Abstract
Eukaryotes contain numerous transposable or mobile elements capable of parasite-like proliferation in the host genome. All known transposable elements in eukaryotes belong to two types: retrotransposons and DNA transposons. Here we report a previously uncharacterized class of DNA transposons called Polintons that populate genomes of protists, fungi, and animals, including entamoeba, soybean rust, hydra, sea anemone, nematodes, fruit flies, beetle, sea urchin, sea squirt, fish, lizard, frog, and chicken. Polintons from all these species are characterized by a unique set of proteins necessary for their transposition, including a protein-primed DNA polymerase B, retroviral integrase, cysteine protease, and ATPase. In addition, Polintons are characterized by 6-bp target site duplications, terminal-inverted repeats that are several hundred nucleotides long, and 5′-AG and TC-3′ termini. Analogously to known transposable elements, Polintons exist as autonomous and nonautonomous elements. Our data suggest that Polintons have evolved from a linear plasmid that acquired a retroviral integrase at least 1 billion years ago. According to the model of Polinton transposition proposed here, a Polinton DNA molecule excised from the genome serves as a template for extrachromosomal synthesis of its double-stranded DNA copy by the Polinton-encoded DNA polymerase and is inserted back into genome by its integrase.
Keywords: ATPase, cysteine protease, DNA polymerase, integrase, transposable elements
Genomes of most eukaryotes are populated by DNA copies of parasitic elements known as transposable elements (TEs) capable of reproducing themselves in the host genome in a non-Mendelian fashion (1, 2). Understanding the biology of transposable elements is of great importance because of their increasingly well documented impact on the host genome (2, 3). Moreover, transposable elements can be used as powerful tools in genetic engineering (4). Despite an enormous diversity of eukaryotic TEs, they belong to only two types, called retrotransposons and DNA transposons. Whereas a retrotransposon is transposed (retroposed) via reverse transcription of its mRNAs, a DNA transposon is transposed via transfer of its genomic copy from one site to another. Each type includes different classes and families of TEs composed of autonomous and nonautonomous elements. Whereas an autonomous element encodes a complete set of enzymes characteristic of its family, a nonautonomous element encodes none, or only some of them, and depends on enzymes encoded by its autonomous relative. Transposition of a retrotransposon is catalyzed by reverse transcriptase and endonuclease (EN) domains of a polyprotein encoded by itself or by other retrotransposons. All retrotransposons can be further divided into two subclasses called LTR and non-LTR retrotransposons (5). In addition to the reverse transcriptase/EN polyprotein, most non-LTR retrotransposons code for a second protein characterized by poorly understood activities, including RNA/DNA binding, chaperone, and esterase. An mRNA molecule expressed during transcription of the genomic non-LTR retrotransposon is reverse transcribed and inserted in the genome (5). LTR retrotransposons, including endogenous retroviruses, represent the most complex TEs in eukaryotes. An LTR retrotransposon may carry three ORFs coding for the gag, env, and pol proteins, the latter is composed of the reverse transcriptase, EN, and aspartyl protease domains (5). The endonuclease domain in LTR retrotransposons is usually called integrase (INT) and is distantly related to the DDE transposases (TPase) encoded by Mariner DNA transposons (6).
DNA transposons identified so far in eukaryotes belong to two classes characterized by the so-called “cut-and-paste” (7) and “rolling-circle” (8) mechanisms of transposition. Unlike retrotransposons, which synthesize their DNA copies by using their own RNA-dependent DNA polymerase (reverse transcriptase), DNA transposons cannot synthesize DNA. Instead, they multiply by using the host replication machinery. A typical autonomous mariner (9), hAT (10), piggyBac (11), P (12), Merlin (13), or Transib (14) DNA transposon encodes only a single protein called transposase, which acts as an endonuclease and catalyses transfer of transposon DNA strands from one genomic site to another. In the En/Spm (10), MuDR (15), Harbinger (16), and Helitron (8) superfamilies, an autonomous transposon usually encodes a TPase and one DNA-binding protein.
Here we report a third class of DNA transposons called Polintons that are widespread in protists, fungi, and animals, including entamoeba, trichomonas, soybean rust, sea urchin, sea anemone, sea squirt, fishes, chicken, lizard, frog, insects, and worms. Autonomous Polintons are typically15–20 kb long and encode up to 10 different proteins, including DNA polymerase B (POLB), retroviral-like integrase, adenoviral-like protease (PRO), and putative ATPase (ATP). They are the most complex DNA transposons in eukaryotes. Based on reported structural and evolutionary characteristics, we propose a model of Polinton transposition. We discuss implications of our findings, including likely origin of Polintons from a linear plasmid and evolution of adenoviruses from an ancient Polinton.
Results and Discussion
Polintons in Vertebrates, Tunicates, and Echinoderms.
