Abstract
We determined the molecular mechanism of cell death response by MutS homologs in distinction to the repair event. Key protein–DNA contacts differ in the interaction of MutS homologs with cisplatinated versus mismatched DNA. Mutational analyses of protein–DNA contacts, which were predicted by molecular dynamics (MD) simulations, were performed. Mutations in suggested interaction sites can affect repair and cell death response independently, and to different extents. A glutamate residue is identified as the key contact with cisplatin-DNA. Mutation of the residue increases cisplatin resistance due to increased non-specific DNA binding. In contrast, the conserved phenylalanine that is instrumental and indispensable for mismatch recognition during repair is not required for cisplatin cytotoxicity. These differences in protein–DNA interactions are translated into localized conformational changes that affect nucleotide requirements and inter-subunit interactions. Specifically, the ability for ATP binding/hydrolysis has little consequence for the MMR-dependent damage response. As a consequence, intersubunit contacts are altered that most likely affect the interaction with downstream proteins. We here describe the interaction of MutS homologs with DNA damage, as it differs from the interaction with a mismatch, and its structural translation into all other functional regions of the protein as a mechanism to initiate cell death response and concomitantly inhibit repair.
INTRODUCTION
Mismatch repair (MMR) proteins contribute to the initiation of cell death in response to DNA damage (1–3). Defects in these proteins significantly increase carcinogenesis, the tolerance to chemotherapy and clonal selection of cancer cells after treatment.
Different hypotheses have been put forward to explain the MMR-dependent cell death pathway (2). ‘Futile repair cycles’ entail repetitive repair attempts of damage-containing DNA that occur on the ‘normal’, undamaged strand. This mechanism requires replication across the damage to generate a mismatch. Abortive repair attempts and persistent DNA damage would result in strand breaks as the factual initiators of cell death. Functional repair activity of the MMR proteins is a prerequisite for this proposed mechanism. In contrast, the damage-signaling hypothesis suggests the recruitment of proapoptotic downstream proteins by MMR proteins and the direct initiation of cell death. This mechanism can be considered repair-independent, though individual protein functions may be overlapping for both pathways. Results from our laboratory and others provide first evidence that the repair function is not required for MMR-dependent cisplatin cytotoxicity (1,4,5). MMR proteins interact with an intricate network of proteins involved in cell cycle checkpoint and apoptotic pathways, among which are p73, ATM, CHK2, BRCA1 and PCNA (6), which provides another piece of evidence suggesting that MMR proteins may play a direct role in drug cytotoxicity.
Both hypotheses for MMR-dependent drug cytotoxicity are based on the ability of the mismatch recognition protein, MutS and its eukaryotic homologs (MSH) to recognize damage-containing DNA, and initiate appropriate responses. For futile repair cycles, the proteins would always recognize a mismatch and initiate repair, while for direct signaling, the MSH proteins would recognize the DNA damage, and, possibly, as a distinction to mismatch binding, initiate cell death. The crystal structures of MutS in complex with mismatched DNA demonstrate that the presence of DNA introduces considerable conformational changes in the protein (7–9). It was proposed that the flexibility of DNA and an induced fit upon binding are pivotal for mismatch specificity. A 60° DNA bend that is introduced at the site of the mismatch, is facilitated by intercalation of a conserved phenylalanine residue. This residue is provided by only one of the subunits (A) of the MutS homodimer (corresponding to MSH6 in the eukaryotic protein) (7–12). Base-stacking interactions of the Phe residue position the mismatched base for hydrogen bond interactions with a conserved glutamate. The phenylalanine and glutamate residues provide the only mismatch-specific contacts between MutS and mismatched DNA. These specific interactions with mismatched DNA introduce conformational changes into the protein as key steps in the initiation of repair. It is likely that distinctly different interaction patterns with DNA damage contribute to and determine the induction of different pathways, such as celldeath.
Defective MMR was shown to result in a 2- to 4-fold increase in tolerance to the chemotherapeutic agent cisplatin, which contributes significantly to the failure of cancer therapy (2,13–15). MMR defects in ovarian cancer have recently been associated with a decreased efficacy of cisplatin therapy (16). A molecular understanding of this correlation is clearly warranted to improve current treatment measures for cancer patients.
We investigated the molecular mechanism of protein–DNA damage interactions as a key element in MMR-dependent drug cytotoxicity. Increased DNA rigidity of cisplatin-adducted DNA (CDDP-DNA) and distinct protein–DNA contacts result in a damage-specific binding pattern by MSH proteins. A conserved glutamate residue is instrumental in the recognition of CDDP-DNA and its cytotoxicity. A mutation in this residue increases overall, non-specific DNA affinity that interferes with cisplatin cytotoxicity. In contrast, a mutation of the phenylalanine demonstrated that this residue is dispensible for cisplatin cytotoxicity, while it was shown to be absolutely required for the interaction with mismatched DNA and proficient repair. We illustrate the biological significance of the individual contacts, and for the first time demonstrate that distinct MSH-CDDP-DNA interactions separate cell death response from repair. These distinct protein–DNA contacts are translated throughout the protein into localized conformational alterations that amplify the distinction between cell death response and repair. Our data significantly impact the mechanistic understanding of MMR-dependent response to chemotherapy.
MATERIALS AND METHODS
Computational modeling
The simulation of the G/T mismatch is based on the X-ray structure of Escherichia coli MutS in complex with heteroduplex DNA (7). Hydrogen atoms were added using the hbuild facility of CHARMM (17). The CHARMM force field was used for the entire complex with additional parameters based on pre-existing cisplatin parameters (17,18). This force field has been extensively parameterized for a wide range of biologically important molecules, including nucleic acids, amino acids, lipids and some small-molecule ligands. The platinated DNA structure was built using the mismatch as a template. The cross-linked structure was fitted into the binding pocket to maximize the structural overlap with the mismatched DNA structure, followed by rotations and translations to minimize the energy of the unrelaxed structure using the coordinate manipulation and energy minimization facilities of CHARMM (17). The structure was fully solvated with TIP3P water in a cubic box using the visual molecular dynamics (VMD) package (19). The fully solvated simulation was performed with a 2.0 fs timestep using SHAKE on all bonds to hydrogen atoms, and with Particle Mesh Ewald as implemented in NAMD (20). The simulation protocol consisted of 100 ps of free solvent equilibration with fixed solute, followed by 400 ps of full unrestrained equilibration, and 1.6 ns of production. During the unrestrained equilibration and the production simulation, all atoms in the complex and the solvent are allowed to move freely. As a result, for example, the DNA can bend freely and the protein side chains can reorient. The angles were calculated with the trajectory analysis tools in VMD using the backbone atoms in the terminal and central amino acids after the equilibration period (19). The conformational analyses were performed using VMD and CHARMM. After 2 ns, the unplatinated mismatch interaction scheme was essentially unaltered from the X-ray, and is not discussed. As a control, an additional simulation was performed of a platinated G/T mismatch using the same protocol. The conclusions from both simulations are similar, although the magnitude of distant conformational changes is smaller in the G/T simulations. Modeling alone at present does not reveal any significant differences between the binding in the G/C and G/T interactions. In the G/T simulations no structure (sampled every 2 ps) has an RMSD of more than 2.1 Å from the X-ray structure, excluding those residues that are undefined in the crystal structure. All the platinated structures are within 2.8 Å of all the structures sampled in the mismatch simulation, including the residues undefined in the X-ray structure (data not shown). The simulation of the mutant MutS-E38A structure was performed using the same methodology after the computational mutagenesis was performed. The mutation was introduced using the wild-type structure with hydrogen atoms as a template. The alanine side chain was built upon the original glutamate side chain, and the entire structure was subjected to 500 steps of steepest descent minimization with an r-dependent dielectric to locally minimize the structure.
Protein purification
The purification of E.coli MutS was performed as described previously (21). Briefly, cells harboring the expression plasmid were lysed by pressure. His-tagged MutS in the supernatant was selectively isolated by ion chelating chromatography. MutS-containing fractions were pooled and further purified by Heparin Sepharose. The protein eluate was concentrated, aliquoted and frozen in 30% glycerol until needed.
DNA substrates
DNA was reacted with cisplatin according to the method of Mello et al. (22). Briefly, a single-stranded DNA oligomer was mixed with 1.2 equivalents of cisplatin in 3 mM NaCl and 1 mM Na2HPO4 (pH7.4) and incubated at 37°C for 16 h. Unreacted cisplatin was removed by extensive dialysis against deionized water. The DNA was then lyophilized and resuspended in deionized water to desired concentration. The sequence of the DNA used in the reaction is 5′-ACCTGTCTGGCAACACTGTCACCGTCGTAT-3′. The oligomer was annealed to the complementary strand to produce homoduplex double-stranded DNA or to the oligonucleotide with the sequence 5′-ATACGACGGTGACAGTGTTGCTAGACAGGT-3′ to produce a single G-T mismatch opposite the site of platination.
