Abstract
Pituitary adenylate cyclase-activating polypeptide (PACAP) is a neuropeptide abundantly expressed in the central nervous system and involved in regulating neurogenesis and neuronal signal transduction. The amino acid sequence of PACAP is extremely conserved across vertebrate species, indicating a strong functional constraint during the course of evolution. However, through comparative sequence analysis, we demonstrated that the PACAP precursor gene underwent an accelerated evolution in the human lineage since the divergence from chimpanzees, and the amino acid substitution rate in humans is at least seven times faster than that in other mammal species resulting from strong Darwinian positive selection. Eleven human-specific amino acid changes were identified in the PACAP precursors, which are conserved from murine to African apes. Protein structural analysis suggested that a putative novel neuropeptide might have originated during human evolution and functioned in the human brain. Our data suggested that the PACAP precursor gene underwent adaptive changes during human origin and may have contributed to the formation of human cognition.
HUMANS diverged from African apes ∼5–7 million years ago (Goodman et al. 1998). Genetically, humans and chimpanzees share nearly 99% DNA sequence similarity (Fujiyama et al. 2002; Hellmann et al. 2003; Shi et al. 2003), which seems to contradict the marked biological divergence between them, e.g., the highly developed cognitive abilities in humans. Therefore, the major challenge in the rapidly progressing human and chimpanzee genome comparison studies is to determine the small subset of sequence differences that have phenotypic significance related to species-specific traits (Enard et al. 2002; Stedman et al. 2004). Recent studies on FOXP2 and myosin demonstrated that the human-specific substitutions in these two genes are related to the phenotypic divergence between humans and nonhuman primates, i.e., the acquiring of language ability and gracilization of masticatory muscles relevant to accelerated encephalization during human evolution (Enard et al. 2002; Zhang et al. 2002; Stedman et al. 2004).
Pituitary adenylate cyclase-activating polypeptide (PACAP) is a well-studied neuropeptide that plays a pivotal role in the central nervous system by acting as a neurohormone and a neurotransmitter (Dicicco-Bloom et al. 1998; Montero et al. 2000; Vaudry et al. 2000; Hamelink et al. 2002; Moretti et al. 2002). In the human genome, the gene encoding the PACAP precursor (ADCYAP1) is located on chromosome 18p11 (Hosoya et al. 1992). This region was shown to be related with holoprosencephaly, the most common hereditary development defect of the forebrain in humans (a single-lobed brain structure with severe skull and facial defects) (Golden 1998). Recent studies showed that PACAP is involved in cerebral cortical neurogenesis by eliciting the transition from proliferation to differentiation in cortical precursors (Dicicco-Bloom et al. 1998; Suh et al. 2001). Gene knockout studies in mouse revealed altered psychomotor behaviors (Hashimoto et al. 2001). The PACAP homolog in Drosophila, amnesiac (amn), was shown to be crucial in adult memory formation (DeZazzo et al. 1999).
The human PACAP precursor gene (ADCYAP1) has five exons encoding a protein of 176 amino acids with PACAP located in the C-terminal region (Figure 1) (Hosoya et al. 1992). PACAP has two forms, the 38-amino-acid form (PACAP38) and the 27-amino-acid form (PACAP27), which are generated through post-translational processing by the precursor convertase (Vaudry et al. 2000). The sequence of PACAP has been remarkably conserved in vertebrates during evolution and no amino acid substitution was observed in the mammal species studied so far (Montero et al. 2000) (Figure 1).
