Abstract
The homeologous Orp1 and Orp2 regions of maize and the orthologous regions in sorghum and rice were compared by generating sequence data for >486 kb of genomic DNA. At least three genic rearrangements differentiate the maize Orp1 and Orp2 segments, including an insertion of a single gene and two deletions that removed one gene each, while no genic rearrangements were detected in the maize Orp2 region relative to sorghum. Extended comparison of the orthologous Orp regions of sorghum and japonica rice uncovered numerous genic rearrangements and the presence of a transposon-rich region in rice. Only 11 of 27 genes (40%) are arranged in the same order and orientation between sorghum and rice. Of the 8 genes that are uniquely present in the sorghum region, 4 were found to have single-copy homologs in both rice and Arabidopsis, but none of these genes are located near each other, indicating frequent gene movement. Further comparison of the Orp segments from two rice subspecies, japonica and indica, revealed that the transposon-rich region is both an ancient and current hotspot for retrotransposon accumulation and genic rearrangement. We also identify unequal gene conversion as a mechanism for maize retrotransposon rearrangement.
THE grasses, including major cereals such as rice, maize, wheat, barley, and sorghum, are the most agronomically and economically important plant species. Despite their fairly recent origin from a common ancestor, the grasses exhibit broad variation in genome size. On the other hand, comparative genetic mapping of rice, maize, wheat, sorghum, and other grasses has revealed extensive conservation of gene content and gene order in all species investigated to date (Gale and Devos 1998), although some large chromosomal rearrangements were also observed (reviewed in Paterson et al. 2000). These studies have provided a foundation for understanding grass genome evolution and have led to the map-based isolation of agronomically important genes (Brueggeman et al. 2002; Feuillet et al. 2003; Yan et al. 2003, 2004; Yahiaoui et al. 2004).
With the near completion of the rice genome sequence (Feng et al. 2002; Sasaki et al. 2002; Rice Chromosome 10 Sequencing Consortium 2003), cross-species sequence comparisons in the grasses become increasingly feasible. So far, several orthologous grass genome segments containing more than one gene have been compared at the level of DNA sequence, including the sh2/a1-homologous regions of maize, sorghum, and rice (Chen et al. 1997); the adh1-homologous regions of maize, sorghum, and rice (Tikhonov et al. 1999; Ilic et al. 2003); the LrK-homologous regions of barley, maize, rice, and wheat (Feuillet and Keller 1999); the genomic regions near Vrn1 and its orthologs in wheat, barley, sorghum, and rice (Ramakrishna et al. 2002a); the Zein gene cluster of maize and its orthologs in sorghum and rice (Song et al. 2002); the Rp1-homologous regions of maize and sorghum (Ramakrishna et al. 2002b); the Rph7-homologous regions of barley and rice (Brunner et al. 2003); and the lg2/lrs1-homeologous regions of maize and its ortholog in rice (Langham et al. 2004). These studies uncovered little or no retention of sequence homology in intergenic spaces but indicate general conservation of gene content and gene order between orthologous genomic segments of grass genomes. In addition, many exceptions to genome microcolinearity such as gene deletion, insertion, duplication, inversion, and translocation were observed (reviewed in Bennetzen and Ma 2003).
Traditional cytological analyses suggested that maize originated from a tetraploid (McClintock 1930), while other genetic and molecular data also indicate that the maize genome contains many duplicated genes and duplicated segments with colinear gene arrangements (Rhoades 1951; Helentjaris et al. 1988; Ahn and Tanksley 1993; Davis et al. 1999). Some duplicated genes in maize have been isolated and sequenced (Gaut and Doebley 1997; Ilic et al. 2003; Langham et al. 2004; Swigoňová et al. 2004). By examining the patterns of sequence divergence among 14 pairs of duplicated genes in maize, Gaut and Doebley (1997) proposed that the modern maize genome originated from an ancient segmental allotetraploid event that occurred between 16.5 and 11.4 million years ago (MYA) after the divergence of sorghum from one of the two maize diploid progenitor lineages that themselves diverged ∼20 MYA. However, by analyzing 11 genes from clearly orthologous segments of maize, sorghum, and rice, Swigoňová et al. (2004) determined that the two maize progenitors and sorghum diverged contemporaneously from a common ancestor ∼11.9 MYA.
