Abstract
The Arabidopsis CAO gene encodes a 52-kDa protein with predicted localization in the plastid compartment. Here, we report that CAO is an intrinsic Rieske iron-sulfur protein of the plastid-envelope inner and thylakoid membranes. Activity measurements revealed that CAO catalyzes chlorophyllide a to chlorophyllide b conversion in vitro and that the enzyme was only slightly active with protochlorophyllide a, the nonreduced precursor of chlorophyllide a. Protein import and organelle fractionation studies identified CAO to be distinct from Ptc52 in the substrate-dependent transport pathway of NADPH:protochlorophyllide oxidoreductase A but instead to be part of a separate translocon complex. This complex was involved in the regulated import and stabilization of the chlorophyllide b-binding light-harvesting proteins Lhcb1 (LHCII) and Lhcb4 (CP29) in chloroplasts. Together, our results provide insights into the plastid subcompartmentalization and evolution of chlorophyll precursor biosynthesis in relation to protein import in higher plants.
Keywords: chlorophyll biosynthesis, light-harvesting proteins, protein import
Chlorophyll (Chl) is one of the most abundant organic molecules on Earth. It is estimated that several million tons of chlorophyll are synthesized and degraded every year. The biosynthetic pathway of Chl is well established in higher plants and algae (1). However, despite considerable progress made over the last three decades toward its understanding, some details of its compartmentalization and regulation remain obscure.
It is generally assumed that Chl a represents the precursor of Chl b and that the pathway leading to Chl b would proceed sequentially (1). The enzyme responsible for Chl b biosynthesis is chlorophyllide a oxygenase (CAO) (2–4). CAO is widespread in higher plants and also occurs in prochlorophytes and chlorophytes (2–4). All CAO genes share high sequence similarity (2–4). Plant CAO expressed in bacteria has been reported to be active as chlorophyllide (Chlide) a oxygenase in vitro but not be active with protochlorophyllide a (Pchlide a) (5). Pchlide a differs from Chlide a by the presence of one double bond between C17 and C18 in ring D of the macrocycle (1). The CAO gene is a single nuclear gene in Chlamydomonas and Arabidopsis thaliana (2–4), and mutants lacking CAO such as chlorina fail to accumulate Chl b (2, 3). At first glance, these findings suggest that there is no redundant way for making Chl b in plants.
According to recent work, this view may be too simple. Phylogenetic analyses have revealed that CAO belongs to a large superfamily of non-heme oxygenases sharing the same conserved Rieske and mononuclear iron binding domains as those found in CAO (6, 7). The other members are choline monooxygenase (8), pheophorbide a oxygenase (PAO) (9), the protein translocon protein Tic55 (6), and a 52-kDa protein associated with the precursor NADPH:protochlorophyllide oxidoreductase A (pPORA) translocon complex, called Ptc52 (10). Pulse-labeling experiments using the isolated Ptc complex and the Pchlide precursor 5-aminolevulinic acid (5-ALA) have shown that Ptc52 operates as Pchlide a oxygenase in the production of Pchlide b, an immediate precursor of Chlide b (10). Xu et al. (11) reported that expression of the AtCAO cDNA in the cyanobacterium Synechocystis, which does not contain a CAO gene in its genome, gave rise not only to Chl b, but also to low levels of Pchlide b and Pchl b. In the present study, the Arabidopsis CAO (AtCAO) cDNA was used to express functional protein and to study its catalytic properties, plastid sublocalization, and role in Chl b precursor (Chlide b and Pchlide b) biosynthesis.
Results
CAO Displays in Vitro Oxygenase Activity with Chlide a and Pchlide a.
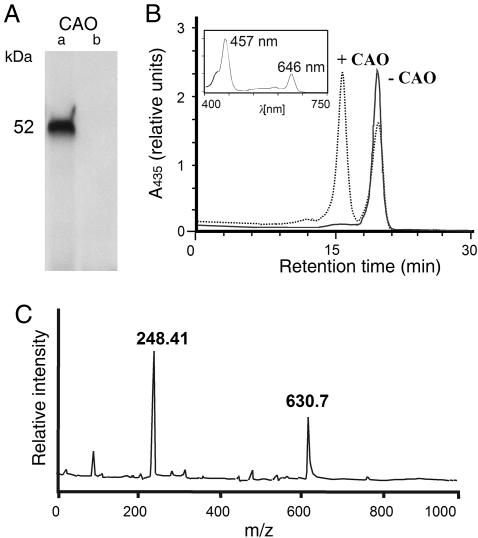
To resolve the issue of substrate specificity of CAO, activity measurements were performed. The A. thaliana At1g44446 gene encodes CAO. A respective cDNA clone was constructed encoding the putative mature CAO protein that lacked the first 36 aa representing the putative chloroplast transit peptide (5). This clone was used for coupled in vitro transcription/translation in a wheat germ lysate and led to the production of a 52-kDa protein (Fig. 1A, lane a). The CAO protein was catalytically active and converted Chlide a to Chlide b in the presence of O2, ferredoxin (Fd), and a Fd-reducing system comprising glucose-6-phosphate, NADPH, glucose-6-phosphate dehydrogenase, and Fd:NADPH oxidoreductase (FNR) (Fig. 1B, dotted line). The identity of Chlide b was established by absorbance measurements of the HPLC-resolved pigment eluting at 16.25 min using a photodiode array detector (Fig. 1B Inset) as well as mass spectrometry (Fig. 1C). When wheat germ extract was programmed with the naked vector DNA lacking the AtCAO insert, neither the CAO protein (Fig. 1A, lane b) nor Chlide b were produced (Fig. 1B, solid line), showing that Chlide a oxygenation to Chlide b is an enzymatic reaction. From the results shown in Fig. 1, we estimate that ≈80% of Chlide a was oxygenated to Chlide b. Analytical tests for substrate specificity revealed that the CAO protein displayed only little activity with Pchlide a (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 1.
