Abstract
The insulator element from the gypsy transposon is a DNA sequence that blocks activation of a promoter by a transcriptional enhancer when placed between them. The insulator contains reiterated binding sites for the Suppressor of Hairy-wing [Su(Hw)] zinc-finger protein. A protein encoded by another gene, modifier of mdg4 [mod(mdg4)], is also required for the enhancer-blocking activity of the Su(Hw) insulator. Here we present evidence that the Su(Hw) insulator activates a weakened yellow promoter at a distance. Deletion of the upstream promoter region (UPR), located close by the TATA box, significantly reduces yellow expression. The Su(Hw) insulator placed at different positions relative to the yellow promoter partially compensates for loss of the UPR. Su(Hw) is able to stimulate yellow expression even if it is located at a 5-kb distance from the promoter. The stimulatory activity depends on the number of Su(Hw)-binding sites. Mutational analysis demonstrates that only the DNA-binding domain and adjacent regions of the Su(Hw) protein are required for stimulation of yellow transcription.
ENHANCER-mediated activation is a fundamental mechanism of gene activation in eukaryotes (Dorsett 1999; West et al. 2002). Enhancers can act over large distances to activate transcription, regardless of their orientation and position relative to the promoter, without affecting adjacent genes. Recently, sequences referred to as insulators have been found in different organisms to prevent activation or repression from extending across them to a promoter (Dorsett 1999; Sun and Elgin 1999; Udvardy 1999; Gerasimova and Corces 2001; Oki and Kamakaka 2002; West et al. 2002; Kuhn and Geyer 2003). The best-studied vertebrate insulator is the chicken β-globin insulator (Bell et al. 1999). Well-characterized insulators in Drosophila include the scs and scs′ sequences found at the boundary of the 87A heat-shock locus (Kellum and Schedl 1991; Zhao et al. 1995), Fab-7 and Fab-8 insulators from the Abd-B region (Hagstrom et al. 1996; Zhou et al. 1996, 1999; Barges et al. 2000) and the Suppressor of Hairy-wing [Su(Hw)] insulator identified in the gypsy retrotransposon (Spana et al. 1988; Mazo et al. 1989).
The properties of an insulator element may be exemplified by the Su(Hw) insulator, which can block diverse enhancers if inserted between an enhancer and a promoter (Holdridge and Dorsett 1991; Geyer and Corces 1992; Geyer and Clark 2002), but does not affect the intrinsic activity of the enhancer (Cai and Levine 1995; Scott and Geyer 1995). The Su(Hw) insulator can also function as a barrier blocking the silencing activity of the Polycomb group response element (Sigrist and Pirrotta 1997; Mallin et al. 1998) and partially protecting a transgene from silencing when inserted into heterochromatin (Roseman et al. 1993; 1995; van der Vlag et al. 2000).
Genetic and molecular approaches have led to identification and characterization of two proteins required for activity of the Su(Hw) insulator. One is Su(Hw), a 12-zinc-finger protein encoded by the su(Hw) gene, which binds to the repeated sequence motifs in the gypsy insulator (Dorsett 1990; Spana and Corces 1990). The enhancer-blocking activity of Su(Hw) requires 9 of its 12 zinc fingers and a domain of ∼150 amino acids including the C-terminal leucine zipper (Harrison et al. 1993; Kim et al. 1996).
Mutations in another gene, modifier of mdg4 [mod(mdg4)], alter the phenotypes of gypsy-induced mutations, indicating that the product of this gene is also involved in the function of the Su(Hw) insulator (Georgiev and Gerasimova 1989; Gerasimova et al. 1995; Georgiev and Kozycina 1996; Cai and Levine 1997; Gdula and Corces 1997). The mod(mdg4) gene, also known as E(var)3-93D, encodes a large set of individual protein isoforms with specific functions in regulating the chromatin structure of different genes (Gerasimova et al. 1995; Buchner et al. 2000). The available genetic data suggest that Mod(mdg4) is required for the enhancer-blocking activity (Gerasimova et al. 1995; Georgiev and Kozycina 1996; Gdula and Corces 1997). Biochemical studies using purified Su(Hw) and Mod(mdg4) proteins indicate that one protein isoform, Mod(mdg4)-67.2, interacts with the enhancer-blocking domain of the Su(Hw) protein (Gause et al. 2001; Ghosh et al. 2001).
Recently it has been found that the Su(Hw) insulator can stimulate transcription from the alcohol dehydrogenase gene (Adh) promoter in a distance-dependent manner (Wei and Brennan 2001). Since the Su(Hw) insulator failed to stimulate the Adh promoter with the GATA-binding site deleted or the white promoter lacking this site in the larval fat body, it was suggested that the Su(Hw) insulator facilitates the access of the GATA transcription factor to the Adh promoter. Here we examined the role of the Su(Hw) insulator in stimulating transcription of the yellow gene. The yellow gene is required for larval and adult cuticle pigmentation (Nash and Yarkin 1974). The temporal and spatial pattern of its expression is controlled by at least five independent, tissue-specific transcriptional enhancers (Geyer and Corces 1987; Martin et al. 1989). The enhancers that control yellow expression in the wings and body cuticle are located in the 5′ upstream region of the yellow gene, whereas the enhancers controlling its expression in the tarsal claw and bristles reside in the intron of the gene.
Previously we found that a particular yellow sequence upstream of the TATA promoter is critical for the yellow transcription during pupal development (Belenkaya et al. 1998). Deletion of the upstream promoter region (UPR) leads to pronounced reduction of yellow expression. Here we show that the Su(Hw) insulator in many transgenic lines partially or completely restores yellow expression in the absence of the UPR. Like a distance-independent enhancer, the Su(Hw) insulator can stimulate the yellow promoter over at least 5 kb. At the same time, the Su(Hw) insulator fails to compensate for deletion of the bristle enhancer; that is, it does not work as a transcriptional enhancer.
MATERIALS AND METHODS
Drosophila strains:
All flies were maintained at 25° on a standard yeast medium. The lines bearing mutations in the su(Hw) gene were obtained from V. Corces. The structure and origin of the su(Hw) mutations were described by Harrison et al. (1993). Drosophila lines carrying combinations of mod(mdg4)u1 with su(Hw)j and su(Hw)v were previously obtained (Georgiev and Kozycina 1996). All other mutant alleles and chromosomes used in this work and all balancer chromosomes are described in Lindsley and Zimm (1992).
DNA constructs:
The 8-kb fragment containing the yellow gene and the cDNA yellow clone were kindly provided by P. Geyer. The 3-kb SalI-BamHI fragment containing the yellow regulatory region (yr) was subcloned into pGEM7 cleaved with BamHI + XhoI (yr plasmid).
The 430-bp gypsy sequence containing the Su(Hw)-binding region was PCR amplified from the gypsy retrotransposon. After sequencing to confirm its identity, the product was inserted in the CaSpeR2 vector (C2-su). The 5-kb BamHI-BglII fragment containing the yellow coding region (yc) was subcloned into CaSpeR3 (C3-yc) or CaSpeR2-su (C3-su-yc).
