Abstract
Background
Gastrointestinal nematodes constitute a major cause of morbidity and mortality in grazing ruminants. Individual animals or breeds, however, are known to differ in their resistance to infection. Gene expression profiling allows us to examine large numbers of transcripts simultaneously in order to identify those transcripts that contribute to an animal's susceptibility or resistance.
Results
With the goal of identifying genes with a differential pattern of expression between sheep genetically resistant and susceptible to gastrointestinal nematodes, a 20,000 spot ovine cDNA microarray was constructed. This array was used to interrogate the expression of 9,238 known genes in duodenum tissue of four resistant and four susceptible female lambs. Naïve animals were used in order to look at genes that were differentially expressed in the absence of infection with gastrointestinal nematodes. Forty one unique known genes were identified that were differentially expressed between the resistant and susceptible animals. Northern blotting of a selection of the genes confirmed differential expression. The differentially expressed genes had a variety of functions, although many genes relating to the stress response and response to stimulus were more highly expressed in the susceptible animals.
Conclusion
We have constructed the first reported ovine microarray and used this array to examine gene expression in lambs genetically resistant and susceptible to gastrointestinal nematode infection. This study indicates that susceptible animals appear to be generating a hyper-sensitive immune response to non-nematode challenges. The gastrointestinal tract of susceptible animals is therefore under stress and compromised even in the absence of gastrointestinal nematodes. These factors may contribute to the genetic susceptibility of these animals.
Background
Grazing ruminants are constantly exposed to natural challenge by gastrointestinal nematodes. Infection by such parasites leads to clinical disease and production losses and is a serious problem in animal husbandry. The economically important gastrointestinal parasites of sheep belong to the Order Strongylida and the family Trichostrongyloidea and include Teladorsagia (Ostertagia), Trichostrongylus, Nematodirus and Haemonchus spp. The host response to parasite challenge is complex and poorly understood due to its polygenic nature. The response varies from sheep breed to sheep breed and from animal to animal [1]. The response can result in rapid or delayed expulsion of parasites and the host response is critical for determining subsequent parasite status [2]. Resistance to nematodes is primarily due to acquired immunity and is largely a Th2 type response [3-8] although innate immunity is also thought to play a role [9]. Sheep lines which have been selected to be resistant to one particular nematode species have been shown to have increased resistance to other nematode species [10-13] primarily due to a non-specific mechanism of parasite expulsion [2]. Historically anthelmintic drenching has been used to control nematode infection, however, the efficacy of this treatment is decreasing as parasite resistance to anthelmintics increases and nematodes resistant to multiple classes of anthlemintics are now found worldwide [14]. The use of chemicals in food production is also increasingly encountering public hostility [15].
An alternative method of helminth control is vaccination of host animals. This strategy has successfully combated many bacterial and viral diseases, however, despite extensive research there is currently no vaccine available against the major gastrointestinal parasitic nematode species [15-17]. This is most likely due to the number of nematode species infecting the host and the complex parasite life cycle which expresses different antigens at each stage.
A natural method of parasite control is breeding for host resistance. Resistance to internal parasites, as measured by faecal egg count (FEC), has moderate heritability (h2 = 0.23–0.41, [1]) and this method has been shown to make significant genetic gains in a variety of sheep breeds [18-24] due to the combination of reduced FEC and reduced pasture contamination. The current method of animal selection is cumbersome and inefficient, however, as animals are selected based on their phenotype. Genetic gain could be accelerated if animals were selected on genotype rather than phenotype.
Lines of Perendale sheep have been divergently selected for parasite resistance and susceptibility at AgResearch, New Zealand since 1986 and differ in FEC by 4.9 fold [24]. The number of adult nematodes in the gastrointestinal tract of animals from the resistant line is also lower than that from the susceptible line [25]. Resistant animals therefore have an increased genetic capability to respond to and subsequently reject parasites when challenged. The identification of genes involved in this process would allow the development of genetic markers, which could be used in marker assisted selection breeding programs. A previous study identified genes differentially expressed between the selection lines in response to parasite challenge, but the use of challenged animals confounded the resistance status of the animals with level of infection. This study also did not distinguish innate from acquired immunity [25]. We therefore chose to look at those genes that were differentially expressed between the selection lines regardless of the level of infection by using naïve animals that had never been exposed to gastrointestinal nematodes. We did this using high-throughput DNA microarray technology. This allowed us to examine the relative expression of thousands of genes in a single experiment and to group genes into expression classes, providing insight into their biological function.
Results
The microarray experiment examined gene expression in duodenum tissue from four resistant and four susceptible naïve Perendale lambs. Poly(A)+ RNA isolated from duodenum tissue was reverse transcribed into cDNA, fluorescently labelled and hybridised to ovine 20 k cDNA microarrays. The experiment design was a factorial dye swap design involving 16 slides, where every animal was compared to every animal in the opposite selection line, as described previously [25]. One hundred ESTs showed differential expression (Figure 1) between the resistant and susceptible animals. Forty ESTs were more highly expressed in the resistant line while 60 were more highly expressed in the susceptible line. These ESTs were resequenced in order to verify the insert sequence. Confirmed ESTs were annotated using BLASTN against the human RefSeq database and a cut-off E value of 1 × 10-18. Thirty of the ESTs more highly expressed in susceptible animals could be verified and annotated in this manner giving 25 unique human RefSeqs. Sixteen of the ESTs more highly expressed in resistant animals could also be annotated. The lists of differentially expressed human RefSeqs is given in Tables 1 and 2.
Figure 1.
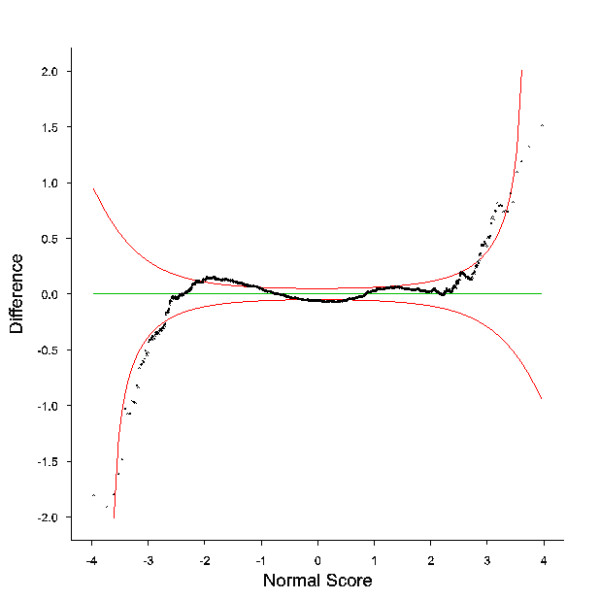
Differenced Normal Q-Q plot for the modified T value of the log ratio of the mean. The expected normal deviate (normal score) is plotted against the difference between the observed order statistic and the expected normal order statistic (95% confidence limits shown in red). Data is combined from all 16 slides of the experiment. ESTs more highly expressed in resistant animals are shown on the right while ESTs more highly expressed in susceptible animals are shown on the left.
Table 1.