DNA binding assays
DNA binding affinities were determined by measuring the change in fluorescence anisotropy of DNA duplexes in which one strand was labeled with fluorescein at the 5′ end. The data were collected using a Safire II microplate reader with a fluorescence anisotropy module (Tecan) with excitation and emission wavelengths of 490 and 525 nm, respectively. To ensure equilibrium binding, the concentration of DNA was kept at least 20-fold below the observed Kd for binding and the concentration of MutS protein was titrated in the range of 5-fold below to 5-fold above the Kd. The dissociation constants were calculated by fitting the data to a model of single site binding (Fbound = [MutS]/Kd + [MutS]). Each experiment was the average of at least four individual measurements.
Genetic assays
Treatment with cisplatin and cell survival assays were performed as described before (1). The IC50 values and 95% confidence limits were obtained by quadratic curve fitting, as described (1,23). Mutation rate determinations based on fluctuation tests have been described (1,12).
RESULTS
The structural model of MutS in complex with CDDP-DNA and prediction of interactions
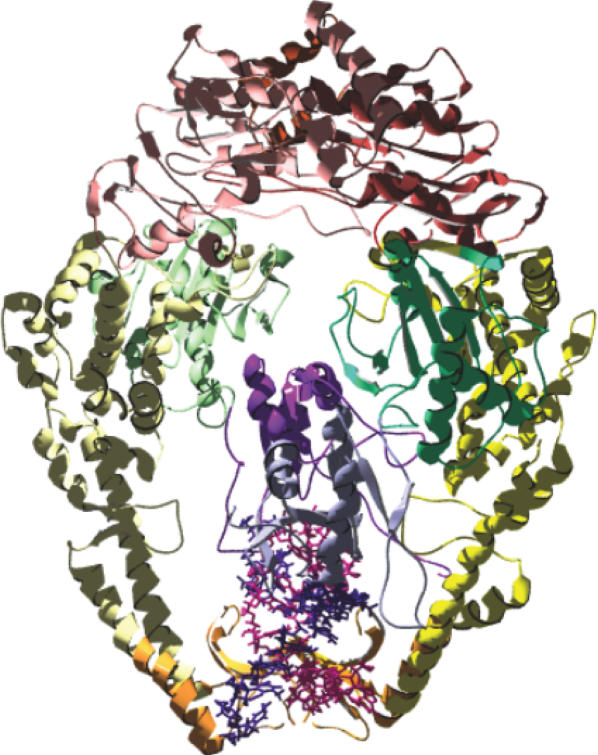
The understanding of MMR-dependent damage-signaling requires knowledge of the structural aspects of damage recognition. No structural information is available for the interaction between MutS homologs with any DNA damage. We therefore performed molecular dynamics (MD) simulations to obtain indications for the structural and molecular aspects of the E.coli MutS-(1,2)GpG cisplatin crosslink complex. The structural model is based on the crystal structure of DNA containing a single (1,2)GpG cisplatin crosslink (24,25) built into the crystal structure of E.coli MutS (7,9). The mismatched DNA in the MutS crystal structure was used as a template. The structural model of MutS in complex with CDDP-DNA is shown in Figure 1.
Figure 1.
The structural model of E. coli MutS—CDDP-DNA. MutS in complex with a 16mer oligonucleotide duplex containing a single (1,2) GpG cisplatin adduct. The adduct cross-links the guanines at position 8 and 9 of one DNA strand. Coloring distinguishes domain structure, with: blue (subunit A)/purple (subunit B): mismatch binding domain I; green: connector domain II; yellow: core domain III; orange: clamp domain IV and red: ATPase/dimerization domain V. DNA is shown in light and dark purple. The symmetry axis of the dimer runs vertical through the structure.
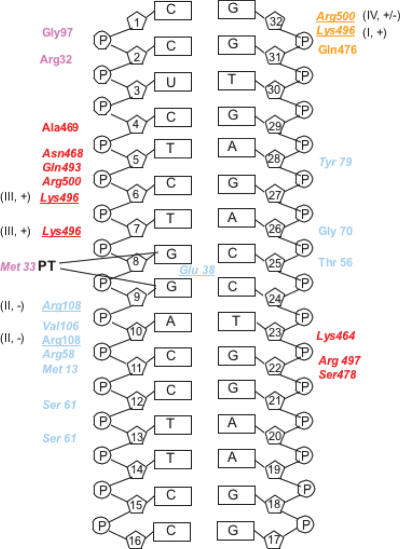
We used this MutS-CDDP-DNA model and snapshots obtained throughout the simulation to predict individual protein–DNA interactions. Figure 2 displays protein–DNA hydrogen bonds that were maintained for more than 90% of the simulation time. The hydrogen bonding pattern is stable, and no significant transient interactions are observed at equilibrium. The structural model with CDDP-DNA demonstrates protein–DNA interactions that resemble the binding pattern described for the individual interactions of MutS with five different mismatches (Figure 2, italics) (9). The principle interactions are hydrogen bonds. Most contacts with CDDP-DNA are provided by domain I of subunit A (corresponding to eukaryotic MSH6) and the clamp domain IV of subunit B (corresponding to MSH2), similar to the interaction with mismatched DNA (7–9). Few unique contacts with CDDP-DNA are identified (subunit A: T56, Y79; subunit B: N468, A469, Q493); none of these newly gained contacts are conserved among species. Other contacts are provided by residues described previously for the interaction with mismatched DNA (7–9), though the location with respect to the DNA damage may differ (Figure 2, e.g. K496, R108, R500).
Figure 2.
Diagram of MutS interactions with CDDP-DNA. Colors distinguish interactions from the individual domains and correspond to Figure 1. Light blue: domain I, subunit A (MSH6), orange: domain IV, subunit A; purple: domain I, subunit B (MSH2), red: domain IV, subunit B. Residues tested herein are underlined. Roman numbers refer to the effect group described in the text. + and − describe whether a mutation in this residue results in a significant effect on cisplatin cytotoxicity (compare to Table 1).
Biological significance of individual backbone contacts between MutS and CDDP-DNA
We performed a mutational and genetic analysis of key protein–DNA contacts predicted to be significant for the interaction with CDDP-DNA. Point mutations at amino acids predicted to contact DNA were introduced into the genes expressing MSH protein(s) (Figure 2, Table 1). For this purpose, the yeast complex of MutS homologs (MSH2/MSH6) was analyzed. The amino acid sequences of MutS homologs are highly conserved among species, which allows the analysis of homologous residues in different systems. The advantage of the eukaryotic MutS homologous proteins is their heterodimeric, rather than homodimeric nature, which allows the separate analysis of each subunit. Results on cell survival after cisplatin exposure were compared to the mutator phenotype associated with the individual mutation (Table 1). The mutation rate, as an indicator for the mutator phenotype, was determined using two well-characterized reporter systems (1). The LYS2-A14 reporter detects frameshift mutations in an A14 nucleotide run. The forward reporter CAN1 primarily monitors base substitutions in a MMR-deficient background. Cells defective in msh2 and msh3 msh6, respectively, display a strong mutator phenotype in both systems (Table 1; msh2: 970- and 32-fold, respectively; msh3msh6: 1600- and 25-fold, respectively). This genetic defect can be complemented by plasmid-based expression of wt genes. As a measure for drug cytotoxicity, dose-dependent cell survival after cisplatin treatment was monitored. The IC50 values were determined by quadratic curve fitting (1). Defects in msh2 and msh3 msh6, respectively, typically result in a 2- to 4-fold increase in resistance to the drug (14,15), which is confirmed by our data [Table 1; msh2: 2.4-fold; msh3msh6: 1.8-fold, non-overlapping confidence limits (CL) with wild-type]. Mutations were introduced into the MSH2 and MSH6 gene, according to predicted protein–cisplatinated DNA interactions (Figure 2), to determine their potential significance for cisplatin cytotoxicity or repair. It has previously been demonstrated (data not shown, 12) that mutant genes are expressed at similar levels as wild-type.
Table 1.
