Abstract
We describe the maternal-effect and zygotic phenotypes of null mutations in the Drosophila gene for the ε-subunit of mitochondrial ATP synthase, stunted (sun). Loss of zygotic sun expression leads to a dramatic delay in the growth rate of first instar larvae and ultimately death. Embryos lacking maternally supplied sun (sun embryos) have a sixfold reduction in ATP synthase activity. Cellular analysis of sun embryos shows defects only after the nuclei have migrated to the cortex. During the cortical divisions the actin-based metaphase and cellularization furrows do not form properly, and the nuclei show abnormal spacing and division failures. The most striking abnormality is that nuclei and spindles form lines and clusters, instead of adopting a regular spacing. This is reflected in a failure to properly position neighboring nonsister centrosomes during the telophase-to-interphase transition of the cortical divisions. Our study is consistent with a role for Sun in mitochondrial ATP synthesis and suggests that reduced ATP levels selectively affect molecular motors. As Sun has been identified as the ligand for the Methuselah receptor that regulates aging, Sun may function both within and outside mitochondria.
ORGANIZATION of the cytoplasm of eukaryotic cells depends largely on the cytoskeleton. The organizational role of the cytoskeleton is particularly dramatic during the cortical syncytial divisions of the early Drosophila embryo (reviewed in Foe et al. 1993; Sullivan and Theurkauf 1995). After 9 divisions in the interior of the embryo, syncytial divisions 10–13 occur in the actin-rich cortex just beneath the plasma membrane. Cortical actin is homogenously distributed prior to the arrival of the nuclei, but undergoes a dramatic redistribution induced by the migrating nuclei and their associated centrosomes. During interphase the actin concentrates into apical caps centered above each cortical nucleus and its apically positioned centrosomes. As the nuclei progress into prophase, the centrosomes migrate toward opposite poles and the actin caps undergo a dramatic redistribution to form an oblong ring outlining each nucleus and its associated separated centrosome pair. The actin rings are structurally and compositionally equivalent to conventional cytokinesis. At metaphase, the furrows invaginate to form a half shell encompassing each spindle. During late anaphase and telophase, the metaphase furrows rapidly regress. Centrosome duplication occurs during late anaphase and newly formed centrosome pairs are again located apically in the next interphase and the actin caps reform (Rothwell and Sullivan 2000).
Mutational analysis has been used to identify components responsible for these cytoskeletal rearrangements (Sullivan et al. 1993). A particularly informative class of mutations are those that develop normally through the precortical divisions, but show extensive errors when the nuclei reach the cortex. The rationale for focusing on this class of mutations was that many are likely to disrupt genes involved in the cytoskeletal rearrangements required for proper furrow formation. During the cortical divisions thousands of nuclei are dividing in a confined monolayer and the furrows are required to prevent collisions between neighboring spindles and nuclei. Eleven mutations in this class have been molecularly characterized. They fall into three major groups: centrosome associated (Li and Kaufman 1996; Rothwell et al. 1998; Stevenson et al. 2001), cortical cytoskeleton components (Zhang et al. 1996; Katayoun et al. 2000), and cell cycle regulators (Hari et al. 1995; Fogarty et al. 1997; Sibon et al. 1997, 1999; Brodsky et al. 2000; Price et al. 2000).
Unexpectedly, disruptions of key metabolic processes produce very specific defects. Mutations in the gene encoding glutamine synthase I have been shown to disrupt chromosome segregation of the cortical divisions (Frenz and Glover 1996). Hypoxia and nitric oxide induce a rapid, reversible metaphase arrest during the syncytial divisions that is accompanied by a depletion of ATP levels (DiGregorio et al. 2001). These examples demonstrate that metabolic processes are important during this time. We report here on the identification and characterization of another gene, stunted (sun), that specifically disrupts the cortical cell divisions. Our analysis of sun shows that it encodes the Drosophila epsilon (ε)-subunit of the mitochondrial ATP synthase. Embyros lacking maternal sun display defects in actin furrow formation, spindle orientation, nuclear divisions, and centrosome positioning in the cortical divisions. We discuss the importance of metabolic processes during these dynamic, rapid divisions and discuss why molecular motors should be particularly sensitive to reduced ATP synthase activity.
MATERIALS AND METHODS
Genetics:
y w v FRT101 and y ovoD1 FRT101/Y; C(1)DX f; F38/F38 stocks were obtained from T.-B. Chou and N. Perrimon. Other stocks were obtained from the Bloomington and Bowling Green stock centers. For EMS mutagenesis, flies were fed 25 mm EMS as a 1% sucrose solution.
Initial screen:
y w v FRT101 males were mutagenized with EMS and mated to runt/FM3 females. y w v FRT101*/FM3 females (* denotes mutagenized chromosome) were mated individually to FM7 males. Offspring were screened for the absence of males, which indicated that an X-linked lethal mutation had been induced. y w v FRT101* females from each line were mated either to FM7 males to generate a reference stock or to ovoD1 FRT101/Y; F38/F38 males (F38 is a heat-shock-inducible FLP recombinase on the second chromosome). Larvae from the latter cross were heat-shocked at 37° for 4 hr to generate germline clones. FRT101*/ovoD1 FRT101 females were selected and mated to X/Y; FG2/FG2 males (FG2 is a lacZ reporter gene at the fushi tarazu locus). The eggs laid by these crosses were screened for cuticular patterning defects or altered patterns of β-galactosidase staining. The reference lines were examined similarly to distinguish between maternal and zygotic effects of the lethal mutations.
The sun1 allele was recovered in this screen. The sun1 chromosome was found to have a mutation in the runt (run) locus. As run is located proximal to the Flipase recombinase target (FRT) on the X chromosome at 19E1–3, it was removed by recombination with a sc cv ct v g f stock to generate a y w v sun1 FRT101* f line. The recombinant line still displayed the maternal-effect phenotype.
F2 screen for further sun alleles:
y w v FRT101 males were mutagenized with EMS and mated to run/FM3 females. y w v FRT101*/FM3 females were recovered and individually mated to two to three y w v sun1 FRT101 f/Dp(1)sdY#3m males. The offspring were scored for the absence of y w v females, indicating the presence of a mutation on the mutagenized chromosome that fails to complement sun1. No lethal alleles of sun were recovered in the 4636 chromosomes screened, although the shrinkled (shk) mutation that shows an incompletely penetrant wing phenotype in trans was found. The shk chromosome was used in an F1 screen: y w v FRT101 males were mutagenized with EMS and mated to shk/shk females. Of 4035 females examined, 118 had a wing phenotype and these were individually remated to y w v sun1 FRT101 f/Dp(1)sdY#3m males. The offspring were scored for the absence of y w v FRT101*/y w v sun1 FRT101 females. Three lines failed to complement sun1 of which two were recovered, sun2 and sun3. The shk stock accumulated modifiers with passaging and was eventually lost.
The zygotic lethal phenotype of the sun locus was mapped to 13F/14A by recombination with the multiply marked sc cv ct v g f and m wy sd oss chromosomes and with X chromosome deficiencies and duplications. Previous mapping within this region had placed the sun lethal complementation group as adjacent and proximal to the D-Myb proto-oncogene (A. Katzen, unpublished results).