Analysis of the recently reported eukaryotic gene family related to retroviral integrases called c-integrases (17), led us to identification of two families of 360- and 369-aa zebrafish c-integrases (INT-1_DR and INT-2_DR). Each family is composed of several copies dispersed in the genome and characterized by 98% intrafamily and 72% interfamily sequence identities. After expansions of the integrase-encoding regions and multiple alignments of the expanded sequences, we derived 18,485-bp and 16,276-bp consensus sequences (called Polinton-1_DR and Polinton-2_DR). Both elements show hallmarks of similarly structured TEs: (i) 344- and 355-bp terminal inverted repeats (TIRs) and (ii) each individual copy is flanked by different 6-bp target-site duplications (TSDs). Although the internal portions of the consensus sequences (958–12,713 in Polinton1_DR and 621–12665 in Polinton2_DR) are 65% identical to each other, the remaining terminal portions are different. Given that both Polinton families include several nearly full-size elements that are 98% identical to each other, they have transposed within the last few million years.
In addition to the integrase, both Polintons contained the same set of seven intronless ORFs coding for DNA polymerase B (1383-aa POLB-1_DR and 1392-aa POLB-2_DR, see DNA polymerase), adenoviral protease (178-aa PRO-1_DR and PRO-2_DR; see Cysteine protease), ATPase (222-aa ATP-1_DR and 231-aa ATP-2_DR; see ATPase in Polintons), and four unclassified proteins (280-aa PX-1_DR and 282-aa PX-2_DR, 435-aa PY-1_DR and PY-2_DR, 151-aa PW-1_DR and 156-aa PW-2_DR, and 256-aa PZ-1_DR and 260-aa PZ-2_DR; see Table 1, which is published as supporting information on the PNAS web site). In both Polintons, order and orientation of all eight proteins are the same (Fig. 1; INT-PX-PW-PY-PRO-PZ-ATP-POLB; proteins encoded by the second strand are underlined). The same order and orientation of these eight proteins also was observed in other fish genomes, including fugu and pufferfish (data not shown).
Fig. 1.
Structure of Polintons. Polintons from different species (DR, zebrafish; CI, sea squirt; XT, frog; CB, nematode; TC, beetle; SP, sea urchin; TV, T. vaginalis protist; GI, G. intraradices fungus) are schematically depicted as rectangles flanked by terminal inverted repeats (gray triangles). Target site sizes are indicated in square brackets. ORFs coding for Polinton-specific proteins are shown as color rectangles: black, INT; red, family B DNA polymerase (POLB); blue, the ATPase (marked by ATP and ATP1); orange, the cysteine PRO. Lavender (PZ), brown (PW), green (PX), yellow (PY), purple (PC1), navy blue (PC2), PTV1-PTV6, PGI 1, and PGI2 rectangles indicate unclassified proteins. Horizontal arrows show orientation of corresponding ORFs. Orientation of different Polintons was defined by direct orientation of their integrases.
Using the zebrafish Polintons-encoded proteins as seeds intblastn searches against sequenced vertebrate genomes, we detected a plethora of diverse Polintons. In the frog genome, Xenopus tropicalis, we reconstructed consensus sequences of two families (Polinton-1_XT and Polinton-2_XT; Fig. 1). Each consensus sequence was derived from several elements ≈95% identical to each other. Despite a low ≈65% identity between the consensus sequences, they are characterized by the same order and orientation of all eight proteins but differing from those in the zebrafish Polintons by inversion of the PZ-ATP-POLB block (Fig. 1). Analogously to the zebrafish TEs, the frog Polintons are also characterized by 6-bp TSDs and 411–677 bp long TIRs.
In addition to fish and amphibians, Polintons were also found in reptiles and birds. We identified one Polinton-1_SPU in the tuatara (Sphenodon punctatus) genome (GenBank accession no. AC153757, position 112,530–100,591). It is flanked by the ATGGCA 6-bp TSD, has ≈800-bp TIRs, and contains remnants of the PY-PRO-PZ-ATP-POLB coding block with order and orientation identical to those in the zebrafish Polintons (Fig. 1). Because the coding regions contain several stop codons, the identified immobile copy of Polinton-1_SPU was inactivated by mutations after its insertion. The missing INT-PX-PW block likely has been deleted from the originally intact element. In the chicken genome, we also detected mutated remnants of Polintons (data not shown). However, we did not find Polintons in mammalian genomes.
Polintons also populate genomes of tunicates and echinoderms. We identified two Polintons in the sea squirt (Ciona intestinalis) genome: 15,061-bp Polinton-1_CI (scaffold_157, position 54,569–39509; TSD: CACAAG) and 13,695-bp Polinton-2_CI (scaffold_257, position 48,367–62,061; TSD: CTCGAC). Although these elements represent two distinct families (they are <65% identical to each other), they are characterized by the same unusual order of the Polinton proteins: PRO-POLB-ATP-PZ-INT-PX-PW-PY (Fig. 1). We identified four families of full-sized Polintons in the echinoderm sea urchin (Strongylocentrotus purpuratus) genome with order and orientation of all eight proteins resembling those in fish Polintons (Fig. 1). None of the sea urchin Polintons is flanked by target site duplications. However, given the small number of copies of these elements and the preliminary assembly of the genome from ≈1,000-bp short shotgun sequences, we cannot rule out a misassembly of highly identical Polinton copies, which could prevent us from observing the TSDs.