The deletion of the regulatory region (yr) between positions −438 and −70 relative to the transcription start site (Geyer et al. 1986) was generated by PCR amplification of the yr plasmid between primers y6, 5′-CATTGGCCTGTCTTCGTCTTCGG-3′, and y7, 5′-CAGGAGGCTCGTGCATAGAATGC-3′. The PCR products were blunted, self-ligated, and used for transformation. One of the successfully mutagenized clones was sequenced to confirm that no unwanted changes had been introduced into the yellow sequence (Δyr).
The four (Sx4) and eight (Sx8) reiterated Su(Hw)-binding sites and five binding sites from the Su(Hw) insulator (Sx5g) were obtained from E. Savitskaya.
(S)dY(S)W:
The Su(Hw) insulator flanked by the frt [frt(su)] sites was inserted in the Δyr plasmid treated with Eco47III. The Δyr-frt(su) fragment was ligated into C2-lox(su)-yc treated with XbaI and BamHI.
(S×8)dY(S)W:
The eight Su(Hw)-binding sites flanked by Flippase recombinase target (FRT) sites [frt(Sx8)] were inserted in the Δyr plasmid treated with Eco47III. The Δyr-frt(Sx8) fragment was ligated into C2-lox(su)-yc treated with XbaI and BamHI.
(Sx4)dYW:
The four reiterated Su(Hw)-binding sites flanked by locus of X-over P (LOX) sites [lox(Sx4)] were inserted in the Δyr plasmid treated with Eco47III. The Δyr-lox(Sx4) fragment was ligated into C3-yc treated with XbaI and BamHI.
(Sg5)(Sx4)dYW:
The five Su(Hw)-binding sites from the gypsy insulator flanked by FRTs [frt(Sg5)] and four reiterated Su(Hw)-binding sites flanked by LOXs [lox(Sx4)] were ligated together. The [frt(Sg5) + lox(Sx4)] fragment was inserted in the Δyr plasmid treated with Eco47III. The Δyr-[frt(Sg5) + lox(Sx4)] fragment was ligated into C3-yc treated with XbaI and BamHI.
To obtain the constructs bearing the intronless yellow gene, the BamHI-BglII fragment containing the cDNA yellow region (Yil) was subcloned into CaSpeR3 (C3-Yil) or CaSpeR2-su (C2-su-Yil). The Su(Hw) insulator was inserted in the yr plasmid treated with Eco47III (yr-su). The yr and yr-su fragments were correspondingly subcloned into C2-su-Yil (YilSW) and C3-Yil (SYilW).
The CaSpeR2 plasmid with the 1.3-kb PstI-to-XhoI fragment containing the su(Hw) gene promoter [pCsu(Hw)Pr] and the plasmids containing cDNAs of the su(Hw) and mod(mdg4) genes (C4Su(Hw)DC and C2-mod(mdg4)-2.2) were obtained from D. Dorsett. The chimeric gene expressing Su(Hw)Mod(mdg4) was constructed by ligation of the EheI-BamHI fragment from C4Su(Hw)DC and the EcoRI-Eco72I DNA fragment from C2-mod(mdg4)-2.2. To obtain the final P transposon with a gene expressing Su(Hw)Mod(mdg4), the BamHI-EcoRI fragment containing parts of the su(Hw) and mod(mdg4) genes was ligated into pCsu(Hw)Pr treated with XhoI and BamHI.
Germline transformation and genetic crosses:
The construct, together with a P element with defective inverted repeats used as a transposase source, P25.7wc (Kares and Rubin 1984), was injected into y ac w1118 preblastoderm embryos (Rubin and Spradling 1982; Spradling and Rubin 1982). The resulting flies were crossed with y ac w1118 flies, and transgenic progeny were identified by their eye color. Chromosome localization of various transgene insertions was determined by crossing the transformants with the y ac w1118 balancer stock containing dominant markers: In(2RL),CyO for chromosome two and In(3LR)TM3,Sb for chromosome three. The transformed lines were examined by Southern blot hybridization (Sambrook et al. 1989) to check for transposon integrity and copy number.
The lines with excisions of the Su(Hw)-binding sites were obtained by crossing flies bearing the transposons with Flp or Cre recombinase-expressing lines w1118; CyO, FLP, ISA/Sco;+ and y1, wi; CyO, P[w+,cre]/Sco; +. All excisions were confirmed by PCR analysis.
To test the effects of Su(Hw) protein on yellow gene expression, lines containing the yellow transposons were crossed into a su(Hw)v/su(Hw)f mutant background. This combination of the su(Hw) alleles reverses the phenotypes associated with gypsy insertions and is female fertile. Su(Hw)v is a deletion of the su(Hw) gene (Harrison et al. 1993), whereas su(Hw)f is a point mutation in the tenth zinc finger finger that retains some ability to bind DNA (Harrison et al. 1993).
The mutations in the su(Hw) and mod(mdg4) genes were combined with P(y) constructs as previously described (Georgiev and Kozycina 1996). Details of the crosses used for genetic analysis and for excision of functional elements are available upon request.
Pigmentation scale:
To determine the yellow phenotype, the extent of pigmentation in bristles of adult flies was estimated visually in 3- to 5-day-old males developing at 25°. The degree of variegation in bristles of the thorax and head was scored using a five-point scale, where 1 denotes loss of pigmentation in all bristles at thorax and head; e-v, extreme variegation (only one to three bristles on the thorax and head are pigmented); m-v, moderate variegation (about half of the bristles are yellow); w-v, weak variegation (only one to three bristles on thorax and head are yellow); and 5, pigmentation of all bristles as in wild-type flies. At least 50 flies were scored independently by two people for each y line.
RESULTS
The Su(Hw) insulator can stimulate yellow transcription when the upstream promoter region is deleted:
As all previous studies (Geyer et al. 1986; Parkhurst and Corces 1986; Geyer and Corces 1992; Georgiev and Kozycina 1996) showed that the Su(Hw) insulator does not activate the wild-type yellow promoter, we used the deletion derivatives of the latter. Belenkaya et al. (1998) showed that the yellow sequence located between positions −146 and −70 relative to the transcription start site is required for the function of the yellow promoter. Deletion of this 77-bp sequence, named the upstream promoter sequence (UPR), strongly reduces yellow expression in the body cuticle, wing blades, and bristles. To further weaken the yellow promoter, we deleted the region from position −438 to −70 (dY). This 368-bp deletion included the UPR and one of the larval enhancers previously mapped to the region between −294 and −92 (Martin et al. 1989). The control construct, dYW (Figure 1), contained the white gene as a marker for selecting successful insertions in the genome of the y ac w1118 strain. All 14 independently obtained transformants had strongly decreased pigmentation of the body cuticle, wing blades, and bristles (Table 1). Flies of 10 independent lines homozygous or heterozygous for the transgene displayed a y1-like phenotype, which suggests almost complete inactivation of yellow. In two homozygous dYW lines, flies had yellow body cuticle and wing blades, and extremely variegated pigmentation of the head and thoracic bristles: only one to three bristles were pigmented. Flies displayed a weak pigmentation of the body cuticle and wing blades, and moderate variegation of bristle pigmentation in only two homozygous dYW lines; about half of the bristles were pigmented. In all dYW lines, flies had yellow-orange eye color, indicating a normal level (euchromatic insertion site) of mini-white expression in the absence of the eye enhancer. Thus, in the dYW lines yellow transcription is strongly repressed in most of the euchromatic insertions.