Non-specific protein–DNA interactions in cisplatin cytotoxicity and MMR
| E.coli | S.cerevisiae | IC50 [µM cisplatin] | Mutation rate in LYS2-A14a | Mutation rate in CAN1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 | CL | -fold | MR 10−6 | CL 10−6 | -fold | MR 10−6 | CL 10−6 | -fold | ||
| Δmsh3msh6 | 430 | 340–510 | 1.8 | 3000 | 860–11 000 | 1600 | 5.0 | 4.6–8.0 | 25 | |
| MSH6 wt | 240 | 220–260 | 1 | 1.9 | 0.7–3.5 | 1 | 0.2 | 0.03–00.3 | 1 | |
| K496 | MSH6-K848A | 380 | 290–470 | 1.6b | 1.4 | 0.8–8.5 | 0.7 | 3.5 | 2.2–4.4 | 18 |
| R108 | MSH6-R412A | 300 | 240–350 | 1.3 | 2800 | 970–3800 | 1500 | 4 | 3–5 | 20 |
| R500 | MSH6-R852A | 300 | 260–340 | 1.3b | 9.1 | 6.5–15 | 5 | 0.9 | 0.5–2.2 | 4.5 |
| Δmsh2 | 360 | 330–390 | 2.4 | 620 | 530–1800 | 970 | 15 | 11–19 | 32 | |
| MSH2 wt | 150 | 130–170 | 1 | 0.64 | 0.4–1.0 | 1 | 0.47 | 0.37–30.66 | 1 | |
| K496 | MSH2-K564A | 230 | 230–240 | 1.5b | 430 | 320–750 | 670 | 6.6 | 5.7–7.5 | 14 |
E.c.: E.coli; IC50: shown are means and standard deviations from at least three independent experiments. MR: mutation rate, CL: 95% confidence limits; -fold: in comparison to wild-type (italics).
aAs described in (1).
bNon-overlapping confidence limits with wild-type; indicates statistically significant difference.
cAs designated from crystal structures with mismatched DNA (9) and predictions from structural model described herein.
Mutations in the putative backbone contacts affect either damage response (as measured by the IC50 for cisplatin exposure) or repair (mutation rate), or both. Several mutants that demonstrate uncorrelated effects on cytotoxicity and repair have been identified:
Cisplatin tolerance, weak effect on repair
Mutation MSH6-K848A conveys no mutator phenotype in the frameshift reporter, and an intermediate mutator phenotype in the base substitution reporter system (18-fold, compared to 25-fold for the knockout). This result per se indicates a functional differences in the repair of different mismatches; the residue appears to have functional significance only for the repair of base substitutions. The K848A mutation affects cisplatin cytotoxicity, with overlapping confidence limits of IC50's with the complete knockout strain (Table 1), and hence increases cisplatin tolerance significantly. The residue is located in the clamp domain of MSH6 (E.coli residue K496). According to the simulation, this residue interacts with the DNA backbone at a considerable distance (7 nt apart), and on the opposite strand to the platinated site (Figure 2). This mutant suggests that contacts distant from the site of damage may gain functional significance when compared to mismatch recognition. We demonstrate here that a point mutation can significantly increase the resistance to cisplatin, independent of its effect on repair.
Small effects on cisplatin cytotoxicity, impaired repair
Residue R108 in MutS (MSH6-R412) affects the repair activity, but displays a weak or intermediate effect on cisplatin sensitivity. This residue is predicted to contact the phosphate backbone immediately 3′ to the cisplatinated GpG crosslink (Figure 2). MSH6-R412A increases cisplatin tolerance 1.3-fold, with confidence limits overlapping with the wild-type and knockout strain, indicating an intermediate or week chemotolerant phenotype (Table 1). In contrast, the mutation results in a mutator phenotype that is indistinguishable from the complete knockout strain (1500-fold increase in LYS2, 20-fold in CAN1, Table 1). This result suggests that the elimination of this putative interaction by mutation is not sufficient to abrogate MMR-dependent damage response entirely. Again, this demonstrates that close vicinity or the distance to the lesion is not indicative of the biological significance of protein-CDDP-DNA interactions. The distance is formally defined as geometric distance between contact and cross-linked site. The R412A demonstrates no obvious correlation between effects on cisplatin cytotoxicity and MMR. In addition to the R412A alteration, a mutation in the nucleotide binding motif of MSH2 (G693A in yeast) shows lack of functional correlation between repair and cisplatin cytotoxicity (1,4). These results demonstrate that the elimination of certain protein–DNA contacts can prevent repair of replication errors, but retain some cisplatin cytotoxicity.
Small effects on both cisplatin cytotoxicity and repair
Mutation MSH6-R852A does not affect damage response or repair significantly. Though both the crystal structure and the structural model suggest that this residue provides a protein–DNA interaction, this contact does not contribute significantly to either repair (5-/4.5-fold increase in mutation rate) or damage response to cisplatin (1.3-fold increase, overlapping confidence limits with wild-type). MMR deficiency results in a relatively weak resistance phenotype to cisplatin in a background where other repair pathways, including those processing cisplatin adducts, are proficient. The IC50 is typically 2- to 4-fold elevated. Though this increase has important implications for the failure of chemotherapy, demonstrating functional significance can be challenging. An effect of mutation MSH6-R852A on cisplatin response cannot entirely be excluded.
Increased cisplatin tolerance, increased repair deficiency
Mutation MSH2-K564A (K496, Domain IV/B, Figure 2) affects both cisplatin cytotoxicity and MMR. This residue is expected to form hydrogen bonds with the phosphate backbone in close vicinity to the adduct (Figure 2). The IC50 for cisplatin cytotoxicity is significantly elevated compared to wild-type (1.5-fold; non-overlapping confidence limits with wild-type), verifying the functional significance of this predicted interaction. On the other hand, the confidence limits are also non-overlapping with the knockout, overall suggesting an intermediate effect on cisplatin cytotoxicity, similar to R412A. The mutation results in a strong mutator phenotype in both reporter systems (Table 1). This result demonstrates that a point mutation can affect both repair activity and drug cytotoxicity.
The identification of mutations that show no correlation between the functional requirements for damage response and repair supports the hypothesis that MMR-dependent cisplatin cytotoxicity is initiated independently of the repair event.
Altered base-specific contacts with CDDP-DNA—Phenylalanine 36
Our structural model of MutS with CDDP-DNA predicts a considerable dislocation of residues that provide indispensable contacts for mismatch recognition (Figure 3). In the structure with mismatched DNA, a conserved phenylalanine residue (E.coli F36/subunit A, MSH6-F337) base-stacks with one base of the mismatch, this is the most important direct interaction with the mismatch. In the interaction with CDDP-DNA, the Phe residue is not parallel to either one of the cross-linked guanine bases. This dislocation prevents base-stacking or other interactions with the CDDP-DNA (Figure 3). The presence of a thymine mismatched with the cross-linked guanines does not alter the location of the Phe significantly (computational data not shown). The dislocation in the protein–DNA complex is hence a result of the DNA damage introduced by CDDP. This prediction that the Phe does not significantly contribute to interactions with CDDP-DNA suggests that a mutation of this residue will not affect CDDP cytotoxicity.
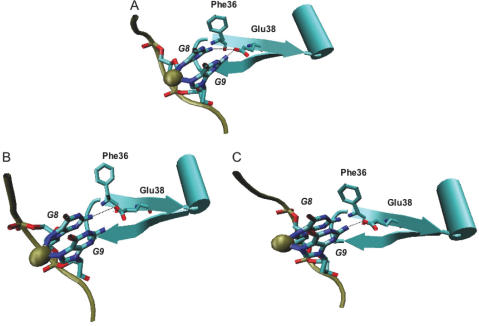
Figure 3.
Base-specific interactions of wild-type MutS with CDDP-DNA. Gold sphere: cisplatinum. Shown are Phe36 and Glu38 (E.coli) in their orientation towards the cross-linked guanines. Hydrogen bonds between Glu38 and the guanines are indicated. (A) Both hydrogen bonds that can be formed between Glu38 and either one of the cross-linked guanines. (B) and (C) The alternate hydrogen bonds formed between Glu38 and one of the cisplatin-cross-linked guanines. The local distortions induced by either conformation are indicated.
The mutational analysis (MSH6-F337A) confirms this prediction and demonstrates differential requirements for the Phe residue in cisplatin cytotoxicity and repair (Table 2). As suggested by the dislocation of the Phe residue from CDDP-DNA (Figure 3), the cisplatin cytotoxicity is not affected by the F337A mutation. In comparison to wild-type MSH6, the MSH6-F337A mutation lowers the IC50 to 180 µM as compared to 240 µM for wild-type. This significant increase in cisplatin sensitivity might be due to the elimination of steric hindrances, and replacement of the side chain with a considerably less bulky one. Alternatively, the impediment of interactions with mismatch may result in an increase in free protein that is available for the interaction with DNA damage, and hence result in the observed increase in cisplatin sensitivity.
Table 2.
Base contacts of protein–DNA interactions in cisplatin cytotoxicity and MMR
| E.coli | S.cerevisiae | IC50 [µM cisplatin] | Mutation rate in LYS2-A14a | Mutation rate in CAN1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 | CL | -fold | MR 10−6 | CL 10−6 | -fold | MR 10−6 | CL 10−6 | -fold | ||
| F36 | MSH6-F337A | 180 | 150–200 | 0.8b | 990 | 610–1600 | 520 | NDc | ||
| E38 | MSH6-E339Q | 400 | 380–410 | 1.7b | 44 | 32–67 | 23 | 3 | 1.4–4.6 | 15 |
E.c.: E.coli; IC50: shown are means and standard deviations from at least three independent experiments. MR: mutation rate, CL: 95% confidence limits; -fold: in comparison to wild-type (italics).
aAs described in (1).
bNon-overlapping confidence limits with wild-type; indicates statistically significant difference.
cThe F337A was shown to result in a strong mutator phenotype in the CAN1 reporter system, indistinguishable from the knockout strain (10). ND: not determined.
