Abstract
We investigated whether the evolution of electric organs and electric signal diversity in two independently evolved lineages of electric fishes was accompanied by convergent changes on the molecular level. We found that a sodium channel gene (Nav1.4a) that is expressed in muscle in nonelectric fishes has lost its expression in muscle and is expressed instead in the evolutionarily novel electric organ in both lineages of electric fishes. This gene appears to be evolving under positive selection in both lineages, facilitated by its restricted expression in the electric organ. This view is reinforced by the lack of evidence for selection on this gene in one electric species in which expression of this gene is retained in muscle. Amino acid replacements occur convergently in domains that influence channel inactivation, a key trait for shaping electric communication signals. Some amino acid replacements occur at or adjacent to sites at which disease-causing mutations have been mapped in human sodium channel genes, emphasizing that these replacements occur in functionally important domains. Selection appears to have acted on the final step in channel inactivation, but complementarily on the inactivation “ball” in one lineage, and its receptor site in the other lineage. Thus, changes in the expression and sequence of the same gene are associated with the independent evolution of signal complexity.
Keywords: animal communication, electric organ, channel inactivation, protein evolution, positive selection
Divergence of animal communication signals accompanies reproductive isolation and speciation (1–3). Because most communication signals are under polygenic control (4), it is difficult to identify the contributions of particular genes to signal evolution. We studied the relationship of gene expression and sequence evolution to signal divergence in weakly electric fish because of the simplicity and species diversity of their electric communication signals and the dependence of those signals on ion currents. Ion currents are generated by ion channels, the genes for which are easily identified.
Two groups of teleosts, the mormyriforms of Africa and the gymnotiforms of South America, have convergently evolved electric organs (EOs) whose weak electrical emissions function as communication signals and for electrolocation. There is a 100-fold difference in electric organ discharge (EOD) duration across species (200 μsec to 20 msec) (ref. 5; Fig. 1), with both lineages showing great diversity in signal waveform. Sexual and natural selection have likely played roles in the evolution of this diversity (6–8). Variation in EOD characteristics, especially pulse duration, forms the basis for detection of species-specific signals (9). This richness in the EOD waveform is based in part on the properties of the ion channels of the cells of the EO, the electrocytes (10). The diversity in membrane excitability of electrocytes contrasts with the simple excitability of vertebrate skeletal muscle from which electrocytes evolved, highlighting ion channel genes as potential candidates for strong evolutionary change in these fishes.
Fig. 1.
Variation in EOD pulses from different species used in this study. (A) Black knifefish, S. macrurus. (B) Brown ghost, A. leptorhynchus, EOD pulse from juvenile, myogenically derived EO. (C) Electric eel, E. electricus. (D) Pintail knifefish, B. pinnicaudatus. (E) The elephant nose mormyrid, G. petersii. All EOD traces are to the same time scale; the EOD of G. petersii is expanded to the right of the trace. Amplitudes are arbitrary. EOD pulses are generated by the summation of action potentials of the electrocytes.
Recent studies have shown remarkable convergent evolution at the morphological level in a number of taxa (11–14). The extent to which morphological convergence is accompanied by convergent molecular change is an interesting question and one that can be profitably addressed by comparing the evolution of ion channel genes underlying the EOD in both lineages of electric fish.
We focused on Na+ channel genes because the EOD in at least one species of gymnotiform is shaped by the properties of the EO Na+ current (10) and because of the wealth of information on channel structure/function from site-directed mutagenesis and point mutations underlying clinical syndromes (15–20).
Results
Convergent Loss of Nav1.4a from Muscle and Gain in Electric Organ.
Previously we cloned portions of six sodium channel genes from the gymnotiform Sternopygus macrurus (21) and found two orthologs (Na6 and Na1, renamed Nav1.4a and Nav1.4b; A. Novak, M. Jost, Y.L., A. Taylor, H.H.Z., A. Ribera, unpublished data) of a single muscle-specific mammalian Na+ channel gene (Nav1.4). Sodium channels are composed of four domains (DI–DIV), each with six membrane-spanning regions (S1–S6). We cloned portions of Nav1.4a and Nav1.4b from two other gymnotiforms (Apteronotus leptorhynchus and Brachyhypopomus pinnicaudatus) and two nonelectric species (Ictalurus punctatus and Danio rerio), all in the superorder Ostariophysa, and in a mormyriform (Gnathonemus petersii) and two nonelectric species (Osteoglossum bicirrhosum and Notopterus chitala), all osteoglossomorphs, the most basal teleost lineage. The widespread occurrence of both genes and their presence in basal teleosts indicates that they duplicated early in teleost evolution, likely the result of a genomewide duplication (22, 23).