AtCAO displays Chlide a oxygenase activity in vitro. (A) SDS/PAGE of 35S-AtCAO (lane a) and lack of 35S-AtCAO in wheat germ extracts programmed with the naked vector DNA missing the AtCAO cDNA insert (lane b). (B) Chlide a oxygenase activity of AtCAO (dotted line) and of the respective control without AtCAO (solid line). The curves show HPLC tracings of acetone-extracted pigments at 435 nm using a photodiode array detector. The pigment eluting at 19.5 min corresponds to Chlide a, whereas the pigment eluting at 16.25 min was identified as Chlide b by absorbance measurements (Inset). (C) Confirmation of the identity of Chlide b eluting at 16.25 min by matrix-assisted laser desorption/ionization mass spectroscopy. The determined molecular mass (630.7 kDa) exactly matched the theoretical isotopic distribution of C35H32N4O6Mg corresponding to Chlide b. The matrix used was terthiophene (molecular mass of 248.4 Da).
A Major Part of CAO Is Localized to the Inner Plastid-Envelope Membrane.
In vitro protein import experiments were conducted to detect the plastid sublocalization of CAO and to test its membrane binding. Wheat germ-translated 35S-CAO precursor (35S-pCAO) was added to isolated, energy-depleted barley or Arabidopsis chloroplasts. Incubations were performed either in the absence or presence of 0.1 or 2 mM Mg-ATP. Whereas binding of 35S-pCAO to the plastids required low, 0.1 mM concentrations of Mg-ATP and was mediated by thermolysin-sensitive receptor components at the outer plastid-envelope membrane, import depended on 2 mM Mg-ATP (Fig. 9A, which is published as supporting information on the PNAS web site). After import and processing, the mature, 52-kDa 35S-CAO attained a thermolysin-resistant but trypsin-sensitive conformation, suggesting that it was largely exposed to the intermembrane space. Whereas thermolysin digests proteins that are exposed to the outer surface of the chloroplast (12), trypsin traverses the outer plastid-envelope membrane and breaks down inner envelope proteins up to their membrane parts (13). Double protease digestions and fractionation experiments showed that 35S-CAO is located both in the inner envelope membrane and thylakoids (Fig. 9 C–E). Salt extraction with either 1 M NaCl or 0.1 M Na2CO3, pH 11 (12), identified 35S-CAO to be tethered to the lipid bilayers of the inner envelope membrane by either one α-helix or β-sheet (Fig. 9D).
CAO Is Not Part of the pPORA Translocon Complex.
In barley and Arabidopsis cotyledons, pPORA import requires Pchlide (10, 14, 15). As a result of the light-induced decline in the level of Pchlide during greening of etiolated barley plants, pPORA accumulates as an envelope-bound import intermediate in planta (16). The precursor is on a productive import pathway from which it can be chased into the plastids in the presence of the Pchlide precursor 5-ALA (16). During membrane passage, it interacts with the components of the Ptc complex and establishes junction complexes between the outer and inner plastid-envelope membranes (10).
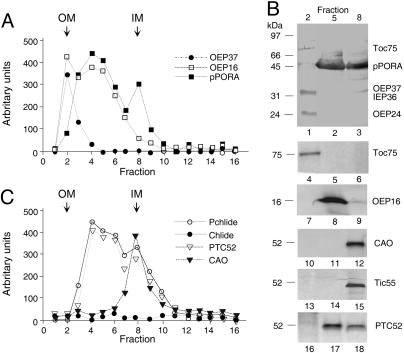
To assess the association of CAO with such junction complexes containing pPORA that would be indicative of redundant roles of CAO and Ptc52, 5-day-old etiolated barley plants were exposed to light for 2 h. Isolated etiochloroplasts bearing pPORA in turn were rapidly disrupted. Crude membranes were fractionated and purified by flotation on a 20–38% sucrose gradient to obtain a light outer membrane fraction (OM), an intermediate density fraction presumably containing pPORA trapped in junction complexes between the outer and inner plastid-envelope membranes (OM–IM), and a slightly denser inner membrane (IM) fraction (17). Immunoblots showed that most of the outer envelope membrane marker proteins Toc75, Oep37, and Oep24 were present in the low density, OM fraction, and most of the inner envelope membrane protein Iep36 was present in the IM fraction (Fig. 2A and B). The intermediate density (OM–IM) fraction contained large amounts of pPORA and therefore consisted of fragments of outer and inner membranes held together by contact sites (17). Significant amounts of precursor were present in the IM fraction (Fig. 2 A and B). The previously identified Oep16 protein, which establishes the translocation pore through which pPORA is imported (18), cofractionated with pPORA in OM–IM junction complexes. In addition, Ptc52 copurified in approximately stoichiometric amounts with pPORA and Pchlide in such complexes, as estimated from the staining intensities on the blots shown in Fig. 2B and the pigment measurements shown in Fig. 2C. No evidence was obtained for an interaction of CAO (Fig. 2 B and C) and PAO (data not shown) with pPORA and Ptc proteins in the OM–IM fraction.
