Abstract
During meiotic recombination in the yeast Saccharomyces cerevisiae, heteroduplex DNA is formed when single-stranded DNAs from two homologs anneal as a consequence of strand invasion. If the two DNA strands differ in sequence, a mismatch will be generated. Mismatches in heteroduplex DNA are recognized and repaired efficiently by meiotic DNA mismatch repair systems. Components of two meiotic systems, mismatch repair (MMR) and large loop repair (LLR), have been identified previously, but the substrate range of these repair systems has never been defined. To determine the substrates for the MMR and LLR repair pathways, we constructed insertion mutations at HIS4 that form loops of varying sizes when complexed with wild-type HIS4 sequence during meiotic heteroduplex DNA formation. We compared the frequency of repair during meiosis in wild-type diploids and in diploids lacking components of either MMR or LLR. We find that the LLR pathway does not act on single-stranded DNA loops of <16 nucleotides in length. We also find that the MMR pathway can act on loops up to 17, but not >19, nucleotides in length, indicating that the two pathways overlap slightly in their substrate range during meiosis. Our data reveal differences in mitotic and meiotic MMR and LLR; these may be due to alterations in the functioning of each complex or result from subtle sequence context influences on repair of the various mismatches examined.
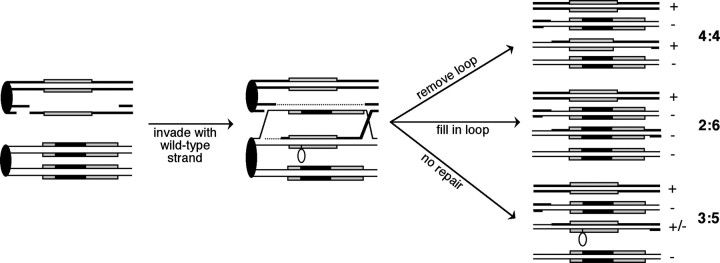
MEIOTIC recombination in the yeast Saccharomyces cerevisiae is a highly regulated process that results in the exchange of DNA sequences between homologous chromosomes. Recombination begins with a double-strand break (DSB) initiated by Spo11p (Keeney et al. 1997), followed by resection of the 5′-ends of the broken DNA molecules. The 3′-ends invade the homologous chromosome to form a heteroduplex DNA molecule (Figure 1) composed of single-stranded DNA from each of the chromosomes. If the DNA sequences included in the heteroduplex region differ, mismatches or unpaired loops will form (Kirkpatrick 1999; Borts et al. 2000). In the AS4/AS13 strain background used in this study, approximately one-half of all diploids initiate a recombination event at HIS4 during meiosis (Nag et al. 1989; White et al. 1993; Fan et al. 1995). This high recombination level leads to a high frequency of mismatch formation in diploids with heterozygous HIS4 alleles.
Figure 1.—
Patterns of aberrant segregation associated with meiotic recombination at HIS4. A double-stranded break initiates recombination on the wild-type chromosome, followed by strand degradation, invasion, repair synthesis, and resolution of crossovers. Recombination initiated on the mutant homolog would follow a similar pattern but result in 5:3, 6:2, or 4:4 segregation. Chromosomes are shown as double-stranded DNA molecules. The HIS4 gene is shown as a shaded rectangle, and the black rectangle within represents a sequence insertion that can form a loop when present in heteroduplex DNA. Dotted lines represent regions of repair synthesis. The segregation pattern of the spore colonies when replica-plated to medium lacking histidine is shown on the far right. +, growth; −, no growth; +/−, sectored colony. The 3:5 tetrads indicate postmeiotic segregation, 2:6 tetrads are gene conversions, and 4:4 tetrads are restorations.
There are several possible fates for the mismatch after it has formed (Figure 1). Repair of the mismatch will either restore normal Mendelian segregation or generate 6:2 or 2:6 gene conversion events, depending on the initiating chromosome. If the mismatch is not detected or repaired, one of the four spores will receive a duplex DNA molecule that contains the mismatch. In the first cell cycle following spore germination, the two alleles composing the mismatch will be replicated and segregated to separate daughter cells. Growth of these cells will lead to a spore colony with a sectored phenotype: a postmeiotic segregation (PMS) that is detected as either 5:3 or 3:5 segregation, depending on the initiating chromosome. Thus, the degree to which a mismatch is recognized and repaired during meiotic recombination is reflected in the ratio of gene conversion (GC) to postmeiotic segregation (PMS) tetrads, with GC tetrads representing repair events and PMS tetrads representing unrepaired mismatches.
In S. cerevisiae, at least three distinct meiotic mismatch repair pathways exist (reviewed in Kirkpatrick 1999; Borts et al. 2000). One pathway is similar to the well-characterized mitotic postreplicative mismatch repair (MMR) pathway (reviewed in Harfe and Jinks-Robertson 2000a) and involves Msh2p, Msh3p, Msh6p, Pms1p, and Mlh1p (Kirkpatrick 1999). In addition, at least two pathways function to repair large loop mismatches. The first large loop repair (LLR) pathway, involving Rad1p, Rad10p, Msh2p, and Msh3p, can repair 26-base loops as well as very large loops up to 5.6 kb in size (Kirkpatrick and Petes 1997; Kearney et al. 2001). These studies indicate that all four of these proteins function in the same repair pathway and that a second large loop repair pathway exists, because repair of large loop mismatches is still seen in strains in which the RAD1-dependent LLR pathway is eliminated. These studies also indicate that LLR in meiosis occurs by a mechanism other than LLR during mitosis, as there is no evidence for a mitotic LLR activity requiring Rad1p, Msh2p, Msh3p, and Rad10p (Tran et al. 1996; Sia et al. 1997b; Harfe et al. 2000; Corrette-Bennett et al. 2001).
Two of the meiotic LLR proteins, Msh2p and Msh3p, are also involved in mitotic MMR. During mitosis, two main multimeric protein complexes function to repair base-base mismatches and small loops that occur as a result of DNA polymerase slippage (reviewed in Harfe and Jinks-Robertson 2000a; Hsieh 2001; Marti et al. 2002). Both complexes contain the MMR proteins Msh2p, Pms1p, and Mlh1p. The first complex also contains Msh6p, while the second contains Msh3p. These two complexes have different substrate specificity for mitotic repair of mismatches (Harfe and Jinks-Robertson 2000a,b; Hsieh 2001; Marti et al. 2002): the Msh6p tetramer recognizes primarily base-base mismatches and single nucleotide insertion/deletion loops, while the Msh3p tetramer recognizes insertion/deletion loops up to 14 or 15 bases in size (Sia et al. 1997b). Two other complexes, in which Pms1p is replaced with Mlh2p or Mlh3p, have lesser roles in MMR. Studies indicate that these minor complexes are involved in repair of some types of frameshift intermediates (Harfe and Jinks-Robertson 2000a,b; Hsieh 2001; Marti et al. 2002). The known DNA mismatch repair proteins that function during mitosis cannot repair loops >14 or 15 bases in length (Sia et al. 1997b).