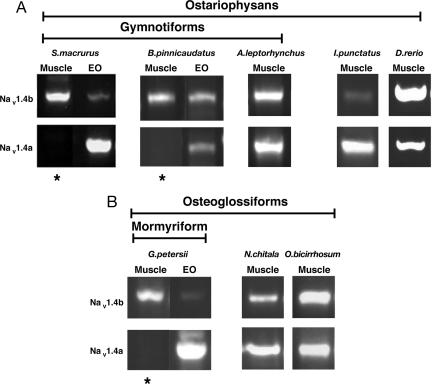
We next determined the expression pattern of these genes in muscle and EO, if present, by RT-PCR. Nav1.4a and Nav1.4b are generally both expressed in muscle in nonelectric species (24), although levels of Nav1.4b are low in muscle from the catfish I. punctatus (Fig. 2). We do not know whether both genes are expressed in all muscle fibers or if they are differentially expressed in different fiber types. In electric fishes, Nav1.4b is expressed in muscle in all four species and in the EO in the gymnotiforms; little or no Nav1.4b is expressed in the EO of G. petersii, a mormyrid. Nav1.4a is expressed solely in the EO in S. macrurus and B. pinnicaudatus, but in the muscle of A. leptorhynchus. Interestingly, A. leptorhynchus possesses a larval myogenic EO that functions only early in life, then degenerates as a neurally derived EO appears (25). We presume that Nav1.4a is expressed in the larval EO in this species but were unable to obtain specimens for analysis. Nav1.4a is expressed only in the EO in G. petersii, a mormyrid. Thus, Nav1.4a has been lost from muscle and expressed exclusively in the EO, an evolutionarily novel structure, in both groups of electric fish.
Fig. 2.
Expression pattern of Nav1.4a and Nav1.4b in muscle and electric organ in two lineages of electric fish and their nonelectric relatives. (A) Three electrogenic gymnotiforms and two nonelectric species, all ostariophysans. (B) An electrogenic mormyriform and two nonelectric species, all osteoglossoforms. Note that Nav1.4a expression is lost from muscle (indicated by the asterisks) in all electric fish except A. leptorhynchus.
Sequence Evolution of Nav1.4a.
To investigate changes in the selective regime acting on the Nav1.4a gene, we used the maximum-likelihood codon models implemented in the codeml program from the paml package (26). Phylogenetic analyses using a number of methods all resulted in the same tree topology (Fig. 3), which resembles the recognized phylogenetic relationships of these species based on other criteria. This topology was fixed for all codeml selection analyses. We applied site models (27) to estimate variation in the pattern of substitution across sites of all 11 taxa and branch-site models (26, 28) to estimate variation in the pattern of substitution across sites on specific lineages identified a priori.
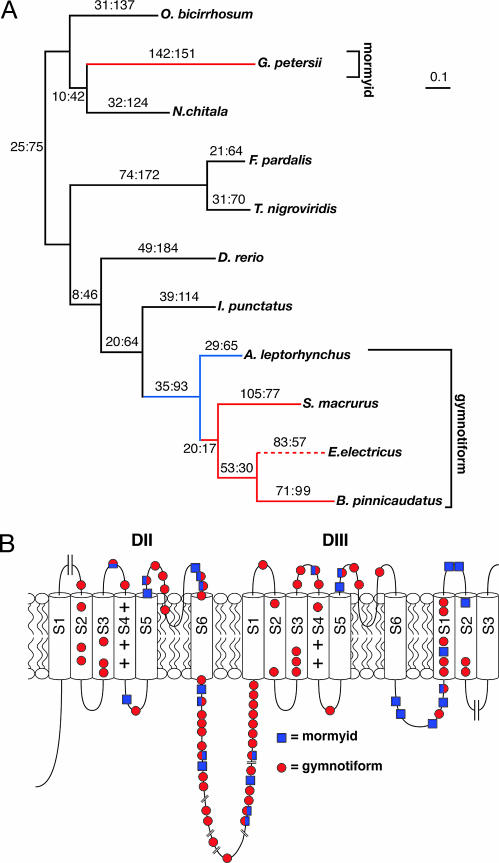
Fig. 3.