Figure 1.—

The structure and protein sequence alignment of the PACAP precursor gene in human and other vertebrate species. The PACAP precursor contains five domains, including the signal peptide (SP), the UD, the PRP, the spacing domain (SD), and PACAP (Vaudry et al. 2000). The numbers indicate relevant amino acid residues. The shaded sites and the solid triangles refer to the fixed substitutions in humans. The arrows indicate the single or paired basic amino acids that can be cleaved by precursor convertase in post-translational processing. The question marks indicate the within-species polymorphic sites (D/G at site 54 in humans, D/H at site 82 in chimpanzees, E/D at site 40 in gorillas, P/L at site 34 in gibbons, and Y/F at site 25, P/Q at site 58, and R/T at site 81 in rhesus monkeys). HUM, human; CHP, chimpanzee; GOR, gorilla; ORA, orangutan; GIB, white-browed gibbon; RHM, rhesus monkey; MOU, mouse; SHP, sheep; CHK, chicken; XEN, Xenopus; and DAN, danio.
In this study, we sought to understand the molecular history of the PACAP precursor gene in primates and other vertebrate species. Our results demonstrate an accelerated evolution of the PACAP precursor in the lineage leading to the emergence of humans, which is likely caused by Darwinian positive selection during human evolution.
MATERIALS AND METHODS
The human DNA samples in this study are from three major continental populations, including 30 Han Chinese (Kunming, China), 10 Europeans (Italy), and 10 Africans (Central African Republic Pygmy and Ethiopian). We also sampled five nonhuman primate species (see Figure 1 for details). All the human and nonhuman primate samples were from the collections of the Kunming Cell Bank, the Chinese Academy of Sciences, and Stanford University.
The PCR primers were designed on the basis of the sequence alignment of human and other available mammalian species (Ensemble genome browser at http://www.ensemble.org and http://pre.ensembl.org/Pan_troglodytes/). The primer sequences are:
NM001117_E1F, 5′-CAAGTGCTGTTCAACTCAGGGA-3′;
NM001117_E1R, 5′-GGCGATGCTAGTAGTCTGGACC-3′;
NM001117_E2F, 5′-GAGACAATTCTCAGCGGAGGAC-3′;
NM001117_E2R, 5′-GCCACTCACTGCCACTCTTTG-3′;
NM001117_E3F, 5′-GGGAGTTATTGGCGAGTTCTG-3′;
NM001117_E3R, 5′-AAAATCTCCAACTGCGACCC-3′;
NM001117_E4F, 5′-GCGGCGATTGAATCTGTGTCTC-3′;
NM001117_E4R, 5′-GAACACATGAGCGATGACTGTTGAG-3′;
NM001117_E4F2, 5′-CTCCAGGGCAGCAGCCAGAC-3′;
NM001117_E4R2, 5′-ACAGCTCGGGCGAATTTTCAA-3′;
NM001117_I2F, 5′-GATCCTATTGCAGCGACAGAAAA-3′;
NM001117_I3R, 5′-CGTGAAGATCCCGTCCGAGTG-3′;
NM001117_I3F, 5′-TGGGGTAAGAGTTTGTGGAAGG-3′.
All the human and nonhuman primate samples were sequenced for the 531-bp coding region of the PACAP precursor gene (exons 2–5; exon 1 is nontranslational). A 400-bp fragment of intron 3 was sequenced in 30 humans (10 samples from each of the three continental populations) and six chimpanzees. The sequences from the other mammalian and vertebrate species were from published data (accession nos. NM_009625, NM_016989, S83511, U67275, AF187877, AF329730; NCBI at http://www.ncbi.nlm.nih.gov/).