Genes more highly expressed in the duodenum of genetically susceptible animals compared to resistant
|
Human Refseq: Genbank accession number |
Gene description | Gene name |
Ensembl human gene Cytogenetic band |
Contig BLAST E value |
Fold increase in expression |
P value |
| NM_003226* | Intestinal trefoil factor 3 | TFF3 | 21q22.3 | 3 × 10-59 | 1.2 | 7.7 × 10-20 |
| NM_006398* | Ubiquitin D | UBD | 6p22.1 | 1 × 10-52 | 1.3 | 1.8 × 10-12 |
| NM_003122* | Serine protease inhibitor, Kazal type 1 | SPINK1 | 5q32 | 2 × 10-58 | 1.3 | 5.5 × 10-10 |
| NM_001009555 | SH3 domain protein D19 | EVE1 | 4q31.3 | 0 | 1.8 | 3 × 10-9 |
| NM_004295 | TNF receptor-associated factor 4 | TRAF4 | 17q11.2 | 0 | 1.1 | 1.3 × 10-8 |
| NM_022342 | Kinesin family member 9 | KIF9 | 3p21.31 | 0 | 1.4 | 1.8 × 10-7 |
| NM_005646 | TAR (HIV) RNA binding protein 1 | TARBP1 | 1q42.2 | 0 | 1.2 | 2 × 10-7 |
| NM_033503 | Bcl2 modifying factor | BMF | 15q15.1 | 1 × 10-18 | 1.2 | 3.7 × 10-7 |
| NM_003392 | Wingless-type MMTV integration site family, member 5A | WNT5A | 3p14.3 | 6 × 10-58 | 1.2 | 4.1 × 10-7 |
| NM_000179 | MutS homologue 6 | MSH6 | 2p16.3 | 0 | 1.2 | 8 × 10-7 |
| NM_001655 | Archain 1 | ARCN1 | 11q23.3 | 0 | 1.2 | 1.4 × 10-6 |
| NM_079423 | Myosin light polypeptide 6 | MYL6 | 12q13.2 | 0 | 1.1 | 1.9 × 10-6 |
| NM_006184 | Nucleobindin 1 | NUCB1 | 19q13.33 | 0 | 1.1 | 3.4 × 10-6 |
| NM_178156 | Fucosyltransferase 8 | FUT8 | 14q23.3 | 0 | 1.3 | 4.5 × 10-6 |
| NM_005533 | Interferon induced protein 35 | IFI35 | 17q21.31 | 3 × 10-77 | 1.1 | 6.7 × 10-6 |
| NM_000062 | Serine (or cysteine) proteinase inhibitor, clade G | SERPING1 | 11q12.1 | 2 × 10-50 | 1.2 | 9 × 10-6 |
| NM_002133 | Decycling heme oxygenase 1 | HMOX1 | 22q12.3 | 0 | 1.1 | 1.1 × 10-5 |
| NM_002816 | Proteasome 26S subunit | PSMD12 | 17q24.2 | 0 | 1.2 | 1.1 × 10-5 |
| NM_002116 | Major histocompatibility complex, class I, A | HLA-A | 6p21.33 | 8 × 10-35 | 1.1 | 1.2 × 10-5 |
| NM_013442 | Stomatin-like 2 | STOML2 | 9p13.3 | 0 | 1.2 | 1.2 × 10-5 |
| NM_201265 | Bone marrow stromal cell-derived ubiquitin-like 7 | BMSC-UbP | 15q24.1 | 0 | 1.3 | 1.5 × 10-5 |
| NM_016248 | A kinase anchor protein 11 | AKAP11 | 13q14.11 | 1 × 10-44 | 1.2 | 1.8 × 10-5 |
| NM_006808 | Sec61 beta subunit | SEC61B | 9q22.33 | 1 × 10-138 | 1.1 | 1.9 × 10-5 |
| NM_032492 | Jagunal homologue 1 | JAGN1 | 3p25.3 | 1 × 10-180 | 1.1 | 2.8 × 10-5 |
| NM_002083 | Glutathione peroxidase 2 | GPX2 | 14q23.3 | 0 | 1.2 | 3.3 × 10-5 |
* More than one EST corresponding to this gene was differentially expressed and their probabilities were combined.
Table 2.
Genes more highly expressed in the duodenum of genetically resistant animals compared to susceptible
|
Human RefSeq: Genbank accession number |
Gene description | Gene name |
Ensembl human gene cytogenetic band |
Contig BLAST E value |
Fold increase in expression |
P value |
| NM_003789 | TNFRSF1A-associated via death domain | TRADD | 16q22.1 | 1 × 10-136 | 1.3 | 2.9 × 10-8 |
| NM_000988 | Ribosomal protein L27 | RPL27 | 17q21.31 | 4 × 10-98 | 1.2 | 5.9 × 10-7 |
| NM_007209 | Ribosomal protein L35 | RPL35 | 9q33.3 | 1 × 10-141 | 1.2 | 2.4 × 10-6 |
| NM_014716 | Centaurin, beta 1 | CENTB1 | 17p13.1 | 7 × 10-62 | 1.3 | 2.9 × 10-6 |
| NM_000477 | Albumin | ALB | 4q13.3 | 1 × 10-151 | 1.2 | 3.4 × 10-6 |
| NM_175571 | IMAP family member 8, GTPase | GIMAP8 | 7q36.1 | 2 × 10-22 | 1.2 | 4.2 × 10-6 |
| NM_006118 | HS1 binding protein | HAX1 | 1q21.3 | 0 | 1.2 | 4.6 × 10-6 |
| NM_005801 | Putative translation initiation factor | SUI1 | 17q21.2 | 0 | 1.1 | 4.7 × 10-6 |
| NM_182752 | Family with sequence similarity 79, member A | FAM79A | 1p36.32 | 1 × 10-117 | 1.2 | 5.3 × 10-6 |
| NM_006827 | Transmembrane trafficking protein | TMP21 | 14q24.3 | 4 × 10-19 | 1.1 | 7.7 × 10-6 |
| NM_001107 | Erythrocyte acylphosphatase 1 | ACYP1 | 14q24.3 | 1 × 10-114 | 1.2 | 9.2 × 10-6 |
| NM_000211 | Integrin, beta 2 | ITGB2 | 21q22.3 | 0 | 1.2 | 1 × 10-5 |
| NM_000613 | Hemopexin | HPX | 11p15.4 | 1 × 10-121 | 1.2 | 1.5 × 10-5 |
| NM_145869 | Annexin A11 | ANXA11 | 10q22.3 | 1 × 10-122 | 1.2 | 2 × 10-5 |
| NM_002872 | Small GTP binding rho family protein | RAC2 | 22q13.1 | 0 | 1.2 | 2 × 10-5 |
| NM_033657 | Death associated protein 3 | DAP3 | 1q22 | 0 | 1.1 | 2.5 × 10-5 |
Northern blotting was carried out to confirm differential expression of a number of genes (Figure 2). TFF3 encodes intestinal trefoil factor 3. This gene is a member of a family of trefoil factors that is involved in mucosal maintenance and repair and is known to be overexpressed during inflammatory processes [26]. In accordance with the microarray results, this gene had a 20% increase in expression in the duodenum of the susceptible animals compared to the resistant (Figure 2A). While the degree of upregulation was not marked it was still significant (P = 0.05).
Figure 2.
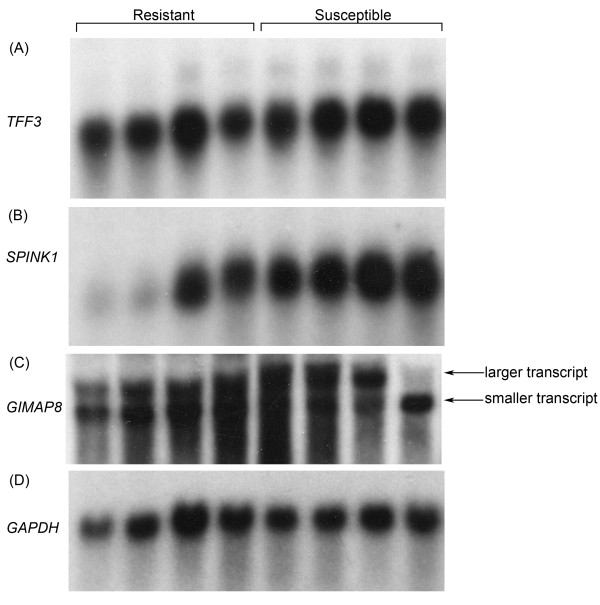
Northern blotting. Northern blots showing differential expression of TFF3 (A), SPINK1 (B), and GIMAP8 (C). mRNA levels were normalised to that of GAPDH (D).