Nav1.4a orthologs in electric fish show higher rates of amino acid replacements than in nonelectric fish species. (A) Gene tree for Nav1.4a with the estimated number of nonsynonymous:synonymous substitutions for each branch. Branches from lineages in which Nav1.4a is exclusively expressed in EO are red. The branch representing the electric eel is a dashed line because Nav1.4a is expressed in the EO and, likely, but not tested, is lacking in muscle. Blue lines represent electric fish lineages in which Nav1.4a is confirmed in muscle in extant species or hypothesized in ancestral lineages. Calibration marker = 0.1 nucleotide substitution per codon. (B) Schematic location of nonconserved amino acid changes in Nav1.4a. The large cut marks at either end indicate where the sequence begins and ends. The small cut marks in LII-III indicate where sequences were edited; LII-III is not drawn to scale. Blue squares, the mormyrid G. petersi; red circles, any gymnotiform. Note that 12 amino acid positions show nonconserved changes in both lineages (indicated by an overlapping red circle and blue square). At the majority of these sites (9 of 12), the amino acids are different in the two lineages. Thus, although there are occasional convergent substitutions of the identical amino acid (3 of 12), this pattern is not common.
The site models suggest that ω (dN/dS or the rate of nonsynonynous substitutions/synonymous substitutions) varies strongly between amino acid sites (M0 vs. M3, P ≪ 0.0001) and that the majority of sites are highly constrained (57% of sites, ω0 = 0.01) (Table 1, which is published as supporting information on the PNAS web site). There was no evidence of sites under positive selection over all lineages (M7 vs. M8, P = 0.56). We applied two branch-site models to test for sites under selection on the individual lineages associated with the EO (five terminal and three internal) by using a correction for multiple tests (α = 0.006). Both branch-site tests agree on the presence of sites under positive selection on the terminal lineages leading to Electrophorus electricus and B. pinnicaudatus, as well as the lineage representing the ancestor of those two species. The test results differed for the lineages leading to S. macrurus and G. petersii, with the model B test suggesting sites under positive selection that were not found by the more conservative modified model A test. Neither test suggested sites under positive selection on the lineage leading to A. leptorhynchus, the lineage ancestral to S. macrurus, B. pinnicaudatus, and E. electricus, or the lineage ancestral to all gymnotiforms. These results suggest that the evolution of the Nav1.4a gene has been shaped to some extent by positive selection in all electric species in this study with the exception of A. leptorhynchus, in which Nav1.4a still resides in muscle.
Convergent Amino Acid Replacements in Parts of the Channel Involved in the Final Step in Inactivation.
Regions critical for fundamental properties of Na+ channels, such as ion selectivity (the pore), voltage dependence (S4), inner lining of the channel (S5 and S6), and cytoskeletal localization (ankyrin-binding motif) are highly conserved in electric fish Nav1.4a. Amino acid replacements tended to occur in S1–S3 or intra- or extracellular loops (Fig. 3B). Because Na+ channel inactivation is likely to have a major influence on electric organ pulse duration, we focus on regions involved in this process.
Inactivation involves a number of distinct domains of the Na+ channel; the final step in fast inactivation, the rapid closing of the channel during a sustained depolarization, is accomplished by the docking of the cytoplasmic loop between DIII and DIV (LIII–IV) with the S4–S5 linkers of DIII and DIV that line the inner mouth of the channel (29–32). This interaction is largely due to hydrophobicity. The LIII–IV loop possesses a highly conserved motif centered on the hydrophobic amino acids isoleucine-phenylalanine-methionine, experimental replacements of which profoundly alter fast inactivation (31). In the mormyrid, which makes extremely brief action potentials, there is a nonconservative replacement (I to C) and the loss of a highly conserved neighboring hydrophilic site (Fig. 4A and D). Such an I to C replacement in the human cardiac Na+ channel gene (Nav1.5) speeds recovery from inactivation (32); this naturally occurring replacement in the mormyrid Na+ channel could be important in maintaining a nondecrementing EOD pulse in the bursts of EOD pulses given during social interactions. Nearby, an S to N replacement eliminates a protein kinase C consensus site (Fig. 4E). Phosphorylation of this site in brain Na+ channels slows inactivation (34); elimination of this consensus sequence would optimize for rapid inactivation.