Polintons in Invertebrates.
Using the protein sequences encoded by vertebrate Polintons as seeds intblastn searches against invertebrate DNA sequences, we identified various 5- to 15-kb regions coding for the Polinton proteins. Based on studies of these regions and their flanks, we identified them as internal parts of Polintons with 200- to 1200-bp TIRs and 6-bp TSDs. We derived consensus sequences of complete Polintons in the nematode (16,633-bp Polinton-1_CB, Caenorhabditis briggsae), red flour beetle (13,486-bp Polinton-1_TC, Tribolium castaneum), and fruit fly (14,782-bp Polinton-1_DY, Drosophila yakuba).
The structure of Polintons in vertebrates, tunicates, and echinoderms is notably closer to those in insects than in nematodes (Fig. 1). Whereas insect Polintons encode the same set of eight proteins found in the vertebrate TEs, although in a different insect-specific order and orientation (ATP-PZ-INT-PX-PW-PY-PRO-POLB), the nematode Polintons code for nematode-specific PC1 and PC2 proteins, instead of PX, PZ and PW, and are characterized by a unique order and orientation of all proteins: INT-PY-PC1-ATP-POLB-PC2-PRO (Fig. 1).
Based on tblastn searches against Trace archives, we also found that the Polinton-specific proteins are encoded in multiple copies by the cnidarian genome (sea anemone Nematostella vectensis) ancestral to insects, worms, and vertebrates.
Polintons in Protists and Fungi.
Although Polintons were not found in plants, we identified them in several other eukaryotic kingdoms, including fungi (soybean rust Phakospora pachyrhizi and Glomus intraradices) and such protists as parabasilids (Trichomonas vaginalis), entamoeba (Entamoeba invadens, E. histolytica, and E. dispar), and heterokonts (Phytophthora infestans). The soybean rust genome is not assembled, but we identified numerous ≈1-kb shotgun sequences encoding Polinton proteins. After analysis of short fragments encoding parts of two different proteins, we conclude that the soybean rust Polintons are characterized by a standard set of eight Polinton proteins: [POLB-ATP], [PX-PW-PY-PRO], and [PZ-INT], where three groups of proteins with known order and orientation are delimited by brackets. Even this incomplete information indicates that the order and orientation of these proteins is unique, although very similar to those in vertebrate and insect Polintons (Fig. 1). We detected one copy of Polinton-1_GI encoding INT and POLB in the G. intraradices fungal genome (GenBank accession no. AC163889, position 5,506–17,459; the CACCTT TSD).
In the T. vaginalis genome, we found a very abundant Polinton-1_TV family characterized by a 20,724-bp consensus sequence (Fig. 1). This family, together with its nonautonomous derivatives, constitutes ≈5% of the genome, which makes T. vaginalis an incubator of Polintons, unlike the other genomes where Polintons constitute only a minor component (<0.2%; data not shown). General properties of Polinton-1_TV are the same as observed in metazoan Polintons: 6-bp TSDs and 160-bp TIR. Although the Polinton-1_TV encodes POLB and INT similar to those in metazoan Polintons, it encodes two different ATPases (ATP and ATP1 in Fig. 1), which are not significantly similar to ATPases from the metazoan Polintons (blast E values >1). Excluding POLB, INT, and ATPases, T. vaginalis Polintons do not code for other proteins found in the metazoan TEs. However, they encode six additional proteins, called PTV1–PTV6 (Fig. 1). Only PTV2 is similar to other known proteins; it contains a 200-aa domain similar to the C-terminal domain of structural proteins involved in assembly of phage tails (Table 1).
The entamoeba genome is the only other sequenced genome coding for PTV6. This protein is encoded by Polinton-1_EI TEs in E. invadens. Although the E. invadens genome was not assembled, we reconstructed the 16,504-bp Polinton-1_EI consensus sequence from ≈1,000-bp short shotgun sequences. This transposon is characterized by the 597-bp TIR and the POLB-ATP1-ATP-PTV6-INT pattern of its proteins.
A 18,398-bp single copy of Polinton-1_PI found in P. infestans is characterized by the POLB-INT-ATP-PRO pattern, 6-bp TSD, and 113-bp TIRs.
The Tetrahymena thermophila ciliate genome also contains Polintons. The genome is not assembled, and we identified several 2-to 3-kb whole genome shotgun sequences coding for Polinton INT and POLB (GenBank accession nos. AAGF01001397, AAGF01001309, and AAGF01001471). INTs encoded by these sequences are ≈50% identical to INT encoded by Tlr transposable elements (18). Moreover, some copies of Tlr encode ATP (18), which is similar to the Polinton ATP. Tlr is characterized by 6-bp TSDs, >100-bp TIRs with the 5′-AGAGA terminus similar to that conserved in Polintons (ref. 18; Fig. 2). Therefore, Tlr elements are nonautonomous derivatives of Polintons.
Fig. 2.