Differential expression of the pancreatic secretory trypsin inhibitor gene (SPINK1) was also confirmed. SPINK1 encodes a gene also known as TATI (tumour associated trypsin inhibitor) that is expressed mainly in the pancreas but is also expressed in the mucosa of the small intestine [27]. One of the major roles of SPINK1 is thought to be the prevention of premature activation of pancreatic proteases. This has the effect of decreasing the rate of mucus digestion by luminal proteases within the stomach and colon. SPINK1 is also known to increase the proliferation of a variety of cell lines and to stimulate cell migration, implying that it may be involved in the healing response following injury [28]. When measured by Northern blot, SPINK1 expression was increased two fold in the susceptible animals compared to the resistant animals (Figure 2B) and this increased expression was highly significant (P = 0.005). In particular two of the resistant animals had very low levels of SPINK1 expression although expression in the other two resistant animals was still lower than expression in any of the susceptible animals.
Differential expression of the immunity associated GTPase, GIMAP8, was also confirmed. This gene is a member of a novel family of GTPases conserved among higher plants and vertebrates [29]. Human GIMAP proteins are known to be expressed most highly in the spleen and lymph nodes, but expression has also been detected in the digestive tract [30]. GIMAP proteins are thought to be involved in the control of cell survival and response to infection and GIMAP8 has been shown to have anti-apoptotic functions. Expression of this gene is known to be decreased in the spleen of mice infected with the protozoan parasite Plasmodium chabaudi [29]. GIMAP8 was expressed 2.6 fold more highly in the intestine of resistant animals compared to susceptible (Figure 2C) as measured by Northern blot. This differential expression is also highly significant (P = 0.0018). It is notable that the fold increase in expression of both GIMAP8 and SPINK1 is markedly higher when quantitated by Northern blot rather than when quantitated by microarray. Differential transcript expression of GIMAP8 was also observed with the four resistant animals predominantly expressing an approximately 3,300 bp transcript, although a larger transcript of approximately 4,100 bp could still be detected. In contrast three of the susceptible animals predominantly expressed the larger transcript although the smaller transcript was detectable, while one susceptible animal expressed the smaller transcript. The precise size of the GIMAP8 transcript is unknown in sheep. Transcript variants of this gene, with sizes of 4,700 bp and 4,200 bp, have been previously reported in human and transcript variants of size 4,200 bp and 2,900 bp have been reported in mouse [29,30].
Gene Ontology (GO) terms significantly associated with the differentially expressed genes were found using EASEonline [31] and are listed in Table 3. A number of GO terms were significantly associated (P < 0.05) with the genes more highly expressed in the intestine of susceptible animals. These terms pertain to an organism's response to stress and stimulus and imply that despite the absence of nematodes from the environment of the susceptible animals, their gastrointestinal tract is still responding to insults or injuries, possibly by other pathogens. The only GO term significantly associated with genes more highly expressed in resistant animals was "cellular process" indicating that the intestine of these animals does not appear to be under stress.
Table 3.
GO terms significantly associated with the differentially expressed genes
| GO term | No. of RefSeqs associated with term | Fisher exact score | |
| Susceptible | Response to biotic stimulus | 6 | 0.00012 |
| Response to stimulus | 7 | 0.00028 | |
| Response to external stimulus | 6 | 0.00067 | |
| Defense response | 5 | 0.00072 | |
| Response to stress | 5 | 0.00124 | |
| Organismal physiological process | 5 | 0.00425 | |
| Immune response | 4 | 0.00486 | |
| Response to pest/pathogen/parasite | 3 | 0.00956 | |
| Cytoskeleton | 3 | 0.0397 | |
| Resistant | |||
| Cellular Process | 9 | 0.0426 |
Promoter regions of the 16 genes more highly expressed in resistant animals and the 25 genes more highly expressed in susceptible animals were analysed for common cis-regulatory motifs. The analysis was performed using the MEME motif prediction program [32]. Only two motifs were significant and both were found in the promoter regions of the genes more highly expressed in susceptible animals. The sequence logo for these motifs is shown in Figure 3. MAST analysis indicated that despite individual motifs not being significant, the combination of motifs identified in promoter regions of both groups were unique to the group in which they were discovered as they were not significantly associated with the opposing group or with all the genes on the array by Fisher's exact test (Table 4). A separate MAST analysis indicated that the motifs identified in this study were also not significantly associated (P > 0.05) with genes previously reported to be differentially expressed in sheep exposed to gastrointestinal nematodes [25] (data not shown).
Figure 3.
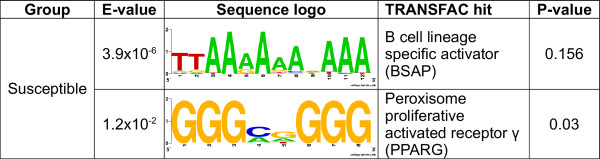
Significant motifs detected in the promoter regions of the differentially expressed genes. The significant motifs found in the promoters of the differentially expressed genes are given along with their sequence logos and top TRANSFAC hit.
Table 4.
MAST results
| Resistant (16) | Susceptible (25) | Array (8809) | Fisher exact score | |
| Proportion of genes with significant combination of resistant motifs | 0.50 | 0.00 | 0.05 | 4.7 × 10-7 |
| Proportion of genes with significant combination of susceptible motifs | 0.06 | 0.44 | 0.08 | 4.6 × 10-6 |
Proportion of promoter regions which contain the combination of motifs detected by MEME with P < 0.0001. Numbers in parenthesis represent the total number of genes in each group. In both cases the combination of motifs is found more frequently in the group in which it was originally identified. The significance of this is given by Fisher's exact score.
In order to identify which transcription factors could bind to the motifs, each motif was compared to the transcription factor binding sites in the TRANSFAC database [33]. Each motif was shuffled 1,000 times and the comparison process repeated in order to establish its significance. Figure 3 lists the significant motifs discovered along with their best TRANSFAC hits. The full list of identified motifs and their TRANSFAC hits are available in Additional files 1 and 2. The two significant motifs found in the promoter regions of the genes more highly expressed in susceptible animals give B cell lineage specific activator protein (BSAP) and Peroxisome proliferative activated receptor gamma (PPARG) as their best hit. BSAP, also known as PAX5 is a transcription factor required for B cell development [34], however, it should be noted that the P value of this hit is not significant and so this motif may be bound by a transcription factor not represented in the TRANSFAC dataset. Interestingly, five of the six genes associated with the GO term "response to biotic stimulus" have this motif in their promoter regions, indicating this motif may be involved in the regulation of this response. Peroxisome proliferative activated receptor gamma (PPARG) is a ligand activated transcription factor which has an important role in adipocyte differentiation [35]. PPARG is known to have anti-inflammatory effects and to play an important role in the maintenance of mucosal integrity in the intestine [36]. This transcription factor may therefore play a role in co-ordinately regulating genes more highly expressed in the intestine of the susceptible animals.
Discussion
This study identified a number of genes differentially expressed between lines of lambs differing genetically in their ability to become resistant to gastrointestinal nematodes. These genes were differentially expressed in the absence of nematode challenge and so represent basal expression differences between the selection lines. It is noteworthy that significant differences were observed in naïve animals as much previous work has shown that animals differing in host resistance primarily differ in the rapidity and strength of their acquired immune response rather than differing in their innate immunity [3,5,6]. To our knowledge this is the first report of constitutive differences in gene expression in naïve sheep differing in their genetic ability to respond to host infection.
