Abstract
Allosteric nucleic acid ligases have been used previously to transform analyte-binding into the formation of oligonucleotide templates that can be amplified and detected. We have engineered binary deoxyribozyme ligases whose two components are brought together by bridging oligonucleotide effectors. The engineered ligases can ‘read’ one sequence and then ‘write’ (by ligation) a separate, distinct sequence, which can in turn be uniquely amplified. The binary deoxyribozymes show great specificity, can discriminate against a small number of mutations in the effector, and can read and recode DNA information with high fidelity even in the presence of excess obscuring genomic DNA. In addition, the binary deoxyribozymes can read non-natural nucleotides and write natural sequence information. The binary deoxyribozyme ligases could potentially be used in a variety of applications, including the detection of single nucleotide polymorphisms in genomic DNA or the identification of short nucleic acids such as microRNAs.
INTRODUCTION
Allosteric nucleic acid enzymes have been generated previously by both design and selection. Enzymes whose activities are modulated by small molecules have been generated by fusing ribozyme domains with allosteric domains [aptamers, yielding aptazymes; (1–10)], while enzymes whose activities are modulated by oligonucleotides have been generated by the strategic insertion of hybridization sites (11–19). In most instances, the modulation of catalytic function has relied on the analyte-dependent re-organization of secondary or tertiary structure. In contrast, a ‘maxizyme’ has been developed in which the modulation of catalytic function relied on the analyte-dependent formation of a specific quarternary structure (20). In this design, two half-ribozymes were brought together by a bridging oligonucleotide in order to form an active hammerhead ribozyme (Figure 1).
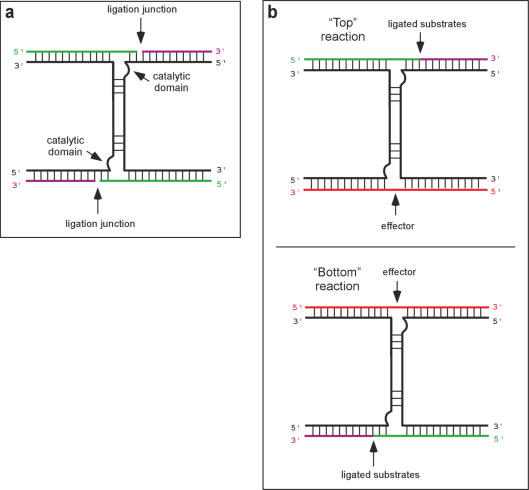
Figure 1.
Design of the bidirectional ligase maxizyme. (a) The DNA ligase was designed to be a binary (two black strands) enzyme with two catalytic domains fused by a common stem structure. When the binary strands associate to form the correct structure, the catalytic domains are formed and are capable of ligating two DNA substrates (green and purple). (b) An effector DNA (red) can specifically base pair with the binary enzyme, stimulating the correct folded structure and catalyzing the ligation of two substrates on the opposite end. In this way the effector oligonucleotide is recoded into a new oligonucleotide ligation product. For convention, we refer to ligation of substrates on the ‘bottom’ and on the ‘top’ of the enzyme.
We have engineered a DNA ligase to function in a manner similar to the maxizyme, in that the half-deoxyribozymes can be activated by a bridging oligonucleotide to carry out a ligation (rather than a cleavage) reaction. The engineered deoxyribozyme can recode nucleic acid information by ‘reading’ one sequence through hybridization and then ‘writing’ a separate sequence by ligation (Figure 1). Since the newly ligated sequence can be a unique template for amplification, the ligase maxizyme can potentially find use in recoding short, hard-to-detect sequences (such as antisense oligonucleotides or microRNAs) into longer templates that can be readily detected by PCR.
As a proof-of-principle, we show that the ligase maxizyme is highly and specifically activated by cognate oligonucleotides, functions faithfully against a background of genomic DNA, and can even read oligonucleotides containing modified nucleotides.