Fig. 2.
CAO is not part of the Ptc complex. (A) Distribution of pPORA, OEP37, and OEP16 in OMs, IMs, and OM–IM junction complexes containing pPORA in barley etiochloroplasts. (B) As in A but showing, in addition, the distribution of Toc75, OEP37, IEP36, and OEP24 in fractions 2, 5, and 8, respectively, on one and the same blot (lanes 1–3) and that of Toc75 (lanes 4–6) and OEP16 (lanes 7–9), as well as CAO (lanes 10–12), Tic55 (lanes 13–15), and Ptc52 (lanes 16–18) on different blots. Each lane contained 10 μg of protein. (C) Pchlide and Chlide levels in relation to Ptc52 and CAO protein levels, respectively, across the gradient.
Identification of Plastid-Envelope Proteins Interacting with CAO During and After Import.
To illuminate the interaction of CAO with other plastid-envelope proteins, chemical crosslinking was carried out by using bacterially expressed and purified pCAO that was derivatized with 125I-labeled N-[4-(p-azidosalicyl-amido)butyl]-3′(2-pyridyldithio)propionamide (125I-APDP) (19, 20). 125I-labeled pCAO was incubated with isolated Arabidopsis chloroplasts either in the absence of added nucleoside triphosphates (used to study energy-independent binding) or in the presence of 0.1 mM Mg-ATP (used to study energy-dependent binding) or 0.1 mM Mg-ATP and 0.1 mM Mg-GTP (used to study the insertion of the precursor into the respective import apparatus). After 15 min in the dark, APDP was activated by exposure to UV light. Chimeric precursor consisting of pSSU of pea and protein A, 125I-APDP-pSSU-protein A (20), and 125I-APDP-pPORB of barley were used as controls. Mixed OM–IM fractions were prepared from lysed chloroplasts and solubilized. Protein was extracted, reduced with DTT, and resolved by SDS/PAGE under reducing conditions. Protein detection was made by autoradiography.
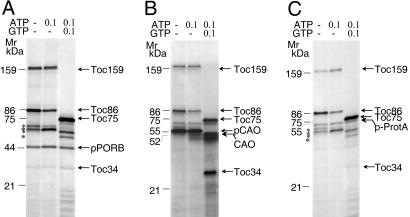
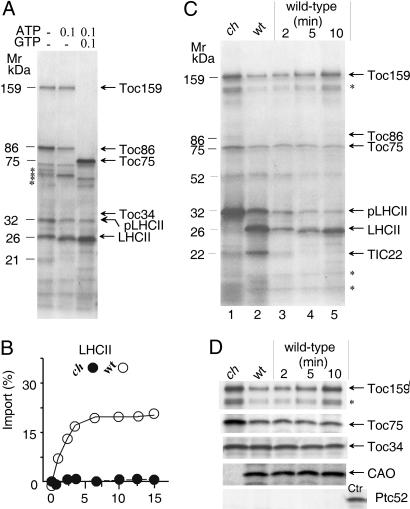
Fig. 3 shows crosslinked products obtained with the different precursors. The results revealed that 125I-APDP-pPORB, 125I-APDP-pCAO, and 125I-APDP-pSSU-protein A produced similar product patterns, comprising major crosslink products of ≈159, ≈86, and ≈75 kDa. The main bands were identified as Toc159 and its major proteolytic product Toc86 as well as Toc75 by immunoprecipitation and protein mass spectrometry (data not shown). In addition, several other bands were obtained, whose labeling intensities were different for the three precursors (Fig. 3). For example, a heavily labeled ≈34-kDa band representing Toc34 was detectable with 125I-APDP-pCAO, but only trace amounts of this band were found for 125I-APDP-pSSU-protein A and 125I-APDP-pPORB (Fig. 3).
Fig. 3.
AtCAO uses the general protein import pathway into chloroplasts. (A–C) Photoaffinity crosslinking of plastid-envelope proteins interacting with 125I-APDP-pPORB (A), 125I-APDP-pCAO (B), and 125I-APDP-pSSU-protein A (C). The identity of the different bands was determined by protein mass spectrometry and immunoprecipitations of the SDS/PAGE-resolved bands (not shown).
To assess whether the imported and processed CAO remained associated with the import apparatus or was sorted out toward other protein complexes, 125I-APDP-derivatized pCAO was imported into isolated Arabidopsis chloroplasts in the presence of 0.1 mM Mg-GTP and 2 mM Mg-ATP. Proteins crosslinked to CAO-(His)6 were purified from isolated, decyl maltoside-solubilized mixed envelope membranes by Ni-NTA agarose chromatography. Proteins adhering to CAO were resolved by SDS/PAGE and blotted onto poly(vinylidene difluoride) membranes, and replicate membranes were used for protein detection by autoradiography, Western blotting, and protein mass spectrometry. Import and crosslinking of 125I-APDP-derivatized pTic55-(His)6 and pPtc52-(His)6 were followed in parallel.