Two of the meiotic LLR proteins, Rad1p and Rad10p, are involved in nucleotide excision repair (NER) during mitotic growth. NER functions to repair bulky DNA lesions, such as thymine dimers and other helix-distorting lesions. During NER the damaged nucleotide is recognized and bound by several NER proteins, and the DNA surrounding the lesion is unwound. The single-stranded DNA containing the lesion is removed by two endonucleases. A heterodimeric complex, consisting of Rad1p and Rad10p, cuts the damaged DNA strand on the 5′-side of the lesion, while Rad2p cuts on the 3′-side of the lesion. These cuts result in the removal of a fragment ∼25–30 nucleotides long. Finally, the single-stranded region undergoes repair synthesis and ligation (Sancar 1996; Prakash and Prakash 2000).
Our favored model for the activities of the proteins in the RAD1-dependent LLR pathway springs from the known enzymatic roles of those proteins during mitotic DNA repair and the observed effects on meiotic recombination and DNA repair upon deletion of the LLR genes (Kirkpatrick and Petes 1997). Given the characterized activities of Rad1/10p and Msh2/3p during DNA repair in mitotic cells, we hypothesize that the Rad1/10p endonuclease functions to cleave the DNA strand opposite the extruded loop during meiotic LLR, while the MSH2 and MSH3 proteins act as loop-recognition factors or confer specificity to the cleavage reaction. Rad1p, Msh2p, Msh3p, and Rad10p have also been shown to interact physically by both yeast two-hybrid and co-immunoprecipitation experiments (Bardwell et al. 1993; Bertrand et al. 1998). Neither study of meiotic LLR determined the size limits for LLR and MMR during meiosis; even very large loops of 5.6 kb are still repaired.
The goal of this study was to define the substrates for the known meiotic DNA repair pathways—the RAD1-dependent LLR pathway and the meiotic MMR pathway. To accomplish this, we used meiotic recombination to generate loop mismatches of various sizes in vivo, and determined the degree to which each was repaired in wild-type strains and in strains lacking a specific repair pathway. We find that for loops above a certain size the efficiency of repair declines as the loop size increases, even in wild-type strains, depending on the sequence of the DNA contained in the loop. Also, the minimum size loop that the RAD1-dependent large loop repair pathway can repair is 16 bases in length. Finally, loss of PMS1 has an effect on the repair of loop sizes up to at least 17 bases, but not >19 bases. Thus, there is an overlap in substrates repaired by the meiotic mismatch repair pathway and RAD1-dependent large loop repair pathway.
MATERIALS AND METHODS
Media, plasmids, and yeast strains:
Standard media were used (Adams et al. 1998). Sporulation plates contained 1% potassium acetate, 0.1% yeast extract, 0.05% glucose, 6 μg of adenine/ml, and 2% agar. Diploids were sporulated at 18° and dissected onto rich growth medium plates (yeast extract-peptone-dextrose). After colonies formed at 30°, the plates were replica plated to omission medium plates to determine the segregation patterns of all heterozygous markers. Postmeiotic segregation (PMS) events at HIS4 were detected as sectored His+/His− colonies by examination under a low-phase microscope (Nikon Eclipse E400 at 30× power).
All strains were derived from the haploid strains AS13 (MATa leu2 ura3 ade6) or AS4 (MATα trp1 arg4 tyr7 ade6 ura3) (Stapleton and Petes 1991). All strains are isogenic except for alterations introduced via lithium acetate transformation.
Plasmids containing his4 alleles with varying length DNA sequence insertions were used to replace the wild-type HIS4 chromosomal sequence in AS13. Each plasmid was constructed by annealing two complementary oligonucleotides and inserting the oligos into the SalI site in HIS4 on pDN9 (Nag et al. 1989) (Table 1). pDN9 is YIp5 (Struhl et al. 1979) with a XhoI-BglII HIS4 fragment. The annealed oligonucleotides could insert into the SalI site in two different orientations: “forward,” with the AG sequence in the transcribed strand, and “reverse,” with the CT sequence in the transcribed strand. In this study we examined alleles with forward insertions. Those insertion lengths that maintain the proper HIS4-reading frame were designed with a stop codon to create a his4 allele (Table 1 and Figure 2). Orientation and sequence of the inserts were confirmed by sequencing with primer 4102403 (+429 into HIS4-reading frame) and/or 4102404 (+610 into HIS4-reading frame).
TABLE 1 .
Oligonucleotides and plasmids
| Name | Sequence | Plasmid |
|---|---|---|
| his4 10mer A | 5′ TCGAGAGGAC | pSLJ009 |
| his4 10mer B | CTCCTGAGCT 5′ | |
| his4 14mer A | 5′ TCGAGAGAGAAGAC | pSLJ010 |
| his4 14mer B | CTCTCTTCTGAGCT 5′ | |
| his4 15mer A | 5′ TCGAGTAGGAGAAGC | pLEJ003 |
| his4 15mer B | CATCCTCTTCGAGCT 5′ | |
| his4 16mer A | 5′ TCGAGAGGAGAAAGAC | |
| his4 16mer B | CTCCTCTTTCTGAGCT 5′ | pLEJ001 |
| his4 17mer A | 5′ TCGAGAGGAGAAGAGAC | pSLJ003 |
| his4 17mer B | CTCCTCTTCTCTGAGCT 5′ | |
| his4 17mer A (random) | 5′ TCGATGGTTGTCTAGGT | pPAJ173 |
| his4 17mer B (random) | ACCAACAGATCCAAGCT 5′ | |
| his4 18mer A | 5′ TCGAGTAGGAGGAAGAGC | pLEJ007 |
| his4 18mer B | CATCCTCCTTCTCGAGCT 5′ | |
| his4 19mer A | 5′ TCGAGAGGAAGAGAAGAGC | pLEJ008 |
| his4 19mer B | CTCCTTCTCTTCTCGAGCT 5′ | |
| his4 20mer A (random) | 5′ TCGAGTCTATGTACTTACAC | pDTK139 |
| his4 20mer B (random) | CAGATACATGAATGTGAGCT 5′ | |
| his4 20mer A | 5′ TCGAGAGGAAGAGAAGAGAC | pSLJ001 |
| his4 20mer B | CTCCTTCTCTTCTCTGAGCT 5′ |
All plasmids were derived from pDN9 and contain an insertion of the indicated DNA sequence within the SalI site in the HIS4 coding sequence. The underlined type indicates stop codons in sequence inserts that maintain the correct reading frame. The insertion duplicates the SalI restriction site. Most alleles contain A and G on one strand; two alleles are a random mix of all four nucleotides, as indicated.
Figure 2.—
Expected configuration of loops forming in heteroduplex DNA. The top DNA strand contains the insertion allele of the indicated size, while the bottom strand is wild-type DNA lacking an insertion. Heteroduplex formation in this region will lead to the extrusion of the extra DNA sequence as a loop. In-frame stop codons are indicated in gray type within the insertion sequences of the 15- and 18-base loops.
To integrate a plasmid-borne his4 insertion allele into the chromosomal HIS4 locus, two-step integration into AS13 was performed following SnaBI plasmid digestion. Ura− derivatives of the initial transformants were isolated after growth on 5-fluoroorotic acid medium (Boeke et al. 1984). Ura− isolates were then screened for a His− phenotype, indicating retention of the his4 insertion allele. The HIS4 region was then sequenced (primer 4102403 and/or 4102404, as above) to confirm the orientation and sequence.