We have performed this experiment and purified the MutS-F36A mutant and performed anisotropy measurements in analogy to the data presented for E38A (Table 3; Figure 4). Surprisingly, the F36A mutant did not exhibit binding to either DNA or cisplatinated DNA. To reconcile these data with our experimental analysis of cisplatin cytotoxicity, we hypothesized that this might be due to the mutant protein being a double mutant, as the E.coli protein consists of a MutS homodimer, rather than the eukaryotic Msh2/Msh6 heterodimer. On the other hand, the bacterial homodimer is a ‘structure–function heterodimer’, in which both subunits are non-equivalent. The phenylalanine residue in subunit B is in no direct interaction with DNA. To determine, if a mutation in this residue in subunit B could nonetheless alter the protein structure sufficiently to explain the lack of any DNA binding activity, we modeled this mutation into the E.coli protein (in analogy to E38A, Figure 4). We note that in the X-ray structure, and the modeled structures, the F36 residue resides in subunit B is in a beta-hairpin structure. This hairpin contains Arg32, which provides one of the few protein–DNA interactions (Figure 2; not conserved in eukaryotic protein). We hypothesized that the F36 in subunit B may play a structural role in maintaining non-specific interactions between subunit B and DNA. Using structures from the ends of both the simulations of mismatched DNA and mismatched DNA with cisplatin, the interaction energies between DNA and each residue of the protein were calculated in a pseudovacuum, and Arg32 and Arg500 had the most important non-specific interactions. The importance of these two interactions was robust. These results prompted simulations of the single and double F36A mutants to see if there were conformational changes consistent with our hypothesis observable on a nanosecond time-scale. The simulations need to start with DNA-bound, and are not capable of observing unbinding of DNA, nor large conformation changes, due to the time-scale of the simulation. However, it is possible to observe local conformational changes, which would be sufficient to test our hypothesis. As a result of this simulation, we observed that within one nanosecond of the simulation, the R32-DNA interaction was mostly lost in the double mutant, but not in the single mutant. This supports our hypothesis that the F36A mutation in the second subunit B (‘Msh2’) of the E.coli protein alters the protein structure sufficiently to destabilize the protein–DNA interaction sufficiently to eliminate all DNA binding, which makes it impossible to biochemically determine the difference between mismatch and cisplatin recognition.
Table 3.
Dissociation constants for the interaction of MutS with mismatched and platinated DNA
| MutS | Kd [µM protein] | |||
|---|---|---|---|---|
| G/C | G/T | CP-GpG/CC | CP-GpG/CT | |
| Wild-type | 2.7 ± 0.087 | 0.1 ± 0.008 | 1.3 ± 0.065 | 0.36 ± 0.046 |
| E38A | 0.61 ± 0.058 | 0.16 ± 0.058 | 0.6 ± 0.078 | 0.2 ± 0.069 |
Numbers are mean values and standard deviations of at least four independent experiments. Lesions present in the DNA are indicated.
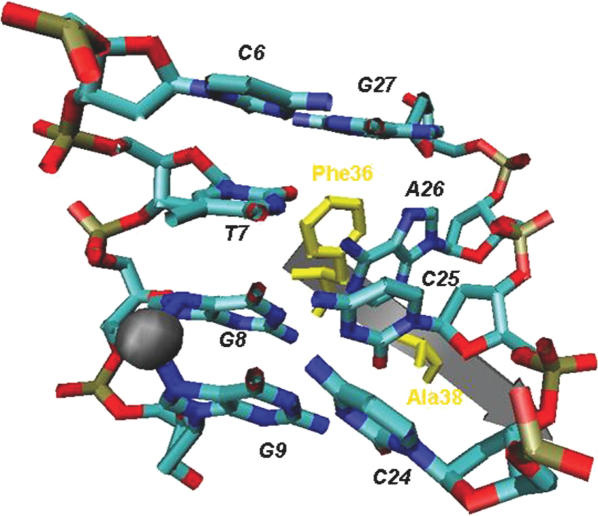
Figure 4.
Base-specific interactions of MutS-E38A with CDDP-DNA. Gold sphere: cisplatinum. The nomenclature for base pairs is the same as shown in Figure 3. Phe36 and the Ala substituting for Glu38 are shown in yellow.
Overall, the computational prediction of a lack of base-stacking interactions is confirmed by the biological experiments. MSH6-F337A results in a strong mutator phenotype that is indistinguishable from a complete knockout in MMR. This result demonstrates the absolute requirement for the Phe residue in MMR. Hence, the contact found to be most important in the interaction with mismatched DNA does not contribute to the interaction with CDDP-DNA and cisplatin cytotoxicity.
Altered base-specific contacts with CDDP-DNA—Glutamate 38
In the structural simulation with CDDP-DNA, a glutamate residue (E.coli E38/subunit A; MSH6-E339) provides significant hydrogen bonds with both cross-linked guanines (Figure 3A). The hydrogen bond containing the glutamate residue switches readily between the two cross-linked guanines in the simulation, although both guanines are always in close contact to the glutamate (Figure 3A). Due to the crosslink, the N7 position of the guanines is not available for hydrogen bonding (26), and a change to the syn conformation that is observed for purine mismatches (9), is prevented. Furthermore, the presence of the positive charge density from the CDDP might be expected to reduce local electrostatic repulsions between the DNA and nearby negatively charged amino acid side chains, e.g. Glu38. Indeed, hydrogen bond formation is predicted between the hydrogens of the N2 position of the guanines and the O-epsilon of Glu38 (Figure 3A). In the interaction with mismatched DNA, the intercalation of the Phe residue re-orients the mismatched base for the interaction with the Glu (9). With the missing Phe base-stacking, no such reorientation of the bases is observed in the simulation studies. The prediction from this observation is that different contacts between the Glu and the CDDP-crosslink are established, which are important for protein–DNA complex formation. To test this prediction, the consequences of a mutation in MSH6-E339 for cisplatin cytotoxicity were analyzed. The mutation confers a resistance phenotype indistinguishable from the complete knockout strain (IC50 of 400 and 430 µM for Δmsh3 msh6, Table 2). The presence of this Glu residue is hence required for cisplatin cytotoxicity. The mutation results in a weak, 23-fold increase in the frameshift mutation rate (compared to 1600-fold for the knockout strain), and a more significant increase in base substitutions (15-fold, compared to 25-fold for the knockout).
To investigate the structural consequences of a mutation in the Glu residue, we simulated the E38A mutation in the MutS-CDDP-DNA model (Figure 4). The number of putative backbone contacts between the mutant MutS and CDDP-DNA increases significantly, so that there are more interactions with MutS in the E39A mutation than in wild-type MutS (computational data not shown). Strikingly, the phenylalanine residue, which is dislocated and functionally dispensable in the MutS wt-CDDP-DNA complex (Figure 3), provides a new protein–DNA interaction in the mutant protein–DNA complex. In the E38A-CDDP-DNA complex, the Phe residue base-stacks with the base 1 nt 3′ to the base pairing cytosine on the strand opposite to the cross-linked guanines (Figure 4). The Phe residue base-stacks with Ade26, which disrupts the A26–T7 base pairing and the base-stacking interactions between A26 and G27. The base pairing of C6 and G27 is preserved (Figure 4). These structural features predict that the mutation of the glutamate residue would enhance DNA binding (due to gained base-stacking interaction and an increased number of contacts with the DNA backbone). This potentially increased affinity should be non-specific for the damage (since the mutation eliminates the direct interactions with the cross-linked bases, but gains backbone contacts).
Purified E.coli MutS-E38A mutant protein was used to directly evaluate the effects on DNA binding by fluorescence polarization (Table 3). The sequence of the 30mer DNA oligomer was chosen to contain only one potential cisplatin site (see Materials and Methods).
The binding affinity of the mutant MutS-E38A was compared to that of wild-type MutS. Increasing amounts of full length wild-type and mutant MutS were incubated with the indicated substrates. Protein–DNA interactions were determined by changes in fluorescence anisotropy and the Kd of interaction determined (Table 3). The wild-type MutS protein recognizes CDDP-DNA; the affinity is about 10-fold weaker than binding to a mismatch (compare Kd hetero of 0.1 µM and Kd CP-GpG/CC of 1.3 µM). The binding affinity for CDDP-DNA is improved when one of the cross-linked guanines is mismatched with a thymine (Kd CP-GpG/CT of 0.36 µM; Table 3). The selectivity for mismatched over matched DNA is significantly weaker when the cisplatin adduct is present (Kd CP-GpG/CC of 1.3 µM and Kd CP-GpG/CT of 0.36 µM; 4-fold difference as compared to 27-fold without cisplatin).