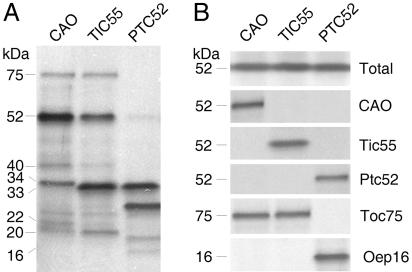
Proteins that were crosslinked after import of pCAO-(His)6, pTic55-(His)6, and pPtc52-(His)6 are indicated in Fig. 4A. Respective Western blots are shown in Fig. 4B. Although CAO and Tic55 likewise crosslinked Toc75, Tic40, and Tic20, they differentially interacted with Tic22 (only found with CAO) and two closely related 33/34-kDa bands. CAO gave rise to an ≈34-kDa band representing Toc34, whereas Tic55 gave rise to a slightly smaller, ≈33-kDa band, which was due to Toc33. Ptc52 crosslinked the same Toc33 band and additionally interacted with Oep16 and a 27-kDa band. No signal was obtained in the 75-kDa range containing Toc75 (Fig. 4 A and B).
Fig. 4.
CAO, Tic55, and Ptc52 establish distinct Tic subcomplexes. (A) 125I-APDP crosslinking of plastid-envelope proteins that interact with CAO, Tic55, and Ptc52, respectively, after import. (B) Western blot of CAO, Tic55, and Ptc52 as well as Toc75 and Oep16 in the different recovered inner envelope membrane complexes. “Total” refers to a blot that was probed with a mixed CAO, Ptc52, and Tic55 antiserum together. Note the differential interaction of CAO and Tic55 with Toc75 and that of Ptc52 with Oep16.
CAO Is Involved in the Regulated Import and Stabilization of Light-Harvesting Chl a/b-Binding Proteins.
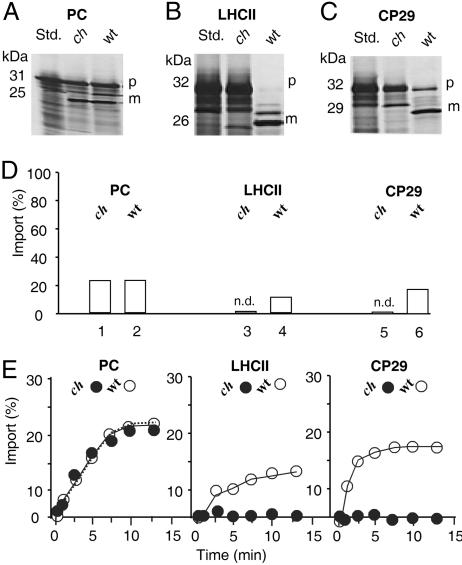
Chlorina mutants (e.g., ch1-3) of Arabidopsis and other plant species and algae lack a functional CAO gene and, for this reason, fail to accumulate Chl b and LHC proteins (2, 3, 21). These mutants seemed ideally suited to perform in vitro import experiments along with in vivo pulse-labeling studies because they would allow direct testing of the involvement of CAO in the import of Chl b-containing LHC proteins. In the first set of experiments, 35S-pLhcb1 (the precursor to the LHCII apoprotein) and 35S-pLhcb4 (the precursor to the pCP29 apoprotein) were synthesized by coupled transcription/translation of respective cDNA clones in a wheat germ lysate and incubated with isolated chloroplasts of wild-type or ch1-3 mutant Arabidopsis plants. Because of a 213-bp deletion that removed part of exon 3, intron 4, and exon 4, and a 5-bp insertion at the site of the deletion, 40 aa are removed from the CAO protein that include most of the mononuclear nonheme Fe-binding site (S334 through T375) and replaced by Val and Ala, respectively (3). As a result, ch1-3 plants lack intact Cao gene and protein. After incubation for 10 min in darkness in the presence of 0.1 mM Mg-GTP and 2 mM Mg-ATP, the chloroplasts were recovered by centrifugation, and the amount of imported and processed precursor, as well as unimported, plastid-bound precursor, was determined. As a control, the plastocyanin precursor was used (35S-pPC).
No difference in import of 35S-pPC by chloroplasts from the two strains could be detected (Fig. 5A and D). Similar amounts of precursor were imported and processed to mature size in wild-type and ch1-3 chloroplasts. By contrast, import of both 35S-pLhcb1 and 35S-pLhcb4 were virtually undetectable with ch1-3 chloroplasts (Fig. 5 B and C; and see Fig. 5D for a quantification of the data). Interestingly, different amounts of precursor were bound to the plastids after their resedimentation (Fig. 5, compare B and C). Some minor bands between the sizes of the precursor and mature proteins may represent nonspecific background signals or partially processed intermediates of pLhcb1 and pLhcb4 that were stuck in the envelopes because of the lack of Chlide b as import trigger. Careful time course analyses confirmed the lack of import of 35S-pLhcb1 and 35S-pLhcb4 as compared with that of 35S-pPC in ch1-3 chloroplasts (Fig. 5E).
Fig. 5.
Ch1-3 mutant chloroplasts are drastically impaired in pLHCII and CP29, but not pPC, import. (A–C) Import of 35S-pPC (A), 35S-pLHCII (B), and 35S-pCP29 (C) into wild-type (wt) and chlorina (ch) 1-3 chloroplasts. P, precursors; m, mature proteins. (D) Quantification of the import data for wild-type and ch1-3 chloroplasts. n.d., not detectable mature LHCII and CP29 protein levels. (E) Time courses of 35S-pPC, 35S-pLHCII, and 35S-pCP29 import into wild-type (open circles) and ch1-3 (filled circles) chloroplasts. Percentages in D and E refer to input radioactivities, set as 100%.