For PMS1 deletions, primers 1305733 and 1305734 were used to amplify the geneticin-resistance gene on the pFA6-KanMX4 plasmid (Wach et al. 1994). The parental strain was transformed with the resulting PCR product and the cells were plated on YPD for 24 hr. Transformants then were replica plated to YPD plates with 100 mg/liter of G418 to select for G418-resistant colonies. Disruption of the PMS1 gene was confirmed by PCR. To delete the RAD1 gene, the appropriate strain was transformed with BamHI-digested pDG18 as previously described (Kirkpatrick and Petes 1997). For pms1 diploid derivatives, the number of generations of growth (∼ 30) between mating and deposition on sporulation medium was minimized to reduce the accumulation of heterozygous lethal mutations; a zero-growth protocol was not used due to the unacceptably low level of sporulation under those conditions in this strain background.
Haploid strains are listed in Table 2. Diploids were generated by mating the AS13-derived strains to AS4 or rad1 or pms1 AS4 as appropriate: MW103 (MW1 × AS4; Nag et al. 1989), DTK257 (TP1011 × DTK256; Kirkpatrick and Petes 1997), DTK510 (DTK509 × AS4), DTK613 (DTK609 × AS4), DTK661 (DTK660 × AS4), DTK664 (DTK623 × TP1011), DTK670 (DTK662 × AS4), DTK680 (DTK677 × TP1011), DTK681 (DTK678 × DNY95), DTK694 (DTK684 × AS4), DTK696 (DTK695 × AS4), DTK698 (DTK697 × AS4), DTK705 (DTK679 × DNY95), DTK711 (DTK691 × TP1011), DTK718 (DTK713 × DNY95), DTK719 (DTK714 × DNY95), DTK720 (DTK715 × DNY95), DTK721 (DTK716 × TP1011), DTK722 (DTK717 × TP1011), DTK737 (DTK727 × TP1011), DTK740 (DTK728 × DNY95), DTK743 (DTK739 × AS4), DTK746 (DTK744 × DNY95), DTK747 (DTK745 × TP1011), DTK748 (DTK731 × DNY95), DTK760 (DTK754 × AS4), DTK768 (DTK766 × DNY95), DTK771 (DTK770 × TP1011), DTK860 (DTK859 × AS4), DTK882 (DTK881 × DNY95), DTK883 (DTK524 × DNY95).
TABLE 2 .
Haploid yeast strains
| Strain | Relevant genotype | Construction details and/or reference |
|---|---|---|
| AS4-derived haploids | ||
| AS4 | Wild type | MATα trp1 arg4 tyr7 ade6 ura3 (Stapleton and Petes 1991) |
| TP1011 | rad1::ura3 | Kirkpatrick and Petes (1997) |
| DNY95 | Δpms1 | Kearney et al. (2001) |
| AS13-derived haploids | ||
| AS13 | Wild type | MATa leu2 ura3 ade6 (Stapleton and Petes 1991) |
| MW1 | his4-Sal | Nag et al. (1989) |
| DTK256 | rad1::ura3 | TST in MW1 with BamHI-digested pDG18 (Kirkpatrick and Petes 1997) |
| DTK509 | his4-F20R | TST in AS13 with SnaBI-digested pDTK139 |
| DTK609 | his4-F16 | TST in AS13 with SnaBI-digested pLEJ001 |
| DTK660 | his4-F15 | TST in AS13 with SnaBI-digested pLEJ003 |
| DTK662 | his4-F20 | TST in AS13 with SnaBI-digested pSLJ001 |
| DTK684 | his4-F17 | TST in AS13 with SnaBI-digested pSLJ003 |
| DTK695 | his4-F10 | TST in AS13 with SnaBI-digested pSLJ009 |
| DTK697 | his4-F14 | TST in AS13 with SnaBI-digested pSLJ010 |
| DTK739 | his4-F19 | TST in AS13 with SnaBI-digested pLEJ008 |
| DTK754 | his4-F18 | TST in AS13 with SnaBI-digested pLEJ007 |
| DTK859 | his4-F17R | TST in AS13 with SnaBI-digested pPAJ173 |
| DTK623 | his4-F16 rad1::ura3 | TST in DTK609 with BamHI-digested pDG18 |
| DTK677 | his4-F15 rad1::ura3 | TST in DTK660 with BamHI-digested pDG18 |
| DTK691 | his4-F20 rad1::ura3 | TST in DTK662 with BamHI-digested pDG18 |
| DTK716 | his4-F10 rad1::ura3 | TST in DTK695 with BamHI-digested pDG18 |
| DTK717 | his4-F14 rad1::ura3 | TST in DTK697 with BamHI-digested pDG18 |
| DTK727 | his4-F17 rad1::ura3 | TST in DTK684 with BamHI-digested pDG18 |
| DTK745 | his4-F19 rad1::ura3 | TST in DTK739 with BamHI-digested pDG18 |
| DTK770 | his4-F18 rad1::ura3 | TST in DTK754 with BamHI-digested pDG18 |
| DTK524 | his4-F20R Δpms1 | TST in DTK509 with BstXI-digested pJH523 (Kearney et al. 2001) |
| DTK678 | his4-F15 pms1::kanMX4 | OST in DTK660 with kanMX4; using primers 1305733 and 1305734 |
| DTK679 | his4-F20 pms1::kanMX4 | OST in DTK662 with kanMX4; using primers 1305733 and 1305734 |
| DTK713 | his4-F10 pms1::kanMX4 | OST in DTK695 with kanMX4; using primers 1305733 and 1305734 |
| DTK714 | his4-F14 pms1::kanMX4 | OST in DTK697 with kanMX4; using primers 1305733 and 1305734 |
| DTK715 | his4-F17 pms1::kanMX4 | OST in DTK684 with kanMX4; using primers 1305733 and 1305734 |
| DTK728 | his4-F16 pms1::kanMX4 | OST in DTK609 with kanMX4; using primers 1305733 and 1305734 |
| DTK731 | his4-F4 pms1::kanMX4 | OST in MW1 with kanMX4; using primers 1305733 and 1305734 |
| DTK744 | his4-F19 pms1::kanMX4 | OST in DTK739 with kanMX4; using primers 1305733 and 1305734 |
| DTK766 | his4-F18 pms1::kanMX4 | OST in DTK754 with kanMX4; using primers 1305733 and 1305734 |
| DTK881 | his4-F17R pms1::kanMX4 | OST in DTK859 with kanMX4; using primers 1305733 and 1305734 |
TST, two-step transplacement; OST, one-step transplacement.
PCR primers:
Primer 4102403 is 5′ CGTACAGACCGTCCTGACGG, and primer 4102404 is 5′ TGGCCATTGCCAGAAGTTTC. Primer 1305733 is 5′ GAACGCGAAAAGAAAAGACGCGTCTCTCTTAATAATCATTATGCGATAAACGTACGCTGCAGGTCGAC, and primer 1305734 is 5′ CTCCCTGTATATAATGTATTTGTTAATTATATAATGAATGAATATCAAAGATCGATGAATTCGAGCTCG.