MATERIALS AND METHODS
Sequences of deoxyribozymes, effectors and substrates
All of the ligase maxizymes were composed of two oligonucleotides, which we designate as the left (L) and right (R) subunits. The sequences of the oligonucleotides are as follows: dR8(L), 5′-CGAAGACAGGTTGTGGCCGCATTAAAA-3′; dR8(R), 5′-AAAAAAACGTTGACCTCTGCTTAGTC-3′; dR3(L), 5′-CGAAGACAGGTTGTGGAGGTTGCCGCATTAAAA-3′; dR3(R), 5′-AAAAAAACGTTGGCTTCCAACACTGCTTAGTC-3′; dR3.1.5(L), 5′-CGAAGACAGGTTGTGTTGGTTGCCGCATTAAAA-3′; and dR3.1.5(R), 5′-AAAAAAACGTTGGCTTCCTTCACTGCTTAGTC-3′. The sequence of the forward effector oligonucleotide, 18N, is 5′-TTTAATGCCGTTTTTTT-3′. The sequence of the reverse effector oligonucleotide, 19N, is 5′-GACTAAGCACCTGTCTTCG-3′. There are two substrate oligonucleotides for the ligation reactions. The 5′ substrates are terminated with a 3′-phosphorothioate (PS) group while the 3′ substrates carried a 5′-iodine (I) group. The sequences of the substrates for the ‘forward’ reaction are KSS2, 5′-TACATGTCTATCGATCTGACTAAGCACC-PS-3′, and 5I.8.c14.m1, 5′-I-TGTCTTCG-3′. The sequences of the substrates for the ‘reverse’ reaction are 3PS.18N.ol1, 5′-TTTTAATGCCG-PS-3′; and 5I.18N.ol2, 5′-I-TTTTTTTACACTTACGAACGT-3′.
The non-natural ligase maxizyme is composed of the oligonucleotides dR8b(L), 5′-CGAAGACAGGTTGTGGCCGCATTiCAAA-3′; and dR8b(R), 5′-AAAiCAAACGTTGACCTCTGCTTAGTC-3′ where iC indicates 5-methyl isocytosine. The sequence of the forward effector for dR8B, 18Nb, is 5′-TTTiGAATGCCGTTTiGTTT-3′, where iG indicates isoguanosine.
All natural and non-natural oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). All phosphorothioate- and iodine-bearing oligonucleotide substrates were synthesized in our laboratory as described in Ref. (3). All oligonucleotides were purified by denaturing PAGE prior to use.
Ligation assays
All ligation reactions were conducted in a volume of 10 μl at 25°C unless otherwise noted. Reactions were assembled by the addition of 10 pmol of each deoxyribozyme subunit, and 20 pmol of DTT treated phosphorothioate-bearing substrate to 1× reaction buffer [500 mM NaCl, 50 mM Tris–HCl (pH 7.4), 10 mM MgCl2 and 500 μM DTT]. The reactions were denatured for 3 min at 70°C and then cooled to 25°C at 0.2°C/s. An aliquot of 10 pmol effector olignonucleotide or H2O (in the effector-independent reactions) was then added, and the reaction was finally initiated by the addition of 20 pmol of the iodine-bearing substrate. Reactions were terminated by the addition of 4 vol of 95% formamide containing bromophenol blue.
Prior to the ligation reaction, the 3′ substrate was radiolabeled using 3′-terminal deoxynucleotidyl transferase (Invitrogen, Carlsbad, CA) and dideoxyadenosine 5′-[α-32P]triphosphate (Amersham Pharmacia Biotech, Piscataway, NJ). After the ligation reaction, the ligated and unligated species were separated on a denaturing 8% polyacrylamide gel containing 7 M urea. Ligation was quantitated using a Phosphorimager (Molecular Dynamics, Sunnyvale, CA) and ImageQuant software.
Reactions were initially linear as a function of time, and pseudo-first order rates were calculated from a best fit line passing through at least three data points taken at <10% total ligation.