In comparison to wild-type MutS, MutS-E38A shows strongly decreased specificity for mismatched DNA. This is due to increased affinity for homoduplex DNA, while the binding affinity for a G/T mismatch is largely unaffected (Kd homo of 0.61 µM and Kd hetero of 0.16 µM). The affinity for homoduplex is only 3.8-fold lower than for heteroduplex DNA. This increase in non-specific binding is consistent with previously published data on the E38A and E38Q mutant (21,27).
The mutant protein binds to CDDP-DNA with an affinity comparable to the one observed for homoduplex DNA (Kd CP-GpG/CC of 0.6 µM and Kd homo of 0.61 µM; Table 3). Similarly, the binding affinity for a G/T mismatch and mismatched CDDP-DNA is roughly the same (Kd CP-GpG/CT of 0.2 µM and Kd hetero of 0.16 µM). The mutant protein shows no specificity for binding to CDDP-DNA.
Conformational differences in the MutS complex with cisplatinated DNA—ATPase domain
The bacterial MutS crystal structures together with experimental data revealed a highly coordinated series of intramolecular conformational changes that is essential for the initiation of repair. These conformational changes are initiated and controlled by the presence of DNA and nucleotide. The presence of DNA is required to structurally order large parts of the protein (7,9); concerted conformational changes that follow, are associated with the consecutive and highly coordinated binding and hydrolysis of ATP by both subunits (28–35). Both of these events affect each other and are tightly controlled. We hypothesized that the distinct protein–DNA interactions described above alter the DNA and nucleotide-dependent intramolecular conformational changes, and that these different, lesion-dependent changes provide the driving force for the initiation of different downstream events. To test the suggestion of a correlation between DNA interaction, conformational changes and the concomitant induction of different pathways, we compared the structure of MutS-mismatched DNA to the model with platinated DNA and identified regions of largest conformational differences between both structures. Based on the structural model (Figure 1), we identified regions of the protein that show largest deviations between the structures with mismatched and platinated DNA, respectively (Table 4). Large scale changes were identified as protein regions in which alpha carbon atoms were moved significantly (>7 Å) in comparison to the crystal structure with mismatched DNA. These changes are summarized in Table 4. Significant structural deviations are very localized and are observed in the ATP binding/hydrolysis motifs (N2-motif, Walker A motif, RH) (7,8), and the variable and disordered loop regions of the MutS protein. The overall functional asymmetry between both subunits is maintained in the conformational changes. Subunit B, corresponding to eukaryotic MSH2, is more severely affected than subunit A (eukaryotic MSH6). While the ATP binding domain (Walker A motif) is affected in subunit B, no alteration in this motif is observed in subunit A (Table 4). The structural model presented here suggests that ATP binding by MSH2 may be impaired in the complex with platinated DNA due to the conformational dislocation of the binding motif. Hence, a mutation in this subunit (G693A in yMSH2) will not affect damage response. This is in consistence with previous data from our laboratory (1) and others (36). In contrast, the N-2 nt binding motif is affected in both subunits. This motif is part of the composite ATPase site, which undergoes conformational changes during the coordinated cycle of ATP binding and hydrolysis performed by both subunits (9,28,31). The N-2 motif is implied to interact with the γ-phosphate of the nucleotide bound to the neighboring subunit, which thereby acts in trans in ATP hydrolysis (9). Mutations in this motif decrease ATP binding and hydrolysis in the bacterial protein [Table 5; (28)]. The conformational changes that are associated with the ATP binding/hydrolysis cycle are amplified throughout the MutS/MSH protein (28,31). We performed mutational analyses of the highly conserved serine residue in the N-2 motif in both eukaryotic subunits, MSH2 and MSH6, and addressed the question of how these mutations affect cisplatin response compared to MMR. The mutation in the MSH2 protein, S742A, does not alter the response to cisplatin, and the IC50 is identical with wild-type response (Table 5). This is consistent with the fact that the ATPase domain of subunit B/MSH2 is primarily affected and structurally altered by binding to cisplatinated DNA, potentially alleviating any functional requirement (Table 4). The homologous mutation in MSH6 (S1036A) results in an intermediate effect, increasing the IC50 1.7-fold above wild-type (Table 5). This result reflects the less pronounced structural changes in this subunit upon interaction with cisplatinated DNA. Mutations in both MSH2 and MSH6 result in an increased mutator phenotype (Table 5). This increase in the mutation rate is likely associated with the reduced ATPase activity of the mutant proteins, as illustrated by the 2-fold reduced Kcat/Km of the single mutants, and the 33-fold reduced activity of the double mutant (Table 5). These data demonstrate that both N-2 motifs contribute to ATP hydrolysis in the eukaryotic MSH2/MSH6 heterodimer, and this activity is required for functional MMR. In contrast, the activity of the composite ATPase sites is largely dispensable for damage response. The missing effect of the MSH2 mutation on damage response reflects and confirms that the ATP binding and hydrolysis motifs of this subunit are structurally altered, preventing nucleotide binding and hydrolysis, while the MSH6 motifs are less affected. This difference between both subunits demonstrates that the overall asymmetry observed in all functional aspects of the MSH proteins is preserved in damage response, though the contribution of each subunit is altered. The weaker effect on the MSH6 subunit may be attributed to the suggestion that the heterodimer exists as ATP-MSH-ADP, suggestively as MSH2-ADP/MSH6-ATP in the absence of DNA (29,37), with ATP already bound to MSH6 prior to the interaction with DNA.
Table 4.
Largest conformational changes when comparing MutS complexes with mismatched and cisplatinated DNA
| Domain | Subunit A (‘MSH6’) | Affected regions | Subunit B (‘MSH2’) | Affected regions |
|---|---|---|---|---|
| III: Core | 287–288 | Variable loop | 283–289 | Variable loop |
| V: ATPase | 668–669 | Next to N-2 motif | 585–596 | RH motif, interdomain |
| Dimerization | 622–623 | ATP binding (Walker A) | ||
| 660–661 | Disordered loop (yD735) | |||
| 668–669 | N-2 motif |
Large conformational changes are defined as Ca RMSD's >7 Å. Numbers depict amino acid residues in E.coli MutS (7, 9s).
Table 5.
Effects of mutations in the N2-motif of the ATPase domains of MSH2 and MSH6
| Allele | IC50 [µM cisplatin] | Mutation rate in LYS2-A14 | Mutation rate in CAN1 | ATPase | ||||
|---|---|---|---|---|---|---|---|---|
| IC50 | -fold | MR 10−6 | -fold | MR 10−6 | -fold | Protein | Kcat/Kma | |
| Msh2-S742A | 170 ± 46 | 1 | 1300 | 2030 | 2.9 | 6.2 | S742A/Msh6wt | 0.5 |
| Msh6-S1036A | 310 ± 96 | 1.7 | 390 | 205 | 1.2 | 6 | Msh2wt/S1036A | 0.6 |
| S742A/S1036A | 0.03 | |||||||
It was previously shown that the MSH-DNA complex dissociates in the presence of ATP and the concomitant absence of other co-factors. This reaction requires only ATP binding, not hydrolysis. We postulated that if the cisplatinated DNA alters the structure such that ATP is not required for cisplatin cytotoxity, the addition of nucleotide will not affect the protein–DNA interaction with the crosslink. The results of a binding study testing this hypothesis are shown in Table 6. The addition of ATP to the G/T mismatch results in the typical dissociation of the complex. In contrast, the nucleotide has no effect on the binding of the cisplatinated matched DNA. Binding to the cisplatinated, mismatched DNA shows a 2-fold weakening of the protein–DNA interaction, which, however, is not as strong as the one observed with the G/T mismatch. These data demonstrate that ATP does not influence the interaction between MutS and cisplatinated DNA, which is in sharp contrast to the interaction with mismatched DNA, and supports the cytotoxicity data from the mutational analysis.
Table 6.
Dissociation constants for the interaction of MutS with mismatched and platinated DNA in the presence and absence of ATP
| MutS | Kd [µM protein] | |||
|---|---|---|---|---|
| G/C | G/T | CP-GpG/CC | CP-GpG/CT | |
| Wild-type | 2.7 ± 0.087 | 0.1 ± 0.008 | 1.3 ± 0.065 | 0.36 ± 0.046 |
| + ATP | 1.08 ± 0.72 | 0.67 ± 0.14 | 1.34 ± 0.58 | 0.74 ± 0.23 |
| Fold change | +2.5 | −6.7 | 1 | −2.1 |
Fold change when comparing dissociation constant with ATP to those without the nucleotide for the same substrate.