The latter results implied that the lack of import of pLhcb1 and pLhcb4 in ch1-3 chloroplasts may be due to the lack of Chlide b. To further test this hypothesis and to directly demonstrate the interaction of the LHCII precursor with CAO, pLhcb1 was expressed in Escherichia coli as a (His)6-containing precursor, purified on a Ni-NTA agarose column, and incubated in the presence of different Mg-GTP and Mg-ATP concentrations with Arabidopsis chloroplasts isolated from either wild-type or ch1-3 plants. After import, the plastids were collected by centrifugation through Percoll, repurified, and lysed. Plastid-envelope proteins bound to the precursor and/or its processed, mature form were isolated from detergent-solubilized mixed envelope membranes by Ni-NTA agarose chromatography, separated by SDS/PAGE, and detected by Coomassie staining.
In Fig. 6A are shown Coomassie-stained envelope proteins that interacted with pLhcb1 in wild-type chloroplasts at different stages of import. Because of the absence or presence of Mg-ATP and Mg-GTP, pLhcb1 differentially bound Toc159 and its proteolytic fragment Toc86, as well as Toc75. These interactions were consistent with previous observations made for other precursors entering the chloroplast through the general protein import site. According to Kouranov and Schnell (20), these interactions can be referred to as energy-independent and energy-dependent binding as well as insertion of pLhcb1 into the respective import machinery. A fraction of pLhcb1 was present in a processed form (Fig. 6A), suggesting that its NH2-terminal presequence was exposed to the stromal processing peptidase before plastid-envelope re-isolation. Time courses of import performed in the presence of 0.1 mM Mg-GTP and 2 mM Mg-ATP suggested that ch1-3 chloroplasts were unable to import pLhcb1 (Fig. 6B). Consistent with this interpretation were crosslinking studies using pLhcb1-(His)6, which revealed a strong correlation between the lack of CAO and the accumulation of unimported pLhcb1 in ch1-3 versus wild-type chloroplasts (Fig. 6 C and D). No evidence was obtained for an interaction of pLhcb1 and its processed form with Ptc52 (Fig. 6D).
Fig. 6.
pLHCII interacts with CAO during import into wild-type but not ch1-3, chloroplasts. (A) Detection of plastid-envelope proteins interacting with pLhcb1/Lhcb1-(His)6 [pLHCII/LHCII-(His)6] in wild-type chloroplasts at different Mg-GTP and Mg-ATP concentrations. (B) Time course of pLhcb1-(His)6 import into wild-type (open circles) and ch1-3 (filled circles) Arabidopsis chloroplasts. Percentages refer to input radioactivities, set as 100%. (C) Purification of plastid-envelope proteins interacting with pLhcb1-(His)6 at different stages of import into wild-type and ch1-3 mutant chloroplasts. Lanes 1 and 2 show products formed after 15 min, whereas lanes 3–5 show products obtained at the indicated time intervals. (D) Toc159, Toc75, Toc34, CAO, and Ptc52 levels in envelopes of ch1-3 mutant and wild-type chloroplasts. Note the lack of Ptc52 in the recovered envelope complexes but its detection in OM–IM junction complexes containing pPORA used as control (Ctr).
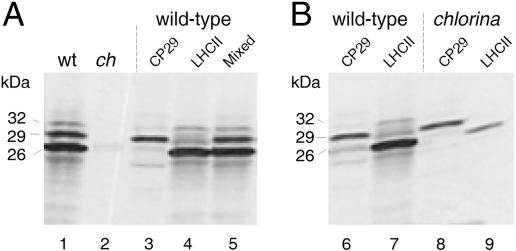
Because isolated envelope membranes were analyzed in the experiment described in Fig. 6, it could not be excluded that the block of pLhcb1 import was not complete and that the coupling of pLhcb1 translocation and CAO-driven Chlide b synthesis was leaky in ch1-3 chloroplasts. For example, Chlide a could replace Chlide b in binding to and triggering pLhcb1 import. To test this hypothesis, pulse-labeling experiments were performed with [35S]methionine in vivo. Leaf tissues from light-grown Arabidopsis wild-type and ch1-3 plants were cut and incubated for 2 h in darkness with the radioisotope. Chloroplasts were isolated on Percoll, and 35S-labeled plastid proteins were extracted with acetone. Lhcb1 and Lhcb4 were resolved electrophoretically and detected by Western blotting or subjected to prior immunoprecipitations by using respective antisera. Lhcb1 and Lhcb4 were below the limit of detection in ch1-3 versus wild-type chloroplasts (Fig. 7, lanes 1 and 2). However, trace amounts of Lhcb1 and Lhcb4 could be immunoprecipitated. These tiny amounts corresponded to ≈0.6% of that amount recovered from wild-type plastids, showing that import of pLhcb1 and pLhcb4 was drastically reduced but not entirely abolished in ch1-3 chloroplasts and that both proteins most likely underwent proteolytic degradation in the absence of Chl(ide) b in planta.
Fig. 7.
The tiny amounts of LHCII and CP29 imported into ch1-3 chloroplasts are degraded. (A) Western blot analysis of Lhcb1 (LHCII) and Lhcb4 (CP29) in wild-type and ch1-3 chloroplasts (lanes 1 and 2) and identification of the various bands by immunoprecipitation with the anti-Lhcb1 (lane 4) and anti-Lhcb4 (lane 3) antisera as well as a mixed antiserum against both proteins (lane 5). (B) Immunoprecipitations of pulse-labeled, mature 35S-Lhcb1 (lanes 7 and 9) and 35S-Lhcb4 (lanes 6 and 8) in chloroplasts prepared from wild-type (lanes 6 and 7) and ch1-3 (lanes 8 and 9) plants. Molecular masses are indicated to the left of the autoradiograms.