Gene Ontology terms associated with the genes more highly expressed in susceptible animals included "response to stimulus", "response to stress", "defence response" and "response to pests, pathogens and parasites". This implies that the gastrointestinal tract of the susceptible animals is responding to stress even in the absence of nematode challenge. This is evidenced by the elevated expression of seven genes that are induced in response to stimulus, HLA-A, MSH6, GPX2, IFI35, UBD, SERPING1 and TFF3. HLA-A encodes an MHC class I heavy chain molecule that, in conjunction with B2M, presents endogenously derived peptides to CD8+ cytotoxic T cells. HLA-A has broad tissue expression and has been shown to be induced by a number of cytokines, in response to infection and in patients with the inflammatory bowel syndrome Crohn's disease [37-39]. MSH6 encodes a MutS homologue protein which, in complex with MSH2, forms the MutSα heterodimer, while MSH2 in complex with MSH3 forms the MutSβ heterodimer. Both these complexes are involved in mismatch repair and repair mutagen-induced lesions in DNA as well as errors in DNA replication [40]. MutSα repairs both DNA mismatches and short 1–2 bp insertions or deletions (indels) while MutSβ repairs longer 2–6 bp indels [41]. A change in expression of either MSH6 or MSH3 can subsequently lead to a change in the MutSα to MutSβ ratio. The MutSα complex is induced at the transcriptional level in response to radiation [42]. Expression of MSH6 may be higher in the duodenum of susceptible animals in order to assist these animals to cope with toxic and mutagenic insults present in ingested food. GPX2 is another stress response gene more highly expressed in the intestine of the susceptible animals. This gene encodes the selenoprotein, gastrointestinal glutathione peroxidase 2, which catalyses the reduction of peroxides by reduced glutathione and protects the cell against oxidative damage. This gene is known to be induced in response to oxidative stress [43]. The interferon inducible gene IFI35 is induced at the transcriptional level in response to interferons, and complexes with the N-Myc interacting protein, NMI, to form a high molecular weight cytosolic complex [44]. The precise function of this complex, however, remains unknown. UBD encodes a small ubiquitin-like modifier protein also known as FAT10. UBD is induced by the cytokines IFNγ and TNFα [45] and can bind proteins and target them for degradation by the proteasome in a cytokine inducible, irreversible, ubiquitin-independent manner [46]. SERPING1, also known as C1 inhibitor (C1-INH), encodes an IFNγ inducible [47] serine protease inhibitor of the complement and contact systems. This protein therefore exhibits an anti-inflammatory effect [48,49]. SERPING1 is known to help prevent endotoxic shock [50] and is under investigation as a clinical treatment for a variety of diseases [51]. TFF3 encodes intestinal trefoil factor 3. Trefoil factors are involved in mucosal protection and healing and are induced during inflammation and in response to gastrointestinal mucosa damage [52]. The increased expression of so many stress response genes in the intestine of the susceptible animals appears to indicate that these animals are responding to gut insult and inflammation. Alternatively, these animals may constitutively express these stress response genes at inappropriate levels in the absence of challenge. Interestingly, stress response genes did not appear to be more highly expressed in the duodenum of susceptible animals after natural challenge with nematodes [25] indicating that these genes may be inappropriately regulated in the gastrointestinal tract of susceptible animals.
The source of stress to the intestinal tract of the susceptible animals remains unknown. Some of the stress response genes more highly expressed in susceptible animals are known to play a role in protecting the cell against oxidative damage. MSH6 is involved in repairing DNA lesions caused by oxidation [53] while GPX2 reduces DNA damaging agents [54]. HMOX1 is also strongly induced by oxidative stress and is involved in heme degradation [55]. Chronic gut inflammation is associated with enhanced production of leukocyte derived oxidants [56]. Therefore the gastrointestinal tract of the susceptible animals may be suffering damage from reactive oxygen species. This could come from a number of sources such as aberrant cellular metabolism or phagocytic leukocytes responding to an infection. Indeed some of the stress proteins more highly expressed in the gastrointestinal tract of the susceptible animals are involved in the immune response and are inducible by pro-inflammatory cytokines. Therefore although free from gastrointestinal nematodes, the susceptible animals may be responding to infection by other viral or bacterial pathogens.
A number of pro-apoptotic genes are more highly expressed in the intestine of the susceptible animals. These genes promote apoptosis in response to stress. Apoptosis is a primary form of defence against infection, stress, damage or injury in the cell. MSH6, TRAF4, UBD and BMF can all induce a protective apoptotic response in cells [45,57-59]. The soluable form of HLA-A has also been shown to trigger apoptosis of CD8+ T cells [60]. Apoptosis can be induced directly by DNA damage or by cytokine signalling.
The genes more highly expressed in intestinal tissue of susceptible animals also included a number that encode proteins involved in protein degradation: these are PSMD12, which encodes a non-ATPase component of the 26S proteasome subunit, UBD which encodes a ubiquitin-like protein that targets proteins for degradation and BMSC-UbP which encodes a bone marrow stromal cell derived ubiquitin-like protein [46,61,62]. Another gene which can play a role in protein degradation is SEC61B, which encodes the beta subunit of the Sec61 protein translocator, which transports proteins across the endoplasmic reticulum. This complex is also known to play a role in retrograde transport of misfolded or degraded proteins back into the cytoplasm for proteasome degradation [63]. Therefore there appears to be increased protein degradation in the intestinal tract of the susceptible animals.
In summary, a number of stress response genes appear to be induced in the intestinal tract of naïve susceptible animals. How the induction of these genes is regulated is unclear at present, however, the identification of two significant motifs in the promoter regions of these genes indicated that transcription may be co-ordinately regulated by the BSAP or PPARG transcription factors. The role of PPARG in regulating genes involved in nematode susceptibility could potentially be examined by treating animals with PPARG agonists in order to modulate PPARG activation. Motifs for BSAP or PPARG binding were not found in the promoter region of genes whose expression was not elevated in the susceptible animals, showing they are unique to these genes.
Only 16 genes were more highly expressed in the intestine of the resistant animals than the susceptible animals. These genes had a variety of biological functions and the only GO term significantly associated with these genes was "cellular process". However two of genes more highly expressed in the resistant animals are vital for maintaining a functioning and healthy immune system. RAC2 plays an important role in response to pathogens as it regulates neutrophil chemotaxis and superoxide production and deficiency of RAC2 leads to impaired host defences and neutrophilia [64,65]. Deficiency of ITGB2, also known as CD18, can also lead to neutrophilia [66] and ITGB2 is also known to be important in innate immunity [67]. Two apoptosis inducing genes are more highly expressed in the intestine of the resistant animals. These genes are DAP3 and TRADD. DAP3, a proposed nucleotide binding protein, is a major positive regulator of apoptosis and has been shown to be critical for anoikis [68]. TRADD encodes a tumour necrosis factor receptor adaptor protein. This protein links the TNF receptor to the caspase pathway initiating apoptosis [69]. Despite a few genes with similar functions being more highly expressed in the intestine of resistant animals, no significant motifs were identified in the promoter regions of these genes indicating that they may not be co-ordinately regulated at a transcriptional level. Alternatively, the small number of genes with significantly elevated expression provided little power to detect motifs responsible for co-ordinate regulation.
A previous study identified genes differentially expressed between the resistant and susceptible lines in response to nematode challenge [25]. Interestingly, there were no genes that were consistently differentially expressed both pre and post-infection. However, the ubiquitin-like modifier gene, UBD, was more highly expressed in the susceptible animals prior to infection but more highly expressed in resistant animals after infection with gastrointestinal nematodes. UBD expression is induced by the cytokines IFNγ and TNFα. Expression of TNFα is known to be induced in intestinal lymph of genetically resistant Romney sheep during primary infection of naïve animals with T. colubriformis [4]. Therefore, while UBD is expressed more highly in naïve susceptible animals, upon infection with nematodes the resistant animals may induce UBD expression to a higher level than the susceptible animals in a TNFα-dependent manner. The previous study also identified smooth muscle function and Major Histocompatibility Complex II expression as important mediators of parasite resistance [25]. The current study did not identify smooth muscle or MHC II genes as differentially expressed between the lines in the absence of challenge and so shows that these genes are induced in response to challenge. Resistance to gastrointestinal nematodes may therefore be mediated, in part, by the ability to induce expression of these genes and this response is generated upon exposure to infection.