Figure 1.—
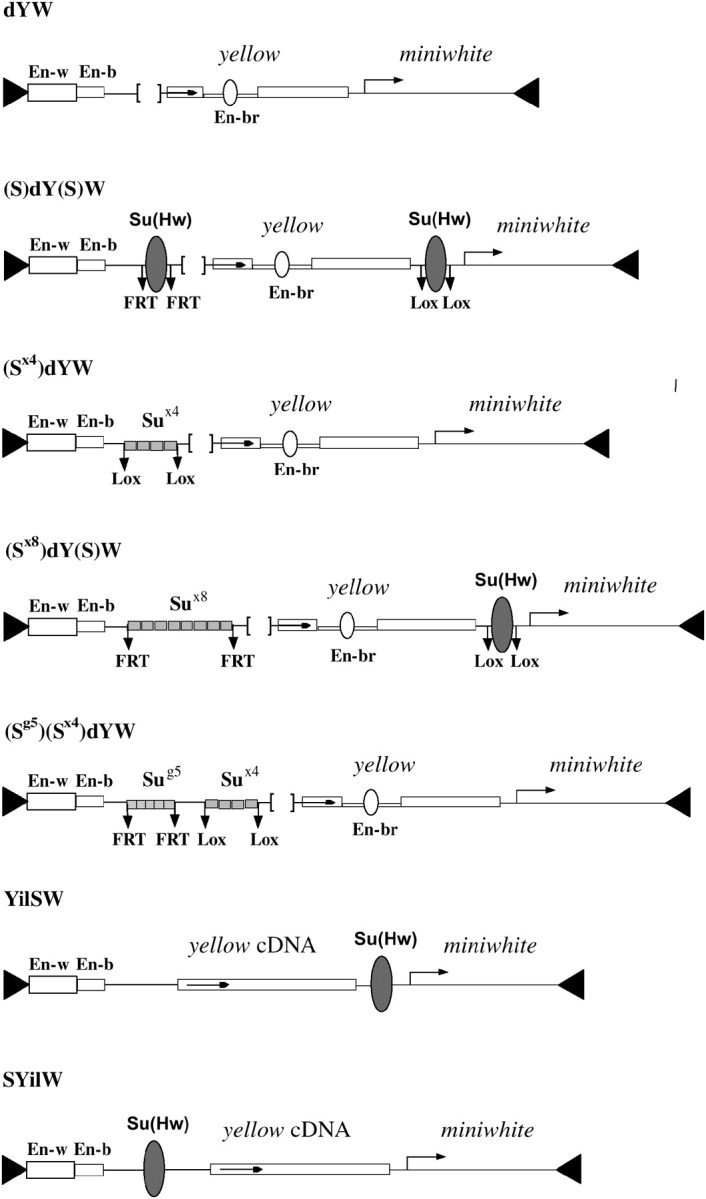
Schematic of transposon constructs. The maps of the constructs (not to scale) show the yellow wing (En-w) and body enhancers (En-b) as partially overlapping open boxes. The bristle enhancer (En-br) is indicated as an open oval in the intron of the yellow gene. The arrows indicate the direction of transcription of the yellow and white genes. Downward arrows labeled FRT or LOX mark the target sites of the Flp or Cre recombinase, respectively. The solid oval represents the Su(Hw) insulator isolated from the gypsy retrotransposon. The synthetic Su(Hw)-binding sites are indicated by the open rectangles.
TABLE 1 .
Summary of phenotypes associated with transgenic dYW, (S)dY(S)W, and (Sx4)dYW lines
| Levels of yellow expression in bristlesb |
|||||||
|---|---|---|---|---|---|---|---|
| Transgenes | Genotype |
N of linesa |
5 | w-v | m-v | e-v | 1 |
| dYW | P/+ | 14 | — | — | — | 2 | 12 |
| P/P | 14(4) | — | — | 2 | 2 | 10 | |
| (S)dY(S)W | P/+ | 23 | 5 | 3 | 3 | 3 | 9 |
| P/P | 14(12) | 2 | 2 | 8 | 2 | ||
| (S)dY(ΔS)W | P/+ | 11(6) | 3 | 2 | 2 | 2 | 2 |
| (ΔS)dY(S)W | P/+ | 11(7) | 2 | 3 | 1 | 1 | 4 |
| (ΔS)dY(ΔS)W | P/+ | 11(11) | — | — | — | 1 | 10 |
| (S×4)dYW | P/+ | 12 | — | 1 | 3 | 3 | 5 |
| P/P | 12(9) | 1 | 3 | 2 | 5 | 1 | |
| (ΔS×4)dYW | P/+ | 12(7) | — | — | — | 1 | 11 |
| P/P | 12(7) | 1 | 1 | 10 | |||
The phenotypes of transgenic lines (P) were examined in males heterozygous (P/+) or homozygous (P/P) for the construct.
Number of tested transgenic lines. Figures in parentheses show the number of lines in which flies acquired a new y phenotype in comparison with flies from the starting line.
Number of flies with similar levels of bristle pigmentation. The degree of variegation in bristles of the thorax and head: 1, loss of pigmentation in all bristles at thorax and head; e-v, extreme variegation (only one to three bristles on thorax and head are pigmented); m-v, moderate variegation (about half of bristles are yellow); w-v, weak variegation (only one to three bristles on thorax and head are yellow); 5, pigmentation of all bristles as in wild-type flies.
To study the assumed stimulatory activity of the Su(Hw) insulator, in (S)dY(S)W (Figure 1) one 340-bp Su(Hw) insulator (S) containing 12 putative Su(Hw)-binding sites (Figure 3A) was inserted at position −525 and another from the 3′ side of the yellow gene at +4964 relative to the yellow transcription start site. The Su(Hw) insulators were flanked by FRT or LOX sites to permit their excision from transgenic flies by crossing the latter with flies expressing either Flp (Golic and Lindquist 1989) or Cre recombinase (Siegal and Hartl 2000). In 23 transgenic lines carrying a single (S)dY(S)W insertion, flies had eyes ranging in color from yellow to dark orange. As the Su(Hw) insulator inserted at −525 blocks the wing and body enhancers, in this and the following experiments we examined yellow expression only in bristles. The bristle enhancer is located in the yellow intron (Geyer and Corces 1987) and thus it is not blocked by the Su(Hw) insulator inserted either upstream or downstream of the yellow gene. In contrast to control dYW transgenic lines, flies heterozygous for the (S)dY(S)W construct in 8 of 23 transgenic lines had wild-type or nearly wild-type levels of bristle pigmentation (Table 1), suggesting substantial activation of the yellow promoter. In 6 (S)dY(S)W lines, flies had moderate or strong variegation of bristle pigmentation and only 9 lines displayed y1-like phenotype. In 12 of 14 transgenic lines, flies homozygous for the construct had more pigmented bristles than did heterozygous ones. To test the contribution of the Su(Hw) protein to transcription stimulation, we crossed flies displaying wild-type or nearly wild-type bristle pigmentation from 7 (S)dY(S)W lines into a su(Hw)v/su(Hw)f mutant background (Table 2). In all tested lines, the level of bristle pigmentation was decreased to nearly the y1-like phenotype. These results suggest that the Su(Hw) insulators stimulate transcription from the weakened yellow promoter in most of the transgenic lines and that the level of activation strongly depends on the site of construct insertion.