Discussion
In the present study, biochemical evidence is provided for a role of CAO in Chlide b synthesis and protein import. We report that CAO is a protein of the inner envelope and thylakoid membranes of Arabidopsis and barley chloroplasts. Protein import and fractionation experiments revealed that CAO is synthesized as a larger precursor that is imported into the intermembrane space via the general protein import machinery comprising Toc159, Toc75, and Toc34. After import and processing, a fraction of the mature CAO accumulated in the inner envelope membrane and remained associated with Toc75 and Toc34. The proteins that interact with thylakoid-associated CAO were not determined in this study. CAO imported into the plastid inner envelope also interacted with Tic40, Tic22, and Tic20 and established a novel Tic subcomplex. This complex was involved in import of Chl(ide) b-binding light-harvesting proteins. Evidence for the existence of distinctive import pathways and respective import machineries in chloroplasts has been provided by genetic and biochemical approaches, highlighting the presence of multiple Toc159- and Toc75-related genes and proteins in Arabidopsis (22, 23). In addition, twin GTP-binding proteins were identified called Toc34 and Toc33 that displayed different expression patterns and precursor specificities in vitro (24, 25). We observed that imported CAO interacted with Toc34, whereas Tic55 and Ptc52 crosslinked Toc33. Except for this difference, the crosslink patterns from CAO and Tic55 were very similar and suggested a modular arrangement of Toc75, Toc33/34, Tic40, and Tic20 as well as Tic55 and CAO. Tic 22, by contrast, interacted only with CAO in the in vitro tests, a result that is at variance with the original report of Kouranov et al. (26) describing the co-isolation of Tic22 and Tic20 in conjunction with pSSU.
Ptc52 generated a crosslink pattern that was distinctive from those obtained for CAO and Tic55. Major products included Toc33 and Oep16. No signal was obtained in the 75-kDa range, consistent with previous results on the roles and interactions of Toc33 and Oep16 in the pPORA translocon, which lacks Toc75 (10, 18, 27). Consistent with these differential interactions of CAO and Ptc52 are their functions in protein import. We found CAO to participate in the chloroplast import of the nucleus-encoded Chl(ide) b-binding light-harvesting proteins pLhcb1 and pLhcb4, whereas Ptc52 operated in the pPORA translocon (10, 27). With isolated chloroplasts of the chl1-3 mutant of Arabidopsis lacking CAO, drastic reductions of import of pLhcb1 and pLhcb4 were observed in vitro and in planta, but no comparable effect was seen for the import of plastocyanin. Bellemare et al. (28) reported that chloroplasts from the chlorina f2 mutant of barley were as capable of importing pLHCP as wild-type chloroplasts. However, the overall amount of imported pLHCII was negligible in either case as compared with that of other precursors. It is not clear whether the prehistory and nature of the precursor and/or source of the plastids (barley versus Arabidopsis) may have affected import. Work is needed to more precisely determine this point.
CAO differs from Tic55, Ptc52, and PAO by its topology and the presence of a large, hydrophilic NH2-terminal domain. Interestingly, this domain contains a binding maquette for Chl(ide) similar to that identified by Eggink and Hoober (29). Yamasato et al. (30) proposed that this motif may provide a sensor for synthesis of Chl(ide) b in chloroplasts and observed that its deletion drastically affected the stability of the enzyme. The NH2 terminus of the CAO from Chlamydomonas also contains a similar Chl(ide)-binding motif (2). Kohorn (31) found that replacement of His in the NH2-terminal Chl-binding motif drastically reduced import of pLhcb1. These findings support a ratchet model of pLhcb1 import in which binding of Chlide a and, more strongly, Chlide b would prevent the precursor from slipping back into the cytosol. Structural data for the crystallized LHCII (32) provoke the hypothesis that the peptidyl carbonyl of Tyr-24 may provide a Chlide b-binding site necessary for Lhcb1 import. Successive binding of Chlide to helix-1 and Tyr-24 thus would favor pLhcb1 import.
We hypothesize that the NH2-terminal pigment sensor domain of CAO may additionally be involved in the recognition of the Chl(ide) b-binding proteins pLhcb1 and pLhcb4. CAO and pLhcb1 copurified in approximately stoichiometric amounts in the envelope of wild-type chloroplasts, and there was good coincidence between the lack of CAO protein and pLhcb1 import in ch1-3 mutant chloroplasts. Because minor traces of mature Lhcb1 and Lhcb4 could nevertheless be detected by pulse-labeling in ch1-3 plants, the coupling between LHC import and CAO-driven Chlide b biosynthesis appears not to be complete. We hypothesize that small amounts of truncated CAO protein lacking the Rieske iron-sulfur center may accumulate in ch1-3 chloroplasts and could provide Chlide a to imported pLhcb1 and pLhcb4 via the NH2-terminal pigment sensor domain. Alternatively, Tic55 could substitute for CAO in ch1-3 mutant chloroplasts. However, because of its unknown substrate specificity and presumed incapacity to bind and oxygenate Chlide a, Tic55 would not be able to functionally replace CAO. As a consequence, imported and processed Lhcb1 and Lhcb4 would be degraded in the stroma. By contrast, Ptc52 was incapable of replacing CAO; otherwise, Chlide b-complexed LHC proteins should have accumulated in ch1-3 chloroplasts, which was not the case. At first glance, this is an unexpected result because etiolated plants accumulate large amounts of Pchlide b that could give rise to Chlide b (33). We hypothesize that Chlide b that cannot be assembled with the nuclear-encoded Chl b-binding proteins during import is converted to Chl(ide) a by virtue of Chlide b reductase (synonymous with 7-formyl reductase) (33). Owing to this mechanism, the risk of photooxidative membrane damage by free, photoexcitable Chlide b would be kept low. In summary, our results provide strong evidence for nonredundant roles of CAO and Ptc52 as part of distinctive protein import complexes in the inner envelope of chloroplasts and highlight the existence of a branched pathway of Chlide b biosynthesis in higher plants.