In vivo fluorescence analysis:
For anti-α-tubulin and propidium iodide staining, embryos were fixed in formaldehyde, devitillinized in methanol, gradually rehydrated, and stained as described previously (Rothwell et al. 1999; Rothwell and Sullivan 2000). Microscopy was performed using an Olympus IMT2 inverted photoscope equipped with a Bio-Rad (Richmond, CA) MRC 600 laser confocal imaging system. In vivo analysis of tubulin behavior was accomplished by microinjection of fluorescently labeled tubulin into embryos and time-lapse images were taken using a fluorescence microscope (Kellogg et al. 1988). In vivo analysis of histones was performed as described previously (Minden et al. 1989). F-actin staining with phalloidin was performed as described previously (Rothwell and Sullivan 2000).
Molecular biology:
Genomic fragments and cDNAs were subcloned into Bluescript.SK+ (Stratagene, La Jolla, CA) using standard techniques. The Drosophila stage 10 library used was constructed by J. Tower using a mixture of random and oligo(dT) priming cloned into the EcoRI site of λ-gt11. DNA probe preparation was with a Prime-It II random primer kit (Stratagene). Sequencing was with the Sequenase 2.0 kit (United States Biochemical, Cleveland) or the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) or the AutoRead kit (Pharmacia, Piscataway, NJ) following the manufacturer's instructions. Sequences were compiled using Geneworks (Intelligenetics, Mountain View, CA) or Lasergene software. PCR for mutant allele sequencing was done with rTth DNA polymerase (Perkin Elmer, Norwalk, CT). PCR products were cloned into the Topo vector (Invitrogen, San Diego). Expressed sequence tags (ESTs) from the Berkeley Drosophila Genome project were used to analyze the transcripts in the sun region. The sequence obtained for EST GM13815 is identical to the cDNA clone sequences RH48911 and RE19513 in GenBank, but is shorter by 7 bp.
Preparation of DNA from larvae:
To sequence mutant alleles, 5–10 mutant larvae were picked by virtue of their retarded development, frozen, crushed in 100 μl buffer A (100 mm Tris/HCl pH 7.6, 100 mm EDTA, 100 mm NaCl, 0.5% SDS), and incubated for 15 min at 65°. The extract was mixed thoroughly with 200 μl of 1.5 m KAc/4.5 m LiCl and chilled for 10 min on ice. The extract was spun at 15,000 rpm for 10 min at room temperature, and the supernatant was transferred to a new tube and respun if necessary. The supernatant was precipitated with 150 μl propanol, and the pellet was washed with 70% ethanol, dried, and resupended in 50 μl TE. Two microliters were used for PCR.
Mitochondrial ATPase assay:
Germline mosaic sun1 and sun3 females were generated following a 1-hr heat-shock, and extracts prepared from their embryos were collected for 3-hr intervals over a 3-day period. No embryos were observed from a non-heat-shock control cross between sun3 females and ovoD1 males. Embryos were homogenized with a plastic pestle in 1.5-ml tubes (Kontes) in ∼10 volumes of 25 mm Tris (pH 7.5), 0.25 m sucrose, 5 mm EDTA, and a protease inhibitor cocktail (10 m benzamidine HCl, 1.2 g/ml phenanthroline, 10 g/ml aprotinin, 10 g/ml leupeptin, and 10 g/ml pepstatin A). Homogenates were centrifuged at 100,000 × g for 1 hr at 4° and supernatant and pellet fractions were collected, frozen in liquid N2, and stored at −80° until needed. The pellet fraction contains membrane-associated ATPases, including mitochondrial, lysosomal, and vacuolar ATPases. Protein concentrations were determined by Bradford assay (Bio-Rad), using BSA as a standard prior to freezing. As controls, protein extracts were also made from embryos collected from wild-type and y w v sun3 FRT101/FM7c stocks. ATPase assays were performed according to Bowman et al. (1978) on 32 and 11 g of total protein from each pellet and supernatant fraction, respectively, in the presence (5 mm) and absence of NaN3. There was no significant difference in ATPase activity in high-speed supernatant fractions between the different embryo preparations.
RESULTS
The stunted maternal-effect locus:
The sun locus was identified in an EMS screen for X-linked zygotic lethals. The mutagenized chromosomes carried an FRT element, allowing rapid and efficient generation of germline clones and analysis of the maternal requirement of the isolated lethals (Chou and Perrimon 1992). Eggs lacking maternal sun (sunmat−; either sun1 or sun3) displayed an almost complete lack of cuticle, and confocal analysis revealed defects in early embryogenesis (see below). An F2 screen was designed to look for new sun alleles, from which we recovered one line that displayed an improperly expanded wing in trans to sun1. Although we were unable to show conclusively that this mutation (shrinkled) was an allele of sun (the allele was later lost), we used it to screen for mutations that are unable to complement its wing phenotype. Two mutations that subsequently failed to complement sun1 were recovered: sun2 and sun3. Mosaic females with germline clones homozygous for either allele produce sunmat− embyros with the same embryonic defects as sun1, indicating that the new mutations are indeed sun alleles, and the same locus is responsible for both zygotic lethality and the maternal phenotype.
We localized the sun locus to 13F by recombination and deficiency mapping. sun lies within Df(1)sd72b, which extends from 13F–14A, an interval previously saturated for lethal mutations (Katzen and Bishop 1996). Complementation analysis showed that sun corresponds to lethal group XV (Katzen and Bishop 1996), which includes two previously recovered alleles. sun4 (synonym EM67) is a zygotic lethal, and attempts to produce germline clones by X-ray-induced mitotic recombination did not lead to any females able to lay eggs. The sun4 chromosome may therefore carry a second lethal. sun5 (synonyms 42-3.0B and EM69) is viable, but displays female sterility in trans to sun1 and sun2 and is lethal in trans to sun3 and Df(1)sd72b. As sun3 is the only allele lethal in trans to sun5, it is probably a strong hypomorph or a null mutation. sun4 was found to give fertile females at a rate of 10% when in trans to sun5, so is probably a weak hypomorph. The maternal effects of sun1, sun2, and sun3 were all found to be temperature sensitive, as rescue of cuticle is observed when mosaic females and eggs are kept at 18° (Kidd 1994). The phenotypic experiments described in the rest of this article were all performed using sun3 and sun1 at 23°–25°.
Molecular characterization of the stunted locus:
The sun locus (complementation group XV) was mapped relative to other complementation groups in Df(1)sd72b and found to lie adjacent to the D-Myb proto-oncogene locus (Katzen et al. 1985, 1998). An 11-kb genomic fragment (Figure 1) was used to generate transgenic flies and found to rescue the zygotic lethal phenotype of sun alleles and no other complementation groups in the region. In addition to D-myb, other transcripts were identified within the rescue fragment by probing maternal mRNA Northern blots with genomic subfragments and by matching public ESTs to genomic sequence. Transgenes containing the novel ubiquitin-like gene UBL3 (Chadwick et al. 1999) under control of its cognate or a heat-shock promoter failed to rescue sun phenotypes. Sequencing of the UBL3 gene in sun mutants also failed to detect any changes. The putative protein-coding region of the second gene, FLI-LRR associated protein-1 (CG8578; Liu and Yin 1998), extends beyond the bounds of the genomic rescue fragment. Expression of CG15914 was not detected on ovarian mRNA Northern blots. Thus we focused on a fourth transcription unit identified by the EST GM13815. Sequencing of GM13815 showed it to encode the 61-amino-acid ε-subunit of the mitochondrial ATP synthase (CG9032-RA; Figure 2). The Drosophila ε-subunit is a small basic protein with 48% identity at the amino acid level to the Arabidopsis ε-subunit, 39% homology to the Saccharomyces cerevisiae ε-subunit, and 34% homology to the bovine ε-subunit. A second ε-subunit, encoded by CG31477, is present in the fly but as it is not represented in any of the public EST collections, it may be expressed only at a low level or in a tissue not yet sampled. The ε-subunit is duplicated in Caenorhabditis elegans and present at three copies in Anopheles; in each case the duplication events appear to have occurred after these species shared a common ancestor with the fly as the ε-subunits are more homologous within each species than between species.