In a recent article, Ilic et al. (2003) presented a detailed genomic sequence comparison of an orthologous segment of the rice, sorghum, and two maize subgenomes. This first comparative sequence analysis involving homeologous segments of maize and corresponding colinear regions in sorghum and rice provides numerous insights into the nature and timing of local genomic rearrangements that occurred in these three important grass lineages. Ilic et al. (2003) identified extensive gene loss by an accumulation of small deletions in the two homeologous segments of maize analyzed, and these two segments seem to be equally unstable compared to the orthologous regions of rice and sorghum. The progressive accumulation of small deletions, most caused by illegitimate recombination, also are responsible for rapid loss of retrotransposons, other intergenic space, and some portions of genes (e.g., introns) in the Arabidopsis, wheat, and rice genomes (Devos et al. 2002; Wicker et al. 2003; Ma et al. 2004; Ma and Bennetzen 2004).
Additional studies are needed to help identify the full spectrum of local genome rearrangement in plants and to determine their frequencies and relative contributions. Here we use comparative sequence analysis to investigate genome structure and change in orthologous Orp regions of maize, sorghum, and rice, thereby uncovering rapid gene movement without gene loss, a hotspot for transposon accumulation, and a propensity for genic rearrangement within a transposon-rich region.
MATERIALS AND METHODS
BAC selection:
An Orp probe was obtained from maize by PCR using primers based on the complete coding sequence of the Orp2 gene (GenBank accession no. M76685). A HindIII BAC library with inserts from cultivar BTx623 sorghum DNA (http://www.tamu.edu/bacindex.html) and a MboI BAC library made from B73 maize DNA (Yim et al. 2002) were screened with the Orp probe as described previously (Song et al. 2002). The positive BAC clones detected in both libraries were fingerprinted with restriction enzyme HindIII and further confirmed by gel-blot hybridization analysis. Meanwhile, the identified BAC clones were digested with 8-bp specificity restriction enzymes AscI, NotI, PacI, and SwaI. The restriction fragments were separated by pulsed-field gel electrophoresis, transferred to nylon membranes, and hybridized with the Orp probe to estimate the BAC insert sizes and to construct restriction maps. This information helped determine the appropriate BACs for sequencing and also experimentally validated the computer sequence assemblies of analyzed BACs.
The largest sorghum BAC, SB18C08, that contained an Orp homolog was sequenced and analyzed. The predicted genes in SB18C08 were used as probes to hybridize with the positive maize BAC clones that we previously identified. Two maize BACs, ZM573L14 and ZM573F08, sharing the greatest genic homology with each other and with sorghum BAC SB18C18, were finally chosen for sequencing.
BAC sequencing:
Shotgun libraries for BACs SB18C08, ZM573L14, and ZM573F08 were constructed as described previously (Dubcovsky et al. 2001; Song et al. 2001). Subclones were sequenced from both directions using ABI PRISM BigDye Terminator Chemistry (Applied BioSystems, Foster, CA) and run on an ABI3700 capillary sequencer. Base calling and quality assessment were done using PHRED (Ewing and Green 1998). Reads were assembled with PHRAP and edited with CONSED (Gordon et al. 1998). Sorghum clone SB18C08 was sequenced at ∼8-fold redundancy, while maize clones ZM573L14 and ZM573F08 were each sequenced at ∼12-fold redundancy. Gaps were filled by a combination of several approaches, as described earlier (Ramakrishna et al. 2002a). The final error frequency estimated by CONSED was less than one base/10 kb. The finished assemblies of BAC sequences were found to agree completely with their restriction maps.
The Orp-orthologous regions in two rice subspecies, japonica (c.v. Nipponbare) (http://rgp.dna.affrc.go.jp/IRGSP/) and indica (c.v. 93-11) (Yu et al. 2002; Zhao et al. 2004), were identified by homology comparisons of the genomic sequences in GenBank deposited by May 2004. Sequence alignments were conducted by using BLASTN (NCBI), BLAST 2.0, BLAST2 (Tatusova and Madden 1999), and CROSS_MATCH (http://www.phrap.org). We considered it an ortholog when a sequence/contig between japonica and indica had a unique match in the japonica genomic sequence and the assembled indica shotgun sequences.