Real-time PCR detection
To accommodate real-time PCR-based detection, the forward substrates were modified. KSS2 was modified to contain a primer-binding site at its 5′ end, followed by a region whose reverse complement could bind to a TaqMan probe (described below), while oligonucleotide 5I.8.c14.m1 was modified to contain a primer-binding site at the 3′ end. The sequence of the resulting 5′ substrate 3PS.RTs1 was 5′-GTGACTTCGTGGAACTATCTAGCGGTGTACGTGAGTGGGCATGTAGCAAGAGGGACTAAGCACC-PS-3′, and the 3′ substrate 5I.RTs2 was 5′-I-TGTCTTCGGTCATCATTCGAATCGTACTGCAATCGGGTATT-3′. The enzyme dR8b(R) was also modified to carry a 3′-amine modification in order to stop nucleotide extension on 5I.RTs2 during PCR. The sequence of the TaqMan probe PLA.TqMnPb was 5′-6FAM-TGTACGTGAGTGGGCATGTAGCAAGAGG-BHQ1-3′ where BHQ1 indicates Black Hole Quencher™1 (IDT, Coralville, IA). The sequences for real-time PCR were adapted from Ref. (21).
For real-time PCR detection, ligation reactions were conducted for 5 min as described above and then directly diluted 1:50 into a real-time PCR mix. Real-time PCR was performed on an MJ DNA Engine Opticon (Bio-Rad, Hercules, CA). The reaction conditions were 20 mM Tris–HCl (pH 8.3), 50 mM KCl, 0.2 mM dNTPs, 500 nM 5′ and 3′ primers, 75 nM PLA.TqMnPb, 0.5× SmartCycler additive [0.1 mg/ml non-acetylated BSA, 75 mM trehalose and 0.1% Tween-20 in 8.5 mM Tris–HCl (pH 8.0)] and 1.5 U of Platinum Taq DNA polymerase (Invitrogen). All real-time PCRs were carried out in a volume of 50 μl. The samples were heated at 92°C for 5 min then cycled 50 times at 92°C for 1 min, 50°C for 1 min and 72°C for 1 min. The fluorescence intensity was measured at the end of each 72°C extension step. Amplification was controlled for using 1 pM full-length template bearing both primer-binding sites and a TaqMan probe-binding region as in Ref. (22).
RESULTS
Design of a binary deoxyribozyme ligase
We have previously used in vitro selection to evolve a deoxyribozyme ligase that can catalyze the formation of internucleotide phosphorothioester linkages (22,23). This enzyme relies upon chemistry pioneered by Xu and Kool (24), in which a 3′ phosphorothioate displaces a 5′ iodide group, resulting in a phosphorothioester. The deoxyribozyme has a small hairpin stem that functions as a catalytic domain, and two single-stranded arms that can base pair with DNA substrates.
At around the same time, the hammerhead ribozyme was re-engineered to act as an allosteric ribozyme or ‘maxizyme’ that could be activated by hybridization to a nucleic acid effector (20). We noted that since the structure of the deoxyribozyme ligase was superficially similar to that of the hammerhead ribozyme it should be possible to engineer a nucleic acid sequence-dependent ligase. The ligase maxizyme was generated by fusing two catalytic domains via a common stem structure (Figure 1a).
The resultant binary deoxyribozyme is composed of two half-molecules, and the catalytic ability of either ligase domain should depend upon the association of the half-molecules. DNAs that bridge the substrate-binding arms should also template the association and folding of the half-molecules, and thus could act as sequence-specific effectors of catalysis. Since there are two catalytic domains, either end of the ligase can bind either substrate or effector DNAs. This duality in turn allows one piece of sequence information to act indirectly upon another, via the intervening enzyme (Figure 1b).
Effector-dependent catalysis
A number of different deoxyribozyme constructs were designed in which the specificity domains (binding arms) accompanying the two catalytic units were different in order to create distinct reactions on each side the maxizyme. For convenience, we will discuss reactions in the ‘bottom’ direction (ligation on the ‘bottom’ of the construct) and in the ‘top’ direction (ligation on the ‘top’) (Figure 1b). The sequence and stability of the stem structure connecting the top and bottom catalytic domains was then systematically varied.
Initially, the deoxyribozymes were assayed for dependence on an effector oligonucleotide that spanned the hybridizing arms on the top side and activated ligation of substrates that bound the bottom side. Of the designs that were assayed, several proved to be constitutively ‘on’ (required no effector), while others were constitutively ‘off’ (could not be activated by effector). For the remainder, activation varied significantly (Supplementary Data). Two constructs (dR8 and dR3) showed strong effector-dependence (Figure 2).