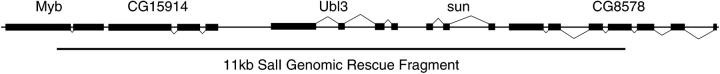
Figure 1.—
Organization of the sun genomic locus. The genomic region around the sun locus deduced from restriction mapping, partial sequencing, and genome project data is shown. Identified genes are shown on the chromosome, with the orientation of the gene shown by the lines linking exons above the line (5′ to 3′ is from left to right) or below the line (5′ to 3′ is from right to left). The 11-kb SalI genomic rescue fragment is shown as a bar below the chromosome.
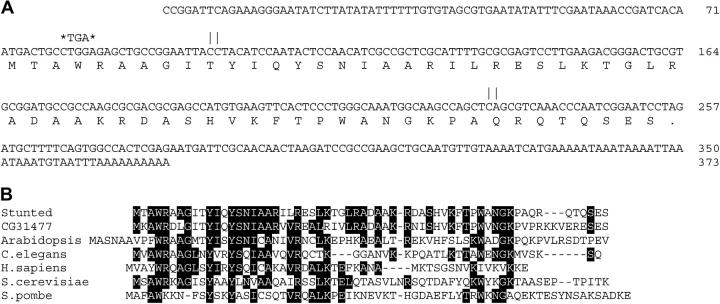
Figure 2.—
Sequence of the sun gene. (A) DNA sequence of the sun gene, with the predicted amino acid sequence of the gene product shown at bottom. The mutation found in sun alleles 1–3 that changes Trp4 to a stop codon is highlighted with asterisks above the DNA sequence. Vertical bars indicate the location of splice sites; the first intron is 129 bp long, and the second is ∼600 bp. The unspliced transcript is predicted to be ∼1.3 kb. (B) Alignment of the Sun amino acid sequence with the ε-subunits of ATP synthase from a representative range of organisms. Residues common to three or more sequences are boxed. The amino acid sequence accession numbers are: Drosophila CG31477, NP_731449; Arabidopsis, Q96253; C. elegans, P34539; Human, NP_008817; S. cerevisiae, NP_015052; and Schizosaccharomyces pombe, NP_596577. The C. elegans and S. pombe sequences are putative proteins identified by analysis of genome projects data.
Sequencing of the ε-subunit gene from three mutant alleles (sun1,2,3) revealed the same nonsense mutation: TGG (Trp4) to TGA (Stop) (Figure 2A). This change is commonly seen in EMS mutagenesis (Ashburner 1989), and as each of the chromosomes displays different complementation characteristics, we believe the three mutations were independent events. The proximity of the nonsense mutation to the start codon indicates that these mutations are null alleles. The differences in complementation characteristics between these mutations are probably due to additional mutations on the chromosomes. This is supported by the ATPase assays described below.
The ATPase activity of mitochondrial ATP synthase is markedly reduced in sun mutants:
Mitochondrial ATP synthase both synthesizes and breaks down ATP. The ATPase activity of mitochondrial ATP synthase is directly correlated to the synthetic enzyme's activity, and so we could assay enzyme activity directly by measuring membrane-associated mitochondrial ATPase activity in 0- to 3-hr embryo extracts. There are multiple sources of ATPase activity, and to identify the mitochondrial ATPase, we took advantage of the fact that sodium azide is a specific and potent inhibitor of mitochondrial ATP synthase. This approach is standard for the field. Extracts enriched for mitochondrial proteins were prepared and assayed in parallel and in duplicate for ATPase activity, in the presence and absence of sodium azide. In wild-type embryo extracts roughly one-half of the total ATPase activity detected is sensitive to inhibition by sodium azide and must be derived from mitochondrial ATP synthase (Figure 3).
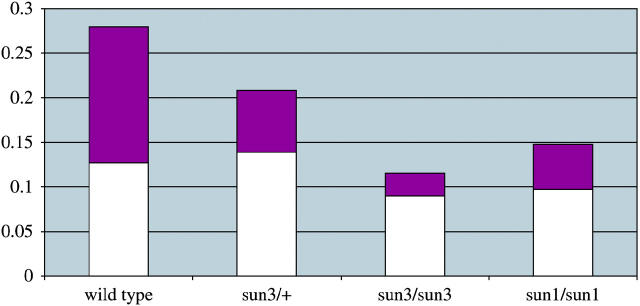
Figure 3.—
ATPase activity in wild-type and sun mutant embryo extracts. Bars indicate ATPase activity in high-speed pellets made from embryos collected from wild-type and sun3/+ stocks or germline mosaic females of sun3 and sun1. Optical density (A660) values and genotypes are indicated along the ordinate and abscissa, respectively. The total ATPase activity derived from the mitochondrial ATP synthase is indicated by the purple portion of each bar. ATPase activity was measured in the absence (purple plus white) and presence (white) of the mitochondrial ATP synthase-specific inhibitor NaN3 (Bowman et al. 1978). Each protein fraction was assayed twice and the average values are shown.
We also assayed extracts made from embryos derived from sun3/+ adult females, which should have half the normal levels of maternally supplied Sun. As expected, we find mitochondrial ATP synthase activity is reduced twofold compared to wild type, indicating that Sun is a limiting component for mitochondrial ATP synthase activity (Figure 3). In embryo extracts derived from sun1 mutant germlines (mosaic females), mitochondrial ATP synthase-specific ATPase activity is reduced sixfold (Figure 3). This major reduction in activity is correlated with the strong sun3 embryonic phenotype. Mitochondrial ATP synthase-specific ATPase activity is also significantly compromised in extracts produced by germline mosaic sun1 females (Figure 3). The higher levels of activity in these extracts are consistent with their weaker embryonic phenotype. These results show that sun activity is a critical component of mitochondrial ATP synthase activity in Drosophila.
Lack of zygotic stunted dramatically reduces larval growth:
All four lethal sun alleles show a zygotic phenotype of greatly delayed larval growth. Mutant sun larvae die stochastically after hatching but some can be maintained in the presence of a yeast food supply for eight days or longer. The mutant larvae undergo little or no growth, and none appear to undergo the first molt to the second instar stage of development. There were no obvious behavioral defects in the larvae, although some individuals can appear sluggish. Maternally supplied sun could be responsible for the partial development observed. The lethal phase probably reflects the point at which the maternal supplies run out. Larval growth is driven by DNA replication, which can be measured by the incorporation of BrdU into DNA (Smith and Orr-Weaver 1991). In wild-type larvae deprived of dietary protein, but supplied with sucrose as an energy source, DNA replication is restricted to mushroom body neuroblasts and the gonad (Britton and Edgar 1998). Larvae lacking zygotic sun have a BrdU incorporation pattern that closely resembled this pattern (M. Garfinkel and B. A. Edgar, personal communication). A P element inserted into the first intron of the α-subunit of mitochondrial ATP synthase also gives a larval growth arrest in the first instar and limited DNA replication (Galloni and Edgar 1999).