Termini of Polintons. Nongapped alignment of 5′ TIRs from 20 families of Polintons is shown. Names of the families are abbreviated as “N_Species,” where “Species” is a two-letter code of a species (CI, C. intestinalis sea squirt; XT, X. tropicalis frog; TC, T. castaneum beetle; CB, C. briggsae nematode; SPU, S. punctatus tuatara; SP, S. purpuratus sea urchin; DR, D. rerio zebra fish; TV, T. vaginalis; EI, E. invadens entamoeba; PI, P. infestans potato blight; GI, G. intraradices fungus) and “N” is a number of the corresponding Polinton family found in this species. Invariantly conserved 5′-AG termini are shaded in black. Terminal repetitions are underlined.
Nonautonomous Polintons.
The best examples of nonautonomous Polintons were found in zebrafish. For instance, a Polinton-1N1_DR family consists of ≈20 recently transposed TEs with ≈97% pairwise identity, including five 11- to 17-kb copies 98% identical to each other and flanked by different 6-bp TSDs (AL953910, position 5,323–19,910; BX897696, 116,625–128,256; AL935200, 105,042–120,037; BX005307, 157,480–142,142; BX842670, 125,298–109,851; BX294179, 82,783–67,292). Remarkably, this transposon is a huge noncoding palindrome composed of 7.3-kb TIRs (>99% identity) and a 0.8-kb internal loop. Only its 1-kb terminus is similar to sequences of autonomous Polintons (85% identity to the Polinton-1_DR terminus). The Polinton-1N1_DR TIRs incorporate one CR1–3_DR non-LTR retrotransposon (position 3,380–3,167 and 12,046–12,259), and one HE1 SINE element (pos. 1,318–1,198 and 14,111–14,231; Fig. 5A, which is published as supporting information on the PNAS web site). Another nonautonomous family, Polinton-2N1_DR, is composed of several copies 98% identical to each other, including five ≈11-kb elements with termini, different 6-bp TSDs, and 2.5- to 5.9-kb TIRs. Its 365-bp termini are 98% identical to the Polinton-2_DR termini, but the remaining portion is not similar to any parts of the autonomous TEs. Moreover, both Polinton-2N1_DR TIRs incorporated a copy of the mariner DNA11TA1_DR transposon (Fig. 5A). Given the observed incorporation of different old TEs into the long TIRs of young nonautonomous Polintons, we propose that originally short (several hundred base pairs) Polinton TIRs have been transformed into very long palindromes by the host replication machinery, after accidental DNA annealing triggered by single-stranded breaks at the stem-loop border (Fig. 5B). Given presence of nonautonomous Polintons with long TIRs in the sea squirt (chr08q, position 910,025–916,953; 2796-bp TIR), and sea urchin genomes (Spur20030922-genome: Contig884, 484–9,563; 3,900-bp TIR), the proposed “palindromization” of internal portions of Polintons may be quite common and may represent a more general model for palindromization of stem-loop structures.
Structural Hallmarks of TIRs.
All identified Polintons are characterized by the universally conserved 5′-AG and CT-3′, in addition to already described TIRs that are several hundred nucleotides long, 6-bp TSDs, and specific sets of proteins, including universally present POLB, INT, and ATP (Fig. 2). This specificity of termini is linked to a structural similarity between the Polinton and retroviral integrases (17). Moreover, LTR retrotransposons that belong to the group I of the Gypsy superfamily are also characterized by the conserved 5′-AG and YT-3′ termini (19). Although there is no significant sequence identity between TIRs from Polintons found in different species, all of them appear to share short simple microsatellite-like terminal repetitions. The 5′-(AGT)2 microsatellite is the most common type of such repetitions; the 5′-(AGA)2, 5′-(AGC)2, and 5′-(AG)3 microsatellites represent less common types of terminal repetitions (Fig. 2).
DNA Polymerase B.
Each autonomous Polinton encodes a 1,000- to 1,400-aa protein whose ≈700-aa C-terminal portion is significantly similar to various protein-primed DNA polymerases that belong to the B family of DNA-dependent DNA polymerases. Protein-primed POLBs constitute a distinctive group of eukaryotic and prokaryotic DNA polymerases encoded by various phages, vertebrate adenoviruses, and linear plasmids from plant and fungal mitochondria. These polymerases display both 3′ to 5′ exonucleolytic and 5′ to 3′ synthetic activities defined by two structurally independent N- and C-terminal domains (20). After inspection of a multiple alignment of the Polinton and POLB polymerases, we found that all known motifs conserved in the protein-primed POLB polymerases are also conserved in the Polinton polymerases (Fig. 6, which is published as supporting information on the PNAS web site). These motifs include the Exo I (defined by the DXE consensus), Exo II (Nx3F/YD), and Exo III (Yx3D) motifs that constitute a catalytic core of the exonuclease conserved in all proofreading POLBs (21–23), and the S/TLx2h motif also conserved in the POLB exonuclease domain (24) (see Fig. 6). The synthetic activities of POLBs are defined by five conserved motifs Dx2SLYP (motif A or 1), Kx3NSxYG (motif B or 2a), Tx2G/AR (motif 2b), YxDTDS (motif C or 3), and KxY (motif 4) (20), which are also well conserved in the Polinton POLBs (Fig. 6).