Fig. 4.
Nonconserved amino acid replacements occur in parts of Nav1.4a involved in inactivation. (A) Schematic illustration of a sodium channel. (B) S4–S5 linker in DII. (C) S4–S5 linker in DIII. (D and E) Inactivation loop in the LIII-IV. Nonconserved amino acid replacements are red. Triangles represent sites at which amino acid replacements are associated with diseases in humans, and the asterisk represents a site at which an amino acid replacement in humans leads to disease and there is also a change in an electric fish gene. Fish sequences used for the paml analyses are below the dashed line and above it, for visual comparison, are sequences from the human Nav1.4 ortholog, a human brain Na+ channel gene (Nav1.1), and the single tunicate Na+ channel gene (TuNa1). Note that tunicate and vertebrate Na+ channel genes diverged ≈500 million years ago, whereas the Nav1.4a orthologs in electric fish diverged from their close relatives depicted here ≈60 million years ago for mormyrids and ≈80 million years ago for gymnotiforms (33).
The inactivation loop pivots around a number of highly conserved proline residues on the C-terminal end. Nonconservative replacements of one of these prolines occur in two gymnotiforms (Fig. 4E). Site-directed mutagenesis of this proline in an expressed rat brain gene (35) perturbs inactivation, confirming its importance. Finally, there are radical replacements of a highly conserved glutamine in the mormyrid (Q to P) and the gymnotiform B. pinnicaudatus (Q to L), both of which generate short EOD pulses. This site has not yet been studied experimentally, but its alteration in the two electric fish taxa with the fastest EOD pulses hints that it may play a pivotal role in influencing the speed of inactivation.
Two gymnotiform species possess a nonconservative amino acid replacement (K or R for N) in the S4–S5 linkers in DIII (Fig. 4C). This linker forms part of the receptor for the inactivation loop and is highly conserved with an N at this site from cnidaria to mammals (H.H.Z., unpublished data); a mutation at this site of the Nav1.5 gene is causal for long QT syndrome (20) (Fig. 4C, asterisk). The effect of a positively charged amino acid in this position is not known; however, site-directed replacement of N with C or S in mammalian Nav1.4 or 1.5 induces incomplete inactivation and slows recovery from inactivation (20, 30). There are no amino acid replacements here in mormyrids.
Amino acid replacements occur in the S4–S5 linker in DII in both groups of electric fish (K to T in mormyrid and G to C in gymnotiform; Fig. 4B). Mutations flanking these sites in the S4–S5 linker DII in human Na+ channel genes disrupt slow inactivation (the propensity for Na+ channel to inactivate after prolonged usage) and are associated with muscle diseases (16, 36).
In sum, despite strong evolutionary conservation of the domains involved in the final step of the inactivation of Na+ channels, there are nonconservative amino acid replacements in these regions in electric fish. Most striking are the amino acid replacements that occur in and around the critical isoleucine-phenylalanine-methionine inactivation motif in mormyrids and in the receptor site for that part of the molecule in gymnotiforms. These replacements suggest that the final step of the inactivation process is under selection in both lineages, but through complementary sides of the process.
Discussion
The main finding of this study is that the evolution of signal diversity in two independently derived lineages of electric fish is accompanied by convergent alteration of expression and changes in sequence within the same or complementary functional domains of the same Na+ channel gene, Nav1.4a. Site-directed mutagenesis allows us to determine how specific amino acid replacements in this gene lead to variations in species-specific communication signals. Because a number of these replacements occur at sites that are otherwise highly conserved (i.e., Q to P or L in the inactivation loop) but previously not singled out for study, our results have also pinpointed a number of amino acid sites of potential functional importance for all Na+ channels.