Data analysis:
Comparisons were performed with Instat 1.12 (GraphPad) for Macintosh, using either chi-square or Fisher's exact variant test. Results are considered statistically significant if P ≤ 0.05. The level of repair was determined by comparison of the number of GC and PMS tetrads in two strains: wild-type and either Δrad1 or Δpms1 derivatives. Significant alterations in the level of aberrant segregation of the his4 insertion allele were determined by comparing the number of tetrads with Mendelian segregation to the number of tetrads with aberrant segregation. The genetic interval between HIS4 and LEU2 was determined by measuring the number of parental ditype (PD), tetratype (T), and nonparental ditype (NPD) tetrads and using the following formula to determine the genetic map distance: cM = 100 × ({0.5 × T + 3 × NPD}/total tetrads). To control for strain-specific variation the results given are the summed total of two independent diploid strains; the only exception is strain DTK746.
RESULTS
Experimental rationale:
To determine the transition point between the MMR and LLR pathways, we constructed strains in which loops of differing sizes were generated during meiotic recombination (Figure 2). To prevent intrastrand pairing in the extruded single-stranded DNA of the loop, the sequence of the insertions was chosen so that when loops formed in heteroduplex DNA, the sequence within the loop would consist of adenine and guanine or cytosine and thymine (Figure 2). We determined the level of recombination and the frequency of loop mismatch repair in wild-type strains and in strains lacking the MMR pathway gene PMS1 or the LLR pathway gene RAD1. As described in Introduction, RAD1 functions specifically in LLR during meiosis, while PMS1 has been demonstrated to function specifically in MMR during meiosis (Kirkpatrick and Petes 1997; Kearney et al. 2001). Comparison of the repair frequencies of each loop allele allowed us to determine when mutations in PMS1 or RAD1 significantly affected repair of a given size loop mismatch (Table 3).
TABLE 3 .
Effects of varying loop size and DNA repair activity on the meiotic segregation patterns of heterozygous markers at theHIS4 locus
| No. of tetrads with various meiotic segregation patternsa |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | HIS4 allele | Loop size |
Repair mutation |
4:4 | 6:2 | 2:6 | 5:3 | 3:5 | Ab 4:4 |
7:1 | 1:7 | 8:0 | 0:8 | Other PMS |
Total tetrads |
% Ab seg |
% unrepaired |
| MW103b | his4-Sal | 4 | WT | 202 | 31 | 50 | 0 | 0 | 0 | 0 | 0 | 2 | 6 | 0 | 291 | 31 | 0 |
| DTK257b | his4-Sal | 4 | rad1 | 249 | 66 | 70 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 0 | 392 | 36 | 0 |
| DTK748 | his4-Sal | 4 | pms1 | 142 | 19 | 15 | 23 | 16 | 2 | 2 | 0 | 1 | 0 | 1 | 221 | 36 | 54* |
| DTK696 | his4-F10 | 10 | WT | 294 | 29 | 49 | 2 | 2 | 0 | 0 | 0 | 0 | 7 | 0 | 383 | 23 | 5 |
| DTK721 | his4-F10 | 10 | rad1 | 185 | 35 | 82 | 1 | 6 | 0 | 0 | 3 | 0 | 2 | 0 | 314 | 41 | 7 |
| DTK718 | his4-F10 | 10 | pms1 | 133 | 29 | 8 | 12 | 19 | 2 | 0 | 0 | 2 | 0 | 2 | 207 | 36 | 47* |
| DTK698 | his4-F14 | 14 | WT | 178 | 22 | 30 | 2 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 237 | 25 | 8 |
| DTK722 | his4-F14 | 14 | rad1 | 167 | 23 | 35 | 3 | 10 | 0 | 0 | 2 | 2 | 1 | 1 | 244 | 32 | 19 |
| DTK719 | his4-F14 | 14 | pms1 | 137 | 28 | 10 | 17 | 25 | 1 | 0 | 2 | 1 | 0 | 0 | 221 | 38 | 52* |
| DTK661 | his4-F15 | 15 | WT | 412 | 99 | 71 | 7 | 6 | 0 | 0 | 0 | 7 | 4 | 0 | 606 | 32 | 7 |
| DTK680 | his4-F15 | 15 | rad1 | 182 | 43 | 33 | 5 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 268 | 32 | 10 |
| DTK681 | his4-F15 | 15 | pms1 | 176 | 25 | 15 | 11 | 10 | 0 | 1 | 1 | 2 | 0 | 0 | 241 | 27 | 34* |
| DTK613 | his4-F16 | 16 | WT | 168 | 22 | 26 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 218 | 23 | 4 |
| DTK664 | his4-F16 | 16 | rad1 | 264 | 56 | 53 | 10 | 12 | 1 | 3 | 3 | 4 | 1 | 0 | 407 | 35 | 18* |
| DTK740 | his4-F16 | 16 | pms1 | 152 | 16 | 7 | 14 | 20 | 1 | 0 | 0 | 0 | 0 | 2 | 212 | 28 | 62* |
| DTK694 | his4-F17 | 17 | WT | 264 | 43 | 30 | 8 | 7 | 1 | 0 | 2 | 0 | 1 | 0 | 356 | 26 | 18 |
| DTK737 | his4-F17 | 17 | rad1 | 200 | 33 | 24 | 29 | 16 | 0 | 1 | 0 | 4 | 0 | 1 | 308 | 35 | 43* |
| DTK720 | his4-F17 | 17 | pms1 | 132 | 11 | 10 | 12 | 27 | 0 | 0 | 0 | 1 | 0 | 1 | 194 | 32 | 65* |
| DTK860 | his4-F17R | 17R | WT | 165 | 32 | 21 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 222 | 26 | 2 |
| DTK882 | his4-F17R | 17R | pms1 | 105 | 27 | 10 | 6 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 152 | 31 | 20* |
| DTK760 | his4-F18 | 18 | WT | 140 | 35 | 9 | 16 | 3 | 0 | 1 | 1 | 4 | 1 | 0 | 210 | 33 | 29 |
| DTK771 | his4-F18 | 18 | rad1 | 146 | 34 | 19 | 24 | 8 | 1 | 6 | 0 | 4 | 0 | 0 | 242 | 40 | 38 |
| DTK768 | his4-F18 | 18 | pms1 | 153 | 30 | 7 | 17 | 6 | 0 | 1 | 1 | 0 | 0 | 0 | 215 | 29 | 39 |
| DTK743 | his4-F19 | 19 | WT | 224 | 31 | 25 | 12 | 4 | 0 | 0 | 0 | 1 | 1 | 0 | 298 | 25 | 22 |
| DTK747 | his4-F19 | 19 | rad1 | 155 | 29 | 22 | 12 | 2 | 1 | 1 | 0 | 1 | 0 | 0 | 223 | 30 | 23 |
| DTK746 | his4-F19 | 19 | pms1 | 83 | 8 | 6 | 2 | 5 | 1 | 0 | 0 | 0 | 0 | 1 | 106 | 22 | 39 |
| DTK670 | his4-F20 | 20 | WT | 311 | 38 | 22 | 17 | 37 | 1 | 1 | 1 | 0 | 1 | 1 | 430 | 28 | 48 |
| DTK711 | his4-F20 | 20 | rad1 | 124 | 37 | 17 | 14 | 36 | 4 | 2 | 0 | 3 | 0 | 4 | 241 | 49 | 50 |
| DTK705 | his4-F20 | 20 | pms1 | 145 | 15 | 14 | 11 | 19 | 1 | 0 | 2 | 0 | 1 | 0 | 208 | 30 | 51 |
| DTK510 | his4-F20R | 20R | WT | 495 | 57 | 40 | 16 | 18 | 2 | 0 | 2 | 3 | 1 | 1 | 635 | 22 | 27 |
| DTK883 | his4-F20R | 20R | pms1 | 171 | 17 | 22 | 4 | 7 | 0 | 2 | 0 | 0 | 0 | 1 | 224 | 24 | 25 |
| DNY27b | his4-lopd | 26 | WT | 252 | 54 | 38 | 11 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 359 | 30 | 12 |
| TP1013b | his4-lopd | 26 | rad1 | 294 | 86 | 28 | 37 | 24 | 3 | 4 | 2 | 3 | 0 | 0 | 481 | 39 | 36* |
| DTK309c | his4-lopd | 26 | pms1 | 238 | 40 | 45 | 6 | 0 | 1 | 0 | 0 | 1 | 2 | 0 | 333 | 29 | 7 |
% Ab seg, the percentage of total tetrads with an aberrant segregation pattern (non-4:4); % unrepaired, the number of unrepaired events (PMS tetrads) divided by the total number of aberrant segregation tetrads (PMS + GC), and expressed as a percentage. WT, wild type.