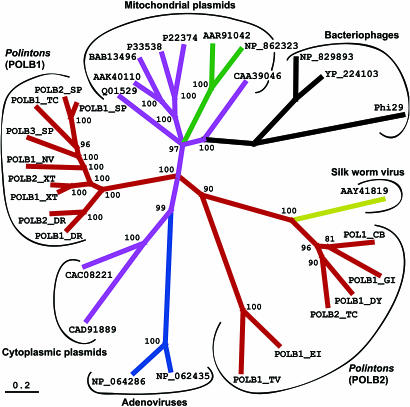
Although all Polinton POLBs cocluster with protein-primed POLBs, they constitute two distinctive clades (Polinton POLB1 and POLB2; Fig. 3). Based on the protein identity, members of the same clade are closer to each other than to members of the other clade. The POLB1 clade includes polymerases encoded by the vertebrate, sea urchin, sea squirt, sea anemone, and some insect (Polinton-1_TC) Polintons. The POLB2 clade includes proteases encoded by the insect, nematode, fungus, and protozoa transposons.
Fig. 3.
Maximum likelihood tree showing phylogenetic relationship of family B DNA polymerases from Polintons, eukaryotic linear plasmids and viruses, and bacteriophages. Polinton polymerases (red branches) are named “POLBN_Species,” where N is a number of the corresponding family of Polintons present in a genome defined by a two-letter species abbreviation (CB, nematode; TC, beetle; NV, sea anemone; SP, sea urchin; XT, frog; DR, zebrafish; CI, sea squirt; GI, fungus; DY, fruit fly; TV, protist T. vaginalis; EI, protist E. invadens). Polymerases from linear plasmids, viruses, and bacteriophages are listed by their GenBank accession numbers. Linear mitochondrial plasmids from fungi (magenta branches): AAK40110, the pMLP2 plasmid of oyster mushroom; BAB13496, the pFV1 plasmid of the basidiomycetous fungus Flammulina velutipes; P22374, the pAI2 plasmid of the filamentous fungus Ascobolus immersus; P33538, the kalilo plasmid of Neurospora intermedia; CAA39046, the maranhar plasmid of Neurospora crassa; Q01529, the pAL2–1 plasmid of Podospora anserina. Linear mitochondrial plasmids from plants (green branches): AAR91042, a plasmid inserted in the Zea mays mitochondrial genome; NP_862323, a plasmid of the Brassica napus rapeseed. Bacteriophages (in black): NP_829893, the Bacillus thuringiensis linear plasmid/prophage pBClin15; YP_224103, the B. thuringiensis phage GIL16c; Phi29, GenBank accession no. P06950, Bacillus phage phi29. Linear cytoplasmic plasmids from yeasts (magenta branches): CAC08221, the pPE1B plasmid from Pichia etchellsii; CAD91889, the pPin1–3 killer plasmid from P. inositovora. Eukaryotic DNA viruses (blue and yellow branches): NP_064286; ovine adenovirus A; NP_062435, frog adenovirus; AAY41819, the Bombyx mori densonucleosis virus type 2 (BmDNV-2).
All POLB1s contain a unique ≈145-aa insertion at the same position between the conserved Exo III and “YxGG” motifs (Fig. 6). The insertion position is ≈15 bp upstream of the YxGG motif (the exact position is not clear because of low identities of POLB1s to other POLBs). The insertion sequence is significantly similar to bacterial ≈140-aa “very-short-patch” DNA repair endonucleases (VSR), which initiate nucleotide excision repair of G:T mismatches introduced by deamination of 5′methyl-cytosines (25). In psi-blast searches against GenBank proteins concatenated with 15 different POLB1 insertions, the VSR proteins were the only proteins, excluding the POLB1 insertions, which were similar to a profile derived from the POLB1 insertions (E <10−5). The catalytic core of VSR consists of Asp-51 and His-69 (26, 27), which are also conserved in the POLB1 insertions (Fig. 6). Although most members of the POLB2 clade do not contain any long insertions between the Exo III and YxGG motifs, we identified several families of Polintons in different species that contain an ≈140-aa specific insertion at a position nearly identical to that in POLB1. Although there is no sequence similarity between these two insertions, the POLB2 insertion is similar to HNH homing nucleases (data not shown). Presence of the VSR and HNH nucleases inserted at the same position between the exonuclease and polymerase domains of the Polinton POLBs appears to be very unusual. We did not find any other POLBs with inclusions of the nucleases, except for several known bacterial POLBs containing the homing nuclease encoded by inteins: analogues of “amino acid self-spliced” introns. In eukaryotes, only a few inteins have been found, in fungi. Although termini of the POLB1 and POLB2 insertions are not perfectly defined and not similar to conserved termini of known bacterial inteins, one cannot rule out an intein-like nature of the POLB1 and POLB2 insertions.
Adenoviral Protease.