Sequence analysis and annotation:
Gene-finding programs FGENESH (http://www.softberry.com/berry.phtml?topic=gfind&prg=FGENES) with the monocot training set, GeneMark.hmm (http://opal.biology.gatech.edu/GeneMark/eukhmm.cgi) with maize and/or rice training sets, and GENSCAN (http://genes.mit.edu/GENSCAN.html) with the maize training set were used to predict potential genes in rice, sorghum, and maize Orp BAC sequences. The genes predicted by these programs and the remaining regions (excluding the identified transposable elements) of these BAC sequences were investigated by BLASTX searches against the GenBank protein database (http://www.ncbi.nlm.nih.gov/BLAST/). Sequences identified as candidate genes by the gene-finding programs were further investigated to determine whether they were actually genes. In our earlier rice genome annotation studies, for instance, we found that >30% of the candidate genes identified by these programs were actually transposons or transposon fragments (Bennetzen et al. 2004). So we used conservation in a distantly related species as an additional criterion for gene certification. Hence, candidate rice genes were used as queries in BlastX searches against the full GenBank database, but were considered likely genes only if they detected homology at an expect value of <e−05 in some species other than rice. The recent release of the genome sequence for maize (Whitelaw et al. 2003) provided a particularly useful data set for this analysis.
Shared genes were detected by orthologous sequence comparisons and multiple sequence alignments using CROSS_MATCH, BLAST2, and ClustalX (Thompson et al. 1997). Genes that were not shared in the orthologous regions were further investigated by BLASTX searches against the Arabidopsis protein database at The Arabidopsis Information Resources (TAIR) (http://www.arabidopsis.org) and against the rice predicted protein database at The Institute for Genomic Research (TIGR) (http://www.tigr.org/tdb/e2k1/osa1/) to determine the copy numbers and distribution of corresponding homologous genes in the Arabidopsis and rice genomes. The genes in sorghum were named in numerical order by their position on the sequenced BAC, while the genes in rice and maize were numbered according to their homology to the shared genes in sorghum. Three unshared genes in rice and one unshared gene in maize were given alphabetical designations.
Transposable elements (transposons and retrotransposons) were identified using a combination of structural analysis of repetitive DNA and homology-based searches against GenBank nucleotide and protein databases and the TIGR cereal repeat database (http://www.tigr.org/tdb/rice/blastsearch.shtml). The programs Repeat and Gap from the Wisconsin Package Version 10.1 (Genetics Computer Group) were used to identify long-terminal-repeat (LTR) retrotransposons as described earlier (Devos et al. 2002; Ma et al. 2004). Newly identified retrotransposons were named according to the retrotransposon nomenclature previously described by SanMiguel et al. (2002). The approximate dates of LTR-retrotransposon insertion and gene duplication in rice were estimated in a manner similar to SanMiguel et al. (1998) and Ramakrishna et al. (2002a), respectively. For dating LTR-retrotransposon insertion times, the molecular clock was set at an average substitution rate of 1.3 × 10−8 mutation/site/year, which we estimated for intergenic regions in rice (Ma and Bennetzen 2004).
RESULTS
Isolation of Orp segments of sorghum, maize, and rice:
The maize genes Orp1 and Orp2, encoding the β-subunit of tryptophan synthase, have been cloned and mapped to the short arms of chromosomes 4 and 10, respectively (Wright et al. 1992). From earlier comparative maps of the cereals (e.g., Gale and Devos 1998), it appears that these two chromosome arms are homeologues. That is, they are orthologous regions descended from two different diploid ancestors of the tetraploid progenitor of maize (Swigoňová et al. 2004). Two contiguous series (contigs) of maize BAC clones that hybridized to the Orp probe were generated by fingerprinting and restriction map analysis. Only one contig of BACs that contain an Orp gene was detected in sorghum. Because the maize genome is primarily composed of large blocks of LTR retrotransposons, often organized in a nested insertion pattern (SanMiguel et al. 1996; Fu and Dooner 2002; Song et al. 2002; Song and Messing 2002, 2003), it was difficult to predict which BACs of maize and sorghum would provide the best alignment of colinear genes. Therefore, we first sequenced an ∼160-kb sorghum BAC, SB18C08, the largest among the overlapping BACs containing the sorghum Orp gene. After analyzing the gene content of BAC SB18C08, probes from 10 additional genes physically linked to the sorghum Orp gene were obtained by PCR and hybridized with the previously identified positive BACs of maize. Two maize BACs, ZM573F08 and ZM573L14, sharing the most genes with the orthologous region of sorghum, were then chosen and completely sequenced.