A genome duplication at the origin of teleosts has been suggested as a substrate for the morphological diversity and extensive speciation characteristic of teleosts (22, 23). Nevertheless, there are few concrete examples of genes whose duplication are directly related to species diversity or the emergence of novel structures in fish. Although the duplication of Nav1.4 in fish is not responsible for the genesis of the EO, it allowed for the compartmentalization of one gene duplicate in the EO, providing a substrate for the evolution and divergence of species-specific signals in two lineages of fishes.
Nav1.4a and Nav1.4b are both expressed in the muscle in nonelectric teleosts. Like mammals, fish have a diversity of muscle fiber types, the properties of some types even varying along the body (37, 38). We do not know whether both Na+ channel genes are redundantly expressed in every muscle fiber or they are differentially expressed in particular groups or functional types of muscle fibers to “tune” the electrical responsiveness of the fiber type to its function.
Multiple lineages of teleost fish have independently evolved sonic communication systems by using a diversity of mechanisms, including the vibration of the swimbladder by specialized muscles at frequencies of hundreds of hertz (39–42). It would be intriguing to determine whether a comparable convergent compartmentalization and specialization of Na+ channels genes occurs in muscles specialized for acoustic communication to allow for rapid or prolonged firing of muscle fibers needed to generate acoustic signals. Interestingly, some mormyrid species generate sounds up to 300–400 Hz, and it is believed that their swimbladder muscles may contract at those rates (40). However, because mormyrids have already “lost” one muscle-expressing Na+ channel gene to the EO, it raises the intriguing possibility that Nav1.4a might also be expressed in the swimbladder muscle of mormyrids, where it supports production of a second communication signal in another modality.
Whatever costs electric fish may have incurred for the loss of this gene from muscle are clearly outweighed by the benefits of its gain in the EO. It will be interesting to see whether there are “compensatory” changes in expression or sequence in the remaining muscle Na+ channel gene (Nav1.4b) or by what alterations in transcriptional mechanisms expression was lost from muscle and gained in an evolutionarily novel structure in both electric lineages.
Na+ channels do not function alone; they are modified by beta subunits and act with other currents, such as K+ currents, to shape electrical activity. It will also be intriguing to see whether any of these other genes show comparable alterations in expression or sequence especially in those species with extremely rapid EOD pulses.
Na+ channel genes are highly constrained, as evidenced by the large number of amino acid replacements that change the properties of the Na+ current and lead to neurological, cardiac, and muscular diseases (15–20). A critical step in the evolution of electric organs is the disabling of excitation-contraction so that the organ does not twitch when it discharges. Even when expressed in a noncontractile EO, amino acid replacements in Nav1.4a would be selected against as long as Nav1.4a is also still expressed in muscle, as occurs in A. leptorhynchus. However, once Nav1.4a was solely expressed in a noncontractile organ, amino acid replacements might occur because of relaxation of negative selection or be selected precisely because these amino acid replacements would have generated variation in Na+ currents and, ultimately, diversity in communication signals. Thus, similar to olfactory receptor or peptide venom genes, spatially restricted expression is permissive for sequence evolution (43, 44).
We noted amino acid replacements in electric fish Nav1.4a at or adjacent to amino acid sites where mutational substitutions in human Na+ channel genes are associated with a disease. Not only will study of electric fish Na+ channel genes be a window on the processes of speciation and evolution of communication signals, but identifying amino acid sites or regions of the Na+ channel that have undergone extensive evolution in electric fish may be useful for predicting amino acids putatively involved in channelopathies or pinpointing functionally important novel regions of Na+ channels.
Methods
Sequences.
We cloned portions of Nav1.4a and Nav1.4b (S2 domain II to S2 domain IV) from the gold-lined black knifefish (S. macrurus: AF378144), the pin-tail knifefish (B. pinnicaudatus: DQ351534), the brown ghost (A. leptorhynchus: DQ351533), the channel catfish (I. punctatus: AY204537), the zebrafish (D. rerio: DQ149506), the elephant nose mormyrid (G. petersii: DQ275142), the arawana (O. bicirrhosum: DQ336343), and the clown knifefish (N. chitala: DQ336344).