In addition to the conserved POLB and INT, Polintons encode an ≈170-aa PRO protein (Fig. 1; see also Fig. 7, which is published as supporting information on the PNAS web site) that is significantly similar to proteases in adenoviruses. The latter, also known as adenain or AVP, belong to a superfamily of cysteine proteases and is characterized by the His-Asn/Glu-Cys catalytic triad (28–31). In addition to the catalytic residues, Gln separated by six amino acids from the catalytic Cys, was also reported as a universally conserved amino acid that is necessary for the protease activity (28, 29). The catalytic triad and Gln are also conserved in the Polinton proteases (Fig. 7). Although Polintons in protozoans do not encode the PRO cysteine protease, it is conserved in metazoan and fungal Polintons. These observations indicate that PRO is necessary for survival of Polintons, which have acquired it in a common ancestor of fungi and animals, after its split from protozoans.
ATPase.
Most Polintons code also for a ≈200-aa ATP protein (Fig. 1; see also Fig. 8, which is published as supporting information on the PNAS web site) characterized by the Walker A and B motifs conserved in ATPases (32, 33). The Polinton ATPase is most similar to a plethora of hypothetical ATPases from dsDNA viruses. Many of these hypothetical ATPases were mistakenly annotated in GenBank as virion-packaging proteins. However, none of these Polinton-like ATPases contain any C-terminal nuclease domain that is present in real virion packaging proteins (34). The Polinton ATPase probably facilitates DNA synthesis by POLB.
Mechanism of Transposition.
Given known distant similarities between retroviral INTs and cut-and-paste TPases, one can expect cut-and-paste transpositions of Polintons catalyzed by their INT. However, some arguments listed below strongly suggest that transposition of Polintons follows a completely different mechanism unseen previously in transposons. First, a perfect conservation of all functional motifs in the extremely diverged POLBs indicates that the DNA-DNA polymerase and proofreading activities are necessary for transposition of Polintons. Second, POLB in Polintons belongs to the group of DNA polymerases that use proteins as primers. This group is composed of polymerases encoded by bacteriophages, linear plasmids, and adenoviruses. Third, all genomes of these objects and Polintons are characterized by TIRs that are usually several hundred base pairs long. Fourth, termini of these genomes and Polintons are composed of short 1- to 3-bp tandem repeats (Fig. 2; figure 6 in ref. 35), which are thought to be necessary for the slide-back mechanism in protein-primed DNA synthesis (35).
Based on all these arguments, we propose that Polintons form a previously uncharacterized class of DNA transposons propagated through protein-primed self-synthesis by their polymerase, according to the model outlined in Fig. 4. First, during host genome replication, the integrase-catalyzed excision of a Polinton element from the host DNA leads to an extrachromosomal single-stranded Polinton that forms a racket-like structure (Fig. 4 A–C). Second, the Polinton POLB replicates the extrachromosomal Polinton (Fig. 4 D–F). Given the arguments listed above, initiation of the replication requires the terminal protein (TP) that binds a free 5′ end of Polinton. It is believed that N-terminal domains of proteins, whose C-terminal parts serve as POLB, encode TP in some linear plasmids (36). Therefore, it is likely the N-terminal 400- to 600-aa domain of the Polinton POLB serves also as TP. After the double-stranded Polinton is synthesized, the INT molecules bind its termini and catalyze its integration in the host genome (Fig. 4 G and H).
Fig. 4.
Model of the Polinton transposition. Polinton single-stranded DNAs are shown in red (those synthesized de novo are shown in orange); their TIRs are in light red and orange. The polymerase, terminal protein, and integrase are depicted as black, blue, and green ovals. Old and new target site duplications are marked by small green and blue rectangles. See Mechanism of Transposition for details.
Evolution of Polintons.
Polintons are present in genomes of species that belong to diverse eukaryotic kingdoms, including opisthokonts (metazoa and fungi), heterokonts (oomycetes), alveolates (ciliates), amoebozoa (entamoeba), and parabasalids. Given the conserved complex structure of Polintons, their monophyletic origin is most likely. Although Polintons are much more complex (up to eight conserved proteins) than known eukaryotic TEs and resemble viruses (adenoviruses and BmDNV-2), we did not find any Polinton protein similar to viral capsid or envelope proteins, which are necessary for the infectious transmission of viruses. Moreover, we are not aware of any viruses capable of spreading over different kingdoms. Most likely, Polintons emerged in a common ancestor of modern species from the eukaryotic crown ≈1 billion years ago. As we reported here, Polintons share their main structural characteristics with “selfish” linear plasmids, bacteriophages, and adenoviruses that multiply by using their protein-primed DNA polymerases. Linear plasmids can be split into two groups: (i), plasmids that exist in mitochondria of plants and fungi and (ii), plasmids that exist in the yeast cytoplasm (37). Although it is likely that mitochondrial linear plasmids evolved from bacteriophages during the evolution of mitochondria from bacteria, understanding the evolution of cytoplasmic plasmids is hampered by different equally plausible scenarios (38, 39). Although Polintons represent a previously unknown link between cytoplasmic plasmids/adenoviruses and mitochondrial plasmids/bacteriophages (Fig. 3), many aspects of evolution of Polintons and cytoplasmic linear plasmids remain unclear. Acquisition of the integrase by a protein-primed replicating genome of an ancient virus or linear plasmid was the most certain stage of the evolution. It has been suggested that the Polinton INT evolved from an INT encoded by an LTR retrotransposon (17). Thus, it might have been acquired after integration of an ancient LTR retrotransposon into the ancestral linear genome. However, we cannot rule out the origin of the Polinton INT from a DNA transposon. For instance, the Tdd-4 transposon from the slime mould Dictyostelium discoideum genome is a DNA transposon characterized by its 145-bp TIRs, 5-bp TSDs, and a TPase that is similar to INTs encoded by LTR retrotransposons (40).