Conformational differences in the MutS complex with cisplatinated DNA—Inter-subunit contacts
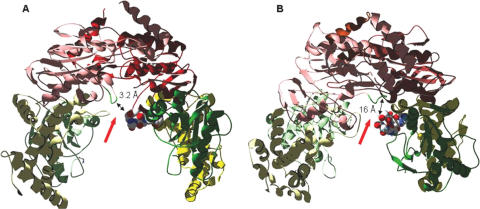
Another set of structural differences between the platinated and mismatched protein–DNA complex involves the disordered loop of subunit B (MSH2) and the variable loop in both subunits. The disordered loop (residues 661–667 in E.coli MutS), which directly precedes the N-2 motif, is not traceable in the structure of the MutS-ADP complex (7,9); this loop becomes ordered and traceable in subunit B (MSH2) in the presence of a non-hydrolyzable ATP analog (31). The same loop region in subunit A (MSH6, the mismatch-interacting subunit) remains disordered. The ordered loop in subunit B communicates the presence of ATP (or its non-hydrolyzable analog) to the variable loop region of domain II in subunit A (Figure 5, left). Both regions, the disordered loop in subunit B/MSH2 and the variable loop in subunit A/MSH6 undergo significant dislocations in the model that replaces the mismatched DNA with platinated DNA (Figure 5). While in the crystal structure of TaqMuts with ADP-BeF, a putative hydrogen bond connects the disordered loop in subunit B with arginine 267 of the variable loop in subunit A, the disordered region in the cisplatin model shows two major conformations. In one conformation, arginine 667 (disordered) and asparagine 289 (variable) are in close enough vicinity to form a hydrogen bond, similar to that observed in the structure with mismatched DNA. However, this conformation exists only for 42% of the simulation. In the second conformation both loop regions are distant, and residues are about 16 Å apart (Figure 5, right). The observation that the MutS-platinated DNA model assumes two different conformations during the simulation—at least one of which preventing inter-subunit interactions—argues that the loop–loop contacts observed with mismatched DNA are not strong enough and disrupted in the complex with cisplatinated DNA. To test the significance of the disordered loop for DNA damage response, we mutated the conserved first aspartate residue in the loop region in both MSH2 and MSH6, and performed cell survival studies. Table 7 demonstrates that a mutation in the conserved aspartate residue in the ‘disordered’ loop region does not affect cell survival after exposure to cisplatin. The IC50 values for both MSH2-D735A (180 µM) and MSH6-D1029A (160 µM) are indistinguishable from the wild-type response (compare to Table 1). In contrast, the mutation rates of Msh2-D735A and Msh6-D1036A mutants are strongly elevated and indistinguishable from a complete knockout (Table 7). This result suggests that the disordered loop region is essential for the repair event, as was implied by studies showing that the nucleotide-dependent structural ‘ordering’ of this loop provides inter-subunit interactions that are essential in the interaction with repair-associated downstream proteins (31). In contrast, our experimental data support the dislocation of this loop region in the complex with cisplatinated DNA, as predicted by the structural model (Figure 5), and the lack or alteration of inter-subunit interactions.
Figure 5.
Schematic presentation of the ‘disordered’ loop of subunit B/MSH2 and the variable loop of subunit A/MSH6 in complexes with mismatched and platinated DNA. The ‘disordered’ loop is shown in green, the variable loop is shown as a space-filling model. (A) Crystal structure of TaqMutS in complex with ADP.beryllium fluoride and mismatched DNA [(31), the only structure with a non-hydrolyzed ATP derivative], showing parts of the connector (green) and core (yellow) domains, and the ATPase/helix–uturn–helix domain (pink). (B) Model of the E.coli MutS in complex with cisplatinated DNA, showing parts of the connector (green) and core (yellow) domains, and the ATPase/helix–uturn–helix domain (pink). Arrows point at the closest distance between the variable loop (space-filling) of subunitA/MSH6 and the ‘disordered’ loop (green) of subunit B/MSH2.
Table 7.
Effects of mutations in the disordered loop of MSH2 and MSH6
| Allele | IC50 [µM cisplatin] | Mutation rate in LYS2-A14 | Mutation rate in CAN1 | |||||
|---|---|---|---|---|---|---|---|---|
| IC50 | -fold | MR 10−6 | CL 10−6 | -fold | MR 10−6 | CL 10−6 | -fold | |
| Msh2-D735A | 180 ± 87 | 1.1 | 950 | 700–1400 | 1500 | 11 | 8.8–33 | 23 |
| Msh6-D1029A | 160 ± 25 | 0.89 | 300 | 140–1100 | 160 | 7.1 | 2.7–19 | 36 |
MR: mutation rate, CL: 95% confidence limits; -fold: in comparison to wild-type (italics).
DISCUSSION
This is the first report to describe distinct structural features of MSH-CDDP-DNA interactions and localized conformational changes that depend on the nature of the lesion, as the mechanism behind the initiation of the MMR protein-dependent cell death, and its autonomy from mismatch recognition and repair. Individual putative interactions between MutS and CDDP-DNA were determined by mutational analysis. Mutational analyses are aided by predictions made from MD simulations (Figure 1). This approach has its limitations, but in the absence of structural information, it is valuable in predicting protein–DNA interactions that can be tested in the experimental setting. Force fields similar to the ones described here have been used to study the structure and dynamics of nucleic acids and nucleic acid–protein complexes in a wide variety of situations, e.g. in protein-mediated base-flipping (38), structural fluctuations in RNA and DNA (39), the energetic analysis of DNA structures (40) and the evaluation of dynamics and energetics of modified DNA (41). The ability of empirical force fields to model atomic level details of biomolecular systems that include proteins, nucleic acids, lipids and carbohydrates has recently been reviewed (42). Results from our experimental data, herein and described previously (1), validate the predictions from the MD simulations for the type of study described herein.
The flexibility of DNA was previously suggested to be a main contributor to the specificity of mismatch recognition. This was supported by the observation that MutS provides very few mismatch-specific interactions, and the majority of contacts occur to the DNA backbone, making a specific ‘readout’ mechanism for each individual mismatch unlikely (8–10). The interaction of MutS with mismatched DNA is rather a general, ‘induced fit’ mode, in which intrinsically destabilized substrates are preferred (21). The presence of a mismatch by itself does not cause any dramatic structural changes in DNA (43–47). It, however, locally destabilizes the DNA, as determined by alterations in the melting temperature (48). This may provide the additional flexibility required for the specific interaction with MutS. In the presence of MutS, a 60° kink is introduced at the site of the mismatch. Recently, it was proposed that a series of DNA bending, kinking and ultimately unbending is required to initiate repair (49). Though the unbending event still requires additional verification, it is clear that the induction of repair requires considerable flexibility of the MutS-DNA complex. In contrast, the 1,2-GpG intrastrand crosslink introduced by cisplatination itself generates a kink in DNA (24,25,50). The structural features of this kink resemble those found for the mismatched DNA in its complex with MutS. Our data demonstrate that the cisplatin adduct leaves the DNA considerably more rigid, and does not allow the flexibility observed for the mismatched DNA (data not shown). The similarity of the cisplatin-induced DNA bend may promote recognition by MutS (Table 3), but the inflexibility of the adducted DNA may impair further downstream events that are directed towards repair. The increased rigidity of CDDP-DNA in complex with MutS may be a determining factor for the initiation of the cell death pathway. The effect of other types of DNA damage that introduce different local distortions remains to be determined and is currently under investigation in our laboratory. In addition, it was recently suggested that the substrate specificity observed for MMR is provided not so much by the initial protein–DNA interactions, but dramatically amplified by downstream events, such as the excision event (51). Whether this is true for the binding to cisplatinated DNA remains to be determined.
The structure of the MutS protein in complex with five different mismatches identified a general binding mode (10). The comparison of the MutS-CDDP-DNA model with the structure containing mismatched DNA reveals most prominent structural differences in the DNA binding domains I and IV (Figure 1). Our functional analysis based on predictions of individual protein contacts with CDDP-DNA (Figure 2) demonstrate that the general binding mode is conserved in the interaction with CDDP-DNA. However, the biological significance of the protein–DNA backbone contacts differs markedly (Table 1). A shift in the backbone interactions with CDDP-DNA is likely to induce different conformational changes throughout the protein (see below).
The location of residues that make specific interactions with mismatches is significantly distorted in the interaction with CDDP-DNA (Figure 3: Table 2). In the structure with mismatched DNA, a conserved Phe residue (F36 of subunit A in E.coli) base-stacks with one of the mismatched bases (8–10). This base-stacking interaction is critical for DNA binding and repair activity; a mutation in this residue abrogates mismatch recognition and repair (10–12,21,52). The intercalation re-orients the mismatched base such that Glu38 (E.coli) can form hydrogen bonds with the base. This conserved residue contacts the N3 of pyrimidines and N7 of purines. Mismatches containing purines enter this interaction in the syn conformation (10). In contrast, no intercalation of the Phe residue is predicted by the model with CDDP-DNA (Figure 3), and experimental data confirm that this residue is not required in cisplatin cytotoxicity (Table 2). Due to this missing base-stacking interaction, no re-positioning of the guanine for hydrogen bonding with the glutamate residue is observed.