Significant (P < 0.05 or better) difference from wild type in the number of PMS tetrads vs. the number of GC tetrads.
For all segregation patterns, the first number represents the wild-type allele and the second, the mutant allele. The segregation patterns include: 4:4 (normal Mendelian segregation), 6:2 and 2:6 (gene conversion), 5:3 and 3:5 (tetrads with a single PMS event), Ab 4:4 (aberrant 4:4; one wild-type, one mutant, and two sectored colonies), 7:1 and 1:7 (tetrads yielding three spore colonies of one genotype and one sectored colony), and 8:0 and 0:8 (tetrads yielding four spores of a single genotype). The “Other PMS” class includes aberrant 6:2 and 2:6 tetrads as well as tetrads with three PMS events.
Data from Kirkpatrick and Petes (1997).
Data from Kearney et al. (2001).
Aberrant segregation of loop alleles during meiosis:
The frequency of aberrant segregation in the wild-type strain varied from a low of 23% (DTK696 and DTK613) to a high of 33% (DTK760) in strains with differing loop sizes (Table 3). No correlation between the level of aberrant segregation and loop size was observed. The Δrad1 derivatives consistently showed an elevated level of aberrant segregation relative to the wild-type control strain. This elevation was statistically significant in DTK721 (his4-F10; P = 0.0001), DTK664 (his4-F16; P = 0.002), DTK737 (his4-F17; P = 0.011), DTK711 (his4-F20; P = 0.0001), and TP1013 (his4-lopd 26 base loop; P = 0.007). The majority of the significant elevations in aberrant segregation frequency in the Δrad1 strains occurred in strains expected to form loops of 16 bases or greater. In contrast, the Δpms1 derivatives exhibited elevated aberrant segregation frequencies in strains expected to form small loops, but not large loops. Recombination was significantly elevated in DTK718 (his4-F10; P = 0.0014) and DTK719 (his4-F14; P = 0.0034).
Meiotic repair of loop mismatches:
We determined the frequency of unrepaired tetrads as a function of the loop size in wild-type strains. In strains with alleles that form small loops, the frequency of PMS events was very low. A 4-base loop was always recognized and repaired (MW103), while a 10-base loop is not recognized or repaired in only 5% of the tetrads exhibiting aberrant segregation (DTK696 his4-F10). A similar percentage of unrepaired loop mismatches were observed for loops up to 16 bases in size. However, as the loop size was increased further, the percentage of unrepaired loops increased significantly (Table 3). In DTK694 (his4-F17), 18% of mismatches formed are not repaired. When we compare this to DTK613, the 16-base loop, the difference is statistically significant at P = 0.019. This decrease in the basal level of repair increases as the loop size increases: at a loop size of 20 bases, half of the aberrant segregation events were not repaired (Table 3, DTK670). Possible reasons for this decline in repair in the wild-type strain are discussed below.
We deleted RAD1 to examine the level of repair in strains lacking the RAD1-dependent LLR pathway. No alteration in repair frequency was detected in rad1 strains with alleles that form loops of <16 bases (Table 3). Both the 16- and 17-base loop strains exhibited a significant increase in PMS tetrads (18% unrepaired in DTK664, P = 0.018, and 43% unrepaired in DTK737, P = 0.0002) compared to the appropriate wild-type parental strain. These data clearly demonstrate that the RAD1-dependent repair pathway can act on loop substrates of 16 bases or larger. Unfortunately, LLR mutant strains with loop alleles >17 bases did not show a significant increase in unrepaired loops due to the high basal level of PMS events detected in those strains, as described above.
We deleted PMS1 to examine the level of repair in strains lacking the PMS1-dependent MMR pathway. All of the pms1 strains up to a loop size of 17 bases showed a significant increase in unrepaired events (Table 3). Strains with loop alleles >17 bases did not show an effect, due to the high basal level of PMS events. These data indicate that meiotic MMR acts on loop substrates up to ∼17 bases in size.
The DNA sequences of the loops, a random mix of adenine and guanine on one strand and thymine and cytosine on the other, were chosen specifically to prevent intrastrand pairing (Figure 2), as loops that form stem-loop structures are poorly repaired (Nag et al. 1989). However, we found that as the loop size increased, the frequency of unrepaired events increased significantly (Table 3). To determine if this effect was due to the loop size or the sequence composition of the loop, we constructed two new loop alleles of 17 and 20 bases (his4-F17R and his4-F20R), whose sequence composition was a random mix of all four nucleotides, arranged to inhibit intrastrand pairing as much as possible (Figure 2).
Tetrad dissection of DTK860 (his4-F17R) and DTK510 (his4-F20R) showed that the frequency of unrepaired events was significantly reduced in the random mix loop allele strains compared to the poly(AG) alleles (Table 3). For 17-base loops, the percentage of unrepaired events dropped from 18% with his4-F17 to 2% with his4-F17R (P = 0.0016), while for 20-base loops the percentage went from 48% with his4-F20 to 27% with his4-F20R (P = 0.0007).
As the random mix loop alleles showed significantly lower frequencies of unrepaired tetrads in the wild-type strains, we deleted the PMS1 gene in both strains to determine if MMR acted on either loop during meiosis. The percentage of unrepaired events went from 2 to 20% with the his4-F17R allele, a highly significant increase (P = 0.005), indicating that loops of 17 bases are acted upon by MMR. Conversely, the percentage of unrepaired events was unchanged with the his4-F20R allele (27 vs. 25%), indicating that MMR may not affect loops of 20 bases or larger during meiosis.