Although both clades of the Polinton polymerase are significantly but distantly coclustered with adenoviruses and cytoplasmic plasmids, POLBs encoded by Polinton-1_CB, Polinton-1_GI, Polinton-1_DY, and Polinton-2_TC from nematode, fungus, fruit fly, and beetle are closest to the POLB encoded by the BmDNV-2 Bombyx mori densonucleosis virus (Fig. 3). This unique virus encodes several structural virion-related proteins, and its POLB was grouped with the adenoviral POLB (21). It remains to be shown whether the BmDNV-2 virus has evolved from a Polinton or from a virus related to Polintons and adenoviruses.
Polintons are characterized by a highly patchy distribution in different species. In insects, Polintons are present in flies and beetles but absent in mosquitoes and bees. In fungi, they are present in basidiomycetes (soybean rust) and glomeromycetes (G. intraradices) but absent in ascomycetes (including the completely sequenced yeast genome). We interpret this patchiness as a frequent loss of Polintons from genomes. Because of the high complexity of Polintons, their transposition may be tightly regulated and may explain their small numbers in most genomes.
While this manuscript was in preparation, Feschotte and Pritham (17) reported that c-integrases in the zebrafish and nematode genomes are encoded by long TEs, called Maverick, characterized by long TIRs and 6-bp TSDs (41). Moreover, Wuitschick et al. reported in 2002 (18) that the T. thermophila genome contains Tlr transposons characterized by the same Maverick-like properties. Because all these authors have not reported the basic enzymatic machinery or mechanism of transposition of Tlr/Maverick TEs, we introduce “Polinton” as a general name of all eukaryotic self-synthesizing DNA transposons.
Materials and Methods
All TEs reported in this work were identified and characterized by using various methods of computational analysis described in our previous papers (8, 14, 16, 19, 42). Significant similarities between distantly related proteins were identified by using psi-blast (43). Multiple alignments of protein sequences were created by using probcons and t_coffee (44, 45). Phylogenetic analysis was performed by using mrbayes 3.0 (46) with the following settings. Rate variation across sites was modeled by using a gamma distribution, with a proportion of sites being invariant (rates = “invgamma”; “mixed” amino acid models). The Markov chain Monte Carlo search was run with four chains for 5 × 105 generations, with trees begin sampled every 1,000 generations (the first 1,000 trees were discarded as “burnin”). The mrbayes input multiple alignment included only the POLB conserved motifs (≈400 amino acids, available upon request). The sequences of TEs reported in this work are deposited in Repbase Update (47).
Supplementary Material
Acknowledgments
We thank Adam Pavlicek and Andrew Gentles for discussions, anonymous reviewers and Margaret Kidwell for helpful suggestions, and Oleksiy Kohany for assistance with computational analysis. This work was supported by National Institutes of Health Grant 5 P41 LM006252-08.
Abbreviations
- INT
integrase
- POLB
family B DNA polymerase
- PRO
protease
- TE
transposable element
- TIR
terminal inverted repeat
- TPase
transposase
- TSD
target-site duplication.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Mobile DNA II. Washington, DC: Am. Soc. Microbiol; 2002. [Google Scholar]
- 2.Kidwell M. G., Lisch D. R. Evolution Int. J. Org. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 3.Fedoroff N. V. Ann. NY. Acad. Sci. 1999;870:251–264. doi: 10.1111/j.1749-6632.1999.tb08886.x. [DOI] [PubMed] [Google Scholar]
- 4.Ivics Z., Izsvak Z. Trends Genet. 2005;21:8–11. doi: 10.1016/j.tig.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Eickbush T. H., Malik H. S. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington DC: Am. Soc. Microbiol; 2002. pp. 1111–1144. [Google Scholar]
- 6.Capy P., Langin T., Higuet D., Maurer P., Bazin C. Genetica. 1997;100:63–72. [PubMed] [Google Scholar]
- 7.Craig N. L. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 8.Kapitonov V. V., Jurka J. Proc. Natl. Acad. Sci. USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plasterk R. H. A., Van Luenen H. G. A. M. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol; 2002. pp. 519–532. [Google Scholar]
- 10.Kunze R., Weil C. F. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol; 2002. pp. 565–610. [Google Scholar]