The (1,2)GpG cisplatin adduct cross-links two adjacent guanines via their N7 positions, leaving this position inaccessible for interactions with the Glu residue. In addition, the crosslink prevents the transition to the syn conformation. The Glu, instead, interacts with the N2 position of the cross-linked guanines (Figure 3). The conservative mutation of this Glu to a glutamine reduces cisplatin cytotoxicity (E339Q, Table 2). A mutant protein bearing an E to Q change in E.coli binds to homoduplex DNA with enhanced affinity (Table 3). At first glance, the increased cisplatin resistance, together with the increased DNA binding activity of the Glu mutant appears counterintuitive. Though differences between the bacterial and eukaryotic proteins cannot be excluded, the DNA binding analysis (Table 3), together with the structural simulation of the mutant protein (Figure 4, Supplementary Figure 1), may explain this discrepancy.
The mutant protein gains base-stacking interactions of Phe36 at a lesion-independent site (Figure 4), and increases the overall number of protein-CDDP-DNA backbone contacts (data not shown). This increased, non-specific binding may be reinforced by the elimination of the repulsive, negative charge. The binding affinity for homoduplex DNA increases disproportionately to the affinity for CDDP-DNA (Table 3). Hence, the mutant protein lacks any specificity for the recognition of CDDP-DNA, and will bind to homoduplex DNA with the same affinity. This lack of selectivity may result in increased recombinational bypass or translesion synthesis (53), which is detected as an increase in cisplatin resistance. Taken together, these data demonstrate that the Glu38 residue plays a significant role in the interaction with CDDP-DNA, the direct interaction with the cross-linked bases and the re-positioning of protein–DNA contacts. Further analysis is required that will determine whether the findings of this report can be transferred to the interaction of MutS with other types of DNA damage. These studies are underway.
As a result of different protein–DNA interactions that are observed in the MutS-platinated DNA complex, distinct conformational changes are transmitted throughout the protein that differ from the complex with mismatched DNA, and are localized to the ATPase domain, the disordered loop of subunit B/MSH2 and the variable loops of both subunits. These conformational changes will prevent ATP binding by MSH2 and hydrolysis by both subunits. This is consistent with data demonstrating that neither function is required for the induction of cell death after cisplatin exposure (1,36) (Tables 5 and 6). The dislocation of the disordered loop in subunit B/MSH2 and the variable loops results in an increased distance between these loop regions, resulting in unstable inter-subunit interactions (Figure 5; Table 7) which prevent signal transmission between the ATPase domains and the DNA binding domains. These missing inter-subunit contacts are predicted to result in altered DNA release patterns and interactions with downstream proteins, likely factors that can lead to the initiation of the cell death pathway, and at the same time hinder repair. Whether this is provoked by the MSH-DNA complex presenting a ‘suicide or dead end complex’ with an increased lifetime that prevents repair and hinders replication, or a dynamic complex, is an important question and remains to be determined. Numerous direct interactions of MMR proteins with proapoptotic proteins suggest a direct role in the initiation of cell death and cell cycle checkpoint control in response to certain DNA damage (54–56).
For a cell to be directed towards cell death response, repair has to be inhibited or be otherwise impossible, and proapoptotic proteins have to be recruited and activated. We here present first experimental data on the signals that preferentially trigger MMR-dependent damage response, and may result in the prevention of repair. We propose a model for MMR-dependent damage response in which altered DNA flexibility and changes in protein–DNA interactions are transmitted across the MSH protein via distinct conformational changes. These structural changes alter the nucleotide requirements, which ultimately affect intermolecular contacts involved in the recruitment of and interaction with downstream proteins. MMR in general is thought to act more rapid than NER (57). Thus, the kinetics of MMR proteins, together with the herein described response to DNA damage may in addition to preventing MMR per se, provide an impediment for other repair proteins, such as NER proteins, to access the side of the lesion. This event would present yet another contribution toward cell death and inhibition of repair. Mutants in all functionally important regions of the protein demonstrate a lack of correlation between the functional requirements for cisplatin cytotoxicity and repair. The identification of these mutants clearly demonstrates that MSH-dependent damage response can operate independently of the repair function. This result argues against the initiation of futile repair cycles, though we cannot exclude that such a mechanism may have an additional contribution to MMR-dependent cell death response. An increasing body of evidence is emerging that suggests a direct role for MMR-dependent apoptosis. Due to their separate functional requirements, MMR mutations that promote carcinogenesis may not predict the response to chemotherapy, and vice versa. We present the first report on the role of DNA flexibility and specific protein–DNA interactions as significant contributors in the MMR-dependent induction of cell death. Apart from a general mechanistic understanding, the knowledge on how certain MMR defects modulate the response to chemotherapeutics will allow a more conscientious choice of cancer treatment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Fred Perrino for helpful discussions and P. Hsieh for the generous gift of the MutS expression plasmid. The computational modeling was performed on the WFU DEAC cluster, supported in part by start-up funds to F.R.S. This work was supported by the CCC of WFUBSM and NIH (CA101829) awarded to K.D.S. and American Cancer Society (RSG-04-187-01-GMC) awarded to T.H. Funding to pay the Open Access publication charges for this article was provided by NIH CA101829.
Conflict of interest statement. None declared.
REFERENCES
- 1.Drotschmann K., Topping R.P., Clodfelter J.E., Salsbury F.R. Mutations in the nucleotide-binding motif of MutS homologs uncouple cell death from cell survival. DNA Repair. 2004;3:729–742. doi: 10.1016/j.dnarep.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Irving J.A., Hall A.G. Mismatch repair defects as a cause of resistance to cytotoxic drugs. Expert Rev. Anticancer Ther. 2001;1:149–158. doi: 10.1586/14737140.1.1.149. [DOI] [PubMed] [Google Scholar]
- 3.Fedier A., Fink D. Mutations in DNA mismatch repair genes: implications for DNA damage signaling and drug sensitivity. Int. J. Oncol. 2004;24:1039–1047. [PubMed] [Google Scholar]
- 4.Lin D.P., Wang Y., Scherer S.J., Clark A.B., Yang K., Avdievich E., Jin B., Werling U., Parris T., Kurihara N., et al. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 5.Yang G., Scherer S.J., Shell S.S., Yang K., Kim M., Lipkin M., Kucherlapati R., Kolodner R.D., Edelmann W. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ. 2001;8:1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- 7.Lamers M.H., Perrakis A., Enzlin J.H., Winterwerp H.H.K., de Wind N., Sixma T.K. The crystal structure of DNA MMR protein MutS binding to a G/T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 8.Obmolova G., Bam C., Hsieh P., Yang W. Crystal structures of MMR protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 9.Natrajan G., Lamers M.H., Enzlin J.H., Winterwerp H.H., Perrakis A., Sixma T.K. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowers J., Sokolsky T., Quach T., Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J. Biol. Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 11.Dufner P., Marra G., Raschle M., Jiricny J. Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutSalpha is abolished by a single mutation in the hMSH6 subunit. J. Biol. Chem. 2000;275:36550–36555. doi: 10.1074/jbc.M005987200. [DOI] [PubMed] [Google Scholar]
- 12.Drotschmann K., Yang W., Brownewell F.E., Kool E.T., Kunkel T.A. Asymmetric recognition of DNA local distortion. Structure-based functional studies of eukaryotic Msh2-Msh6. J. Biol. Chem. 2001;276:46225–46229. doi: 10.1074/jbc.C100450200. [DOI] [PubMed] [Google Scholar]
- 13.Li G.-M. The role of MMR in DNA damage-induced apoptosis. Oncol. Res. 1999;11:393–400. [PubMed] [Google Scholar]
- 14.Fink D., Nebel S., Aebi S., Zheng H., Cenni B., Nehme A., Christen R.D., Howell S.B. The role of DNA MMR in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 15.Drummond J.T., Anthoney A., Brown R., Modrich P. Cisplatin and adriamycin resistance are associated with MutLalpha and MMR deficiency in an ovarian tumor cell line. J. Biol. Chem. 1996;21:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 16.Marcelis C.L.M., van der Putten H.W.H.M., Tops C., Lutgens L.C.H.W., Moog U. Chemotherapy resistant ovarian cancer in carriers of an hMSH2 mutation? Fam. Cancer. 2001;1:107–109. doi: 10.1023/a:1013865323890. [DOI] [PubMed] [Google Scholar]