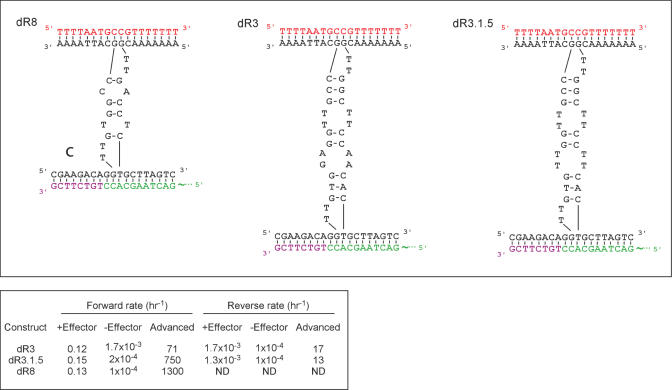
Figure 2.
Secondary structures of deoxyribozymes dR8, dR3 and dR3.1.5. The two oligonucleotides composing the enzymes (black) fold into the active structure (shown) in the presence of the effector (18N, red) in order to bind two substrates (green and purple, as in Figure 1). This orientation is designated as the bottom orientation, and is the standardized presentation of the ligase maxizyme.
While it should be possible for the ligase maxizymes to be activated in either the bottom or top direction (i.e. with a full-length effector oligonucleotide at either end), activation was found to occur predominantly in one direction, the bottom direction. Activation required a full-length effector oligonucleotide; the two oligonucleotide substrates which compose the effector did not stabilize the maxizyme structures (data not shown).
The construct dR8 had a relatively fast rate of bottom ligation (0.13 h−1) and showed the largest effector-dependence (1300-fold). This construct, however, showed no detectable ligase activity in the top direction. Construct dR3 showed approximately the same rate of ligation as dR8 in the bottom direction, but also had a higher rate of effector-independent ligation. This resulted in an overall lower effector-dependence (71-fold). Construct dR3 also displayed modest effector-dependent ligation in the top direction (0.0017 h−1; 17-fold activation).
We hypothesized that the differences between ligation rates in the bottom and top directions might be due to the inherent asymmetry of the maxizymes, and that a more symmetrical construct might be activated in both directions. Therefore, all bulge sequences in the internal stem structure of dR3 were mutated to T–T mispairs, creating dR3.1.5. This modification had no significant effect on top ligation, but did increase activation in the bottom direction from 71- to 740-fold. The background activity of dR3.1.5 was also significantly lower than dR3, presumably because T–T base-pairing is inherently less favorable than A–A or A–G base pairing.
We attempted to measure substrate turnover by dR3 and dR8. When monitoring ligation as a function of time, no accumulation of ligation products was observed in the absence of effector, consistent with previous results. In the presence of the effector, some accumulation of ligation products was observed, but it was extremely slow, yielding a maximum turnover number of ∼4 at >100 h (data not shown). The lack of turnover in our system can most likely be attributed to the increased stability of base pairing between the ligated bottom product and the deoxyribozyme (19 bp with the ligation product, as opposed to 8 and 11 bp with the corresponding substrates). In an attempt to optimize turnover, we shortened the lengths of the hybridizing arms of dR3 and dR8. However, even the deletion of 1 bp completely abolished effector-dependent ligation. Conversely, increasing the number of base pairs between the hybridizing arms and substrates resulted in higher overall rates of catalysis but also less effector-dependent activation, owing to the greater stability of the effector-independent enzyme-substrate complex at room temperature. These results confirm the extraordinary sensitivity of the ligase maxizyme to even small changes in binding energies.