Examination of the ratio of 6:2 and 2:6 gene conversion events in the wild-type strains revealed an interesting trend. In MW103, which forms a 4-base loop, more 2:6 than 6:2 GC events were detected (Table 3). As the loop size increased, however, the bias was reversed, until in DNY27, which forms a 26-base loop, significantly more 6:2 than 2:6 events were seen (P = 0.0095, 6:2 vs. 2:6 in MW103 and DNY27). This trend is slightly accentuated in the repair deficient strains—the Δpms1 and Δrad1 strains show a bias at lower loop sizes than do the wild-type strains or a stronger directionality to the bias. However, the effect on bias is not evident in Δpms1 strains with large loops that are not affected by loss of PMS1 (his4-F20R or his4-lopd) or in Δrad1 strains with small loops that similarly are not affected by loss of RAD1 (his4-Sal, his4-F10, and his4-F14). This correlation indicates that the alteration in bias is likely to be dependent on the repair pathway acting on the loop. In agreement with this interpretation, previous genetic data (Kirkpatrick and Petes 1997; Kearney et al. 2001) indicated that Rad1p cleaved the DNA strand opposite the extruded loop, rather than acting to remove the loop. Loss of this activity leads to a decrease in the number of 2:6 tetrads, consistent with the data reported here for the Δrad1 strains.
HIS4-LEU2 crossovers in wild-type and mutant strains:
As a second measure of recombination, we monitored the level of intergenic recombination between HIS4 and LEU2. There was no statistically significant difference in crossover frequency between any of the wild-type or mutant strains. The genetic map distance between HIS4 and LEU2 averaged 32 cM in wild-type strains, 33 cM in rad1 strains, and 29 cM in pms1 strains. These data demonstrate that the observed alterations in the level of HIS4 aberrant segregation and repair frequency in strains in this study are a localized effect, rather than occurring genomewide.
Spore viability in DNA repair mutants:
Spore viability was monitored for each strain to ensure that changes in the number of tetrads in the various aberrant segregation classes were not due to elevated loss of a certain class of tetrad. No significant deviations were observed within each strain type (wild type, Δrad1, or Δpms1). The wild-type strains had an average overall spore viability of 88% (18,401 viable spores of 20,852 deposited), while the average was 81% (14,163 of 17,480 total) for the rad1 strains and 67% (14,141 of 21,148) for the pms1 strains.
The number of viable spores per tetrad was also determined. The distribution of the viability classes was similar in the wild-type and Δrad1 strains, although the Δrad1 strains had a higher percentage of inviable spores in each category (wild type—4:0–69%, 3:1–19%, 2:2–10%, 1:3–2%, 0:4–1%; Δrad1—4:0–51%, 3:1–28%, 2:2–15%, 1:3–5%, 0:4–1%). However, the distribution in strains lacking PMS1 was significantly different. The classes with two inviable spores (2:2) and no viable spores (0:4) were elevated relative to those classes in the wild-type and Δrad1 strains (Δpms1—4:0–38%, 3:1–16%, 2:2–29%, 1:3– 2%, 0:4–6%). There are two explanations for this pattern of spore viability: segregation of heterozygous recessive lethal mutations and an increase in nondisjunction during the first meiotic division. Loss of PMS1 leads to an increase in the basal rate of mutation (a mutator phenotype), and thus we favor the first explanation for the altered spore viability distribution.
DISCUSSION
A number of conclusions can be drawn from this study. First, even in wild-type strains the efficiency of repair declined as the loop size increased (Table 3). This decline is apparently related to the sequences composing the loops. Second, the minimum size loop that is repaired by RAD1-dependent LLR is 16 bases in length (Table 3). Third, loss of PMS1 affects the repair of loop sizes up to ∼17, but not >19 bases. Thus, there is an overlap in substrates repaired by the meiotic MMR pathway and the RAD1-dependent LLR pathway.
Repair declines as loop size increases:
Our initial loop alleles were constructed with adenine and guanine on one strand to minimize intrastrand pairing (Figure 2). We found that as the size of these poly(AG) loops was increased, the degree to which they were repaired declined, even in the wild-type strain. For loop mismatches up to 16 bases, there was a gradual decrease in the repair frequency. From 16 to 20 bases there was a more rapid decline. Nearly one-half of the mismatches formed in the strain with the his4-F20 allele were not repaired (Table 3). We constructed 17- and 20-base loop alleles (his4-F17R and his4-F20R) whose DNA sequences consisted of all four nucleotides randomly distributed to reduce the likelihood of intrastrand pairing. These alleles exhibited significantly fewer unrepaired tetrads compared to the poly(AG) loops of the same size (Table 3).
The decline in repair observed in the poly(AG) loops could be explained in several ways. It is unlikely that there is a simple correlation between loop size and degree of repair, given the difference in repair efficiencies of the randomized and poly(AG) 17- and 20-base loop alleles. In another study performed in this strain background, a 26-base loop (Kirkpatrick and Petes 1997) exhibited 12% unrepaired mismatches (Table 3). Also, very large insertions (up to 5.6 kb) are capable of undergoing gene conversion repair; increasing inefficiency of repair as a function of increasing loop size predicts that very large insertions would be very poorly repaired.
Alternatively, there could be unexpected secondary structure forming in the larger poly(AG) loop mismatches. Nag et al. (1989) showed that palindromic loop mismatch sequences are not as well repaired as nonpalindromic mismatches of identical length, suggesting that hairpin-loop formation affects the efficiency of repair. Similarly, another study found that loops containing triplet repeats capable of forming hairpin structures were less well repaired (Moore et al. 1999). It was suggested that the hairpin structures are not detected or are protected from repair due to the binding of a structure-specific protein(s) (Nag and Petes 1991; Nag and Kurst 1997). For our poly(AG) loop alleles, some form of unconventional intrastrand base pairing might allow formation of a hairpin. Nag and Petes found that the minimum length of an inverted repeat required to form a hairpin structure was 14 bases (Nag and Petes 1991): at this size insert there was a dramatic increase in the percentage of PMS tetrads. If there is unusual base pairing in the longer inserts used in this study, such structures may not form until the insert reaches ≥18 bp in length, as that is the smallest loop size to exhibit elevated PMS events.
Another explanation is that the primary sequence is affecting the repair of loops. If this is the case, the poly(AG) sequence is escaping repair while the random nucleotide mix sequences are not. Also, this model implies that the sequence of the shorter poly(AG) loops is insufficient to affect repair; however, the sequence in longer size loops does influence repair. Although almost all loop mismatches that show high levels of PMS are predicted to form secondary structure in the looped out sequence, there is one example in which the loop sequence was nonpalindromic and showed a high level of PMS (White et al. 1985, 1988). The authors suggested that a protein binding to the base of the loop mismatch prevented repair. The loop sequences and the sequences at the junction formed at the base of the loops used in our study were examined, and no canonical protein binding sites were detected (data not shown).