- 17.Brooks B.R., Bruccoleri R.D., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 18.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins and nucleic acids. J. Phys. Chem. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 19.Humphrey W., Dalke A., Schulten K. VMD—visual molecular dynamics. J. Molec. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 20.Kale L., Skeel R., Bhandarkar M., Brunner R., Gursoy A., Krawetz N., Phillips J., Shinozaki A., Varadarajan K., Schulten K. NAMD2: greater scalability for parallel molecular dynamics. J. Comp. Phys. 1999;151:283–312. [Google Scholar]
- 21.Schofield M.J., Brownewell F.E., Nayak S., Du C., Kool E.T., Hsieh P. The Phe-X-Glu DNA binding motif of MutS. J. Biol. Chem. 2001;276:45505–45508. doi: 10.1074/jbc.C100449200. [DOI] [PubMed] [Google Scholar]
- 22.Mello J.A., Acharya S., Fishel R., Essigmann J.M. The mismatch-repair protein hMSH2 binds selectively to DNA adducts of the anticancer drug cisplatin. Chem. Biol. 1996;3:579–589. doi: 10.1016/s1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 23.Clodfelter J.E., Gentry M.B., Drotschmann K. MSH2 missense mutations alter cisplatin cytotoxicity and promote cisplatin-induced genome instability. Nucleic Acids Res. 2005;33:3323–3330. doi: 10.1093/nar/gki646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahara P.M., Rosenzweig A.C., Frederick C.A., Lippard S.J. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 25.Takahara P.M., Frederick C.A., Lippard S.J. Crystal structure of the anticancer drug cisplatin bound to duplex DNA. J. Am. Chem. Soc. 1996;118:12309–12321. [Google Scholar]
- 26.Fichtinger-Schepman A.M., van der Veer J.L., den Hartog J.H., Lohman P.I., Reedijk J. Adducts of the antitumor drug cis-diamminedichloroplatinum (II) with DNA: formation, identification, and quantitation. Biochemistry. 1985;24:707–713. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]
- 27.Junop M.S., Yang W., Funchain P., Clendenin W., Miller J.H. In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA Repair. 2003;2:387–405. doi: 10.1016/s1568-7864(02)00245-8. [DOI] [PubMed] [Google Scholar]
- 28.Junop M.S., Obmolova G., Rausch K., Hsieh P., Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 29.Bjornson K.P., Modrich P. Differential and simultaneous adenosine di- and triphosphate binding by MutS. J. Biol. Chem. 2003;278:18557–18562. doi: 10.1074/jbc.M301101200. [DOI] [PubMed] [Google Scholar]
- 30.Blackwell L.J., Bjornson K.P., Allen D.J., Modrich P. Distinct MutS DNA-binding modes that are differentially modulated by ATP binding and hydrolysis. J. Biol. Chem. 2001;276:34339–34347. doi: 10.1074/jbc.M104256200. [DOI] [PubMed] [Google Scholar]
- 31.Alani E., Lee J.Y., Schofield M.H., Kijas A.W., Hsieh P., Yang W. Crystal structure and biochemical analysis of the MutS-ADP-Beryllium Fluoride complex suggests a conserved mechanism for ATP interactions in mismatch repair. J. Biol. Chem. 2003;278:16088–16094. doi: 10.1074/jbc.M213193200. [DOI] [PubMed] [Google Scholar]
- 32.Lamers M.H., Winterwerp H.H.K., Sixma T.K. The alternating ATPase domains of MutS control DNA mismatch repair. EMBO J. 2003;22:746–756. doi: 10.1093/emboj/cdg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iaccarino I., Marra G., Palombo F., Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gradia S., Subramanian D., Wilson T., Acharya S., Makhov A., Griffith J., Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 35.Antony E., Hingorani M.M. Asymmetric ATP binding and hydrolysis activitiy of the Thermus aquaticus MutS dimer is key to modulation of its interactions with mismatched DNA. Biochem. 2004;43:13115–13128. doi: 10.1021/bi049010t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin D.P., Wang Y., Scherer S.J., Clark A.B., Yang K., Avdievich E., Jin B., Werling U., Parris T., Kurihara N., et al. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 37.Martik D., Baitinger C., Modrich P. Differential specificities and simultaneous occupancy of human MutSalpha nucleotide binding sites. J. Biol. Chem. 2004;279:28402–28410. doi: 10.1074/jbc.M312108200. [DOI] [PubMed] [Google Scholar]
- 38.Huang N., Banavali N.K., MacKerell A.D. Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase. Proc. Natl Acad. Sci. USA. 2003;100:68–73. doi: 10.1073/pnas.0135427100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y.P., MacKerell A.D. Altered structural fluctuations in duplex RNA versus DNA: a conformational switch involving base pair opening. Nucleic Acids Res. 2003;31:7131–7140. doi: 10.1093/nar/gkg941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foloppe N., Nilsson L., MacKerell A.D. Ab initio conformational analysis of nucleic acid components: intrinsic energetic contributions to nucleic acid structure and dynamics. Biopolymers. 2001;61:61–76. doi: 10.1002/1097-0282(2001)61:1<61::AID-BIP10047>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 41.Banavali N.K., MacKerell A.D. Re-examination of the intrinsic, dynamic and hydration properties of phosphoramidate DNA. Nucleic Acids Res. 2001;29:3219–3230. doi: 10.1093/nar/29.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacKerell A.D. Empirical force fields for biological macromolecules: overview and issues. J. Comput. Chem. 2004;25:1584–1605. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 43.Hunter W.N., Brown T., Anand N.N., Kennard O. Structure of an adenine-cytosine base pair in DNA and its implications for mismatch repair. Nature. 1986;320:552–555. doi: 10.1038/320552a0. [DOI] [PubMed] [Google Scholar]
- 44.Hunter W.N., Brown T., Kneale G., Anand N.N., Rabinovich D., Kennard O. The structure of a guanosine-thymidine mismatches in B-DNA at 2.5 Å resolution. J. Biol. Chem. 1987;262:9962–9970. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- 45.Skelly J.V., Edwards K.J., Jenkins T.C., Neidle S. Crystal structure of an oligonucleotide duplex containing G.G base pairs: influence of mispairing on DNA backbone conformation. Proc. Natl Acad. Sci. USA. 1993;90:804–808. doi: 10.1073/pnas.90.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter W.N., Brown T., Kennard O. Structural features and hydration of a dodecamer duplex containing two C.A mispairs. Nucleic Acids Res. 1987;15:6589–6606. doi: 10.1093/nar/15.16.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peyret N., Seneviratne P.A., Allawi H.T., SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A,C.C,G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 48.Werntges H., Steger G., Riesner D., Fritz H.J. Mismatches in DNA double strands: thermodynamic parameter and their correlation to repair efficiencies. Nucleic Acids Res. 1986;14:3773–3790. doi: 10.1093/nar/14.9.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H., Yang Y., Schofield M.J., Du C., Fridman Y., Lee S.D., Larson E.D., Drummond J.T., Alani E., Hsieh P., et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl Acad. Sci. USA. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gelasco A., Lippard S.J. NMR solution structure of a DNA dodecamer duplex containing a cis-diammineplatinum (II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin. Biochemistry. 1998;37:9230–9239. doi: 10.1021/bi973176v. [DOI] [PubMed] [Google Scholar]
- 51.Hays J.B., Hoffman P.D., Wang H. Discrimination and versatility in mismatch repair. DNA Repair. 2005;4:1463–1474. doi: 10.1016/j.dnarep.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto A., Schofield M.J., Biswas I., Hsieh P. Requirement for Phe36 for DNA binding and mismatch repair byEscherichia coli MutS protein. Nucleic Acids Res. 2000;28:3564–3569. doi: 10.1093/nar/28.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bassett E., King N.M., Bryant M.F., Hector S., Pendyala L., Chaney S.G., Cordeiro-Stone M. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res. 2004;64:6469–6475. doi: 10.1158/0008-5472.CAN-04-1328. [DOI] [PubMed] [Google Scholar]
- 54.Shimodaira H., Yoshioka-Yamashita A., Kolodner R.D., Wang J.Y.J. Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc. Natl Acad. Sci. USA. 2003;100:2420–2425. doi: 10.1073/pnas.0438031100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S.J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 56.Mac Partlin M., Homer E., Robinson H., McCormick C.J., Crouch D.H., Durant S.T., Matheson E.C., Hall A.G., Gillespie D.A., Brown R. Interactions of the DNA mismatch repair proteins MLH1 and MSH2 with c-MYC and MAX. Oncogene. 2003;22:819–825. doi: 10.1038/sj.onc.1206252. [DOI] [PubMed] [Google Scholar]
- 57.Wang H., Hoffman P.D., Lawrence C., Hays J.B. Testing Excision models for responses of mismatch-repair systems to UV photoproducts in DNA. Environ. Mol. Mutagen. 2006;47:296–306. doi: 10.1002/em.20206. [DOI] [PubMed] [Google Scholar]
- 58.Drotschmann K., Yang W., Kunkel T.A. Evidence for sequential action of two ATPase active sites in yeast Msh2-Msh6. DNA Repair. 2002;1:743–753. doi: 10.1016/s1568-7864(02)00081-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.