Recognition of non-natural nucleotides
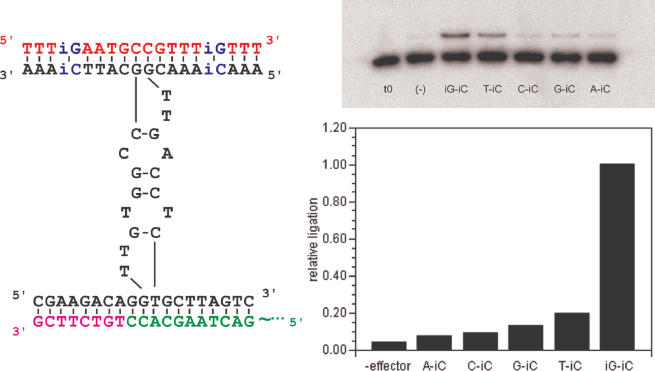
In order to expand the range of information that could be recoded by the ligase maxizyme we incorporated two non-natural nucleotides, 5-methyl-isocytidine (isoMeC) and isoguanosine (isoG) (25) into the effector oligonucleotide and the substrate-binding arms of the maxizyme. A variant of construct dR8 was first synthesized that included isoMeC residues at two positions in its substrate-binding arms. The resultant construct, dR8b, was assayed for its ability to specifically recognize an effector oligonucleotide that contained two complementary isoG substitutions (18Nb). While construct dR8b was highly activated by the non-natural effector 18Nb, it was much less active with effectors bearing residues that were predicted to form mismatches, such as A–isoMeC, C–isoMeC, G–isoMeC and T–isoMeC (Figure 3). The extent of activation correlated well with the previously observed stabilities of the mispairs (26), except for a greater than expected activation by the T–isoMeC mispairs.
Figure 3.
Expanded base pairing in an allosteric deoxyribozyme. Construct dR8 was re-engineered to carry a non-natural nucleotide, 5-methyl-isocytidine (isoMeC), at two positions (blue), generating dR8b. The effector was then synthesized with compensatory isoguanosine (isoG) substitutions, generating 18Nb. The adjacent gel shows the ligation activity of dR8b with different substrates. The lower band on the gel is a radiolabeled, unligated substrate, while the higher band is the ligated product. The lane labeled ‘t0’ is time 0 of the reaction and all subsequent lanes are 3.5 h reactions with effector variants engineered to make the indicated base-pairings. The extent of ligation is normalized to the most active pairing, iG–iC.
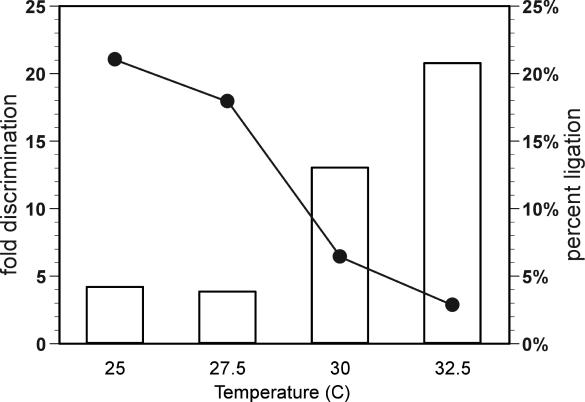
To increase the fidelity of recognition against the T–isoMeC mispair, we increased the temperature of the reaction. At 25°C, dR8b showed 5-fold discrimination for its cognate effector versus the effector bearing the T–isoMeC mispair, while at 32.5°C the discrimination increased to 21-fold (Figure 4). The increase in discrimination was accompanied by an expected decline in the rate of ligation, since the originally selected (22) ligase was most active near room temperature. The decrease in activity at higher temperatures was also consistent with the previous finding that the deoxyribozyme was so finely poised that the deletion of even a single base pair led to loss of activation.
Figure 4.
Temperature optimization of base pair discrimination. Temperature was increased to aid discrimination by dR8b for the correct effector, 18Nb versus the most active competitor, 18N. Fold discrimination (bars) indicates the ratio of ligated product in the presence of 18Nb to 18N. Circles indicate percent ligation of substrates in the presence of 18Nb. Ligation reactions were allowed to proceed for ∼16 h.
Detection of recoded sequence information
While we have demonstrated that a deoxyribozyme can be used to recode sequence information, the utility of this method will rely on being able to detect the transformed product. To this end, we attempted to recode short oligonucleotide sequences that could not be readily amplified by PCR into different, longer amplicons. Moreover, we wished to recode and detect one sequence against a background of non-specific sequence information. As a proof-of-principle, we used the ligase maxizyme dR8b that recognizes non-natural base pairs to detect non-natural oligonucleotides obscured within vast amounts of genomic DNA.