- 11.Sarkar A., Sim C., Hong Y. S., Hogan J. R., Fraser M. J., Robertson H. M., Collins F. H. Mol. Genet. Genomics. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- 12.Rio D. C. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol; 2002. pp. 484–518. [Google Scholar]
- 13.Feschotte C. Mol. Biol. Evol. 2004;21:1769–1780. doi: 10.1093/molbev/msh188. [DOI] [PubMed] [Google Scholar]
- 14.Kapitonov V. V., Jurka J. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walbot V., Rudenko G. N. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol; 2002. pp. 533–564. [Google Scholar]
- 16.Kapitonov V. V., Jurka J. DNA Cell Biol. 2004;23:311–324. doi: 10.1089/104454904323090949. [DOI] [PubMed] [Google Scholar]
- 17.Gao X., Voytas D. F. Trends Genet. 2005;21:133–137. doi: 10.1016/j.tig.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Wuitschick J. D., Gershan J. A., Lochowicz A. J., Li S., Karrer K. M. Nucleic Acids Res. 2002;30:2524–2537. doi: 10.1093/nar/30.11.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapitonov V. V., Jurka J. Proc. Natl. Acad. Sci. USA. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco L., Salas M. J. Biol. Chem. 1996;271:8509–8512. doi: 10.1074/jbc.271.15.8509. [DOI] [PubMed] [Google Scholar]
- 21.Bernad A., Blanco L., Lazaro J. M., Martin G., Salas M. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 22.Morrison A., Bell J. B., Kunkel T. A., Sugino A. Proc. Natl. Acad. Sci. USA. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 24.de Vega M., Lazaro J. M., Salas M., Blanco L. J. Mol. Biol. 1998;279:807–822. doi: 10.1006/jmbi.1998.1805. [DOI] [PubMed] [Google Scholar]
- 25.Lieb M., Bhagwat A. S. Mol. Microbiol. 1996;20:467–473. doi: 10.1046/j.1365-2958.1996.5291066.x. [DOI] [PubMed] [Google Scholar]
- 26.Tsutakawa S. E., Muto T., Kawate T., Jingami H., Kunishima N., Ariyoshi M., Kohda D., Nakagawa M., Morikawa K. Mol. Cell. 1999;3:621–628. doi: 10.1016/s1097-2765(00)80355-x. [DOI] [PubMed] [Google Scholar]
- 27.Tsutakawa S. E., Morikawa K. Nucleic Acids Res. 2001;29:3775–3783. doi: 10.1093/nar/29.18.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding J., McGrath W. J., Sweet R. M., Mangel W. F. EMBO J. 1996;15:1778–1783. [PMC free article] [PubMed] [Google Scholar]
- 29.McGrath W. J., Ding J., Didwania A., Sweet R. M., Mangel W. F. Biochim. Biophys. Acta. 2003;1648:1–11. doi: 10.1016/s1570-9639(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 30.Weber J. M. Acta Microbiol. Immunol. Hung. 2003;50:95–101. doi: 10.1556/AMicr.50.2003.1.9. [DOI] [PubMed] [Google Scholar]
- 31.Tihanyi K., Bourbonniere M., Houde A., Rancourt C., Weber J. M. J. Biol. Chem. 1993;268:1780–1785. [PubMed] [Google Scholar]
- 32.Walker J. E., Saraste M., Runswick M. J., Gay N. J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorbalenya A. E., Koonin E. V. Nucleic Acids Res. 1989;17:8413–8440. doi: 10.1093/nar/17.21.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanamaru S., Kondabagil K., Rossmann M. G., Rao V. B. J. Biol. Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 35.Mendez J., Blanco L., Esteban J. A., Bernad A., Salas M. Proc. Natl. Acad. Sci. USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim E. K., Jeong J. H., Youn H. S., Koo Y. B., Roe J. H. Curr. Genet. 2000;38:283–290. doi: 10.1007/s002940000157. [DOI] [PubMed] [Google Scholar]
- 37.Schaffrath R., Meinhardt F., Meacock P. A. FEMS Microbiol. Lett. 1999;178:201–210. doi: 10.1111/j.1574-6968.1999.tb08678.x. [DOI] [PubMed] [Google Scholar]
- 38.Kempken F., Hermanns J., Osiewacz H. D. J. Mol. Evol. 1992;35:502–513. doi: 10.1007/BF00160211. [DOI] [PubMed] [Google Scholar]
- 39.Rohe M., Schrunder J., Tudzynski P., Meinhardt F. Curr. Genet. 1992;21:173–176. doi: 10.1007/BF00318478. [DOI] [PubMed] [Google Scholar]
- 40.Wells D. J. Nucleic Acids Res. 1999;27:2408–2415. doi: 10.1093/nar/27.11.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feschotte C., Pritham E. J. Trends Genet. 2005;21:551–552. doi: 10.1016/j.tig.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Kapitonov V. V., Jurka J. Genetica. 1999;107:27–37. [PubMed] [Google Scholar]
- 43.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Do C. B., Mahabhashyam M. S., Brudno M., Batzoglou S. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Notredame C., Higgins D. G., Heringa J. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 46.Ronquist F., Huelsenbeck J. P. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 47.Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.