Figure 3.—
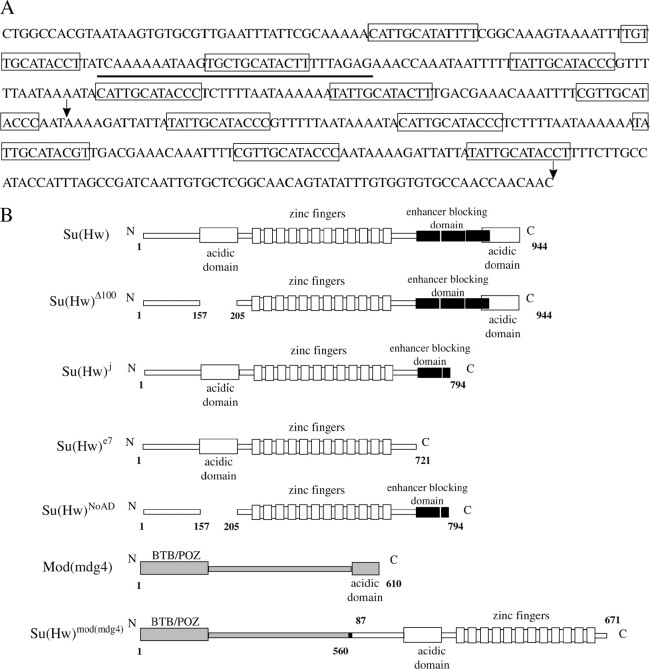
(A) The sequence of the Su(Hw) insulator isolated from the gypsy retrotransposon (Marlor et al. 1986). The 12 core binding sites are boxed. The underlining indicates the sequence of the nucleotide used to produce the synthetic Su(Hw)-binding regions. The consensus for the Su(Hw)-binding site was taken from Scott et al. (1999). The arrows indicate the sequence of the 8–12 Su(Hw)-binding sites. (B) Schematic of the Su(Hw) and Mod(mdg4)-67.2 proteins in mutations and constructions used in this study.
TABLE 2 .
Influence of thesu(Hw) mutations onyellowexpression in bristles
| Levels of yellow expression in bristles |
|||||||
|---|---|---|---|---|---|---|---|
| Transgenes | Genotype |
N of lines |
5 | w-v | m-v | e-v | 1 |
| dYW | su(Hw)+ | 4 | — | — | — | 2 | 2 |
| su(Hw)− | 4 | — | — | — | 2 | 2 | |
| (S)dY(S)W | su(Hw)+ | 7 | 5 | 2 | — | — | — |
| su(Hw)− | 7(7) | — | — | — | 1 | 6 | |
| (Sx4)dYW | su(Hw)+ | 4 | — | 1 | 3 | — | — |
| su(Hw)− | 4(4) | — | — | — | — | 4 | |
| (Sx8)dY(ΔS)W | su(Hw)+ | 6 | 4 | 2 | — | — | — |
| su(Hw)− | 6(6) | — | — | — | — | 6 | |
| SYilW | su(Hw)+ | 4 | — | — | 2 | 1 | 1 |
| su(Hw)− | 4 | — | — | 2 | 1 | 1 | |
| YilSW | su(Hw)+ | 5 | — | 1 | 2 | 1 | 1 |
| su(Hw)− | 5 | — | 1 | 2 | 1 | 1 | |
The levels of bristle pigmentation were examined in heterozygotes (P/+) for the transgene males in the su(Hw)+ (su(Hw)+/su(Hw)+) or su(Hw)− (su(Hw)v/su(Hw)f) background. Other designations are as in Table 1.
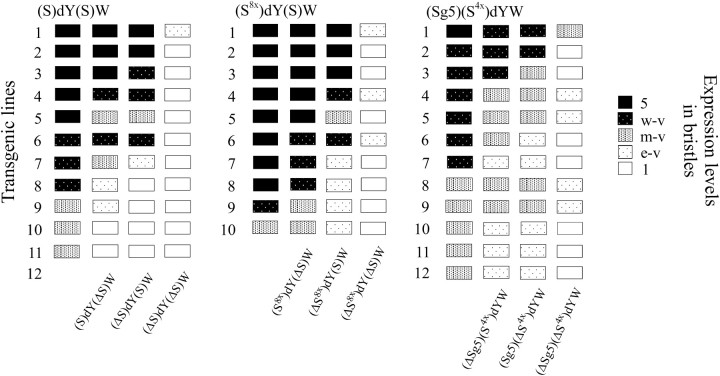
To assess the contribution of each Su(Hw) insulator to transcription stimulation, we deleted either the upstream ((ΔS)dY(S)W) or the downstream [(S)dY(ΔS)W] Su(Hw) insulator or both [(ΔS)dY(ΔS)W] from 11 transgenic lines in which flies had pigmented bristles (Table 1). In 5 transgenic lines, deletion of either Su(Hw) insulator did not significantly change bristle pigmentation, while deletion of both Su(Hw) insulators almost completely abolished it (Figure 2). This finding suggests that the Su(Hw) insulator does not stimulate yellow expression just as a neutral boundary that prevents spreading of the negative effects of surrounding chromatin. In contrast, the Su(Hw) insulator appears to be an active stimulator of the weakened yellow promoter.
Figure 2.—
Summary of phenotypes associated with selected transgenic (S)dY(S)W, (Sg5)(Sx4)dYW, and (Sx8)dY(S)W lines and their derivatives. All transgenic lines were numbered. For each line, pigmentation levels reflecting expression of the yellow gene in bristles are indicated by boxes using a five-level scale. Open boxes indicate a y1-like phenotype and solid boxes indicate a wild-type level of bristle pigmentation.
In the other six transgenic lines, deletion of either Su(Hw) insulator partially reduced or completely eliminated bristle pigmentation (Figure 2). In 4 of 11 cases, deletion of the upstream Su(Hw) insulator had a more pronounced effect, suggesting that the Su(Hw) insulator located upstream from the yellow promoter is more stimulatory.
The level of transcriptional stimulation directly correlates with the number of the Su(Hw)-binding sites:
The natural Su(Hw) insulator consists of 12 degenerate Su(Hw)-binding sites (Figure 3A), which have different affinity to the Su(Hw) protein (Spana and Corces 1990; Kim et al. 1996; Scott et al. 1999). It is possible that other proteins in addition to Su(Hw) bind with the 12-bp core sequence (consensus, 5′-PyPuTTGCATACCPy-3′) and are also involved in transcription stimulation. To examine this possibility, we used synthetic binding regions with 4 and 8 sites for Su(Hw), generated by concatemerization of a 31-unit oligonucleotide corresponding to the third Su(Hw)-binding site reported as the most effective one (Spana and Corces 1990; Kim et al. 1996).