For phylogenetic analyses, we used additional published sequences for the electric eel (E. electricus: M22252), the green spotted pufferfish (Tetraodon nigroviridis: CAAE01014976), and the Japanese pufferfish (Takifugu pardalis: AB030482).
Cloning and Sequencing.
Tissue was homogenized and RNA was isolated by the guanidinium thiocyanate method with STAT-60 (Ambion, Austin, TX). RNA was quantitated by absorbance readings, and quality was confirmed by gel electrophoreses. RNA was reverse transcribed and amplified by PCR. The PCR products were cloned into the TOPO TA vector (Invitrogen) and grown in bacteria. Inserts were sequenced in both directions with an ABI 3100 sequencer (Applied Biosystems).
Tissue Expression.
One microgram of total mRNA from dorsal trunk muscle (taken from behind the head) in all species and EO from electric fish was reverse transcribed to cDNA with specific primers for each channel. Specific primers targeted intracellular loop DII-DIII and produced fragments of ≈700–1,000 bp, depending on the species. Temperature was optimized for each primer set. PCR was carried out for 10, 20, 30, or 40 cycles. Thirty cycles was chosen as the standard for comparison because it produced a strong signal without saturation of the reaction. The use of primer sets for both Nav1.4a and Nav1.4b in both tissues served as an internal positive control for the quality of mRNA and the robustness of the primers.
Codon Maximum Likelihood Analyses.
Sequences were edited and aligned. The nucleotides used corresponded to the following nucleotides from the Electrophorus sequences (starting at initial ATG): 2,149–2,882; 2,896–2,917; 2,998–3,028; 3,038–3,084; 3,151–3,195; and 3,232–4,531. Phylogenetic trees were estimated with paup* (45).
Analyses were performed by using the codeml program from paml version 3.14 (26–28, 46–49). For a given tree and codon model, codeml finds the set of parameter values that maximize the probability of observing the data, i.e., the likelihood score. The likelihood scores of two nested models are compared via a likelihood ratio test. A significant likelihood score difference between two models that differ in the allowance of a particular feature (such as the presence of positive selection) is taken as evidence for that feature. All models were run under the F3 × 4 setting of codeml in which the equilibrium codon frequencies are taken to be the product of empirical nucleotide frequencies at each codon position. Models M0, M3, M7, and M8 were as described in the literature (27) and are used to detect specific sites under positive selection across all lineages in a data set.
Branch-site models require a division of lineages into foreground branches (branches of interest specified a priori) and background branches (the remaining branches) and test for evidence of positive selection on specific foreground branches, each in a separate test.
Branch-site model B allows for sites that were constrained to some degree on the background branches to evolve under positive selection on the foreground branches. This model may be compared via a likelihood ratio test to site model M3 with two site categories as a test for sites under positive selection on the foreground lineage. This test was applied as in ref. 28. It has been suggested that this test can result in false positives when sites are released from evolutionary constraint on the foreground branch but none experience positive selection (48).
A recently introduced, more conservative branch-site test was also used, now termed “the branch-site test of positive selection” (49). The test uses a modified form of branch-site model A in which ω0 is estimated and may take any value between zero and one. This model is compared via a likelihood ratio test to a restriction of the same model (in which ω1 is fixed at 1) with 1 df. This test is less likely to mistake relaxation of constraint for positive selection and has been shown to have excellent false-positive rates but relatively low power.
Branch-site models are also able to identify individual sites on the foreground branches with a high probability of being positively selected by using an empirical Bayesian approach. It has been shown in the context of site models that the accuracy of these site predictions is low when the number of sequences in the data set is small (<15 sequences) (50). Because our data set is small at 11 sequences, we do not present the Bayesian probability data for individual sites.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Theodore Holmes Bullock, pioneer in comparative biology, mentor, and friend (1915–2005). We thank Michael V. Bennett and Ziheng Yang for helpful discussions; Maggie Patay for fish care; Philip Stoddard (Florida International University, Miami) for providing B. pinnicaudatus; MingMing Wu and Sukant Khurana for providing unpublished sequences; and Marianna Grenadier for artwork. This work was funded by National Institutes of Health Grant R01 NS025513 (to H.H.Z. and Y.L.) and National Science Foundation Integrative Graduate Education and Research Traineeship Program DGE-0114387 (to D.J.Z. and D.M.H.).