Oligonucleotide substrates were prepared for dR8b that, upon ligation, would yield an amplicon that could be readily detected by real-time PCR by extending the 5′ and 3′ substrates so that they contained primer and TaqMan probe-binding sites (Figure 5a). Ligation reactions were carried out with the oligonucleotide effector, 18Nb, in the presence of a 1000-fold mass excess of genomic DNA (Materials and Methods). When dR8b-mediated ligation was allowed to proceed for only 5 min, the presence of the effector 18Nb in the genomic DNA resulted in a 12 cycle threshold (CT) real-time PCR advantage for the ligated product relative to amplification in the absence of the effector (Figure 5b). This is a huge CT value relative to typical, diagnostically relevant real-time PCR signals (21), and corresponds to a nearly 4100-fold activation of the non-natural enzyme by its cognate effector. Importantly, the presence of 1000-fold excess genomic DNA also decreased the extent of effector-independent ligation (5 cycles or 32-fold). This effect is likely due to genomic DNA sequestering either substrate or half-enzyme molecules (by non-specific hybridization) or magnesium (by chelation). Either mechanism would decrease effective reactant concentrations, increase the overall stringency of the reaction and thereby increase reaction specificity.
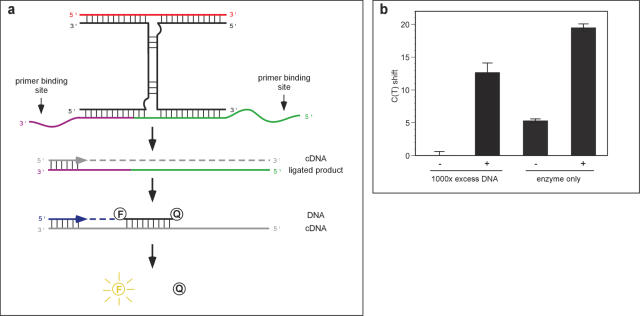
Figure 5.
Real-time PCR detection of the ligation product formed by dR8b. (a) Each of the substrates for dR8b was extended to carry one primer-binding site. When ligated, the product becomes a template for PCR. The complementary DNA strand (cDNA, gray) formed by the first primer extension bears a site complementary to a Taqman probe. When the cDNA strand is replicated the Taqman probe is digested, liberating the fluorescent molecule from the quencher. The fluorescent readout is quantified by real-time PCR. (b) Quantification of the ligation product of dR8b by real-time PCR. dR8b was allowed to react with the modified substrates for 5 min and the product was detected as in (A). Minus and plus indicate the absence and presence of the effector molecule, 18Nb in the reaction. CT shift indicates the decrease in number of cycles required to reach exponential amplification relative to the effector-independent background reaction plus 1000× excess DNA (left-most bar), which had an absolute CT value of 37 ± 0.15. Error bars represent 2 SDs.
DISCUSSION
The ligase maxizyme is a catalyst that recodes nucleic acid sequence information: it ‘reads’ one sequence and ‘writes’ another. There are a variety of potential applications for such a simple reagent. For example, in many instances the detection of specific alleles by PCR suffers from background problems due to primer binding to closely related alleles or to other related sequences in genomic DNA (27). The ligase maxizyme could transform individual alleles into amplicons that were unrelated to most sequences in a genome, and thereby reduce problems associated with background amplification.
Nucleic acid enzymes have been engineered previously to detect biologically relevant nucleic acids such as microRNAs and regulatory untranslated regions (10,18,19). As the ligase maxizyme converts short oligonucleotide sequences into longer amplicons, it could also prove useful as a tool for detecting therapeutic nucleic acids such as antisense, aptamers or siRNA molecules by real-time PCR. Such a technique might be especially useful if the therapeutic contained modified or non-standard nucleotides that were difficult to detect or amplify by normal methods. In addition, short, natural sequences, such as microRNA molecules that are usually difficult to detect, could be transformed into amplicons and then read by PCR.