The DNA sequences were aligned using DNASTAR (DNASTAR, Madison, WI), and checked manually. The amino acid sequences were aligned using CLUSTAL X (Jeanmougin et al. 1998). The phylogenetic relationships among the primate species tested were based on the published data (Goodman et al. 1998). The ancestral sequences (internal nodes) in the phylogenetic tree (Figure 2) were inferred by using the ANC-GENE program developed by Zhang et al. (1998). With the use of MEGA2.0 program (Kumar et al. 2001), the Pamilo-Bianchi-Li method was employed to estimate the nonsynonymous and synonymous substitution ratios (Ka/Ks), in which the transition and transversion bias was taken into account (Li 1993; Pamilo and Bianchi 1993). On the basis of results from the Pamilo-Bianchi-Li analysis, which suggested that the human lineage has an accelerated evolution, we elected to run a likelihood-ratio test to compare two alternative models of sequence evolution: a model in which rates of dN/dS (ω) are constrained to be equal vs. a model in which the human lineage is assumed to have a different rate from the other lineages (Yang 1998). The relative rate test was used to detect amino acid substitution rate variations in different primate lineages (Tajima 1993). The McDonald-Kreitman test was conducted to detect positive selection by comparing the between-species nonsynonymous/synonymous substitution ratios with the within-species nonsynonymous/synonymous polymorphism ratios (McDonald and Kreitman 1991). The protein structure analysis was conducted by utilizing the tools (the PROSITE SCAN package) from the ExPASy Molecular Biology Server hosted by the North Carolina Supercomputing Center (http://us.expasy.org/).
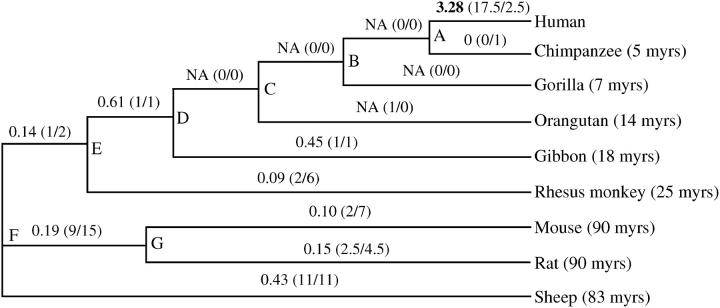
Figure 2.—
The Ka/Ks ratios of different evolutionary lineages. The calculations were based on the data from the UD and PRP domains only. The Pamilo-Bianchi-Li method was used in estimating the Ka/Ks ratios (Li 1993; Pamilo and Bianchi 1993). The numbers in parentheses refer to the numbers of nonsynonymous and synonymous substitutions. The divergence times between human and other mammal species are indicated (myrs, million years) (Hedges and Kumar 1999).
RESULTS AND DISCUSSION
In the effort to identify the fast-evolving genes in the human genome, we observed rapid amino acid substitutions between humans and chimpanzees in the PACAP precursor gene. A total of 17 amino acid substitutions (9.7%, 17/176) were detected in the human PACAP precursor, which is much higher than the average protein sequence divergence (<1.0%) between humans and chimpanzees (Fujiyama et al. 2002; Hellmann et al. 2003; Shi et al. 2003) (Figure 1). Surprisingly, when the human and chimpanzee protein sequences were aligned with those of other mammalian species, 11 of the 17 amino acid substitutions were found conserved in mouse, rat, and sheep (Figure 1), indicating that the accelerated evolution of the PACAP precursor occurred only in the human lineage. The same conserved substitution pattern was observed when we sequenced four other nonhuman primate species (one sample from each species), including gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), gibbon (Hylobates hoolock), and rhesus monkey (Macaca mulatta), which have varied degrees of divergence from humans (7–25 million years) (Goodman et al. 1998) (Figures 1 and 2).