The sun maternal effect specifically disrupts the arrangement of nuclei of the syncytial blastoderm:
sunmat− embryos show defective cuticles indicative of an early requirement for sun activity. Preliminary examination of mutant embryos showed that they are already abnormal by the blastoderm stage, so we stained 0- to 4-hr embryos using the nuclear dye propidium iodide (PI; Figure 4). Nuclear migration during cycles 4–10 (axial expansion and cortical migration) appears normal (Table 1). The early cortical divisions (nuclear cycles 10 and 11) are indistinguishable from those in wild-type embryos. However, sunmat− embryos display highly irregular nuclear spacing during the late cortical divisions (Figure 4; Table 1). An increasing tendency of the nuclei to cluster was observed, coincident with the occurrence of nuclear fusion, which is rarely seen in wild-type embryos.
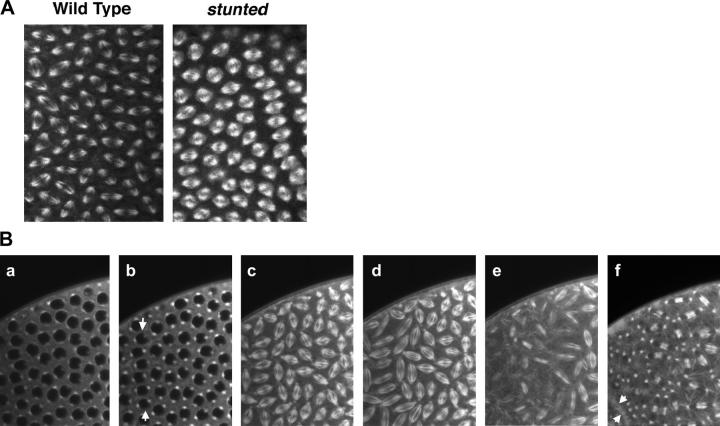
Figure 4.—
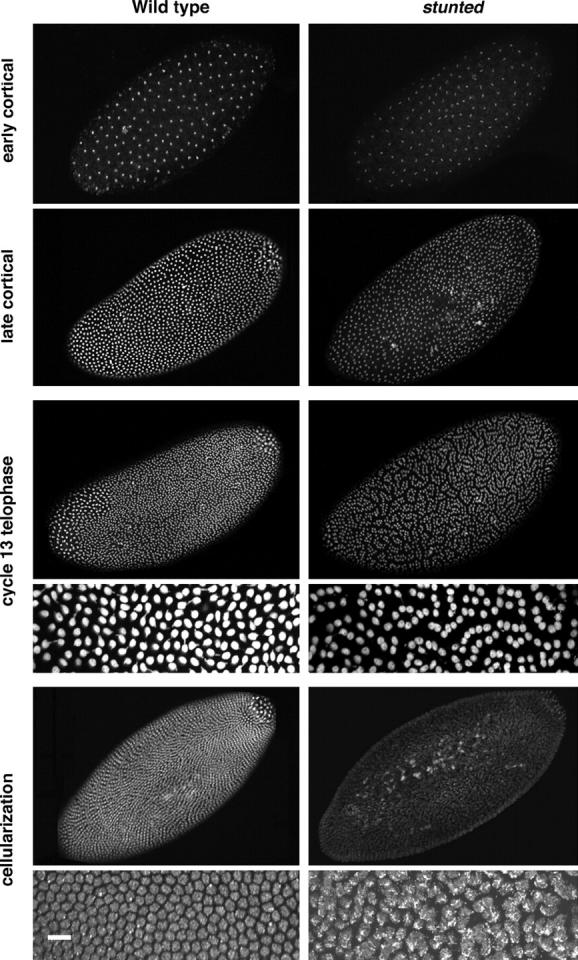
Nuclear behavior of wild-type and stunted embryos. Wild-type (left column) and sunmat− embryos (stunted, right column) were stained with the nuclear dye propidium iodide. During the early cortical divisions, slight abnormalities are observed in sunmat− embryos. During the late cortical divisions, areas in which large numbers of nuclei have fallen back into the center of the embryo are observed and spacing between nuclei has become irregular. At telophase of cycle 13, the spacing defects are readily apparent as nuclei come closer together than in wild type, forming lines of nuclei arranged in clumps in a manner reminiscent of the “paisley” pattern. The 60× insets reveal many nuclei are in contact with their neighbors and form lines of touching nuclei. At cellularization, nuclear fusion has occurred throughout the sunmat− embryos; this is apparent in the 60× inset. Bar, 10 μm.
TABLE 1.
Nuclear behavior in wild-type andsun embryos
|
sunmat−
|
sun/FM7
|
|||
|---|---|---|---|---|
| Cycle | % abnormal | N | % abnormal | N |
| 0–9 | 0 | 62 | 0 | 21 |
| 10 | 6 | 15 | 0 | 9 |
| 11 | 15 | 34 | 11 | 7 |
| 12 | 50 | 24 | 14 | 9 |
| 13 | 86 | 22 | 0 | 7 |
| 14 | 97 | 33 | 0 | 15 |
| Cellularization | 97 | 34 | 0 | 9 |
The nuclei of sunmat− and sunmat−/FM7 control embryos were stained with propidium iodide and examined for defects in morphology, spacing between nuclei, and position relative to the plasma membrane. The sun3 allele was used for this analysis.
To identify a basis for these phenotypes, we examined nuclear dynamics in sunmat− embryos by injecting fluorescently labeled histone into living embryos (Figure 5). A time-lapse movie revealed that numerous nuclear fusions between nonsister nuclei occur at telophase of cycle 13 (Figure 5, D–F). After fusion, these nuclei drop back into the yolk, often leaving behind free centrosomes (Kidd 1994; R. Abu-Shumays and W. Sullivan, unpublished data). A downstream consequence is the formation of large multinucleate cells just prior to cellularization (cycle 14; Figures 4–6). Syncytial nuclear fusions are the direct result of a failure to properly form metaphase furrows that are generated by the actin cytoskeleton (Schweisguth et al. 1990; Simpson and Wieschaus 1990; Sullivan et al. 1993).
Figure 5.—
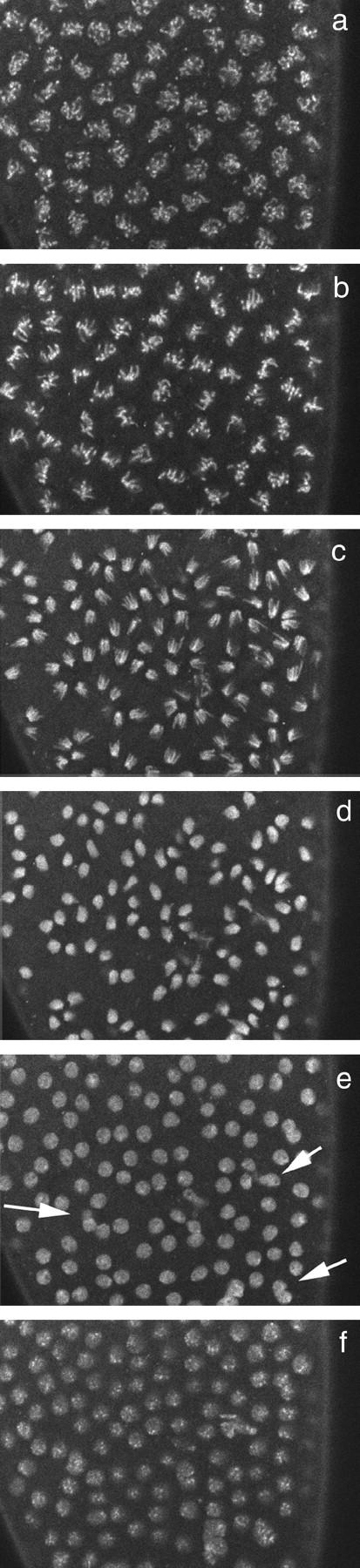
Nuclear dynamics in a sunmat− embryo. Images of living sunmat embryos injected with fluorescently labeled histones at nuclear cycle 13 are shown. Prophase (a), metaphase (b), anaphase (c), telophase (d and e), and interphase (f) are shown. Fusions between dividing nuclei occur at telophase (see arrows in e).