Materials and Methods
Isolation of Plastid-Envelope Proteins Interacting with CAO.
Chemical crosslinking was performed according to Ma et al. (15) and Kouranov and Schnell (16). Briefly, 125I-APDP-derivatized, hexahistidine [(His)6]-tagged CAO, Tic55, and Ptc52, respectively, were imported into the inner envelope membrane of isolated, energy-depleted Arabidopsis wild-type or ch1-3 chloroplasts in the presence of 2 mM Mg-ATP and 0.1 mM Mg-GTP. After 15 min in darkness, the samples were exposed to UV light. Plastids in turn were lysed, and mixed envelopes were separated on a sucrose gradient. After their solubilization with 1.3% decyl maltoside (6), proteins adhering to CAO, Tic55, and Ptc52, respectively, were purified by Ni-NTA agarose chromatography, eluted, resolved by electrophoresis, and detected by Coomassie staining, Western blotting using respective antisera, and/or autoradiography. Control blots were probed with antisera against Toc75 and Oep16. Import and chemical crosslinking of 125I-APDP-pLhcb1 were performed essentially in the same manner.
Isolation and fractionation of barley etiochloroplasts containing pPORA trapped in junction complexes between the outer and inner envelope membranes is described in refs. 10 and 16.
Measurement of CAO Activity.
Activity measurements were performed with CAO protein that had been expressed from a clone corresponding to At1g44446 in wheat germ lysates or produced in E. coli strain SG13009 (Qiagen). Bacterially expressed precursor was recovered from inclusion bodies by dissolution in 8 M urea containing 20 mM imidazole-HCl, pH 8.0, as described in ref. 18. Final protein preparations were ≈85% pure. Enzyme assays were conducted in 50-μl assays containing the following supplements: 2 μM Chlide a or Pchlide a dissolved in dimethyl sulfoxide, 10 μg of Fd (Sigma), and a Fd-reducing system [2 mM glucose-6-phosphate/1 mM NADPH/50 milliunits of glucose-6-phosphate dehydrogenase/10 milliunits of Fd-NADPH-oxidoreductase (Sigma)] (modified after refs. 5 and 9). In the case of the bacterially expressed and denatured protein, wheat germ extract was added to the assays and the functional protein reconstituted during a preincubation. To this end, the final protein was dissolved in assay buffer to give rise to a final concentration of 1–2 mg/ml. Appropriate aliquots then were withdrawn and diluted into 1 ml of wheat germ extract (1.2 mg/ml protein concentration) such that the protein concentration was 60 μg/ml with respect to the CAO protein. Enzyme assays were carried out at 23°C in darkness. Pigments were then extracted with 100% acetone and separated by HPLC on either C18 reverse-phase columns (Hypersil ODS, Shandon; 5 μm; used for Pchlides a and b) or C30 reverse-phase columns (YMC, Wilmington, NC; 250 × 4.6 mm, 5 μm; used for Chlides a and b) (33). Detection and quantification of pigments were made by absorbance measurements using synthetic standards (33). Mass spectroscopy of HPLC-separated pigments was performed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Voyager-DE STR biospectrometry work station; Applied Biosystems). Before analysis, 10 μg of either Chlide b or Pchlide b was mixed with terthiophene (used as matrix) dissolved in acetone.
Miscellaneous.
Pulse-labeling of proteins with [35S]methionine and immunoprecipitations with respective anti-LHCII and anti-CP29 antisera were carried out according to Scharf and Nover (34). Protein analysis was made by SDS/PAGE and Western blotting using the indicated antisera (10). Protein mass spectrometry was done according to Ferro et al. (35).
Supplementary Material
Acknowledgments
For gifts of cDNA clones and antisera, we thank D. J. Schnell (University of Massachusetts, Amherst), F. Kessler (Université de Neuchātel, Neuchātel, Switzerland), and J. Soll (University of Munich, Munich); and for typing of the manuscript, we thank L. Reinbothe.