The ligase maxizyme can also be viewed as a simple machine or ‘part’ that would have a unique function in the nascent fields of DNA computation and synthetic biology. To date most of the various implementations of nucleic acid computation have involved what can be described as ‘hybridization logic,’ assembling and analyzing answers based upon pre-encoded patterns of hybridization (14–17,28–32). As might be expected from the error-prone nature of DNA hybridization and polymerization, large-scale nucleic acid computation based on hybridization logic is inherently infeasible (33). For example, the best nucleic acid computer built to date demonstrated an error rate of ∼1 in 2500 (29), far greater than even a poor silicon device. However, the use of nucleic acid enzymes as ‘silicomimetic’ devices that can interface with biology and make decisions has been demonstrated brilliantly by Stojanovic and Stefanovic, who encoded an algorithm for playing Tic-Tac-Toe into a series of deoxyribozymes (16). More recently, Shapiro and co-workers have extended these ideas to the creation of self-diagnosing and self-actuating nucleic acid therapeutics (34). A deoxyribozyme that can transform information might be extremely useful in the design and implementation of such therapeutic nanomachines, especially since the unidirectional activation observed with most of the constructs examined would allow ‘reading’ and ‘writing’ to be uniquely specified.
Irrespective of the application, this is a further example of how allosteric nucleic acid enzymes can be generated by engineering at the level of secondary structure, and stands as one of the first demonstrations that DNA can act not only as a molecule to carry information but as a machine that can recode information.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
This work was supported by the United States Department of Defense, University of Florida (UF03010), the National Institute of Health and the Texas Higher Education Coordinating Board, TDT-003658-0611-2003. Funding to pay the Open Access publication charges for this article was provided by Texas Higher Education Coordinating Board.
Conflict of interest statement. None declared.
REFERENCES
- 1.Battiste J.L., Mao H., Rao N.S., Tan R., Muhandiram D.R., Kay L.E., Frankel A.D., Williamson J.R. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide–RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 2.Legault P., Li J., Mogridge J., Kay L.E., Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 3.Levy M., Ellington A.D. ATP-dependent allosteric DNA enzymes. Chem. Biol. 2002;9:417–426. doi: 10.1016/s1074-5521(02)00123-0. [DOI] [PubMed] [Google Scholar]
- 4.Jose A.M., Soukup G.A., Breaker R.R. Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res. 2001;29:1631–1637. doi: 10.1093/nar/29.7.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koizumi M., Soukup G.A., Kerr J.N., Breaker R.R. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nature Struct. Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- 6.Soukup G.A., Breaker R.R. Design of allosteric hammerhead ribozymes activated by ligand-induced structure stabilization. Structure. 1999;7:783–791. doi: 10.1016/s0969-2126(99)80102-6. [DOI] [PubMed] [Google Scholar]
- 7.Soukup G.A., Emilsson G.A., Breaker R.R. Altering molecular recognition of RNA aptamers by allosteric selection. J. Mol. Biol. 2000;298:623–632. doi: 10.1006/jmbi.2000.3704. [DOI] [PubMed] [Google Scholar]
- 8.Tang J., Breaker R.R. Rational design of allosteric ribozymes. Chem. Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 9.Tang J., Breaker R.R. Mechanism for allosteric inhibition of an ATP-sensitive ribozyme. Nucleic Acids Res. 1998;26:4214–4221. doi: 10.1093/nar/26.18.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Najafi-Shoushtari S.H., Famulok M. Competitive regulation of modular allosteric aptazymes by a small molecule and oligonucleotide effector. RNA. 2005;11:1514–1520. doi: 10.1261/rna.2840805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komatsu Y., Yamashita S., Kazama N., Nobuoka K., Ohtsuka E. Construction of new ribozymes requiring short regulator oligonucleotides as a cofactor. J. Mol. Biol. 2000;299:1231–1243. doi: 10.1006/jmbi.2000.3825. [DOI] [PubMed] [Google Scholar]