Figure 6.—
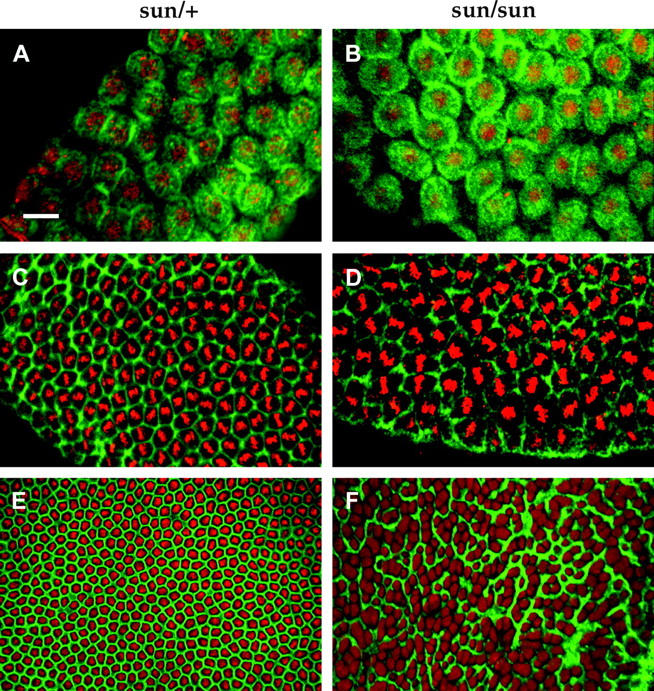
Actin distribution in sunmat− embryos. Wild-type (sunmat−/FM7c) and sunmat− (stunted) embryos were stained with fluroscein-labeled phalloidin and propidium iodide. The actin caps appear to form normally in sunmat− (stunted) embryos (A and B). However, the metaphase furrows are frequently absent in sunmat− embryos (D). By cellularization, the normal orderly outline of cells (E) is severely disrupted (F). Bar, 10 μm.
The actin cytoskeleton displays defects in sunmat− embryos:
In sunmat− embryos, actin caps form normally above interphase nuclei (Figure 6, A and B). However, during metaphase, gaps in the metaphase furrows are common and most frequently found at regions of the metaphase plate most distant from the centrosomes (Figure 6D). This furrow defect is similar to that observed in nuf mutant embryos (Rothwell et al. 1998). At the onset of cellularization, the normally regular actin network is highly disorganized, and in some areas completely absent, resulting in the formation of multinucleate cells (Figure 6F; Kidd 1994; R. Abu-Shumays and W. Sullivan, unpublished data). A number of zygotic and maternal mutants have this phenotype, e.g., serendipity, nullo, and nuf (Schweisguth et al. 1990; Simpson and Wieschaus 1990; Schejter and Wieschaus 1993).
Abnormal centrosome positioning:
Previous studies have demonstrated that proper centrosome duplication, segregation, and position are essential for normal furrow formation and nuclear division (Rothwell and Sullivan 2000). To understand the metaphase furrow and nuclear division defects in sunmat− embryos, we examined centrosome behavior during the cortical divisions.
Living sunmat− embryos were injected with fluorescently labeled tubulin to follow centrosome dynamics (Figure 7) (Rothwell and Sullivan 1999). In the late syncytial divisions of wild-type embryos, centrosome duplication occurs during telophase and the sister centrosomes separate to opposite poles in early interphase. Analysis of sunmat− embryos indicates that centrosome duplication and separation occur normally. This is evidenced by the fact that all interphase nuclei contain centrosomes normally positioned at opposite poles (Figure 7B, a). However, the relative position of centrosome pairs on neighboring nuclei is abnormal in mutant embryos. During the late syncytial divisions of wild-type embryos, mitotic spindles are evenly spaced in an array of orientations that maximize the distance between centrosomes (Valdes-Perez and Minden 1995). In contrast, in metaphase sunmat− embryos, the mitotic spindles are often found in parallel arrays (Figure 7, A and B). The parallel arrays arise from the abnormal positioning of neighboring nonsister centrosomes in early interphase (Figure 7B, b, arrows). Since repulsion between overlapping astral microtubules is thought to be the primary factor in positioning neighboring centrosomes, this astral-based process may be compromised in sunmat− embryos (see discussion). The abnormal interphase centrosome orientation foreshadows the abnormal orientation of neighboring spindles at metaphase (Figure 7B, c). The abnormal orientation of the neighboring spindles could be due to defects in furrow formation, centrosome positioning, or a combination of both. The variability in the sun phenotype made distinguishing between the two possibilities inconclusive.
Figure 7.—
Analysis of microtubule behavior in a sunmat− embryo. (A) Fixed analysis of wild-type vs. sunmat− embryos. (B) Time-lapse sequence of a sunmat− syncytial blastoderm embryo injected with fluorescently labeled tubulin. (a and b) Prophase. The syncytial nuclei can be seen by an absence of tubulin staining due to exclusion by the nuclear envelope. The position of the centrosomes can be determined from increased density of tubulin on either side of the nuclei. On the left, a line of nuclei (indicated by arrows) in which the centrosomes have aligned can be seen. (c) During metaphase, the mitotic spindles corresponding to the aligned centrosomes can be seen on the left. (d) During late metaphase, irregular mitotic figures become apparent, with inappropriate interactions between neighboring spindles, notably at the bottom left and right corners. (e) Anaphase. A lack of mitotic coordination becomes apparent as some spindles break down more quickly than others, and midbodies form (accumulation of microtubules at the spindle equator; these are more apparent in f). (f) Telophase. The position of the centrosomes can be deduced from small local concentrations of tubulin; midbody formation has occurred at the remaining spindles. A group of tightly apposed abnormal midbodies can be seen in the bottom left (indicated by arrows).