In the (Sx4)dYW construct (Figure 1), four Su(Hw)-binding sites (Sx4) were inserted at position −525 relative to the transcription start in the yellow gene carrying the 368-bp deletion (dY). The Sx4 fragment was flanked by LOX sites. In 7 of 12 transgenic lines heterozygous for the (Sx4)dYW construct and in 11 of 12 lines homozygous for (Sx4)dYW, flies had partially pigmented bristles (Table 1). Thus, four Su(Hw)-binding sites are able to stimulate yellow expression in most of genomic sites of the construct insertion. Deletion of the Sx4 fragment eliminated bristle pigmentation in most of the lines, confirming the role of the Su(Hw)-binding sites in yellow stimulation. To verify the role of the Su(Hw) protein in transcription stimulation, we crossed flies with pigmented bristles from four (Sx4)dYW lines into a su(Hw)v/su(Hw)f mutant background (Table 2). Inactivation of Su(Hw) led to a y1-like phenotype, supporting the significance of Su(Hw) in transcription stimulation in (Sx4)dYW lines.
In the (Sx8)dY(S)W construct (Figure 1), eight Su(Hw) binding sites (Sx8) flanked with FRTs were inserted at −525. The Su(Hw) insulator flanked with LOXs was inserted at the 3′ side of the yellow gene. In 14 of 25 lines heterozygous for the (Sx8)dY(S)W construct, flies had detectable bristle pigmentation (Table 3, Figure 2). Deletion of the Su(Hw) insulator (ΔS) from the 3′ side of yellow did not significantly reduce yellow expression: flies in 8 transgenic lines heterozygous for (Sx8)dY(ΔS)W had nearly wild-type levels of bristle pigmentation (Figure 2). Additional deletion of the Sx8 fragment led to complete repression of yellow in 21 of 25 tested (ΔSx8)dY(ΔS)W derivative lines. To confirm the role of Su(Hw), yellow expression was examined on the su(Hw)− background in 6 (Sx8)dY(ΔS)W lines in which flies had nearly wild-type bristle pigmentation (Table 2). In all cases, inactivation of the Su(Hw) protein led to almost complete yellow repression in bristles. Comparison of bristle pigmentation of flies carrying the construct with deletion of either the Su(Hw) insulator [(Sx8)dY(ΔS)W] or the eight Su(Hw) binding sites [(ΔSx8)dY(S)W] demonstrated that the Su(Hw) binding sites inserted at −525 stimulated yellow expression more efficiently than the Su(Hw) insulator inserted at the 3′ side of the yellow gene (Figure 2). As in the (S)dY(S)W lines, the upstream Su(Hw) insulator had a more pronounced stimulatory effect than the downstream one; we suggest that the Su(Hw) insulator and the eight Su(Hw)-binding sites stimulate transcription with comparable effectiveness.
TABLE 3 .
Summary of phenotypes associated with the (Sg5)(Sx4)dYW, (Sx8)dY(S)W, SyilW, and YilSW transgenic lines
| Levels of yellow expression in bristles |
||||||
|---|---|---|---|---|---|---|
| Transgenes | N of lines | 5 | w-v | m-v | e-v | 1 |
| (Sx8)dY(S)W | 25 | 8 | 1 | 1 | 4 | 11 |
| (Sx8)dY(ΔS)W | 25(9) | 5 | 3 | 2 | 2 | 13 |
| (ΔSx8)dY(S)W | 25(12) | 3 | 2 | 1 | 4 | 16 |
| (ΔSx8)dY(ΔS)W | 25(14) | — | — | — | 4 | 21 |
| (Sg5)(Sx4)dYW | 23 | 1 | 6 | 5 | 3 | 8 |
| (ΔSg5)(Sx4)dYW | 23(11) | — | 3 | 5 | 5 | 10 |
| (Sg5)(ΔSx4)dYW | 23(12) | — | 2 | 5 | 6 | 10 |
| (ΔSg5)(ΔSx4)dYW | 23(15) | — | — | 1 | 4 | 18 |
| SYilW | 11 | — | — | 2 | 1 | 8 |
| YilSW | 14 | — | 1 | 2 | 2 | 9 |
All designations are as in Table 1.
As flies in the (Sx8)dY(ΔS)W lines had more pigmented bristles than flies from the (Sx4)dYW lines, we decided to further examine the correlation between the number of Su(Hw)-binding sites and their ability to stimulate transcription. In the (Sg5)(Sx4)dYW construct (Figure 1), a DNA fragment including four Su(Hw)-binding sites flanked with LOXs (Sx4) and five Su(Hw)-binding sites (8–12) from the Su(Hw) insulator (Sg5) (Figure 3A), flanked with FRTs, was inserted at −525. In 15 of 23 transgenic lines, flies heterozygous for the construct displayed detectable bristle pigmentation (Table 3). Deletion of either four (S×4) or five (Sg5) Su(Hw)-binding sites partially reduced bristle pigmentation, while deletion of all Su(Hw)-binding sites completely eliminated bristle pigmentation in most transgenic lines. These results further confirm that the efficiency of yellow stimulation directly correlates with the number of Su(Hw)-binding sites.
The Su(Hw) insulator does not compensate the deletion of the yellow enhancer:
As the Su(Hw) insulator stimulates the yellow expression at a large distance, it is possible that the Su(Hw) insulator acts as an enhancer. To test the ability of the Su(Hw) insulator to activate yellow expression in the absence of the bristle enhancer, we made two constructs bearing an intronless yellow gene and the Su(Hw) insulator inserted either at −893 bp (ESYilW, Figure 1) or at the 3′-end of the yellow gene (YilSW, Figure 1). As shown previously (Geyer and Corces 1987; Martin et al. 1989), flies bearing an intronless yellow gene produced yellow bristles.
In 11 SYilW lines and 14 YilSW lines, flies had yellow-colored bristles (Table 3). The bristle pigmentation was unchanged in the su(Hw)v/su(Hw)f background in flies from four SYilW lines and five YilSW lines (Table 2). These results indicate that the Su(Hw) insulator is unable to functionally substitute for the bristle enhancer.
Structural and functional analysis of Su(Hw) domains with regard to the insulator activity:
Su(Hw) has two acidic domains located at the amino- and carboxy-termini of the protein and an enhancer-blocking region located between 737 and 880 aa that is most important for insulation (Harrison et al. 1993; Kim et al. 1996; Gdula and Corces 1997). To address the role of individual Su(Hw) protein domains in yellow expression, different su(Hw) mutations (Figure 3B) were crossed into selected (S)dY(S)W, (Sx4)dYW, and (Sx8)dY(S)W lines and their derivatives carrying constructs on the X or second chromosome. In all selected transgenic lines, flies had wild-type or nearly wild-type levels of bristle pigmentation (Table 4).
TABLE 4 .