- 12.Burke D.H., Ozerova N.D., Nilsen-Hamilton M. Allosteric hammerhead ribozyme TRAPs. Biochemistry. 2002;41:6588–6594. doi: 10.1021/bi0201522. [DOI] [PubMed] [Google Scholar]
- 13.Penchovsky R., Breaker R.R. Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes. Nat. Biotechnol. 2005;23:1424–1433. doi: 10.1038/nbt1155. [DOI] [PubMed] [Google Scholar]
- 14.Stojanovic M.N., Mitchell T.E., Stefanovic D. Deoxyribozyme-based logic gates. J. Am. Chem. Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]
- 15.Stojanovic M.N., Semova S., Kolpashchikov D., Macdonald J., Morgan C., Stefanovic D. Deoxyribozyme-based ligase logic gates and their initial circuits. J. Am. Chem. Soc. 2005;127:6914–6915. doi: 10.1021/ja043003a. [DOI] [PubMed] [Google Scholar]
- 16.Stojanovic M.N., Stefanovic D. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 17.Stojanovic M.N., Stefanovic D. Deoxyribozyme-based half-adder. J. Am. Chem. Soc. 2003;125:6673–6676. doi: 10.1021/ja0296632. [DOI] [PubMed] [Google Scholar]
- 18.Hartig J.S., Grune I., Najafi-Shoushtari S.H., Famulok M. Sequence-specific detection of microRNAs by signal-amplifying ribozymes. J. Am. Chem. Soc. 2004;126:722–723. doi: 10.1021/ja038822u. [DOI] [PubMed] [Google Scholar]
- 19.Najafi-Shoushtari S.H., Mayer G., Famulok M. Sensing complex regulatory networks by conformationally controlled hairpin ribozymes. Nucleic Acids Res. 2004;32:3212–3219. doi: 10.1093/nar/gkh643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuwabara T., Warashina M., Tanabe T., Tani K., Asano S., Taira K. A novel allosterically trans-activated ribozyme, the maxizyme, with exceptional specificity in vitro and in vivo. Mol. Cell. 1998;2:617–627. doi: 10.1016/s1097-2765(00)80160-4. [DOI] [PubMed] [Google Scholar]
- 21.Pai S., Ellington A.D., Levy M. Proximity ligation assays with peptide conjugate ‘burrs’ for the sensitive detection of spores. Nucleic Acids Res. 2005;33:e162. doi: 10.1093/nar/gni150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy M., Ellington A.D. Selection of deoxyribozyme ligases that catalyze the formation of an unnatural internucleotide linkage. Bioorg. Med. Chem. 2001;9:2581–2587. doi: 10.1016/s0968-0896(01)00033-5. [DOI] [PubMed] [Google Scholar]
- 23.Levy M., Ellington A.D. In vitro selection of a deoxyribozyme that can utilize multiple substrates. J. Mol. Evol. 2002;54:180–190. doi: 10.1007/s00239-001-0066-1. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y., Kool E.T. High sequence fidelity in a non-enzymatic DNA autoligation reaction. Nucleic Acids Res. 1999;27:875–881. doi: 10.1093/nar/27.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Switzer C.Y., Moroney S.E., Benner S.A. Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry. 1993;32:10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- 26.Geyer C.R., Battersby T.R., Benner S.A. Nucleobase pairing in expanded Watson–Crick-like genetic information systems. Structure (Camb.) 2003;11:1485–1498. doi: 10.1016/j.str.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Mir K.U., Southern E.M. Sequence variation in genes and genomic DNA: methods for large-scale analysis. Ann. Rev. Genomics Hum. Genet. 2000;1:329–360. doi: 10.1146/annurev.genom.1.1.329. [DOI] [PubMed] [Google Scholar]
- 28.Adleman L.M. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 29.Braich R.S., Chelyapov N., Johnson C., Rothemund P.W., Adleman L. Solution of a 20-variable 3-SAT problem on a DNA computer. Science. 2002;296:499–502. doi: 10.1126/science.1069528. [DOI] [PubMed] [Google Scholar]
- 30.Faulhammer D., Cukras A.R., Lipton R.J., Landweber L.F. Molecular computation: RNA solutions to chess problems. Proc. Natl Acad. Sci. USA. 2000;97:1385–1389. doi: 10.1073/pnas.97.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee C.M., Kim S.W., Kim S.M., Sohn U. DNA computing the Hamiltonian path problem. Mol. Cells. 1999;9:464–469. [PubMed] [Google Scholar]
- 32.Lipton R.J. DNA solution of hard computational problems. Science. 1995;268:542–545. doi: 10.1126/science.7725098. [DOI] [PubMed] [Google Scholar]
- 33.Li D., Huang H., Li X., Li X. Hairpin formation in DNA computation presents limits for large NP-complete problems. Biosystems. 2003;72:203–207. doi: 10.1016/s0303-2647(03)00145-x. [DOI] [PubMed] [Google Scholar]
- 34.Benenson Y., Gil B., Ben-Dor U., Adar R., Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.