DISCUSSION
The effect of reduced ATP levels in the early embryo:
We have described the maternal and zygotic effects of null mutations in the sun locus. sun encodes the ε-subunit of the mitochondrial ATP synthase, the universal enzyme for cellular ATP synthesis (reviewed in Boyer 1997). In yeast the ε-subunit is a nonessential gene required for dimerization and oligomerization of ATP synthase and is involved in generating the inward foldings of the inner mitochondrial membrane, the cristae (Lai-Zhang et al. 1999; Pawnard et al. 2003). The ε-subunit is also a potential binding site or target of the natural inhibitor protein IF1 that serves to prevent ATP hydrolysis (Solaini et al. 1997; Minauro-Sanmiguel et al. 2002). The ε-subunit appears necessary for the maximum efficiency of the ATP synthase complex and it has been proposed to be a molecular clutch regulating the coupling of ATP synthesis to proton flow (Lai-Zhang et al. 1999). Expression of the bovine ε-subunit can rescue the growth defect of S. cerevisiae carrying a deletion of the ε-subunit gene (Lai-Zhang and Mueller 2000). This result suggests that the molecular function of the ε-subunit within the mitochondrial ATP synthase complex has been conserved throughout eukaryotic evolution. Thus we interpret the sun mutant phenotypes as a consequence of reduced levels of ATP. This is consistent with the increased severity of the defects seen at higher temperatures in the sun mutants, because ATP requirements and oxygen consumption increase at higher temperatures. Reducing ATP levels in the embryo by inhibiting oxidative phosphorylation with cyanide or azide induces a cell cycle arrest (DiGregorio et al. 2001; W. Sullivan, unpublished observations). We saw no evidence of cell cycle arrests in sun maternal-effect embryos.
The sun maternal effect is most dramatic during the late cortical cycles, presumably reflecting a greater energetic load at these stages. Lack of sun activity disrupts alignment of neighboring spindles and formation of the metaphase and cellularization furrows. A direct consequence of this is fusions of sister nuclei. Computational studies of these mitoses indicate that the even spacing results from interactions of each nucleus with its neighbors (Valdes-Perez and Minden 1995). These interactions most likely arise from centrosome-based astral microtubules repelling one another (de Saint Phalle and Sullivan 1998). The force of their repulsion is inversely proportional to the distance between neighboring centrosomes. The abnormal arrangement of spindles in sun embryos is not due to a failure to form astral microtubules, as we do not see a difference between wild-type and sun astral microtubules in the light microscope.
Motor proteins and the sun phenotype:
One interpretation of the sun phenotype is that the activity of the motor proteins during the cortical divisions places extra energetic demands on the embryo. In sun embryos, while the reduced ATP levels are sufficient for the early divisions, they are insufficient for the cortical divisions. On the basis of the sun maternal-effect phenotype, we propose that maintaining regular spacing of the closely packed nuclei is the most ATP-demanding process in the syncytial embryo. The inability to keep nuclei apart leads to inappropriate microtubule interactions, nuclear fusion, and dropping of nuclei back toward the yolk.
The repulsive force between anti-parallel microtubules is generated by microtubule-based motor proteins. Given the number of molecular motors already described, it is highly likely that several contribute to spindle positioning (Scholey et al. 2003). The sun mutant phenotype could be a failure of the motor proteins to provide this repulsive force. For example, the motor protein KLP61F acts on anti-parallel microtubules to maintain separation of sister centrosomes (Sharp et al. 1999). In addition, embryos lacking the motor protein Ncd display centrosomal defects and microtubule spurs between mitotic spindles (Endow et al. 1994). Figure 4 of Endow and Komma (1996) shows three aligned spindles reminiscent of the sun maternal effect. A further candidate motor is the Drosophila homolog of MKLP1 kinesin-like protein, which transports oppositely oriented microtubules relative to one another (Schmid and Tautz 1998; Sharp et al. 1999).
Intracellular circulation of ATP:
One might have expected decreased levels of ATP to have highly pleiotropic effects, and so it is surprising that sun has such a specific effect on the syncytial mitoses. Many other mutations affecting the syncytial mitoses turn out to be centrosomal or key regulators of the cell cycle, e.g., nuclear fallout (Rothwell et al. 1998). The sun locus is distinctive by encoding a component of a central metabolic enzyme. Mutations in another essential metabolic enzyme, Glutamine synthetase 1 (Gs1), also specifically affect the syncytial mitoses (Frenz and Glover 1996). Gs1 catalyzes the amination of glutamate in an ATP-dependent manner to produce glutamine, which is required for amino acid, purine, and pyrimidine biosynthesis. Analysis of Gs1 mutants suggests that delays in syncytial cell cycle progression lead to nuclei being discarded due to a reduction in amino acid and nucleotide availability (Frenz and Glover 1996). Surprisingly, given the requirement of Gs1 for ATP, the sun and Gs1 mutant phenotypes are almost reciprocal, with Gs1 affecting nuclear events during S phase and sun affecting cytoplasmic events during M phase, with little or no effect on DNA segregation.
Why are the sun and Gs1 mutant phenotypes so reciprocal? Gs1 may function at lower ATP concentrations than required for the molecular motors to maintain spindle separation. In the sun mutant, there may be sufficient ATP to carry out the functions of Gs1, but not those of the microtubule-associated motors. There may also be distinct biochemical pools from which Gs1 and the motors obtain ATP. Under most conditions, intracellular circulation keeps the ATP concentration perfectly homeostatic, meaning the concentration does not change even when ATP-dependent work is being performed (Hochachka 2003). The sun mutant syncytium may resemble a fatigued cell, with local differences in intracellular circulation of ATP to the metabolic pools containing molecular motors and biosynthetic enzymes.
The role of the ε-subunit in multicellular organisms:
S. cerevisiae deleted for the ε-subunit grow slowly on medium with glycerol as the carbon source, indicating that the ε-subunit is not an essential gene (Lai-Zhang et al. 1999). In contrast to S. cerevisiae, the ε-subunit of ATP synthase is essential for survival of Drosophila. As the bovine ε-subunit can rescue the yeast ε-deletion mutant (Lai-Zhang and Mueller 2000), we believe that the phenotypic differences originate in differences in ATP homeostasis between unicellular and multicellular organisms. No other eukaryotic ε-subunit mutants have been published. Our phenotypic and biochemical observations indicate that ATP levels are reduced but not eliminated, supporting the hypothesis that the ε-subunit is required for maximal efficiency of ATP synthase. The presence of maternal ATP in sun mutants allows growth until an energetically demanding process is encountered. In the early embryo, the first defects are seen in the cortical divisions, but the embryos continue to grow and cellularize, albeit abnormally. Embryos lacking maternal sun fail to gastrulate (T. Kidd and D. Ish-Horowicz, unpublished observations), suggesting the dynamic cell movements are incompatible with the reduced ATP levels. In the larva, the energetically demanding processes of DNA replication and protein synthesis normally drive a 200-fold increase in mass over 4 days (Galloni and Edgar 1999). The absence of zygotic sun causes a larval growth arrest before any significant growth has occurred. Interestingly, the same phenotype is seen for a mutation, colibri, in the α-subunit of ATP synthase (Galloni and Edgar 1999). The α-subunit, known as bellwether in Drosophila, should be absolutely required for ATP synthesis; a series of alleles have been characterized as recessive lethal, but the exact lethal phase was not determined (Jacobs et al. 1998). The colibri allele of bellwether is a P-element insertion in an intron and is most probably a hypomorphic allele allowing some synthesis of ATP in a manner similar to that of sun alleles. Mutant clones of colibri in the wing show a severe size reduction whereas mutant clones in the eye survive well (Galloni and Edgar 1999), suggesting different energetic requirements in the two tissues. We believe that ε-subunit mutations will be essential in all multicellular organisms, but the effects will vary from tissue to tissue.
The ε-subunit as a mitochondrial subunit and as an extracellular ligand:
Sun was unexpectedly identified as the ligand for the Drosophila G-protein-coupled receptor (GPCR), Methuselah (Mth; Cvejic et al. 2004). Mutations in methuselah (mth) extend life span, and the protein is required in motor neurons where it regulates neurotransmitter release (Lin et al. 1998; Song et al. 2002). A Mth-GFP fusion localizes to the plasma membrane of the presynaptic terminals, although it has not conclusively been shown to be exposed to the extracellular environment (Song et al. 2002). Analysis of life span in sun mutants (using the alleles generated in this study) revealed extended life span (Cvejic et al. 2004).