To see if the amino acid substitutions are fixed in human populations, we sequenced 50 human individuals (100 chromosomes) from the major continental populations, including 30 Han Chinese, 10 Europeans, and 10 Africans. All the amino acid substitutions observed between human and nonhuman primates were conserved in the human populations. There are three polymorphic sites observed in humans; two are silent changes and one a missense (Asp54Gly) (Figure 1). We conducted a phylogenetic analysis to reveal the detailed mutation patterns in different evolutionary lineages (Figure 2). The synonymous (silent) and nonsynonymous (missense) substitution rates (Ks and Ka, respectively) were calculated following Pamilo-Bianchi-Li's method (Li 1993; Pamilo and Bianchi 1993). As shown in Figure 2 and Table 1, when the unidentified domain (UD) and PACAP-related peptide (PRP) (Vaudry et al. 2000) regions were considered, the human lineage had a large Ka/Ks ratio (Ka/Ks = 3.28, P = 0.026, one-tailed z-test). Similar results were observed when calculating the ω (dN/dS) values using the maximum-likelihood method developed by Yang (1998)(data not shown). The likelihood-ratio test indicated that the human lineage has a significantly larger ω-value than the nonhuman primate lineages (2ΔLnL = 9.14, P = 0.003; Yang 1998), reflecting a rapid evolution of the human PACAP precursor. In contrast, all the other lineages, including that of the chimpanzee, had very small Ka/Ks ratios with most of them following the expectation of negative selection (functional constraint) (Ka/Ks < 1). When the entire coding region of the PACAP precursor gene is considered, the Ka/Ks ratio in the human lineage is 1.37, which is still significantly larger than that of the nonhuman primate lineages according to the likelihood-ratio test (2ΔLnL = 7.90, P = 0.005; Yang 1998), again indicating the rapid amino acid substitution rate in the human lineage (Figure 1 and Table 1). The amino acid substitution rate in the human lineage is at least seven times faster than those of the other mammalian lineages (3.4 vs. 0–0.45 amino acid substitutions/gene/million years), and the relative rate test based on protein sequences showed a significant rate difference between humans and chimpanzees (P < 0.001) (Tajima 1993).
TABLE 1.
The calculation of theKa/Ks ratios in the human and chimpanzee lineages
| Human node A
|
Chimpanzee node A
|
|||||
|---|---|---|---|---|---|---|
| Gene domain | Ka/Ks |
z-test positive |
z-test negative |
Ka/Ks |
z-test positive |
z-test negative |
| UD | 2.61 | 0.085 | 1.000 | 0 | 1.000 | 0.176 |
| PRP | 11.35 | 0.067 | 1.000 | NA | NA | NA |
| UD and PRP | 3.28 | 0.026* | 1.000 | 0 | 1.000 | 0.166 |
| PACAP | 0 | 1.000 | 0.104 | 0 | 1.000 | 0.198 |
| Entire gene | 1.37 | 0.261 | 1.000 | 0 | 1.000 | 0.040* |
Node A refers to the common ancestor of humans and chimpanzees (Figure 2). The z-test was used to detect whether the ratios of Ka/Ks deviate statistically from 1, the neutral expectation. The probabilities (P) of one-tailed z-tests for positive and negative selection (z-test positive and z-test negative) are listed. *Significant P-values (P < 0.05). NA, not applicable (Ka = 0).
The rapid evolution of the human PACAP precursor can be due to either Darwinian positive selection or relaxation of functional constraint. To further test whether positive selection or relaxation of functional constraint is responsible for the observed rapid evolution of the PACAP precursor in the human lineage, we conducted the neutrality test following McDonald and Kreitman (1991). To obtain enough polymorphic sites in human populations (only three polymorphic sites were observed in the coding region), we sequenced the upstream segment of intron 3 (400 bp) of the PACAP precursor gene in 30 humans (10 samples from each continental population) and six chimpanzees. A total of seven polymorphic sites were observed in humans while no polymorphism was found in the chimpanzees. If there is positive selection in the human lineage, the ratio of nonsynonymous/synonymous substitutions between human and chimpanzee (17.5/20.5) should be significantly higher than the ratio of nonsynonymous/synonymous polymorphisms within human populations (1/9), which turned out to be the case (P = 0.039, one-tailed Fisher's exact test) when the intron sequences were treated as synonymous sites. Hence, the accelerated amino acid substitution rate in the human lineage was likely caused by adaptive evolution instead of relaxation of functional constraint. The substitution rate between human and chimpanzee at the synonymous sites (Ks = 0.056) is slightly larger than that of the intron region (Ki = 0.036), but not statistically significant (P > 0.05, two-tailed Fisher's exact test); therefore, the use of intron sequences would not bring much bias in the McDonald and Kreitman test.