Influence of varioussu(Hw) alleles onyellow expression in bristles
| Genotypes |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| + | v/f | v/2 | j | j | Δ100 | e7 | NoAD | Su-M | ||
| Transgene | Levels of yellow expression in bristles: |
+ | + | m | + | m | + | + | + | + |
| dYW | ev | ev | ev | ev | ev | ev | ev | ev | ev | |
| (S)dY(S)W-1 | 5 | ev | ev | 5 | 5 | 5 | 5 | wv | 5 | |
| (S)dY(ΔS)W-1 | 5 | ev | ev | 5 | 5 | 5 | 5 | mv | 5 | |
| (ΔS)dY(S)W-1 | 5 | ev | ev | 5 | 5 | 5 | 5 | ev | wv | |
| (ΔS)dY(ΔS)W-1 | ev | ev | ev | ev | ev | ev | ev | ev | ev | |
| (S)dY(S)W-4 | 5 | 1 | 1 | 5 | 5 | 5 | 5 | wv | 5 | |
| (S)dY(S)W-5 | 5 | 1 | 1 | 5 | 5 | 5 | 5 | mv | 5 | |
| (Sx4)dYW | wv | 1 | 1 | wv | wv | wv | mv | 1 | mv | |
| (ΔSx4)dYW | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| (Sx8)dY(ΔS)W-1 | 5 | ev | ev | 5 | 5 | 5 | 5 | mv | 5 | |
| (Sx8)dY(S)W-2 | 5 | 1 | 1 | 5 | 5 | 5 | 5 | wv | 5 | |
| (Sx8)dY(ΔS)W-2 | 5 | 1 | 1 | 5 | 5 | 5 | wv | ev | 5 | |
| (ΔSx8)dY(S)W-2 | 5 | 1 | 1 | 5 | 5 | 5 | wv | ev | wv | |
| (ΔSx8)dY(ΔS)W-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| (Sx8)dY(ΔS)W-5 | 5 | 1 | 1 | 5 | 5 | 5 | wv | ev | wv | |
+, su(Hw)+ or mod(mdg4)+; v/f, su(Hw)v/su(Hw)f; v/2, su(Hw)v/su(Hw)2; m, mod(mdg4)u1/mod(mdg4)u1; j, su(Hw)j/su(Hw)j; Δ100, su(Hw)Δ100/su(Hw)Δ100; su(Hw)v/su(Hw)2; e7, su(Hw)e7/su(Hw)v; NoAD, su(Hw)v su(Hw)NoAD/su(Hw)2 su(Hw)NoAD; Su-M, su(Hw)Mod(mgd4)/su(Hw)Mod(mdg4); su(Hw)v/su(Hw)f. Italics indicate cases in which the su(Hw) mutations change yellow expression. The selected transgenic lines have the same numbers as in Figure 2. Other designations are described in the legend of Table 1.
The Su(Hw) protein (Figure 3B) contains a large acidic domain in the amino-terminal region and a second minor one in the carboxy terminus (Harrison et al. 1993). We have used the su(Hw)Δ100 allele to address the question whether the amino-terminal acidic domain is involved in yellow activation. The su(Hw)Δ100 mutation has an in-frame deletion of the 48 amino acids that constitute the amino-terminal acidic domain (Harrison et al. 1993). Flies heterozygous for the transposon and homozygous for the su(Hw)Δ100 allele had the same phenotype as those heterozygous for only the yellow transposon (Table 4). This result suggests that the N-terminal acidic domain of Su(Hw) is not important for yellow stimulation.
To address the effect of the Su(Hw) carboxy-terminal domain on yellow expression, the su(Hw)j allele was crossed into flies heterozygous for the yellow transposons. The Su(Hw) protein encoded by this allele lacks the 149 terminal residues, including the carboxy-terminal acidic domain and a part of the enhancer-blocking domain (Harrison et al. 1993; Kim et al. 1996; Gdula and Corces 1997). Similarly to su(Hw)Δ100, su(Hw)j does not influence yellow expression in transgenic lines (Table 4). Thus, the carboxy-terminal portion of the Su(Hw) protein is also not required for yellow activation. Because in the Su(Hw)j protein the domain interacting with the Mod(mdg4)-67.2 protein is only partially deleted (Gause et al. 2001; Ghosh et al. 2001), we examined the role of Mod(mdg4)-67.2 in yellow stimulation by the Su(Hw) insulator. The mod(mdg4)u1 mutation, which is known to affect the interaction between the Mod(mdg4)-67.2 isoform and Su(Hw) (Gause et al. 2001; Ghosh et al. 2001), was combined with the su(Hw)j allele (Georgiev and Kozycina 1996). Combination of the su(Hw)j and mod(mdg4)u1 mutations did not influence bristle pigmentation in the tested transgenic lines (Table 4). Thus, the Mod(mdg4)-67.2 protein is not required for yellow activation by the Su(Hw) insulator.
The su(Hw)e7 mutation leads to the loss of 223 amino acids from the carboxy-terminal end of the Su(Hw) protein (Harrison et al. 1993). This mutation only slightly affects the yellow phenotype in some transgenic lines (Table 4). Thus, the Su(Hw) protein lacking the domain responsible for enhancer blocking and the C-terminal acidic domain is still able to stimulate yellow expression when the Su(Hw) insulator is located at either the 5′- or the 3′-end of the yellow gene.
The Su(Hw)NoAD protein lacks the amino- and carboxy- terminal acidic domains and the part of the enhancer blocking domain (Harrison et al. 1993; Gdula and Corces 1997). The su(Hw)NoAD mutation partially relieved the mutant phenotype of the transgenic lines (Table 4). However, yellow repression is considerably less prominent than in the su(Hw)− background. This result might suggest that simultaneous deletion of both acidic domains and the enhancer-blocking domain partially affects the activating capacity of the Su(Hw) protein. Alternatively, the inability of Su(Hw)NoAD to effectively stimulate yellow expression could be explained by the instability of the truncated protein or less effective interaction with the Su(Hw) insulator.
Next we obtained two transgenic lines expressing the chimeric protein Su(Hw)Mod(mdg4) under the control of the Su(Hw) promoter as described in Kim et al. (1996). Su(Hw)Mod(mdg4) contains only the DNA-binding domain and the amino-terminal acidic domain that is joined to the C-terminal end of the truncated Mod(mdg4)-67.2 protein with deletion of the C-terminal domain required for interaction with Su(Hw) (Figure 3B). In all tested transgenic lines and their derivatives, the Su(Hw)Mod(mdg4) protein efficiently stimulated yellow transcription at the level of the Su(Hw)e7 protein (Table 4). As Mod(mdg4)-67.2 is not required for yellow activation, we suggest that the DNA-binding region and adjacent regions of Su(Hw) are sufficient for the transcriptional stimulation mediated by the Su(Hw) insulator.