Conclusion
Despite divergent selection over many years relatively few genes were differentially expressed between the selection lines in the absence of nematode challenge. Many more genes were found to be differentially expressed between the lines in response to natural parasite challenge [25] however, the genes identified in the present study may contribute to an animals innate resistance or susceptibility. The genes more highly expressed in resistant animals had a variety of functions, but some were involved in maintaining a healthy immune system, while some were pro-apoptotic genes. A number of genes more highly expressed in the susceptible animals were related to cellular response to stress and infection indicating that the susceptible animals may have a compromised gastrointestinal tract, even in the absence of nematode infection, and this may contribute to their innate susceptibility. It is notable that the human orthologs of the differentially expressed genes are located on a number of different human chromosomes (Tables 1 and 2). It is not known whether the polymorphisms that give rise to the differences in expression reported here are due to cis or trans mutations. Although it is possible that a mutation in one or a small number of genes may give rise to the expression differences, the diversity of the actions and pathways of the genes differentially expressed is consistent with genetic resistance to nematodes being due to many genes with small effects rather than a mutation in a single locus. The recent release of the bovine genome [70], a closely related species, will greatly aid mapping and resequencing of sheep genes responsible for parasite resistance.
Methods
Selection lines and tissue collection
Resistant and susceptible lines of Perendale sheep have been selected based on faecal egg count (FEC) since 1986. These lines now differ in faecal egg count by 4.9 fold [24]. Pregnant ewes were adjusted to concentrate feed, treated with anthelmintic, brought indoors and subsequently lambed indoors. The lambs were raised indoors to insure their immune system remained naïve with respect to gastrointestinal nematodes. During this period the ewes and lambs had access to commercially formulated sheep nuts and chopped Lucerne hay ad libitum. All animals were faecal sampled periodically to ensure no parasitic nematodes were present. Four female lambs per selection line were chosen and at an average age of 84 days (standard deviation, 6.8 days) these animals were sacrificed and duodenum tissue promptly collected. The tissue was frozen in liquid nitrogen and stored at -80°C. No adult nematodes could be detected in the abomasums or intestines of the animals confirming their naïve status. The average live weight of the lambs pre-slaughter was 23.4 kg (standard deviation, 3.8 kg). No significant differences were observed between the weights of the animals in the two lines at any stage. All procedures were approved by the AgResearch Invermay Animal Ethics Committee, formally constituted under the New Zealand Animal Welfare Act.
RNA preparation
Total RNA was isolated from the duodenum of each animal using TRIzol (Invitrogen) and was cleaned using an RNeasy kit (Qiagen). RNA integrity was confirmed by denaturing agarose gel electrophoresis and RNA was quantitated using a NanoDrop® ND-1000 Spectrophotomoter (NanoDrop Technologies). First-strand cDNA was made from 25 μg of total RNA by anchored oligo(dT)20-primed reverse transcription incorporating amino-modified dNTPs, and was subsequently labelled indirectly by fluorescent coupling of Cy™3 and Cy™5 mono-reactive dyes (Amersham) using the SuperScript™ Indirect cDNA Labeling System (Invitrogen) according to the manufacturer's instructions.
Array preparation
Ovine cDNA libraries were prepared from 27 tissues by MWG Biotech (Germany). These libraries were single-pass sequenced from the 5' end generating expressed sequence tags (ESTs). The inserts, representing ovine expressed sequences, were then amplified in 96 well plates using the universal primers SP6 and T7 in 50 μl reactions (1.5 mM MgCl2, 2 mM each dNTP, 45 pmol primers and 2.5 units AB Red Hot Taq polymerase). The reactions were denatured at 94°C for 3 minutes then cycled 36 times at 94°C for 45 sec, 55°C for 45 sec and 72°C for 60 sec. Finally products were extended at 72°C for 5 min. The PCR products were verified by agarose gel electrophoresis, precipitated and resuspended in printing solution as described previously [25]. At the University of Otago Genomics Facility an ovine microarray consisting of 19,968 spots was printed onto poly-L-lysine coated glass slides using an ESI array robot with 32 split pinheads depositing 0.6 nl with a 100 μm spot size. After printing the slides were UV irradiated to cross-link the DNA to the polylysine coating.
Slide hybridisation, scanning and normalisation
Slides were prehybridised by incubation for 45 min at 42°C in 50 ml of 5 X SSC, 0.1% SDS, 1% BSA. They were then rinsed twice in deionised water, once in isopropanol and dried. The labelled cDNA was denatured by heating at 95°C and then combined with 50 μl SlideHyb #1 (Ambion) and applied to slides. Slides were hybridised at 54°C for 16 hours in humidified chambers (Monterey). Post-hybridisation the slides were washed in the dark at 54°C for 10 min in 2 X SSC, 0.1% SDS, 5 min in 1 X SSC and 10 min in 0.1 X SSC. All buffers were filtered through 0.22 μm filters. The slides were dried and scanned in a ScanArray 5000 (Packard Biosciences) and the dual images collected in TIFF format. The combination and processing of the images was carried out using GenePix Pro (Axon Instruments). The data for each slide was normalised following the procedure of Baird et al., [71]. For each EST on the array, the normalised data from all 16 slides was combined and a number of average statistics calculated. ESTs were excluded from further analysis if they had more than 6/16 bad spots. The remaining ESTs were sorted based on the modified T value [72] for the log ratio of the mean [Additional file 3]. ESTs where the tail of the probability plot was more extreme than the 95% confidence limit [73] were counted as differentially expressed (Figure 1). All the microarray information has been submitted into the National Centre for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) website [74]. The accession number for the experiment series is GSE3738.
Northern blotting
20 μg of total RNA from duodenum tissue was separated on a 1% formaldehyde agarose gel and transferred to Hybond nylon membrane (Amersham) by capillary transfer. Probes were generated by amplification from the original EST using the gene specific primers listed in Table 5. The PCR products were verified by sequencing, labelled with α32P-dCTP (Amersham) using the RadPrime DNA labelling system (Invitrogen) and unincorporated radioactivity removed using the High Pure PCR product purification kit (Roche). Membranes were prehybridised for 45 min at 42°C in 10 ml ULTRAhyb (Ambion). The probes were diluted 1:10 in 10 mM EDTA and denatured by boiling for 10 min followed by cooling on ice for 5 min. After addition of 0.5 ml of ULTRAhyb, the probe was added to the prehyb solution and incubated overnight at 42°C. The membranes were then washed twice for 5 min at room temperature in 2 X SSC, 0.1% SDS followed by 2 X 15 min in 0.1 X SSC, 0.1% SDS at 42°C. The membranes were then exposed to BioMax XAR film (Kodak). The images were scanned using an ImageScanner (Amersham) and the resulting images quantitated using ImageQuant TL (Amersham) and normalised to GAPDH mRNA levels. This gene was not differentially expressed in the microarray experiment making it a suitable choice of housekeeping gene for normalisation.
Table 5.
Primers used for Northern blot probe amplification
| Primer Name | Primer Sequence 5'-3' |
| GAPDHF | TGAAGGTCCGTGTGAACGGATTTGGC |
| GAPDHR | CATGTAGGCCATGAGGTCCACCAC |
| GIMAP8F | TGCATACCTTTCCCTCTTCG |
| GIMAP8R | GCCTAGCCGTAAATAGGAACC |
| SPINK1F | CGGTGCAGTTTTCAACTGAG |
| SPINK1R | CCAAGCACGCATTGTAGTGT |
| TFF3F | TACGGTCCGGATTCCGGG |
| TFF3R | CCTCATGCTGAGCACGGG |
Microarray data interpretation
All ESTs in the ovine libraries, along with all ovine ESTs deposited in NCBI were assembled into contigs using CAP3 [75] after an initial clustering step using BLAST. ESTs on the array were annotated by finding the human RefSeq (RefSeq release as at 11/4/2005) corresponding to the contig to which they belonged using BLASTN [76] and the following options: -e 0.01 -v 5 -b 5. Each EST was annotated with the top human RefSeq hit. In the case of the differentially expressed ESTs, a more stringent E value cutoff of 1 × 10-18 was applied. In cases where the EST matched more than one transcript variant of a gene then the top hit is listed. All analysis hereafter refers to the annotated RefSeq genes. Gene Ontology (GO) terms significantly associated with the differentially expressed genes were found using the Expression Analysis Systematic Explorer (EASE). The list of differentially expressed genes was submitted to EASEonline [77]. The background list submitted included all human RefSeqs on the array. EASE calculates overrepresented functional gene categories compared to all the genes on the array [31].