The human-specific substitutions are located in the central region of the PACAP precursor, in contrast to the highly conserved PACAP region (Figure 1). The fixed substitutions in humans resulted in three potential protein motif changes by gaining an α-amidation site (Gly79) and a N-myristoylation site (Gly122) while losing a protein kinase C phosphorylation site (Gly102) (Table 2). The gain of the α-amidation site is particularly intriguing because previous studies showed that the C-terminal α-amidation is a critical determinant of biologic activity for neuropeptides, including PACAP (Eipper et al. 1992), and a putative novel peptide would be expected in humans due to the gain of the α-amidation site. There are seven potential post-translational processing sites in the human PACAP precursor, which are single or pairs of basic amino acids (lysine and/or arginine) that can be cleaved by precursor convertase (Seidah et al. 1998; Vaudry et al. 2000) (Figure 1). The mutation at site Gly79 provided a novel putative α-amidation site in humans, which may create a bioactive peptide of 41 amino acids (sites 38–78, Figure 1) with the following sequence: PEEEAYGEDGNPLPDFD(G)GSEPPGAGSPASAPRAAAAWYRPA-NH2.
TABLE 2.
Protein motif comparison between human and other mammalian species
| Location | Motif | Human | Nonhuman primates |
Mouse | Rat | Sheep |
|---|---|---|---|---|---|---|
| α-Amidation site | ||||||
| 78–81 | AGRR | Yes | No | No | No | No |
| Protein kinase C phosphorylation site | ||||||
| 100–102 | SAR | No | Yes | Yes | Yes | Yes |
| N-Myristoylation site | ||||||
| 118–123 | GGGAGD | Yes | No | No | No | No |
The underlined sites are those human-specific substitutions causing protein motif changes. The protein structure analysis was conducted by utilizing the tools from the ExPASy Molecular Biology Server hosted by the North Carolina Supercomputing Center (http://us.expasy.org/).
The α-amidation of neuropeptides can protect them from enzymatic degradation (half-life) and increase binding affinity to the receptors (Eipper et al. 1992). Therefore, as only humans have the potential α-amidation site, the 41-aa putative novel peptide may exist and function only in humans. When compared with chimpanzees, the putative novel peptide in humans acquired 12 amino acid substitutions (29.3%, 12/41), which is consistent with the proposed strong positive selection on the UD region that led to the creation of a novel peptide during human evolution. However, functional data need to be generated to confirm the existence and the biological function of the proposed novel neuropeptide in the human brain.
The PRP region also showed a strong signal of positive selection in humans although the statistical test was not significant (Ka/Ks = 11.35, P = 0.067, one-tailed z-test). The two fixed substitutions of PRP (Gly102 and His104) in humans are conserved in all the other vertebrate species including chicken, Xenopus, and fish (Figure 1). As described above, the Gly102 resulted in the potential loss of a protein kinase C phosphorylation site in humans, implying that the human PRP might acquire a modified function during human evolution.
It is generally accepted that human intelligence is the product of adaptive evolution (Dunbar 1998). The enlarged brain and highly developed cognitive abilities is one of the fundamental differences that sets us apart from our close relatives, the nonhuman primates. The UD and PRP regions of the PACAP precursor underwent strong positive selection within the human lineage, resulting in marked protein divergence between humans and nonhuman primates. We postulate that since the putative novel peptide and PRP are coexpressed with PACAP in the human central nervous system (Vaudry et al. 2000), it is likely that they might play important roles in human brain development and function. The 41-aa putative novel peptide in humans might interact with PACAP in regulating the biological processes in the human brain, e.g., neurogenesis and/or signal transduction. A similar evolutionary mechanism was reported in a Xenopus species in which the recently evolved peptide acted as the antagonist of the original peptide encoded by the same gene (Chen et al. 2003).