DISCUSSION
The Su(Hw) insulator does not notably stimulate yellow transcription when the yellow promoter is functional (Geyer and Corces 1992). However, the Su(Hw) protein can behave as an activator of the yellow promoter if the upstream activator region is deleted. The level of yellow activation directly correlates with the number of the Su(Hw)-binding sites. The promoter stimulation activity of the Su(Hw) insulator is not restricted to the yellow promoter. Previously it was found that the Su(Hw) insulator stimulates the alcohol dehydrogenase promoter (Wei and Brennan 2001). The Su(Hw) protein also may be an activator of the weak gypsy promoter, as levels of gypsy RNA considerably decrease in su(Hw) mutants (Parkhurst and Corces 1986; Smith and Corces 1995). It seems that the Su(Hw) insulator can strengthen weak promoters but its effect is not visible in the case of a strong promoter.
Like a distance-independent enhancer, the Su(Hw) insulator can stimulate the yellow promoter over at least 5 kb. At the same time, the Su(Hw) insulator fails to compensate the deletion of the bristle enhancer, suggesting that the Su(Hw) insulator does not work as an enhancer. The long-distance effect of the Su(Hw) insulator cannot be explained by the boundary activity. As we found in many genomic sites, stimulation of transcription requires only one copy of the Su(Hw) insulator located either upstream or downstream from the yellow promoter. If only boundary function is important for the transcriptional stimulation, the location of the Su(Hw) insulator relative to the promoter must be crucial. As the transcriptional stimulation by the Su(Hw) insulator could be observed when the yellow promoter was partially inactivated by deletion of UPR, we suggest that Su(Hw) facilitates the assembling of a transcriptional complex at the yellow promoter.
The Su(Hw) insulator completely lost the ability to stimulate yellow transcription on the Su(Hw)− background, suggesting the main role of the Su(Hw) protein in this activity. Previous studies showed that the Su(Hw) protein has several different activities in the regulation of transcription. The enhancer-blocking activity mainly depends on the conserved domain located between the DNA-binding and carboxy-terminal acidic domains of the Su(Hw) protein (Harrison et al. 1993; Kim et al. 1996). The Mod(mdg4)-67.2 protein interacts with the enhancer-blocking domain of the Su(Hw) protein and contributes to the insulator activity (Gause et al. 2001; Ghosh et al. 2001). Inactivation of the Mod(mdg4)-67.2 protein in the mod(mdg4)u1 mutant converts the Su(Hw) insulator to a promoter-specific silencer (Gerasimova et al. 1995; Georgiev and Kozycina 1996; Cai and Levine 1997; Wei and Brennan 2001). It is likely that in the absence of the Mod(mdg4)-67.2 protein, Su(Hw) can directly interfere with the transcription complex at a promoter (Georgiev and Kozycina 1996; Cai and Levine 1997). Genetic analysis of the su(Hw) mutations involving deletions of particular domains of the Su(Hw) protein showed that the carboxy-terminal acidic domain is responsible for direct repression of the yellow promoter in the absence of Mod(mdg4)-67.2 (Georgiev and Kozycina 1996; Gdula and Corces 1997).
Here we found that deletion of either the acidic domain or the enhancer-blocking domain does not affect the ability of Su(Hw) to stimulate the weakened yellow promoter. However, deletion of both acidic domains and the enhancer-blocking domain in the Su(Hw)NoAD protein affects the activating capacity of the Su(Hw) insulator. Previously it was found that interaction with Mod(mdg4)-67.2 facilitates the binding of Su(Hw) to insulator sequences in vivo (Gerasimova and Corces 1998). As Su(Hw)NoAD fails to interact with Mod(mdg4)-67.2, we suggest that the deletion of the acidic and enhancer-blocking domains decreases DNA-binding affinity of the truncated Su(Hw)NoAD protein. As the level of transcriptional stimulation directly correlates with the number of the Su(Hw)-binding sites, the reducing of DNA-binding affinity of Su(Hw)NoAD might lead to the inability of the Su(Hw) insulator to efficiently stimulate transcription.
The chimeric Su(Hw)Mod(mdg4) protein consisting of the Su(Hw) DNA-binding domain, amino-terminal acidic domain, and Mod(mdg4)-67.2 can effectively stimulate transcription. As the Mod(mgd4)-67.2 protein and the amino-terminal acidic domain of Su(Hw) are not required for transcriptional stimulation by the Su(Hw) insulator, we suggest that the DNA-binding domain of Su(Hw) fulfills the main role in activation of the weak yellow promoter. Thus, different domains of Su(Hw) are required for enhancer blocking, promoter repression, and transcriptional stimulation. Interestingly, the enhancer-blocking and acidic domains of Su(Hw) are also not required for the boundary function of the Su(Hw) insulator in preventing gene repression by centric or telomeric heterochromatin (Geyer and Clark 2002).
As only DNA-binding domain and adjacent regions of Su(Hw) are required for long-distance transcriptional stimulation, we suggest that these domains form an entry site for the modification complexes. Recently, it was found (Torigoi et al. 2000) that the DNA-binding domain of Su(Hw) interacts with Chip that has been proposed to be a facilitator protein required for communication between an enhancer and a promoter (Morcillo et al. 1997; Dorsett 1999). Since only some of the 12 zinc fingers are required for DNA binding (Kim et al. 1996), other zinc fingers can be involved in recruiting protein complexes. The long-distance transcriptional stimulation might be explained by spreading of modification complexes to the yellow promoter. Consistent with this possibility, Chen and Corces (2001) showed that the Su(Hw) insulator increases the long-distance accessibility of the DNA to nucleases independently of the transcriptional status of the yellow gene. As a result of chromatin modifications, general transcription factors would gain access to the promoter region with a higher probability. Alternatively, the Su(Hw) insulator can directly interact with the yellow promoter by looping out the intervening DNA. The ability of the Su(Hw) insulator to repress yellow transcription in the absence of Mod(mdg4)-67.2 supports the possibility of direct interactions between proteins bound to the yellow promoter and the Su(Hw) insulator. Further study is required to understand the mechanism of the long-distance transcriptional stimulation of the yellow promoter by the Su(Hw) insulator.
Acknowledgments
We thank A. V. Galkin for critical reading and correction of the manuscript. We also thank D. Dorsett, E. Savitskaya, and Y. Schwartz for the plasmids and V. Corces for the su(Hw) mutants. This work was supported by the Molecular and Cellular Biology program of the Russian Academy of Science, by a stipend from the Center for Medical Studies, University of Oslo, to A.G., and by an International Research Scholar award from the Howard Hughes Medical Institute to P.G.