BLASTN searches against the nonredundant database at GenBank using the predicted genes in sorghum as queries were conducted to identify potential homologous segments of rice. A contig of five overlapping finished BAC sequences (GenBank accession nos. AP003896, AP005620, AP005618, AP005250, and AP004591) from the japonica cultivar Nipponbare were found to contain most of the genes homologous to the genes predicted on sorghum clone SB18C08, defining this contig as an orthologous Orp region in rice. Therefore, a 313-kb contiguous Orp region in rice was selected for further analysis.
Sequence organization of the Orp regions of sorghum, maize, and rice:
The complete sequence of the Orp segment in sorghum clone SB18C08 is 159,669 bp (GenBank accession no. AF466200). With our criteria for gene identification (see materials and methods), we identified 22 sorghum genes on this BAC (Table 1). The average gene density is one gene/7.3 kb, similar to that previously observed in the sorghum sh2/a1 region (Chen et al. 1997), the sorghum adh region (Tikhonov et al. 1999), and the region near the Vrn1 ortholog of sorghum (Ramakrishna et al. 2002a), but higher than the density of one gene/10.8 kb in the 215-kb region comprising the kafirin gene (Song et al. 2002). No intact transposable elements were annotated in the sorghum region, but two non-LTR retrotransposon fragments (−f) and one DNA transposon fragment (TNP2-f) were detected by homology-based searches (Figure 1).
TABLE 1 .
Identified genes inOrp regions of maize, sorghum, and rice
| Homology |
|||||||
|---|---|---|---|---|---|---|---|
| Gene | Rice Orp | Sorghum Orp |
Maize Orp2 |
Maize Orp1 |
Protein products | Accession no. |
E-value |
| 1 | Present | Expressed protein (Arabidopsis) | NP_188808 | 2e-6 | |||
| 2 | Present | Present | Present | Homeobox protein (Arabidopsis) | NP_193906 | 0 | |
| 3 | Present | Present | Present | Present | Trytophan synthase β-subunit (Arabidopsis) |
NP_194437 | 2e-87 |
| 4 | Present | F box protein (Arabidopsis) | NP_191482 | 2e-6 | |||
| 5 | Present | Present | Present | Hypothetical protein (Arabidopsis) | NP_188494 | 2e-14 | |
| 6 | Present | Hypothetical protein (Arabidopsis) | NP_200360 | 2e-24 | |||
| 7 | Present | Present | Endonuclease/exonuclease/phosphatase family (Arabidopsis) |
NP_566904 | 2e-24 | ||
| 8 | Present | Present | Present | Expressed protein (Arabidopsis) | NP_192387 | 1e-9 | |
| 9 | Present | Present | Present | Glycosyl hydrolase family 17 (Arabidopsis) | NP_181895 | 5e-21 | |
| 10 | Present | Present | Copine-related protein (Arabidopsis) | NP_565206 | 2e-98 | ||
| 11 | Present | Present | Hypothetical protein (Arabidopsis) | NP_187362 | 6e-19 | ||
| 12 | Present | Present | Transporter related protein (Arabidopsis) | NP_566487 | 1e-70 | ||
| 13 | Present | Hypothetical protein (Arabidopsis) | NP_201419 | 6e-13 | |||
| 14 | Present | Present | Phospholipid/glycerol acyltransferase family (Arabidopsis) |
NP_181346 | 4e-89 | ||
| 15 | Present | Protein phosphatase 2C (Arabidopsis) | NP_194903 | 5e-20 | |||
| 16 | Present | Hypothetical protein (Arabidopsis) | NP_173847 | 4e-47 | |||
| 17 | Present | Present | Galactosyltransferase family (Arabidopsis) |
NP_177618 | 9e-56 | ||
| 18 | Present | Present | Hypothetical protein (Arabidopsis) | NP_175765 | e-155 | ||
| 19 | Present | Present | Cytochrome P450 protein (Arabidopsis) | NP_171635 | 0 | ||
| 20 | Present | Present | Putative lip transfer proteion precuror (Arabidopsis) |
NP_179109 | 1e-13 | ||
| 21 | Present | Present | Phototropic response protein family (Arabidopsis) |
NP_174332 | e-151 | ||
| 22 | Present | Present | Expressed protein (Arabidopsis) | NP_193039 | 8e-17 | ||
| a | Present | Expressed protein (Arabidopsis) | NP_564055 | 1e-24 | |||
| b | Present | Copine BONZAI1 (BON1) (Arabidopsis) | NP_568944 | 3e-11 | |||
| c | Present | Putative protein (Arabidopsis) | NP_198805 | 5e-19 | |||
| d | Present | Tubby-like protein (Arabidopsis) | NP_849975 | 2e-11 | |||
“Present” indicates that the genes listed are present in the corresponding regions of maize, rice, and/or sorghum.