So far, only a handful of genes involved in human brain function have been reported to have undergone positive selection, e.g., the FOXP2 gene for human speech and language ability and the ASPM gene and microcephalin gene for human brain size (Enard et al. 2002; Zhang et al. 2002; Zhang 2003; Evans et al. 2004a,b; Kouprina et al. 2004; Wang and Su 2004). The rapid evolution of the human PACAP precursor gene provides a new clue to a better understanding of the genetic basis of human cognition and human origin.
Acknowledgments
We thank Xiao-na Fan, Yi-chuan Yu, and Reshmi Indugula for technical help. We also thank M. Borchers for his critical reading of this article. This study was supported by a grant from the Chinese Academy of Sciences (KSCX2-SW-121), the National Natural Science Foundation of China (30370755), and the Natural Science Foundation of Yunnan Province of China.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AY592934, AY592935, AY592936, AY592937, AY592938, AY592939, AY592940, AY592941, AY592942, AY592943, AY592944, AY592945, AY592946, AY592947, AY592948, AY592949, AY592950, AY592951, AY592952, AY592953.
References
- Chen, T., M. O'Rourke, D. F. Orr, D. J. M. Coulter, D. G. Hirst et al., 2003. Kinestatin: a novel bradykinin B2 receptor antagonist peptide from the skin secretion of the Chinese toad, Bombina maxima. Regul. Pept. 116: 147–154. [DOI] [PubMed] [Google Scholar]
- DeZazzo, J., S. Xia, J. Christensen, K. Velinzon and T. Tully, 1999. Developmental expression of an amn1 transgene rescues the mutant memory defect of amnesiac adults. J. Neurosci. 19: 8740–8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicicco-Bloom, E., N. Lu, J. E. Pintar and J. W. Zhang, 1998. The PACAP ligand/receptor system regulates cerebral cortical neurogenesis. Ann. NY Acad. Sci. 865: 274–289. [DOI] [PubMed] [Google Scholar]
- Dunbar, D. M., 1998. The social brain hypothesis. Evol. Anthropol. 6: 178–189. [Google Scholar]
- Eipper, B. A., D. A. Stoffers and R. E. Mains, 1992. The biosynthesis of neuropeptides: peptide α-amidation. Annu. Rev. Neurosci. 15: 57–85. [DOI] [PubMed] [Google Scholar]
- Enard, W., M. Przeworski, S. E. Fisher, C. S. L. Lai, V. Wiebe et al., 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418: 869–872. [DOI] [PubMed] [Google Scholar]
- Evans, P. D., J. R. Anderson, E. J. Vallender, S. S. Choi and B. T. Lahn, 2004. a Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum. Mol. Genet. 13: 1139–1145. [DOI] [PubMed] [Google Scholar]
- Evans, P. D., J. R. Anderson, E. J. Vallender, S. L. Gilbert, C. M. Malcom et al., 2004. b Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum. Mol. Genet. 13: 489–494. [DOI] [PubMed] [Google Scholar]
- Fujiyama, A., H. Watanabe, A. Toyoda, T. D. Taylor, T. Itoh et al., 2002. Construction and analysis of a human-chimpanzee comparative clone map. Science 295: 131–134. [DOI] [PubMed] [Google Scholar]
- Golden, J. A., 1998. Holoprosencephaly: a defect in brain patterning. J. Neuropathol. Exp. Neurol. 57: 991–999. [DOI] [PubMed] [Google Scholar]
- Goodman, M., C. A. Porter, J. Czelusniak, S. L. Page, H. Schneider et al., 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9: 585–598. [DOI] [PubMed] [Google Scholar]
- Hamelink, C., O. Tjurmina, R. Damadzic, W. S. Young, E. Weihe et al., 2002. Pituitary adenylate cyclase-activating polypeptide is a sympathoadrenal neurotransmitter involved in catecholamine regulation and glucohomeostasis. Proc. Natl. Acad. Sci. USA 99: 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, H., N. Shintani, K. Tanaka, W. Mori, M. Hirose et al., 2001. Altered psychomotor behaviors in mice lacking pituitary adenylate cyclase-activating polypeptide (PACAP). Proc. Natl. Acad. Sci. USA 98: 13355–13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, S. B., and S. Kumar, 1999. Divergence times of eutharian mammals. Science 285: 2031. [Google Scholar]