References
- Barges, S., J. Mihaly, M. Galloni, K. Hagstrom, M. Muller et al., 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulated iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127 779–790. [DOI] [PubMed] [Google Scholar]
- Belenkaya, T., K. Barseguyan, H. Hovhannisyan, I. Biryukova, E. Z. Kochieva et al., 1998. P element sequences can compensate for a deletion of the yellow regulatory region in Drosophila melanogaster. Mol. Gen. Genet. 259 79–87. [DOI] [PubMed] [Google Scholar]
- Bell, A. C., A. G. West and G. Felsenfeld, 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98 387–396. [DOI] [PubMed] [Google Scholar]
- Buchner, K., P. Roth, G. Schotta, V. Krauss, H. Saumweber et al., 2000. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics 155 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, H., and M. Levine, 1995. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 376 533–536. [DOI] [PubMed] [Google Scholar]
- Cai, H., and M. Levine, 1997. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 16 1732–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., and V. G. Corces, 2001. The gypsy insulator of Drosophila affects chromatin structure in a directional manner. Genetics 159 1649–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett, D., 1990. Potentiation of a polyadenylation site by a downstream protein DNA interaction. Proc. Natl. Acad. Sci. USA 87 4373–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett, D., 1999. Distant liaisons: long range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 9 505–514. [DOI] [PubMed] [Google Scholar]
- Gause, M., P. Morcillo and D. Dorsett, 2001. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of Hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol. 21 4807–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula, D. A., and V. G. Corces, 1997. Characterization of functional domains of the su(Hw) protein that mediate the silencing effect of mod(mdg4) mutations. Genetics 145 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev, P. G., and T. I. Gerasimova, 1989. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 220 121–126. [DOI] [PubMed] [Google Scholar]
- Georgiev, P., and M. Kozycina, 1996. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 142 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova, T. I., and V. G. Corces, 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92 511–521. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., and V. G. Corces, 2001. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 35 193–208. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., D. A. Gdula, D. V. Gerasimov, O. B. Simonova and V. G. Corces, 1995. A Drosophila protein that impacts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82 587–597. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., and I. Clark, 2002. Protecting against promiscuity: the regulatory role of insulators. Cell. Mol. Life Sci. 59 2112–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1 996–1004. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6 1865–1873. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., C. Spana and V. G. Corces, 1986. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 5 2657–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, D., T. I. Gerasimova and V. G. Corces, 2001. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 20 2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, K. G., and S. Lindquist, 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59 499–509. [DOI] [PubMed] [Google Scholar]
- Hagstrom, K., M. Muller and P. Schedl, 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10 3202–3215. [DOI] [PubMed] [Google Scholar]
- Harrison, D. A., D. A. Gdula, R. S. Coyne and V. G. Corces, 1993. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 7 1966–1978. [DOI] [PubMed] [Google Scholar]
- Holdridge, C., and D. Dorsett, 1991. Repression of hsp70 heat shock gene transcription by the suppressor of Hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kares, R. E., and G. M. Rubin, 1984. Analysis of P transposable element functions in Drosophila. Cell 38 135–146. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and P. Schedl, 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64 941–950. [DOI] [PubMed] [Google Scholar]
- Kim, J., B. Shen, C. Rosen and D. Dorsett, 1996. The DNA-binding and enhancer-blocking domains of the Drosophila suppressor of Hairy-wing protein. Mol. Cell. Biol. 16 3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, E. J., and P. K. Geyer, 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell. Biol. 15 259–265. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992 The Genome of Drosophila melanogaster. Academic Press, New York.
- Mallin, D. R., J. S. Myung, J. S. Patton and P. K. Geyer, 1998. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 148 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor, R. L., S. M. Parkhurst and V. G. Corces, 1986. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol. Cell. Biol. 6 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M., Y. B. Meng and W. Chia, 1989. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. Mol. Gen. Genet. 218 118–126. [DOI] [PubMed] [Google Scholar]
- Mazo, A. M., L. J. Mizrokhi, A. A. Karavanov, Y. A. Sedkov, A. A. Krichevskaya et al., 1989. Suppression in Drosophila: su(Hw) and su(f) gene products interact with a region of gypsy (mdg4) regulating its transcriptional activity. EMBO J. 8 903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo, P., C. Rosen, M. K. Baylies and D. Dorsett, 1997. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes. Dev. 11 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash, W. G., and R. J. Yarkin, 1974. Genetic regulation and pattern formation: a study of the yellow locus in Drosophila melanogaster. Genet. Res. 24 19–26. [DOI] [PubMed] [Google Scholar]
- Oki, M., and R. T. Kamakaka, 2002. Blockers and barriers to transcription: Competing activities? Curr. Opin. Cell Biol. 14 299–304. [DOI] [PubMed] [Google Scholar]
- Parkhurst, S., and V. G. Corces, 1986. Interactions among the gypsy element and the yellow and suppressor of Hairy-wing loci in Drosophila melanogaster. Mol. Cell. Biol. 6 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R. R., V. Pirrotta and P. K. Geyer, 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi et al., 1995. A P element containing suppressor of Hairy-wing binding region has novel properties for mutagenesis in Drosophila melanogaster. Genetics 141 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218 348–353. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989 Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Scott, K. S., and P. K. Geyer, 1995. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 14 6258–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, K. S., A. D. Taubman and P. K. Geyer, 1999. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics 153 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal, M. L., and D. L. Hartl, 2000. Application of Cre/loxP in Drosophila. Site-specific recombination and transgene co-placement. Methods Mol. Biol. 136 487–495. [DOI] [PubMed] [Google Scholar]
- Sigrist, C. J. A., and V. Pirrotta, 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P.A., and V. G. Corces, 1995. The suppressor of Hairy-wing protein regulates the tissue-specific expression of the Drosophila gypsy retrotransposon. Genetics 139 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana, C., and V. G. Corces, 1990. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev. 4 1505–1515. [DOI] [PubMed] [Google Scholar]
- Spana, C., D. A. Harrison and V. G. Corces, 1988. The Drosophila melanogaster Suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2 1414–1423. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., and G. M. Rubin, 1982. Transposition of cloned P elements into germline chromosomes. Science 218 341–347. [DOI] [PubMed] [Google Scholar]
- Sun, F.-L., and S. C. R. Elgin, 1999. Putting boundaries on silence. Cell 99 459–462. [DOI] [PubMed] [Google Scholar]
- Torigoi, E., I. M. Bennani-Baiti, C. Rosen, K. Gonzalez, P. Morcillo et al., 2000. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc. Natl. Acad. Sci. USA 97 2686–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag, J., J. L. den Blaauwen, R. G. Sewalt, R. van Driel and A. P. Otte, 2000. Transcriptional repression mediated by polycomb group proteins and other chromatin-associated repressors is selectively blocked by insulators. J. Biol. Chem. 275 697–704. [DOI] [PubMed] [Google Scholar]
- Wei, W., and M. D. Brennan, 2001. The gypsy insulator can act as a promoter-specific transcriptional stimulator. Mol. Cell. Biol. 21 7714–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, A. G., M. Gaszner and G. Felsenfeld, 2002. Insulators: many functions, many mechanisms. Genes Dev. 16 271–288. [DOI] [PubMed] [Google Scholar]
- Udvardy, A., 1999. Dividing the empire: boundary chromatin elements delimit the territory of enhancers. EMBO J. 18 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., C. M. Hart and U. K. Laemmli, 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81 879–889. [DOI] [PubMed] [Google Scholar]
- Zhou, J., S. Barolo, P. Szymanski and M. Levine, 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 10 3195–3201. [DOI] [PubMed] [Google Scholar]
- Zhou, J., H. Ashe, C. Burks and M. Levine, 1999. Characterization of the transvection mediating region of the Abdominal-B locus in Drosophila. Development 126 3057–3065. [DOI] [PubMed] [Google Scholar]