This diverse function might indicate that the sun gene does not encode a genuine homolog of mitochondrial ATP synthase ε-subunits from other species. However, this view appears unlikely. The results from our study strongly suggest Sun indeed participates in ATP synthesis (Figure 3). Rather, the genetic and biochemical analyses suggest that Sun is a bifunctional protein. Such a dual function is reminiscent of another mitochondrial protein, cytochrome C, which functions within the respiratory electron transport chain and is released from the mitochondria to participate in apoptosis (Li et al. 2004). A better understanding of Sun function awaits the development of antibody reagents to visualize Sun localization, particularly in Drosophila models of aging. For the moment, there are some intriguing hints as to how Sun might be regulated: sun transcription has been shown to be downregulated by oxidative stress, which is thought to limit life span in multicellular organisms (Finkel and Holbrook 2000; Girardot et al. 2004). Sun has been shown to bind the regulatory subunit of cAMP protein kinase, suggesting it may be a substrate for phosphorylation (Pka-R1; Giot et al. 2003), although it remains to be confirmed in vivo.
Acknowledgments
We are grateful to Helen Francis-Lang for recognizing the sun maternal effect as a morphogenesis defect, for suggesting the name stunted, and for coordinating research interactions between the Ish-Horowicz and Sullivan laboratories. We thank Ze'ev Paroush, Wendy Rothwell, and Kristina Yu for technical assistance and advice, as well as other members of the Ish-Horowicz and Sullivan laboratories. We thank Michelle Garfinkel and Bruce Edgar for examining sun mutant larvae. We thank Corey Goodman in whose laboratory some of this work was carried out. We are especially grateful to T.-B. Chou and N. Perrimon for supplying stocks before publication and to Barry Bowman for his help with the ATPase assays. This work was supported by the Imperial Cancer Research Fund (now Cancer Research UK), by a grant from the Howard Hughes Medical Institute International Research Scholars Program (to D.I.-H.), and by grants from the National Institutes of Health (R01 GM68961 to A.K. and R01 GM46409) and the University of California Cancer Coordinating Committee to W.S.
References
- Ashburner, M., 1989 Drosophila: A Laboratory Handbook, pp. 345–346. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bowman, B. J., S. E. Mainzer, K. E. Allen and C. W. Slayman, 1978. Effects of inhibitors on the plasma membrane and mitochondrial adenosine triphosphatases of Neurospora crassa. Biochim. Biophys. Acta 512: 13–28. [DOI] [PubMed] [Google Scholar]
- Boyer, P. D., 1997. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 66: 717–749. [DOI] [PubMed] [Google Scholar]
- Britton, J. S., and B. A. Edgar, 1998. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125: 2149–2158. [DOI] [PubMed] [Google Scholar]
- Brodsky, M. H, J. J. Sekelsky, G. Tsang, R. S. Hawley and G. M. Rubin, 2000. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 14: 666–678. [PMC free article] [PubMed] [Google Scholar]
- Chadwick, B. P., T. Kidd, D. Ish-Horowicz and A. Frischauf, 1999. Cloning, mapping and expression of HCG1, a novel ubiquitin-like gene. Gene 233: 189–195. [DOI] [PubMed] [Google Scholar]
- Chou, T. B., and N. Perrimon, 1992. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131: 643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvejic, S., Z. Zhu, S. J. Felice, Y. Berman and X.-Y. Huang, 2004. The endogenous ligand Stunted of the GPCR Methuselah extends lifespan in Drosophila. Nat. Cell Biol. 6: 540–546. [DOI] [PubMed] [Google Scholar]
- de Saint Phalle, B., and W. Sullivan, 1998. Spindle assembly and mitosis without centrosomes in parthenogenetic Sciara embryos. J. Cell Biol. 141: 1383–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGregorio, P. J., J. A. Ubersax and P. H. O'Farrell, 2001. Hypoxia and nitric oxide induce a rapid reversible cell cycle arrest of the Drosophila syncytial divisions. J. Biol. Chem. 276: 1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow, S. A., and D. J. Komma, 1996. Centrosome and spindle function of the Drosophila Ncd microtubule motor visualized in live embryos using Ncd-GFP fusion proteins. J. Cell Sci. 109: 2429–2442. [DOI] [PubMed] [Google Scholar]
- Endow, S. A., R. Chandra, D. J. Komma, A. H. Yamamoto and E. D. Salmon, 1994. Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle pole defects in mitosis. J. Cell Sci. 107: 859–867. [DOI] [PubMed] [Google Scholar]
- Finkel, T., and N. J. Holbrook, 2000. Oxidants, oxidative stress and the biology of aging. Nature 408: 239–247. [DOI] [PubMed] [Google Scholar]
- Foe, V. E., G. M. Odell and B. A. Edgar, 1993 Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint, pp. 149–300 in The Development of Drosophila melanogaster, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Fogarty, P., S. D. Campbell, R. Abu-Shumays, B. S. Phalle, K. R. Yu et al., 1997. The Drosophila grapes gene is related to checkpoint gene chk1/rad27 and is required for late syncytial division fidelity. Curr. Biol. 7: 418–426. [DOI] [PubMed] [Google Scholar]
- Frenz, L. M., and D. M. Glover, 1996. A maternal requirement for glutamine synthetase 1 for the mitotic cycles of syncytial Drosophila embryos. J. Cell Sci. 109: 2649–2660. [DOI] [PubMed] [Google Scholar]
- Galloni, M., and B. A. Edgar, 1999. Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 126: 2365–2375. [DOI] [PubMed] [Google Scholar]
- Giot, L., J. S. Bader, C. Brouwer, A. Chaudhuri, B. Kuang et al., 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736. [DOI] [PubMed] [Google Scholar]
- Girardot, F., V. Monnier and H. Tricoire, 2004. Genome wide analysis of common and specific stress responses in adult Drosophila melanogaster. BMC Genomics 5: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari, K. L., A. Santerre, J. J. Sekelsky, K. S. McKim, J. B. Boyd et al., 1995. The mei-41 gene of D. melanogaster is a structural and functional homologue of the human ataxia telangiectasia gene. Cell 82: 815–821. [DOI] [PubMed] [Google Scholar]
- Hochachka, P. W., 2003. Intracellular convection, homeostasis and metabolic regulation. J. Exp. Biol. 206: 2001–2009. [DOI] [PubMed] [Google Scholar]
- Jacobs, H., R. Stratmann and C. Lehner, 1998. A screen for lethal mutations in the chromosomal region 59AB suggests that bellwether encodes the alpha subunit of the mitochondrial ATP synthase in Drosophila. Mol. Gen. Genet. 259: 383–387. [DOI] [PubMed] [Google Scholar]
- Katayoun, A., B. Stuart and S. A. Wasserman, 2000. Functional analysis of the Drosophila Diaphanous FH protein in early embryonic development. Development 127: 1887–1897. [DOI] [PubMed] [Google Scholar]
- Katzen, A. L., and J. M. Bishop, 1996. myb provides an essential function during Drosophila development. Proc. Natl. Acad. Sci. USA 93: 13955–13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen, A. L., T. B. Kornberg and J. M. Bishop, 1985. Isolation of the proto-oncogene c-myb from D. melanogaster. Cell 41: 449–456. [DOI] [PubMed] [Google Scholar]
- Katzen, A. L., J. Jackson, B. P. Harmon, S. M. Fung, G. Ramsay et al., 1998. Drosophila myb is required for the G2/M transition and maintenance of diploidy. Genes Dev. 12: 831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, D. R., T. J. Mitchison and B. M. Alberts, 1988. Behavior of microtubules and actin filaments in living Drosophila embryos. Development 103: 675–686. [DOI] [PubMed] [Google Scholar]
- Kidd, T., 1994 Genetic and molecular studies of early embryogenesis in Drosophila. Ph.D. Thesis, Oxford University, Oxford.