- Hellmann, I., S. Zöllner, W. Enard, I. Ebersberger, B. Nickel et al., 2003. Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Res. 13: 831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya, M., C. Kimura, K. Ogi, S. Ohkubo, Y. Miyamoto et al., 1992. Structure of the human pituitary adenylate cyclase-activating polypeptide (PACAP) gene. Biochim. Biophys. Acta 1129: 199–206. [DOI] [PubMed] [Google Scholar]
- Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins and T. J. Gibson, 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23: 403–405. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., A. Pavlicek, G. H. Mochida, G. Solomon and W. Gersch, 2004. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2: 0653–0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001 MEGA2: Molecular Evolutionary Genetics Analysis Software. Arizona State University, Tempe, AZ. [DOI] [PubMed]
- Li, W. H., 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36: 96–99. [DOI] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Montero, M., L. Yon, S. Kikuyama, S. Dufour and H. Vaudry, 2000. Molecular evolution of the growth hormone-releasing hormone/pituitary adenylate cyclase-activating polypeptide gene family: functional implication in the regulation of growth hormone secretion. J. Mol. Endocrinol. 25: 157–168. [DOI] [PubMed] [Google Scholar]
- Moretti, C., C. Mencacci, G. V. Frajese, M. Cerilli and G. Frajese, 2002. Growth hormone-releasing hormone and pituitary adenylate cyclase-activating polypeptide in the reproductive system. Trends Endocrinol. Metab. 13: 428–435. [DOI] [PubMed] [Google Scholar]
- Pamilo, P., and N. O. Bianchi, 1993. Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Mol. Biol. Evol. 10: 271–281. [DOI] [PubMed] [Google Scholar]
- Seidah, N. G., R. Day, M. Marcinkiewicz and M. Chretien, 1998. Precursor convertase: an evolutionary ancient, cell-specific, combinatorial mechanism yielding diverse bioactive peptides and proteins. Ann. NY Acad. Sci. 839: 9–24. [DOI] [PubMed] [Google Scholar]
- Shi, J., H. Xi, Y. Wang, C. Zhang, Z. Jiang et al., 2003. Divergence of the genes on human chromosome 21 between human and other hominoids and variation of substitution rates among transcription units. Proc. Natl. Acad. Sci. USA 100: 8331–8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman, H. H., B. W. Kozyak, A. Nelson, D.M. Thesier, L. T. Su et al., 2004. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature 428: 415–418. [DOI] [PubMed] [Google Scholar]
- Suh, J., N. Lu, A. Nicot, I. Tatsuno and E. DiCicco-Bloom, 2001. PACAP is an anti-mitogeni signal in developing cerebral cortex. Nat. Neurosci. 4: 1–2. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1993. Simple methods for testing molecular clock hypothesis. Genetics 135: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudry, D., B. J. Gonzalez, M. Basille, L. Yon, A. Fournier et al., 2000. Pituitary adenylate cyclase-activating polypeptide and its receptors: from structure to functions. Pharmacol. Rev. 52: 269–324. [PubMed] [Google Scholar]
- Wang, Y. Q., and B. Su, 2004. Molecular evolution of microcephalin, a gene determining human brain size. Hum. Mol. Genet. 13: 1131–1137. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML, a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 15: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573. [DOI] [PubMed] [Google Scholar]
- Zhang, J., 2003. Evolution of the human ASPM gene, a major determinant of brain size. Genetics 165: 2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., H. F. Rosenberg and M. Nei, 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95: 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., D. M. Webb and O. Podlaha, 2002. Accelerated protein evolution and origins of human-specific features: FOXP2 as an example. Genetics 162: 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]