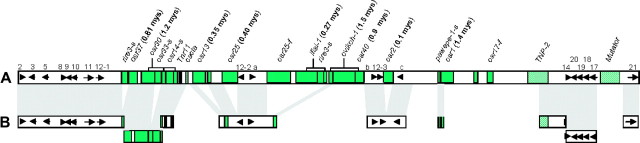
Figure 1.—
Sequence organization and comparison of orthologous Orp regions of sorghum, maize, and rice. Arrows represent identified genes and their transcriptional orientations. The orthologous or homeologous genes shared by different regions are connected by gray lines. The organizations of retrotransposons and transposons are illustrated by gray-shaded boxes and cross-hatched boxes, respectively. Dotted lines in rice connect contiguous sequences. f, truncated transposon fragment; s, solo LTR.
The complete sequence of the Orp1 segment in maize clone ZM573F08 is 181,627 bp (GenBank accession no. AY555142). It contains four identified genes (Table 1). The average gene density is one gene/45.4 kb. LTR retrotransposons in this region are more abundant than in the average sequenced regions of maize (SanMiguel et al. 1996; Fu and Dooner 2002; Ramakrishna et al. 2002b; Song et al. 2002; Ilic et al. 2003). A total of 14 LTR retrotransposons, one solo LTR, and three retrotransposon fragments were identified (Table 1), constituting ∼138 kb of DNA (∼76% of the region). The majority of the retrotransposons in this region are organized in typical nested fashion (SanMiguel et al. 1996). The four predicted genes are separated into two gene pairs by the largest (∼53 kb) retrotransposon block.
The complete sequence of the Orp2 segment in maize clone ZM573L14 is 144,792 bp (GenBank accession no. AY555143) and contains four apparent genes (Table 1). The average gene density is one gene/36 kb. Three of these four genes are clustered together and separated from the other gene by a cluster of intact retrotransposons, retrotransposon fragments, and a newly identified CACTA-like transposon, fanal-1, which inserted into retrotransposon milt-1. The transposable elements on this BAC include six retrotransposons, five retrotransposon fragments, one DNA transposon (fanal-1), and two DNA transposon fragments (TNP-f and Tam3-f), together accounting for ∼50% of this region.
The 313-kb rice genomic sequence contains 19 identified genes, including a triplication of one locus (genes 12-1, 12-2, and 12-3). The average gene density is 1 gene/16.5 kb, much lower than estimated for the whole rice genome (1 gene/7–9 kb; Feng et al. 2002; Goff et al. 2002; Sasaki et al. 2002; Song et al. 2002; Yu et al. 2002; Rice Chromosome 10 Sequencing Consortium 2003). This low gene density is mainly due to the presence of a large cluster of repetitive DNA that harbors only four predicted genes (Figure 1). This repetitive domain is predominantly composed of LTR retrotransposons, including nine intact elements, five solo LTRs, and five truncated fragments, three of which (ifisi, ovikoh, and pawepe) are discovered and named in this study. These elements constitute ∼105 kb of DNA, accounting for ∼55% of the retrotransposon-rich area or 33% of the whole region investigated. We also identified four DNA transposons and/or fragments in the rice region, constituting 22 kb of DNA.
Sequence comparison of colinear Orp regions of sorghum, maize, and rice:
Three genes, 3, 8, and 9, are shared among rice, sorghum, and maize Orp1 regions, distributed across 25 kb in rice, 53 kb in sorghum and 68 kb in maize. The maize Orp2 region also shares three predicted genes, 2, 3, and 5, with rice and sorghum. These genes are distributed across 17 kb in rice, 35 kb in sorghum, and 18 kb in maize, respectively. In addition to genes 2, 3, and 5, one more gene (gene 7) is shared between sorghum and the maize Orp2 region. Several genes are missing from one or more of the four otherwise colinear segments (Figure 1). This dramatic variation of gene organization and intergenic distance is due to both the variable amount of intergenic repetitive DNAs and the local genic rearrangements, such as deletions or insertions of genes. The maize Orp1 and Orp2 regions share only gene 3, the Orp loci. Hence, as observed at adh1 and lg2/lrs1 loci (Ilic et al. 2003; Langham et al. 2004), most of the duplicated genes present from the tetraploidization of the maize ancestor ∼11.9 MYA have been reduced by deletion to a near-diploid state (Swigoňová et al. 2004). The colinearity of the Orp genes and other genes shared between maize and sorghum and between maize and rice (Figure 1) does indicate that the maize Orp1 and Orp2 regions are homeologous segments derived from the two diploid progenitors of maize.