A region of 1,500 bp upstream of the transcription start site of each of the RefSeq genes on the array was retrieved from the human genome browser at University of California-Santa Cruz [78] and sequences were masked for repeats through the retrieval process. The choice of promoter length was based on the report that approximately 75% of human core promoters lie within 1,500 bp of the transcription start site [79]. Of the 9,238 unique RefSeqs on the array, promoter regions for 8,809 RefSeqs were retrieved. Motifs in the promoter regions of the differentially expressed RefSeq genes were detected using the MEME motif detection program [32]. The motif lengths ranged between 6 and 12 bases, any number of repetitions of the motif was permitted, the reverse complement was allowed and the top 15 motifs were identified in each group The identified motifs were also compared to the transcription factor database, TRANSFAC release 8.1, [33] using the method described in Aerts et al., [80]. Each motif was permuted 1,000 times and best matches and their level of significance are reported. The full list of motifs identified in the promoter regions of the genes more highly expressed in susceptible and resistant animals, and their top TRANSFAC hits are available in Additional files 1 and 2 respectively. In order to carry out a MAST analysis [81] motifs with a Pearson's correlation coefficient greater than 0.6 were identified and excluded from further analysis. Promoter regions of both groups of differentially expressed genes and all RefSeq genes on the array were subsequently screened for the presence of all unique motifs using MAST. Once again, reverse complement orientation of the motifs was allowed and the results obtained were analysed using Fisher's exact test based on the number of RefSeq promoters with a combined motif significance of P < 0.0001 in each of the groups.
Authors' contributions
OMK extracted all RNA, carried out the microarray experiments, Northern blotting and Gene Ontology analysis as well as preparing the manuscript. AZ carried out the MEME and MAST analysis. TW and DLH designed, constructed and validated the ovine cDNA arrays. GJG was responsible for all animal care, handling and experimentation. DBB developed the microarray normalisation procedure and carried out statistical analysis, AFM curated the EST database, assembled the contigs and provided bioinformatics support. AMC and JCM conceived the study, participated it its design, coordination, analysis and writing. All authors read and approved the final manuscript.
Supplementary Material
This file contains all motifs detected by MEME in the promoter regions of genes more highly expressed in susceptible animals.
This file contains all motifs detected by MEME in the promoter regions of genes more highly expressed in resistant animals.
This file ranks the ESTs on the array according to their differential expression and contains statistical information for each EST on the array.
Acknowledgments
Acknowledgements
We thank Roger Wheeler, Wendy Bain, Chris Morris, Kevin Knowler, Mary Wheeler and Nadia McLean for assistance with maintenance of the Perendale selection lines, animal handling and slaughter. This work was funded by the Ovita Consortium Ltd.
Contributor Information
Orla M Keane, Email: orla.keane@agresearch.co.nz.
Amonida Zadissa, Email: amonida@sanger.otago.ac.nz.
Theresa Wilson, Email: theresa.wilson@agresearch.co.nz.
Dianne L Hyndman, Email: dianne.hyndman@agresearch.co.nz.
Gordon J Greer, Email: gordon.greer@agresearch.co.nz.
David B Baird, Email: david.baird@agresearch.co.nz.
Alan F McCulloch, Email: alan.mcculloch@agresearch.co.nz.
Allan M Crawford, Email: allan.crawford@agresearch.co.nz.
John C McEwan, Email: john.mcewan@agresearch.co.nz.
References
- Dominik S. Quantitative trait loci for internal nematode resistance in sheep: a review. Genet Sel Evol. 2005;37 Suppl 1:S83–96. doi: 10.1051/gse:2004027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balic A, Bowles VM, Meeusen EN. The immunobiology of gastrointestinal nematode infections in ruminants. Adv Parasitol. 2000;45:181–241. doi: 10.1016/s0065-308x(00)45005-0. [DOI] [PubMed] [Google Scholar]
- Gill HS. Genetic control of acquired resistance to haemonchosis in Merino lambs. Parasite Immunol. 1991;13:617–628. doi: 10.1111/j.1365-3024.1991.tb00557.x. [DOI] [PubMed] [Google Scholar]
- Pernthaner A, Cole SA, Morrison L, Hein WR. Increased expression of interleukin-5 (IL-5), IL-13, and tumor necrosis factor alpha genes in intestinal lymph cells of sheep selected for enhanced resistance to nematodes during infection with Trichostrongylus colubriformis. Infect Immun. 2005;73:2175–2183. doi: 10.1128/IAI.73.4.2175-2183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney CH, Parker CF, McClure KE, Herd RP. Resistance of exotic and domestic lambs to experimental infection with Haemonchus contortus. Int J Parasitol. 1985;15:101–109. doi: 10.1016/0020-7519(85)90107-9. [DOI] [PubMed] [Google Scholar]
- Stear MJ, Strain S, Bishop SC. Mechanisms underlying resistance to nematode infection. Int J Parasitol. 1999;29:51–6; discussion 73-5. doi: 10.1016/S0020-7519(98)00179-9. [DOI] [PubMed] [Google Scholar]
- McClure SJ, Emery DL, Wagland BM, Jones WO. A serial study of rejection of Trichostrongylus colubriformis by immune sheep. Int J Parasitol. 1992;22:227–234. doi: 10.1016/0020-7519(92)90106-U. [DOI] [PubMed] [Google Scholar]
- Bisset SA, Vlassoff A, Douch PG, Jonas WE, West CJ, Green RS. Nematode burdens and immunological responses following natural challenge in Romney lambs selectively bred for low or high faecal worm egg count. Vet Parasitol. 1996;61:249–263. doi: 10.1016/0304-4017(95)00836-5. [DOI] [PubMed] [Google Scholar]
- Aumont G, Gruner L, Hostache G. Comparison of the resistance to sympatric and allopatric isolates of Haemonchus contortus of Black Belly sheep in Guadeloupe (FWI) and of INRA 401 sheep in France. Vet Parasitol. 2003;116:139–150. doi: 10.1016/S0304-4017(03)00259-0. [DOI] [PubMed] [Google Scholar]
- Woolaston RR, Barger IA, Piper LR. Response to helminth infection of sheep selected for resistance to Haemonchus contortus. Int J Parasitol. 1990;20:1015–1018. doi: 10.1016/0020-7519(90)90043-M. [DOI] [PubMed] [Google Scholar]
- Gray GD, Barger IA, Le Jambre LF, Douch PG. Parasitological and immunological responses of genetically resistant Merino sheep on pastures contaminated with parasitic nematodes. Int J Parasitol. 1992;22:417–425. doi: 10.1016/0020-7519(92)90142-8. [DOI] [PubMed] [Google Scholar]
- Sreter T, Kassai T, Takacs E. The heritability and specificity of responsiveness to infection with Haemonchus contortus in sheep. Int J Parasitol. 1994;24:871–876. doi: 10.1016/0020-7519(94)90013-2. [DOI] [PubMed] [Google Scholar]