- Lai-Zhang, J., and D. M. Mueller, 2000. Complementation of deletion mutants in the genes encoding the F1-ATPase by expression of the corresponding bovine subunits in yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267: 2409–2418. [DOI] [PubMed] [Google Scholar]
- Lai-Zhang, J., Y. Xiao and D. M. Mueller, 1999. Epistatic interactions of deletion mutants in the genes encoding the F1-ATPase in yeast Saccharomyces cerevisiae. EMBO J. 18: 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K., and T. C. Kaufman, 1996. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell 85: 585–596. [DOI] [PubMed] [Google Scholar]
- Li, P., D. Nijhawan and X. Wang, 2004. Mitochondrial activation of apoptosis. Cell 116: 57–59. [DOI] [PubMed] [Google Scholar]
- Lin, Y. J., L. Seroude and S. Benzer, 1998. Extended lifespan and stress resistance in the Drosophila mutant methuselah. Science 282: 943–946. [DOI] [PubMed] [Google Scholar]
- Liu, Y. T., and H. L. Yin, 1998. Identification of the binding partners for flightless I, a novel protein bridging the leucine-rich repeat and the gelsolin superfamilies. J. Biol. Chem. 273: 7920–7927. [DOI] [PubMed] [Google Scholar]
- Minauro-Sanmiguel, F., C. Bravo and J. J. Garcia, 2002. Cross linking of the endogenous inhibitor protein (IF1) with rotor (γ, ε) and stator (α) subunits of the mitochondrial ATP synthase. J. Bioenerg. Biomembr. 34: 433–443. [DOI] [PubMed] [Google Scholar]
- Minden, J. S., D. A. Agard, J. W. Sedat and B. M. Alberts, 1989. Direct cell lineage analysis in Drosophila melanogaster by time lapse three dimensional optical microscopy of living embryos. J. Cell Biol. 109: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawnard, P., J. Vaillien, B. Coulary, J. Schaeffer, V. Soubannier et al., 2003. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 21: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, D., S. Rabinovitch, P. H. O'Farrell and S. D. Campbell, 2000. Drosophila wee1 has an essential role in the nuclear divisions of early embryogenesis. Genetics 155: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell, W. F., and W. Sullivan, 1999 Fluorescent analysis of Drosophila embryos, pp. 141–157 in Drosophila Protocols, edited by W. Sullivan, A. Ashburner and R. S. Hawley. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rothwell, W. F., and W. Sullivan, 2000. The centrosome in early Drosophila development. Curr. Top. Dev. Biol. 49: 409–447. [DOI] [PubMed] [Google Scholar]
- Rothwell, W. F., P. Fogarty, C. M. Field and W. Sullivan, 1998. Nuclear-fallout, a Drosophila protein that cycles from the cytoplasm to the centrosomes, regulates cortical microfilament organization. Development 125: 1295–1303. [DOI] [PubMed] [Google Scholar]
- Rothwell, W. F., C. X. Zhang, C. Zelano, T. S. Hsieh and W. Sullivan, 1999. The Drosophila centrosomal protein Nuf is required for recruiting Dah, a membrane associated protein, to furrows in the early embryo. J. Cell Sci. 112: 2885–2893. [DOI] [PubMed] [Google Scholar]
- Schejter, E. D., and E. Wieschaus, 1993. Functional elements of the cytoskeleton in the early Drosophila embryo. Annu. Rev. Cell Biol. 9: 67–99. [DOI] [PubMed] [Google Scholar]
- Schmid, K. J., and D. Tautz, 1998. Sequence and expression of DmMKLP1, a homolog of the human MKLP1 kinesin-like protein from Drosophila melanogaster. Dev. Genes Evol. 208: 474–476. [DOI] [PubMed] [Google Scholar]
- Scholey, J. M., I. Brust-Mascher and A. Mogilner, 2003. Cell division. Nature 442: 746–752. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F. C., J.-A. Lepesant and A. Vincent, 1990. The serendipity-alpha gene encodes a membrane associated protein required for the cellularization of the Drosophila embryo. Genes Dev. 4: 922–931. [DOI] [PubMed] [Google Scholar]
- Sharp, D. J., K. R. Yu, J. C. Sisson, W. Sullivan and J. M. Scholey, 1999. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1: 51–54. [DOI] [PubMed] [Google Scholar]
- Sibon, O. C., V. A. Stevenson and W. E. Theurkauf, 1997. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388: 93–97. [DOI] [PubMed] [Google Scholar]
- Sibon, O. C, A. Laurencon, R. Hawley and W. E. Theurkauf, 1999. The Drosophila ATM homologue Mei-41 has an essential checkpoint function at the midblastula transition. Curr. Biol. 9: 302–312. [DOI] [PubMed] [Google Scholar]
- Simpson, L., and E. Wieschaus, 1990. Zygotic activity of the nullo locus is required to stabilize the actin myosin network during cellularization in Drosophila. Development 110: 851–863. [DOI] [PubMed] [Google Scholar]
- Smith, A. V., and T. L. Orr-Weaver, 1991. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development 112: 997–1008. [DOI] [PubMed] [Google Scholar]
- Solaini, G., A. Baracca, E. Gabellieri and G. Lenaz, 1997. Modification of the mitochondrial F1-ATPase ε subunit, enhancement of the ATPase activity of the IF1–F1 complex and IF1 binding dependence of the conformation of the ε subunit. Biochem. J. 327: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W., R. Ranjan, K. Dawson-Scully, P. Bronk, L. Marin et al., 2002. Presynaptic regulation of neurotransmission in Drosophila by the g protein-coupled receptor methuselah. Neuron 36: 105–119. [DOI] [PubMed] [Google Scholar]
- Stevenson, V., J. Kramer, J. Kuhn and W. E. Theurkauf, 2001. Centrosomes and the Scrambled protein coordintate microtubule-independent actin reorganization. Nat. Cell Biol. 3: 68–75. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., and W. E. Theurkauf, 1995. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr. Opin. Cell Biol. 7: 18–22. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., P. Fogarty and W. Theurkauf, 1993. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development 118: 1245–1254. [DOI] [PubMed] [Google Scholar]
- Valdes-Perez, R. E., and J. S. Minden, 1995. Drosophila melanogaster syncytial nuclear divisions are patterned: time-lapse images. Hypothesis and computational evidence. J. Theor. Biol. 175: 525–532. [DOI] [PubMed] [Google Scholar]
- Zhang, C., M. P. Lee, A. D. Chen, S. D. Brown and T.-S. Hsieh, 1996. Isolation and characterization of a Drosophila gene essential for early embryonic development and formation of cortical cleavage furrows. J. Cell Biol. 134: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]