Among the orthologous regions (from gene 2 to gene 9) shared by sorghum, rice, and maize, several gene rearrangements can be attributed to specific lineages because we can compare four chromosomal segments: (1) Gene 5 adjacent to Orp1 was deleted in maize; (2) genes 4 and 6 were inserted into the sorghum region after the divergence of sorghum and maize ancestors; and (3) gene d was acquired by the maize Orp1 region after the divergence of sorghum and maize ancestors (Figure 1). In addition, 3′ to Orp1 in maize, genes 1 and 2 were found to be deleted by analyzing the next maize BAC that is downstream of Orp1 and contains the Fie1 locus (Lai et al. 2004).
Extended comparison of colinear Orp regions of rice and sorghum:
The sorghum Orp segment was compared with the continuous 313-kb orthologous region of rice. We found numerous alterations in gene content, order, and orientation. A total of 14 predicted genes were found to be shared, distributed across 313 kb in rice and 159 kb in sorghum, whereas 13 additional genes were not in orthologous locations. This includes 8 genes (1, 4, 6, 7, 13, 15, 16, and 22) present in this region of sorghum but absent in the orthologous region of rice, and 5 genes (a, b, c, 12-2, and 12-3) present only in the rice region. Inspection of the adjacent BACs to the rice contig that we analyzed in this study indicated no copies homologous to genes 1 and 22. For most of the nonorthologous genes, on the basis of comparative analysis of two species we do not know whether they were gained or deleted in sorghum or in rice. However, for genes 4, 6, and 7 (present in sorghum or maize but not in rice), the simplest explanation suggests that they inserted in these locations in the lineage that gave rise to sorghum and/or maize.
An inversion of a cluster of four predicted genes (genes 17, 18, 19, and 20) was detected between rice and sorghum. These four genes are arranged in the indica genome in the same order as present in japonica (Figure 2), but it is not clear whether the inversion occurred in an ancestor of rice or sorghum.
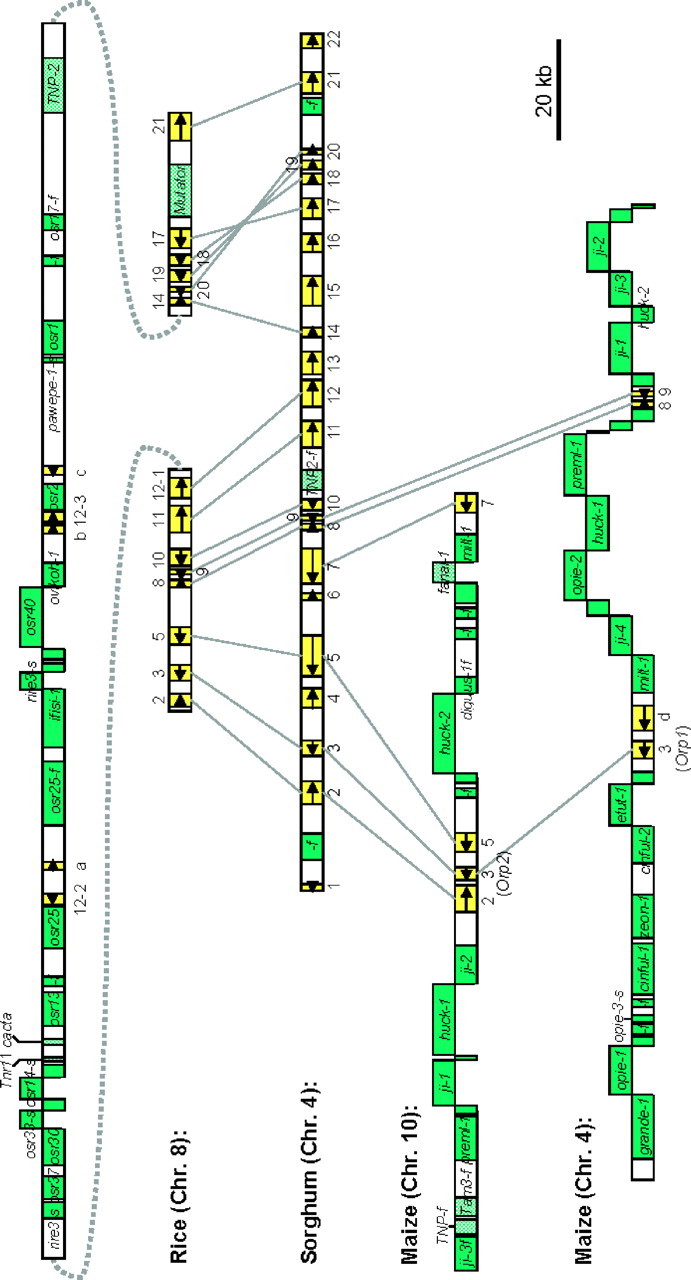
Figure 2.—
Comparison of orthologous regions of (A) japonica and (B) indica. Arrows represent predicted genes. Gray-shaded boxes and cross-hatched boxes represent retrotransposons and DNA transposons, respectively. The shaded regions connecting the japonica and indica sequences outline the conserved regions between these two subspecies. Because the indica sequence is fragmentary, we cannot compare the order or orientation of the sequences on different indica fragments with the ordered sequence for japonica. mys, million years since insertion.