We favor the anomalous secondary structure model to explain the decrease in repair efficiency in large loops containing poly(AG) sequences. However, our genetic data cannot rule out some variations of the other models presented here.
Substrates of meiotic LLR and MMR:
Loss of RAD1 specifically affects the RAD1-dependent LLR pathway (Kirkpatrick and Petes 1997; Kearney et al. 2001). For those loop sizes that require the RAD1-dependent LLR pathway, we expected to see an increase in unrepaired mismatches when the RAD1 gene is deleted. We found that the lower limit of loop size recognized by the RAD1-dependent LLR pathway occurs at 16 bases, as the 16-base loop size is the first to show a statistically significant difference between the wild-type and rad1 strain (P = 0.017). We also saw a difference at the 17-base loop size (P = 0.0002).
PMS1 is a component of the MMR pathway, and so we expected to see an increase in unrepaired mismatches for loop sizes that require the MMR pathway when the PMS1 gene was deleted. The last point at which the loss of PMS1 results in a statistically significant effect is at the 17-base loop size (P < 0.0001 with the poly(AG) insertion, and P = 0.005 for the randomized insertion). No effect on the repair of the randomized 20-base loop is detected in a Δpms1 derivative, indicating that the upper limit for meiotic MMR is either 18 or 19 bases. Data from mitotic studies indicate that the upper limit of loop mismatches repaired by MMR during mitosis is 14–15 bases (Sia et al. 1997b). This limit is larger than that for mitotic MMR, demonstrating a difference between mitotic and meiotic mismatch repair.
Our data show that there is overlap in substrate specificity between MMR and LLR. Correction of both the 16- and 17-base loop sizes is affected by loss of LLR or MMR, and the overlap between the two pathways may also extend to loops of 18 or 19 bases. A similar overlap in repair pathways is seen in mitotic MMR, where both MSH6 and MSH3 function in the repair of very small (1 base) loop mismatches (Sia et al. 1997b). Overlap between repair pathways may further ensure that a mismatch is recognized and repaired, especially at the limits of the substrate range for the repair activities, where the frequency of repair may be decreased due to a decreased ability to detect the lesion.
Mismatch repair in meiosis vs. mitosis:
The results presented in this report, in combination with prior studies, demonstrate several differences between MMR and LLR in meiosis and mitosis. First, the mitotic MMR pathway functions in the repair of mismatches ≤14 bases in length (Sia et al. 1997b). During meiosis, however, MMR functions to repair mismatches <18 bases in length. Second, in meiosis, RAD1 is involved in the repair of mismatches ≥16 bases. A mitotic study showed that a rad1 mutation had no effect on the repair of a 16-base loop (Corrette-Bennett et al. 2001). Third, repair of large loops during meiosis is affected by loss of MSH2 and MSH3 (Kirkpatrick and Petes 1997; Kearney et al. 2001). However, several studies have shown that repair of large loops during mitosis is independent of MSH2 and MSH3 (Tran et al. 1996; Sia et al. 1997b; Corrette-Bennett et al. 1999, 2001).
There are also similarities in MMR and LLR during meiosis and mitosis. Loss of MMR affects the repair of mismatches ≤15 bases in both meiosis and mitosis. Second, LLR is not dependent on PMS1 (Tran et al. 1996; Harfe and Jinks-Robertson 1999; Corrette-Bennett et al. 1999, 2001). Also, loops that form secondary structure are not well repaired during mitosis (Corrette-Bennett et al. 2001) and meiosis (Nag et al. 1989; Nag and Petes 1991; Moore et al. 1999). One study found a mitotic LLR activity that required both MSH3 and RAD1 to repair loops of ∼100 bases formed during frameshift reversion and that neither PMS1 nor MSH2 was involved in loop repair (Harfe and Jinks-Robertson 1999). However, another study found that a PMS1 and MSH2 dependent pathway functions to repair very large loops (>2 kb) during HO endonuclease-initiated mitotic recombination (Clikeman et al. 2001). To date, this is the only study showing involvement of PMS1 in LLR; the differences in mitotic repair activities may reflect differences in the manner in which loop formation is initiated.
One caveat to our observed differences between meiotic and mitotic MMR or LLR is the influence of sequence context on the repair activities. In the various studies discussed above, the primary sequence of DNA surrounding the mismatch, the sequence of the mismatch, and the manner of their formation differ significantly; these factors may contribute to the observed differences. Given the difficulties inherent in forming mismatches on demand during mitotic cell cycles, it is unlikely that this issue will be quickly resolved.
Broader implications of meiotic DNA repair:
Repetitive tracts usually are divided into classes based on repeat unit size. Microsatellites contain repeats ranging from a single base pair to ∼14 bp in length (Sia et al. 1997a). Minisatellites have repeat units ranging from ∼15 to 100 bp (Bois and Jeffreys 1999; Jauert et al. 2002). Microsatellites primarily destabilize during mitotic growth (Sia et al. 2001), while minisatellites become unstable during meiosis. However, some tracts that have been labeled as minisatellites show a high level of mitotic instability, and mutation of replication factors such as RAD27 can affect the stability of some minisatellites (Kokoska et al. 1998; Lopes et al. 2002). Meiotic instability of minisatellites is most likely due to recombination events in which misalignment can result in loop mismatches (Jeffreys et al. 1998; Bois and Jeffreys 1999; Bishop and Schiestl 2000). These loops may be substrates for repair by LLR proteins. In support of this idea, meiotic expansions in the overall length of a human HRAS1 minisatellite tract inserted into the yeast genome were significantly reduced in a strain lacking RAD1 (Jauert et al. 2002). The data presented here on the substrate range of meiotic LLR may be useful in distinguishing “microsatellite”-type tracts from “minisatellite”-type tracts.
Several human disease phenotypes have been associated with alterations in minisatellites, possibly due to alterations in the expression of nearby genes (reviewed in Bois and Jeffreys 1999). For example, rare alleles of the minisatellite adjacent to the HRAS1 gene have been associated with several types of cancer (Krontiris et al. 1993). In addition to cancer, other diseases such as insulin-dependent diabetes mellitus (Bennett et al. 1995; Kennedy et al. 1995) and progressive myoclonus epilepsy (Lafreniere et al. 1997; Virtaneva et al. 1997) have been correlated with allelic variation in minisatellites. Increased understanding of how mismatches are recognized during meiosis and which meiotic repair activities are involved will allow us to better understand the initiation and progression of diseases of this type.
Acknowledgments
We thank Selina Jaman for technical assistance during the course of this work. This research was supported by a Grant-in-Aid of Research, Artistry and Scholarship from the Graduate School, University of Minnesota.