- Gruner L, Bouix J, Brunel JC. High genetic correlation between resistance to Haemonchus contortus and to Trichostrongylus colubriformis in INRA 401 sheep. Vet Parasitol. 2004;119:51–58. doi: 10.1016/j.vetpar.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Kaplan RM. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Knox DP, Redmond DL, Newlands GF, Skuce PJ, Pettit D, Smith WD. The nature and prospects for gut membrane proteins as vaccine candidates for Haemonchus contortus and other ruminant trichostrongyloids. Int J Parasitol. 2003;33:1129–1137. doi: 10.1016/S0020-7519(03)00167-X. [DOI] [PubMed] [Google Scholar]
- Meeusen EN, Piedrafita D. Exploiting natural immunity to helminth parasites for the development of veterinary vaccines. Int J Parasitol. 2003;33:1285–1290. doi: 10.1016/S0020-7519(03)00162-0. [DOI] [PubMed] [Google Scholar]
- Newton SE, Meeusen EN. Progress and new technologies for developing vaccines against gastrointestinal nematode parasites of sheep. Parasite Immunol. 2003;25:283–296. doi: 10.1046/j.1365-3024.2003.00631.x. [DOI] [PubMed] [Google Scholar]
- McEwan JC, Dodds KG, Watson TG, Greer GJ, Hosking BC, Douch PGC. Selection for resistance to roundworms by the New Zealand sheep breeding industry; the WormFec service. Proc Aust Assoc Anim Breed Genet. 1995;11:70–73. [Google Scholar]
- Albers GA, Gray GD. Breeding for worm resistance: a perspective. Int J Parasitol. 1987;17:559–566. doi: 10.1016/0020-7519(87)90132-9. [DOI] [PubMed] [Google Scholar]
- Windon RG. Selective breeding for the control of nematodiasis in sheep. Rev Sci Tech. 1990;9:555–576. doi: 10.20506/rst.9.2.496. [DOI] [PubMed] [Google Scholar]
- Bisset SA, Morris CA, McEwan JC, Vlassoff A. Breeding sheep in New Zealand that are less reliant on anthelmintics to maintain health and productivity. N Z Vet J. 2001;49:236–246. doi: 10.1080/00480169.2001.36238. [DOI] [PubMed] [Google Scholar]
- McEwan JC, Bisset SA, Morris CA. The selection of sheep for natural resistance to internal parasites. In: G.K B, editor. Sustainable Control of Internal Parasites in Ruminants. Canterbury, New Zealand , Lincoln University Press; 1997. pp. 161–182. [Google Scholar]
- Woolaston RRPLR. Selection of Merino sheep for resistance to Haemonchus contortus: genetic variation. Anim Sci. 1996;62:451–460. [Google Scholar]
- Morris CA, Wheeler M, Watson TG, Hosking BC, Leathwick DM. Direct and correlated responses to selection for high or low faecal nematode egg count in Perendale sheep. N Z J Agric Res. 2005;48:1–10. [Google Scholar]
- Diez-Tascon C, Keane OM, Wilson T, Zadissa A, Hyndman DL, Baird DB, McEwan JC, Crawford AM. Microarray analysis of selection lines from outbred populations to identify genes involved with nematode parasite resistance in sheep. Physiol Genomics. 2005;21:59–69. doi: 10.1152/physiolgenomics.00257.2004. [DOI] [PubMed] [Google Scholar]
- Okada H, Kimura MT, Tan D, Fujiwara K, Igarashi J, Makuuchi M, Hui AM, Tsurumaru M, Nagase H. Frequent trefoil factor 3 (TFF3) overexpression and promoter hypomethylation in mouse and human hepatocellular carcinomas. Int J Oncol. 2005;26:369–377. [PMC free article] [PubMed] [Google Scholar]
- Bohe H, Bohe M, Lundberg E, Polling A, Ohlsson K. Production and secretion of pancreatic secretory trypsin inhibitor in normal human small intestine. J Gastroenterol. 1997;32:623–627. doi: 10.1007/BF02934111. [DOI] [PubMed] [Google Scholar]
- Marchbank T, Freeman TC, Playford RJ. Human pancreatic secretory trypsin inhibitor. Distribution, actions and possible role in mucosal integrity and repair. Digestion. 1998;59:167–174. doi: 10.1159/000007485. [DOI] [PubMed] [Google Scholar]
- Krucken J, Epe M, Benten WP, Falkenroth N, Wunderlich F, Schroetel RM, Muller IU, Saidani N, Marinovski P, Stamm O. Malaria-suppressible expression of the anti-apoptotic triple GTPase mGIMAP8: Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. J Cell Biochem. 2005;341:291–304. [Google Scholar]
- Krucken J, Schroetel RM, Muller IU, Saidani N, Marinovski P, Benten WP, Stamm O, Wunderlich F. Comparative analysis of the human gimap gene cluster encoding a novel GTPase family. Gene. 2004;341:291–304. doi: 10.1016/j.gene.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis GJ, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Shapiro-Shelef M, Tunyaplin C, Calame K. Regulatory events in early and late B-cell differentiation. Mol Immunol. 2005;42:749–761. doi: 10.1016/j.molimm.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- Dubuquoy L, Dharancy S, Nutten S, Pettersson S, Auwerx J, Desreumaux P. Role of peroxisome proliferator-activated receptor gamma and retinoid X receptor heterodimer in hepatogastroenterological diseases. Lancet. 2002;360:1410–1418. doi: 10.1016/S0140-6736(02)11395-X. [DOI] [PubMed] [Google Scholar]
- Koretz K, Momburg F, Otto HF, Moller P. Sequential induction of MHC antigens on autochthonous cells of ileum affected by Crohn's disease. Am J Pathol. 1987;129:493–502. [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Osvath SR, Hall RA, King NJ, Sedger LM. Regulation of antigen processing and presentation molecules in West Nile virus-infected human skin fibroblasts. Virology. 2004;324:286–296. doi: 10.1016/j.virol.2004.03.036. [DOI] [PubMed] [Google Scholar]
- Johnson DR. Locus-specific constitutive and cytokine-induced HLA class I gene expression. J Immunol. 2003;170:1894–1902. doi: 10.4049/jimmunol.170.4.1894. [DOI] [PubMed] [Google Scholar]
- Young LC, Thulien KJ, Campbell MR, Tron VA, Andrew SE. DNA mismatch repair proteins promote apoptosis and suppress tumorigenesis in response to UVB irradiation: an in vivo study. Carcinogenesis. 2004;25:1821–1827. doi: 10.1093/carcin/bgh191. [DOI] [PubMed] [Google Scholar]
- Mark SC, Sandercock LE, Luchman HA, Baross A, Edelmann W, Jirik FR. Elevated mutant frequencies and predominance of G:C to A:T transition mutations in Msh6(-/-) small intestinal epithelium. Oncogene. 2002;21:7126–7130. doi: 10.1038/sj.onc.1205861. [DOI] [PubMed] [Google Scholar]
- Li Z, Xia L, Lee LM, Khaletskiy A, Wang J, Wong JY, Li JJ. Effector genes altered in MCF-7 human breast cancer cells after exposure to fractionated ionizing radiation. Radiat Res. 2001;155:543–553. doi: 10.1667/0033-7587(2001)155[0543:egaimh]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kelner MJ, Bagnell RD, Montoya MA, Lanham KA. Structural organization of the human gastrointestinal glutathione peroxidase (GPX2) promoter and 3'-nontranscribed region: transcriptional response to exogenous redox agents. Gene. 2000;248:109–116. doi: 10.1016/S0378-1119(00)00137-2. [DOI] [PubMed] [Google Scholar]
- Chen J, Shpall RL, Meyerdierks A, Hagemeier M, Bottger EC, Naumovski L. Interferon-inducible Myc/STAT-interacting protein Nmi associates with IFP 35 into a high molecular mass complex and inhibits proteasome-mediated degradation of IFP 35. J Biol Chem. 2000;275:36278–36284. doi: 10.1074/jbc.M006975200. [DOI] [PubMed] [Google Scholar]