In rice, we discovered three copies of gene 12 (12-1, 12-2, and 12-3) that were not tandemly arrayed. Gene 12-1 remains intact, while genes 12-2 and 12-3 are truncated at their N termini when compared with rice gene 12-1 and sorghum gene 12. If the divergence time for rice and sorghum ancestors is 60 MYA (Wolfe et al. 1989; Kellogg 2001), we roughly estimate that the first duplication of rice gene 12 homologs occurred ∼25 MYA. However, because only gene 12-1 appears to be intact, the truncated genes 12-2 and 12-3 may be evolving more rapidly than functional loci that usually follow standard molecular clocks. Hence, this first duplication may have occurred much <25 MYA, and, similarly, the second duplication (yielding genes 12-2 and 12-3) may have taken place more recently than the 8 MYA that we calculated. Gene 12-2 is arranged in inverted orientation relative to 12-1 and 12-3 in rice and gene 12 in sorghum, an event that probably occurred after the second duplication. In addition, putative genes a and b were found between genes 12-1 and 12-2 and between genes 12-2 and 12-3, respectively. The extra three genes (a, b, and c) in rice are also truncated. Altogether, these data indicate a high frequency of several different types of genic rearrangement in this specific region of rice.
Chromosomal locations of homologs in rice and Arabidopsis:
Nearly complete genomic sequence and comprehensive sequence annotation of the Arabidopsis and rice genomes allowed us to investigate the nature of some local gene rearrangements at the whole-genome level. All of the genes predicted in the Orp regions of sorghum and/or maize but not shared with the orthologous region of rice were used as queries to search against nucleotide databases and protein databases of the rice genome at TIGR (http://www.tigr.org/tdb/e2k1/osa1/) and the Arabidopsis genome at TAIR (http://www.tigr.org/servlets/sv). The rice and Arabidopsis homologs closest to the corresponding sorghum genes and their chromosomal locations in individual genomes are summarized in Table 2.
TABLE 2 .
Chromosomal distribution of homologs of investigated genes in rice and Arabidopsis
| In rice |
In Arabidopsis |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Best match |
Best match |
||||||||
| Gene |
Orp region |
Copy no. | Chromosome | Locus | E-value | Copy no. | Chromosome | Locus | E-value |
| 1 | Sorghum | 1 | 7 | 2009.t00001 | 4e-4 | 1 | 3 | At3g21710.1 | 2e-7 |
| 4 | Sorghum | >2 | 6 | 3460t00003 | 3e-120 | >2 | 3 | At3g59230.1 | 5e-8 |
| 6 | Sorghum | 2 | 9 | 5014t00010 | 8e-104 | 1 | 5 | At5g55490.1 | 3e-24 |
| 7 | Sorghum | 1 | 12 | 5048.t00006 | 8e-125 | 1 | 3 | At3g48425.1 | 3e-26 |
| 13 | Sorghum | 1 | 2 | 5875t00005 | 8e-127 | 1 | 5 | At5g66180.1 | 2e-14 |
| 15 | Sorghum | >2 | 2 | 5021t00003 | 2e-46 | >2 | 4 | At4g31750.1 | 1e-21 |
| 16 | Sorghum | >2 | 4 | 5483t00011 | 3e-87 | >2 | 1 | At1g24370.1 | 5e-49 |
| 22 | Sorghum | 1 | 5 | 6505t00025 | 7e-74 | 1 | 4 | At4g13030.1 | 3e-14 |
| d | Maize | >2 | 2 | 4877t00020 | 1e-15 | >2 | 2 | At2g18280.1 | 6e-13 |
All of the identified genes (1, 4, 6, 7, 13, 15, 16, 22, and d) absent in the rice Orp region were found to have homologs in both the rice and the Arabidopsis genome protein databases (Table 2). These homologs (the best matches) are distributed along several different chromosomes, including chromosomes 2, 4, 5, 6, 7, 9, and 12 in rice and chromosomes 1, 2, 3, 4, and 5 in Arabidopsis (Table 2). None of these genes were closely linked to each other on any rice or Arabidopsis chromosome (data not shown). Genes 1, 7, 13, and 22 have only single copies in both rice and Arabodopsis genomes, suggesting but not proving that these loci are orthologous and further suggesting that numerous independent rearrangements involving these genes must have occurred after the divergence of sorghum and rice lineages. Multiple copies were observed for genes 4, 15, 16, and d in both rice and Arabidopsis.