Abbreviations
- EO
electric organ
- EOD
electric organ discharge.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Boughman J. Nature. 2001;411:944–948. doi: 10.1038/35082064. [DOI] [PubMed] [Google Scholar]
- 2.Pomiankowski A., Iwasa Y. Proc. Natl. Acad. Sci. USA. 1998;95:5106–5111. doi: 10.1073/pnas.95.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan M. Science. 1998;281:1999–2003. doi: 10.1126/science.281.5385.1999. [DOI] [PubMed] [Google Scholar]
- 4.Hoy R., Hahn J., Paul R. Science. 1977;195:82–84. doi: 10.1126/science.831260. [DOI] [PubMed] [Google Scholar]
- 5.Hopkins C. J. Exp. Biol. 1999;202:1217–1228. doi: 10.1242/jeb.202.10.1217. [DOI] [PubMed] [Google Scholar]
- 6.Crampton W. J. Fish Biol. 1998;53:307–330. [Google Scholar]
- 7.Hopkins C. D. Nature. 1973;424:268–270. [Google Scholar]
- 8.Stoddard P. Nature. 1999;400:254–256. doi: 10.1038/22301. [DOI] [PubMed] [Google Scholar]
- 9.Hopkins C. D., Bass A. H. Science. 1981;212:85–87. doi: 10.1126/science.7209524. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari M. B., McAnelly M. L., Zakon H. H. J. Neurosci. 1995;15:4023–4032. doi: 10.1523/JNEUROSCI.15-05-04023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossuyt F., Milinkovitch M. C. Proc. Natl. Acad. Sci. USA. 2000;97:6585–6590. doi: 10.1073/pnas.97.12.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Losos J., Jackman T., Larson A., Queiroz K., Rodriguez-Schettino L. Science. 1998;279:2115–2118. doi: 10.1126/science.279.5359.2115. [DOI] [PubMed] [Google Scholar]
- 13.Albertson R. C., Streelman J. T., Kocher T. D. Proc. Natl. Acad. Sci. USA. 2003;100:5252–5257. doi: 10.1073/pnas.0930235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kocher T., Conroy J., McKaye K., Stauffer J. Mol. Phylogenet. Evol. 1993;2:158–165. doi: 10.1006/mpev.1993.1016. [DOI] [PubMed] [Google Scholar]
- 15.Berkovic S., Heron S., Giordano L., Marini C., Guerrini R., Kaplan R., Gambardella A., Steinlein O., Grinton B., Dean J., et al. Ann. Neurol. 2004;55:550–557. doi: 10.1002/ana.20029. [DOI] [PubMed] [Google Scholar]
- 16.Bendahhou S., Cummins T., Kula R., Fu Y., Ptacek L. Neurology. 2002;58:1266–1272. doi: 10.1212/wnl.58.8.1266. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara T., Sugawara T., Mazaki-Miyazaki E., Takahashi Y., Fukushima K., Watanabe M., Hara K., Morikawa T., Yagi K., Yamakawa K., et al. Brain. 2003;126:531–546. doi: 10.1093/brain/awg053. [DOI] [PubMed] [Google Scholar]
- 18.Jurkat-Rott K., Lerche H., Lehmann-Horn F. J. Neurol. 2002;249:1493–1502. doi: 10.1007/s00415-002-0871-5. [DOI] [PubMed] [Google Scholar]
- 19.Tan H., Bezzina C., Smits J., Verkerk A., Wilde A. Cardiovasc. Res. 2003;57:961–973. doi: 10.1016/s0008-6363(02)00714-9. [DOI] [PubMed] [Google Scholar]
- 20.Tian X., Yong S., Wan X., Wu L., Chung M., Tchou P., Rosenbaum D., Van Wagoner D., Kirsch G., Wang Q. Cardiovasc. Res. 2004;61:256–267. doi: 10.1016/j.cardiores.2003.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopreato G., Lu Y., Southwell A., Atkinson A., Hillis D., Wilcox T., Zakon H. Proc. Natl. Acad. Sci. USA. 2001;98:7588–7592. doi: 10.1073/pnas.131171798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaillon O., Aury J., Brunet F., Petit J., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., et al. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 23.Amores A., Force A., Yan Y.-L., Joly L., Amemiya C., Fritz A., Ho R., Langeland K., Prince V., Wang Y.-L., et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 24.Venkatesh B., Lu S. Q., Dandona N., See S. L., Brenner S., Soong T. Curr. Biol. 2005;15:2069–2072. doi: 10.1016/j.cub.2005.10.068. [DOI] [PubMed] [Google Scholar]