- Raasi S, Schmidtke G, Groettrup M. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J Biol Chem. 2001;276:35334–35343. doi: 10.1074/jbc.M105139200. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G. FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol Cell Biol. 2005;25:3483–3491. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Scafidi J, Prada AE, Zahedi K, Davis AE. Nuclear phosphatases and the proteasome in suppression of STAT1 activity in hepatocytes. Biochem Biophys Res Commun. 2002;299:574–580. doi: 10.1016/S0006-291X(02)02694-3. [DOI] [PubMed] [Google Scholar]
- Cai S, Davis AE. Complement regulatory protein C1 inhibitor binds to selectins and interferes with endothelial-leukocyte adhesion. J Immunol. 2003;171:4786–4791. doi: 10.4049/jimmunol.171.9.4786. [DOI] [PubMed] [Google Scholar]
- Caliezi C, Wuillemin WA, Zeerleder S, Redondo M, Eisele B, Hack CE. C1-Esterase inhibitor: an anti-inflammatory agent and its potential use in the treatment of diseases other than hereditary angioedema. Pharmacol Rev. 2000;52:91–112. [PubMed] [Google Scholar]
- Liu D, Cramer CC, Scafidi J, Davis AE. N-linked glycosylation at Asn3 and the positively charged residues within the amino-terminal domain of the c1 inhibitor are required for interaction of the C1 Inhibitor with Salmonella enterica serovar typhimurium lipopolysaccharide and lipid A. Infect Immun. 2005;73:4478–4487. doi: 10.1128/IAI.73.8.4478-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AE. Biological effects of C1 inhibitor. Drug News Perspect. 2004;17:439–446. doi: 10.1358/dnp.2004.17.7.863703. [DOI] [PubMed] [Google Scholar]
- Lafontaine PO, Arnal M, Buron N, Solary E, Bron AM, Westley BR, May FE, Bara J, Gespach C, Creuzot-Garcher C. Trefoil factor family mRNA and protein expression in pterygium. Int J Oncol. 2005;27:997–1003. [PubMed] [Google Scholar]
- Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Chu FF, Esworthy RS, Chu PG, Longmate JA, Huycke MM, Wilczynski S, Doroshow JH. Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res. 2004;64:962–968. doi: 10.1158/0008-5472.CAN-03-2272. [DOI] [PubMed] [Google Scholar]
- Lee YM, Cheng PY, Hong SF, Chen SY, Lam KK, Sheu JR, Yen MH. Oxidative stress induces vascular heme oxygenase-1 expression in ovariectomized rats. Free Radic Biol Med. 2005;39:108–117. doi: 10.1016/j.freeradbiomed.2005.02.033. [DOI] [PubMed] [Google Scholar]
- Kimura H, Hokari R, Miura S, Shigematsu T, Hirokawa M, Akiba Y, Kurose I, Higuchi H, Fujimori H, Tsuzuki Y, Serizawa H, Ishii H. Increased expression of an inducible isoform of nitric oxide synthase and the formation of peroxynitrite in colonic mucosa of patients with active ulcerative colitis. Gut. 1998;42:180–187. doi: 10.1136/gut.42.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann M, Tomicic MT, Kaina B. Phosphorylation of mismatch repair proteins MSH2 and MSH6 affecting MutSalpha mismatch-binding activity. Nucleic Acids Res. 2002;30:1959–1966. doi: 10.1093/nar/30.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Villunger A, O'Reilly LA, Beaumont JG, Coultas L, Cheney RE, Huang DC, Strasser A. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293:1829–1832. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- Sax JK, El-Deiry WS. Identification and characterization of the cytoplasmic protein TRAF4 as a p53-regulated proapoptotic gene. J Biol Chem. 2003;278:36435–36444. doi: 10.1074/jbc.M303191200. [DOI] [PubMed] [Google Scholar]
- Puppo F, Contini P, Ghio M, Indiveri F. Soluble HLA class I molecules/CD8 ligation trigger apoptosis of CD8+ cells by Fas/Fas-ligand interaction. ScientificWorldJournal. 2002;2:421–423. doi: 10.1100/tsw.2002.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Liu S, Yu Y, An H, Li N, Lin N, Wang W, Zhang W, Wan T, Cao X. Cloning and identification of a novel ubiquitin-like protein, BMSC-UbP, from human bone marrow stromal cells. Immunol Lett. 2003;86:169–175. doi: 10.1016/S0165-2478(03)00004-X. [DOI] [PubMed] [Google Scholar]
- Kalies KU, Allan S, Sergeyenko T, Kroger H, Romisch K. The protein translocation channel binds proteasomes to the endoplasmic reticulum membrane. Embo J. 2005;24:2284–2293. doi: 10.1038/sj.emboj.7600731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Kim C, Zhen L, Lowe JB, Kapur R, Petryniak B, Spaetti A, Pollock JD, Borneo JB, Bradford GB, Atkinson SJ, Dinauer MC, Williams DA. Deficiency of the hematopoietic cell-specific Rho family GTPase Rac2 is characterized by abnormalities in neutrophil function and host defense. Immunity. 1999;10:183–196. doi: 10.1016/S1074-7613(00)80019-9. [DOI] [PubMed] [Google Scholar]
- Carstanjen D, Yamauchi A, Koornneef A, Zang H, Filippi MD, Harris C, Towe J, Atkinson S, Zheng Y, Dinauer MC, Williams DA. Rac2 regulates neutrophil chemotaxis, superoxide production, and myeloid colony formation through multiple distinct effector pathways. J Immunol. 2005;174:4613–4620. doi: 10.4049/jimmunol.174.8.4613. [DOI] [PubMed] [Google Scholar]
- Forlow SB, Schurr JR, Kolls JK, Bagby GJ, Schwarzenberger PO, Ley K. Increased granulopoiesis through interleukin-17 and granulocyte colony-stimulating factor in leukocyte adhesion molecule-deficient mice. Blood. 2001;98:3309–3314. doi: 10.1182/blood.V98.12.3309. [DOI] [PubMed] [Google Scholar]
- Lee SH, Corry DB. Homing alone? CD18 in infectious and allergic disease. Trends Mol Med. 2004;10:258–262. doi: 10.1016/j.molmed.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Shen M, Fujikura D, Tosa N, Kim HR, Kon S, Uede T, Reed JC. Functional role of death-associated protein 3 (DAP3) in anoikis. J Biol Chem. 2004;279:44667–44672. doi: 10.1074/jbc.M408101200. [DOI] [PubMed] [Google Scholar]
- Dempsey PW, Doyle SE, He JQ, Cheng G. The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev. 2003;14:193–209. doi: 10.1016/S1359-6101(03)00021-2. [DOI] [PubMed] [Google Scholar]
- Bovine Genome Project [http://www.hgsc.bcm.tmc.edu/projects/bovine/]
- Baird D, Johnstone P, Wilson T. Normalization of microarray data using a spatial mixed model analysis which includes splines. Bioinformatics. 2004;20:3196–3205. doi: 10.1093/bioinformatics/bth384. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Michael JR. The stabilized probability plot. Biometrika. 1983;70:11–17. [Google Scholar]
- Gene Expression Omnibus [http://www.ncbi.nlm.nih.gov/geo/]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- EASEonline [http://david.niaid.nih.gov/david/]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XT, Qian MP, Tang HX. Predicting polymerase II core promoters by cooperating transcription factor binding sites in eukaryotic genes. Acta Biochim Biophys Sin (Shanghai) 2004;36:250–258. doi: 10.1093/abbs/36.4.250. [DOI] [PubMed] [Google Scholar]
- Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains all motifs detected by MEME in the promoter regions of genes more highly expressed in susceptible animals.
This file contains all motifs detected by MEME in the promoter regions of genes more highly expressed in resistant animals.
This file ranks the ESTs on the array according to their differential expression and contains statistical information for each EST on the array.