Abbreviations
- 125I-APDP
125I-labeled N-[4-(p-azidosalicyl-amido)butyl]-3′(2-pyridyldithio)propionamide
- Fd
ferredoxin
- IM
inner membrane
- OM
outer membrane.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.von Wettstein D., Gough S., Kannangara C. G. Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka A., Ito H., Tanaka R., Tanaka N. K., Yoshida K., Okada K. Proc. Natl. Acad. Sci. USA. 1998;95:12719–12723. doi: 10.1073/pnas.95.21.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Espineda C. E., Linford A. S., Devine D., Brusslan J. A. Proc. Natl. Acad. Sci. USA. 1998;96:10507–10511. doi: 10.1073/pnas.96.18.10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomitani A., Okada K., Miyashi H., Matthijs H. C., Ohno T., Tanaka A. Nature. 1999;400:159–162. doi: 10.1038/22101. [DOI] [PubMed] [Google Scholar]
- 5.Oster U., Tanaka R., Tanaka A., Rüdiger W. Plant J. 2000;21:305–310. doi: 10.1046/j.1365-313x.2000.00672.x. [DOI] [PubMed] [Google Scholar]
- 6.Caliebe A., Grimm R., Kaiser G., Lübeck J., Soll J., Heins L. EMBO J. 1997;16:7342–7350. doi: 10.1093/emboj/16.24.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray J., Wardzala E., Yang M., Reinbothe S., Haller S., Pauli F. Plant Mol. Biol. 2004;54:39–54. doi: 10.1023/B:PLAN.0000028766.61559.4c. [DOI] [PubMed] [Google Scholar]
- 8.Rathinasabapathi B., Burnet M., Russell B. L., Gage D. A., Liao P.-C., Nye G. J., Scott P., Goldbeck J. H., Hanson A. D. Proc. Natl. Acad. Sci. USA. 1997;94:3454–3458. doi: 10.1073/pnas.94.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruzinska A., Tanner G., Anders I., Boca M., Hörtensteiner S. Proc. Natl. Acad. Sci. USA. 2003;100:15259–15264. doi: 10.1073/pnas.2036571100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinbothe S., Quigley F., Gray J., Schemenewitz A., Reinbothe C. Proc. Natl. Acad. Sci. USA. 2004;101:2197–2202. doi: 10.1073/pnas.0307284101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu H., Vavilin D., Vermaas W. Proc. Natl. Acad. Sci. USA. 2001;98:14168–14173. doi: 10.1073/pnas.251530298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cline K., Werner-Washburne M., Andrews J., Keegstra K. Plant Physiol. 1984;75:675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler F., Blobel G. Proc. Natl. Acad. Sci. USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinbothe S., Runge S., Reinbothe C., van Cleve B., Apel K. Plant Cell. 1995;7:161–172. doi: 10.1105/tpc.7.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim C., Apel K. Plant Cell. 2004;16:88–98. doi: 10.1105/tpc.015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinbothe C., Lebedev N., Apel K., Reinbothe S. Proc. Natl. Acad. Sci. USA. 1997;94:8890–8894. doi: 10.1073/pnas.94.16.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell D. J., Kessler F., Blobel G. Science. 1994;266:1007–1012. doi: 10.1126/science.7973649. [DOI] [PubMed] [Google Scholar]
- 18.Reinbothe S., Quigley F., Springer A., Schemenewitz A., Reinbothe C. Proc. Natl. Acad. Sci. USA. 2004;101:2203–2208. doi: 10.1073/pnas.0301962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y., Kouranov A., LaSala S. E., Schnell D. J. J. Cell Biol. 1996;134:315–327. doi: 10.1083/jcb.134.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouranov A., Schnell D. J. J. Cell Biol. 1997;139:1677–1685. doi: 10.1083/jcb.139.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Wettstein D., Kahn A., Nielsen O. F., Gough S. Science. 1974;184:800–802. doi: 10.1126/science.184.4138.800. [DOI] [PubMed] [Google Scholar]
- 22.Bauer J., Hiltbrunner A., Kessler F. Cell. Mol. Life Sci. 2001;58:420–433. doi: 10.1007/PL00000867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin A., Wardle A., Patel R., Dudley P., Park S. K., Twell D., Inoue K., Jarvis P. Plant Physiol. 2005;138:715–733. doi: 10.1104/pp.105.063289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvis P., Chen L.-J., Li H.-m., Peto C. A., Fankhauser C., Chory J. Science. 1998;282:100–103. doi: 10.1126/science.282.5386.100. [DOI] [PubMed] [Google Scholar]
- 25.Gutensohn M., Schulz B., Nicolay P., Flügge U.-I. Plant J. 2000;23:771–783. doi: 10.1046/j.1365-313x.2000.00849.x. [DOI] [PubMed] [Google Scholar]
- 26.Kouranov A., Chen X., Fuks B., Schnell D. J. Cell Biol. 1998;143:991–1002. doi: 10.1083/jcb.143.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinbothe S., Pollmann S., Springer A., Johari James R., Tichtinsky G., Reinbothe C. Plant J. 2005;42:1–12. doi: 10.1111/j.1365-313X.2005.02353.x. [DOI] [PubMed] [Google Scholar]
- 28.Bellemare G., Bartlet S. G., Chua N.-H. J. Biol. Chem. 1982;257:7762–7767. [PubMed] [Google Scholar]
- 29.Eggink L. L., Hoober J. K. J. Biol. Chem. 2000;275:9087–9090. doi: 10.1074/jbc.275.13.9087. [DOI] [PubMed] [Google Scholar]
- 30.Yamasato A., Nagata N., Tanaka R., Tanaka A. Plant Cell. 2005;17:1585–1597. doi: 10.1105/tpc.105.031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohorn B. Plant Physiol. 1990;93:339–342. doi: 10.1104/pp.93.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 33.Reinbothe S., Pollmann S., Reinbothe C. J. Biol. Chem. 2003;278:800–806. doi: 10.1074/jbc.M209737200. [DOI] [PubMed] [Google Scholar]
- 34.Scharf K.-D., Nover L. Cell. 1982;30:427–437. doi: 10.1016/0092-8674(82)90240-9. [DOI] [PubMed] [Google Scholar]
- 35.Ferro M., Salvi D., Brugière S., Miras S., Kowalski S., Louwagie M., Garin J., Joyard J., Rolland N. Mol. Cell. Proteomics. 2003;25:325–345. doi: 10.1074/mcp.M300030-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.