- 25.Kirschbaum F. Naturwissenschaftern. 1977;64:387–388. [Google Scholar]
- 26.Yang Z. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z., Nielsen R., Goldman N., Pedersen A. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Z., Nielsen R. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 29.Smith M., Goldin A. Biophys J. 1997;73:1885–1895. doi: 10.1016/S0006-3495(97)78219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popa M. O., Alekov A. K., Bail S., Lehmann-Horn F., Lerche H. J Physiol. 2004;561:39–51. doi: 10.1113/jphysiol.2004.065912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellenberger S., West J., Scheuer T., Catterall W. J. Gen. Physiol. 1997;109:589–605. doi: 10.1085/jgp.109.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deschenes I., Trottier E., Chahine M. Cardiovasc. Res. 1999;42:521–529. doi: 10.1016/s0008-6363(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 33.Alves-Gomes J. J. Exp. Biol. 1999;202:1167–1183. doi: 10.1242/jeb.202.10.1167. [DOI] [PubMed] [Google Scholar]
- 34.Numann R., Catterall W. A., Scheuer T. Science. 1991;254:115–118. doi: 10.1126/science.1656525. [DOI] [PubMed] [Google Scholar]
- 35.Kellenberger S., West J., Catterall W., Scheuer T. J. Gen. Physiol. 1977;109:607–617. doi: 10.1085/jgp.109.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kearney J., Plummer N., Smith M., Kapur J., Cummins T., Waxman S., Goldin A., Meisler M. Neuroscience. 2001;102:307–317. doi: 10.1016/s0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
- 37.Swank D. M., Zhang G., Rome L. C. J. Exp. Biol. 1997;200:1297–1307. doi: 10.1242/jeb.200.9.1297. [DOI] [PubMed] [Google Scholar]
- 38.Buckingham S. D., Ali D. W. J. Exp. Biol. 2004;207:841–852. doi: 10.1242/jeb.00839. [DOI] [PubMed] [Google Scholar]
- 39.Rome L. C., Syme D. A., Hollingworth S., Lindstedt S. L., Baylor S. M. Proc. Natl. Acad. Sci. USA. 1996;93:8095–8100. doi: 10.1073/pnas.93.15.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawford J. D., Huang X. J. Exp. Biol. 1999;202:1417–1426. doi: 10.1242/jeb.202.10.1417. [DOI] [PubMed] [Google Scholar]
- 41.Ladich F., Bass A. H. Brain Behav. Evol. 1998;51:315–330. doi: 10.1159/000006545. [DOI] [PubMed] [Google Scholar]
- 42.Bass A. H., Marchaterre M. A., Baker R. J. Neurosci. 1994;14:4025–4039. doi: 10.1523/JNEUROSCI.14-07-04025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duda T. F., Jr, Palumbi S. R. Proc. Natl. Acad. Sci. USA. 1999;96:6820–6823. doi: 10.1073/pnas.96.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niimura Y., Nei M. Gene. 2005;346:23–28. doi: 10.1016/j.gene.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 45.Swofford D. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods. Sunderland, MA: Sinauer Associates; 1998. Version 4. [Google Scholar]
- 46.Goldman N., Yang Z. Mol. Biol. Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z., Bielawski J. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J. Mol. Biol. Evol. 2004;21:1332–1339. doi: 10.1093/molbev/msh117. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J., Nielsen R., Yang Z. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 50.Anisimova M., Bielawski J., Yang Z. Mol. Biol. Evol. 2002;19:950–958. doi: 10.1093/oxfordjournals.molbev.a004152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.