References
- Adams, A., D. E. Gottschling, C. A. Kaiser and T. Stearns, 1998 Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Bardwell, A. J., L. Bardwell, D. K. Johnson and E. C. Friedberg, 1993. Yeast DNA recombination and repair proteins Rad1 and Rad10 constitute a complex in vivo mediated by localized hydrophobic domains. Mol. Microbiol. 8 1177–1188. [DOI] [PubMed] [Google Scholar]
- Bennett, S. T., A. M. Lucassen, S. C. Gough, E. E. Powell, D. E. Undlien et al., 1995. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat. Genet. 9 284–292. [DOI] [PubMed] [Google Scholar]
- Bertrand, P., D. X. Tishkoff, N. Filosi, R. Dasgupta and R. D. Kolodner, 1998. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc. Natl. Acad. Sci. USA 95 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A. J., and R. H. Schiestl, 2000. Homologous recombination as a mechanism for genome rearrangements: environmental and genetic effects. Hum. Mol. Genet. 9 2427–2434. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., F. LaCroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197 345–346. [DOI] [PubMed] [Google Scholar]
- Bois, P., and A. J. Jeffreys, 1999. Minisatellite instability and germline mutation. Cell. Mol. Life Sci. 55 1636–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts, R. H., S. R. Chambers and M. F. Abdullah, 2000. The many faces of mismatch repair in meiosis. Mutat. Res. 451 129–150. [DOI] [PubMed] [Google Scholar]
- Clikeman, J. A., S. L. Wheeler and J. A. Nickoloff, 2001. Efficient incorporation of large (>2 kb) heterologies into heteroduplex DNA: Pms1/Msh2-dependent and -independent large loop mismatch repair in Saccharomyces cerevisiae. Genetics 157 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrette-Bennett, S. E., B. O. Parker, N. L. Mohlman and R. S. Lahue, 1999. Correction of large mispaired DNA loops by extracts of Saccharomyces cerevisiae. J. Biol. Chem. 274 17605–17611. [DOI] [PubMed] [Google Scholar]
- Corrette-Bennett, S. E., N. L. Mohlman, Z. Rosado, J. J. Miret, P. M. Hess et al., 2001. Efficient repair of large DNA loops in Saccharomyces cerevisiae. Nucleic Acids Res. 29 4134–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Q., F. Xu and T. D. Petes, 1995. Meiosis-specific double-strand DNA breaks at the HIS4 recombination hot spot in the yeast Saccharomyces cerevisiae: control in cis and trans. Mol. Cell. Biol. 15 1679–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 1999. Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 4766–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. a DNA mismatch repair and genetic instability. Ann. Rev. Genet. 34 359–399. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. b Mismatch repair proteins and mitotic genome stability. Mutat. Res. 451 151–167. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., B. K. Minesinger and S. Jinks-Robertson, 2000. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 10 145–148. [DOI] [PubMed] [Google Scholar]
- Hsieh, P., 2001. Molecular mechanisms of DNA mismatch repair. Mutat. Res. 486 71–87. [DOI] [PubMed] [Google Scholar]
- Jauert, P. A., S. N. Edmiston, K. Conway and D. T. Kirkpatrick, 2002. RAD1 Controls the Meiotic Expansion of the Human HRAS1 Minisatellite in Saccharomyces cerevisiae. Mol. Cell. Biol. 22 953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A. J., D. L. Neil and R. Neumann, 1998. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J. 17 4147–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney, H. M., D. T. Kirkpatrick, J. L. Gerton and T. D. Petes, 2001. Meiotic recombination involving heterozygous large insertions in Saccharomyces cerevisiae: formation and repair of large, unpaired DNA loops. Genetics 158 1457–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375–384. [DOI] [PubMed] [Google Scholar]
- Kennedy, G. C., M. S. German and W. J. Rutter, 1995. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat. Genet. 9 293–298. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, D. T., 1999. Roles of the DNA mismatch repair and nucleotide excision repair proteins during meiosis. Cell. Mol. Life Sci. 55 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick, D. T., and T. D. Petes, 1997. Repair of DNA loops involves DNA mismatch and nucleotide excision repair proteins. Nature 387 929–931. [DOI] [PubMed] [Google Scholar]
- Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin et al., 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting Okazaki fragment processing (rad27) and DNA polymerase ∂ (pol3-t). Mol. Cell. Biol. 18 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krontiris, T. G., B. Devlin, D. D. Karp, N. J. Robert and N. Risch, 1993. An association between the risk of cancer and mutations in the HRAS1 minisatellite locus. N. Engl. J. Med. 329 517–523. [DOI] [PubMed] [Google Scholar]
- Lafreniere, R. G., D. L. Rochefort, N. Chretien, J. M. Rommens, J. I. Cochius et al., 1997. Unstable insertion in the 5′ flanking region of the cystatin B gene is the most common mutation in progressive myoclonus epilepsy type 1, EPM1. Nat. Genet. 15 298–302. [DOI] [PubMed] [Google Scholar]
- Lopes, J., H. Debrauwere, J. Buard and A. Nicolas, 2002. Instability of the human minisatellite CEB1 in rad27Delta and dna2–1 replication-deficient yeast cells. EMBO J. 21 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, T. M., C. Kunz and O. Fleck, 2002. DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol. 191 28–41. [DOI] [PubMed] [Google Scholar]
- Moore, H., P. W. Greenwell, C.-P. Liu, N. Arnheim and T. D. Petes, 1999. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl. Acad. Sci. USA 96 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, D. K., and A. Kurst, 1997. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics 146 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, D. K., and T. D. Petes, 1991. Seven-base-pair inverted repeats in DNA form stable hairpins in vivo in Saccharomyces cerevisiae. Genetics 129 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, D. K., M. A. White and T. D. Petes, 1989. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature 340 318–320. [DOI] [PubMed] [Google Scholar]
- Prakash, S., and L. Prakash, 2000. Nucleotide excision repair in yeast. Mutat. Res. 451 13–24. [DOI] [PubMed] [Google Scholar]
- Sancar, A., 1996. DNA excision repair. Annu. Rev. Biochem. 65 43–81. [DOI] [PubMed] [Google Scholar]
- Sia, E. A., S. Jinks-Robertson and T. D. Petes, 1997. a Genetic control of microsatellite stability. Mutat. Res. 383 61–70. [DOI] [PubMed] [Google Scholar]
- Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell and T. D. Petes, 1997. b Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17 2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia, E. A., M. Dominska, L. Stefanovic and T. D. Petes, 2001. Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol. 21 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton, A., and T. D. Petes, 1991. The Tn3 beta-lactamase gene acts as a hotspot for meiotic recombination in yeast. Genetics 127 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl, K., D. T. Stinchcomb, S. Scherer and R. W. Davis, 1979. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc. Natl. Acad. Sci. USA 76 1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, H. T., D. A. Gordenin and M. A. Resnick, 1996. The prevention of repeat-associated deletions in Saccharomyces cerevisiae by mismatch repair depends on size and origin of deletions. Genetics 143 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva, K., E. D'Amato, J. Miao, M. Koskiniemi, R. Norio et al., 1997. Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat. Genet. 15 393–396. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- White, J. H., K. Lusnak and S. Fogel, 1985. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature 315 350–352. [DOI] [PubMed] [Google Scholar]
- White, J. H., J. F. DiMartino, R. W. Anderson, K. Lusnak, D. Hilbert et al., 1988. A DNA sequence conferring high postmeiotic segregation frequency to heterozygous deletions in Saccharomyces cerevisiae is related to sequences associated with eucaryotic recombination hotspots. Mol. Cell. Biol. 8 1253–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M. A., M. Dominska and T. D. Petes, 1993. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]