A rapidly evolving retrotransposon block in rice:
The rice interval contains a transposable element-rich region, composed mainly of LTR retrotransposons (∼105 kb of DNA). This regions occupies ∼190 kb of DNA, but contains only five genes, including two that are duplicated (Figure 1). This segment contains a high percentage (∼55%) of LTR retrotransposons, similar to that recently observed in the centromeric region of rice chromosome 8 (Wu et al. 2004).
The assembled whole-genome shotgun sequence generated from indica cultivar 93-11 (Yu et al. 2002; Zhao et al. 2004) was used in this study to investigate the timing and lineage specificities of the dramatic accumulation of retrotransposons and genic rearrangements identified in the Orp region of japonica rice. By sequence homology searches and sequence alignments, we identified nine assembled contiguous segments (accession nos. AAAA01000069, AAAA01004112, AAAA01006364, AAAA01008548, AAAA01009470, AAAA01009525, AAAA009834, AAAA01013675, and AAAA01023118) from indica that have unique matches in both the japonica genomic sequence and the indica whole-genome shotgun sequences, suggesting that these segments are orthologous (Figure 2).
We found eight LTR retrotransposons or fragments uniquely present in the Orp region of japonica, although seven LTR retrotransposons or fragments were shared by indica and japonica in the comparable regions (Figure 2). For all LTR retrotransposons that are relatively intact, we employed LTR divergence as a tool to date approximate times of insertion (SanMiguel et al. 1998). We found that all intact LTR retrotransposons uniquely present in japonica were younger than 0.44 MY (the estimated divergence time of indica and japonica, Ma and Bennetzen 2004) and that all shared intact elements had inserted >0.44 MYA (Figure 2). Hence, it appears that this retrotransposon block has been continuously and independently expanding in both indica and japonica lineages by insertion of LTR retrotransposons.
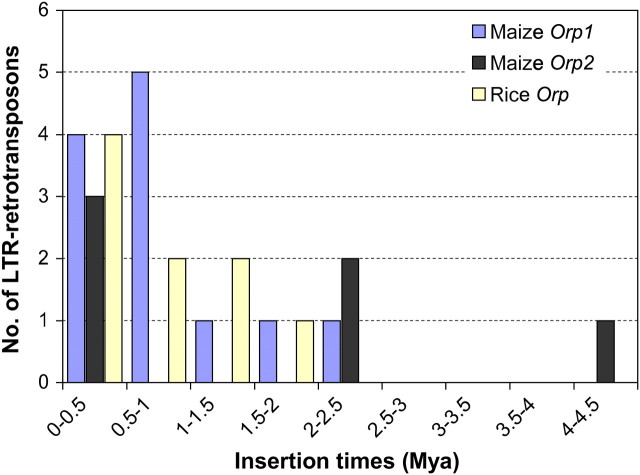
The relatively intact LTR retrotransposons found in the maize Orp1 and Orp2 regions are all recent insertions. The majority of intact elements inserted <2 MYA (Figure 3). Our estimate is consistent with the previous dating of LTR retrotransposons in maize (SanMiguel et al. 1998; Swigoňová et al. 2004).
Figure 3.—
Estimated insertion times of identified LTR retrotransposons in the Orp regions of maize and rice. The dates of LTR retrotransposon insertions are shown in brackets. Mya, million years ago.
The structure of a rearranged retrotransposon:
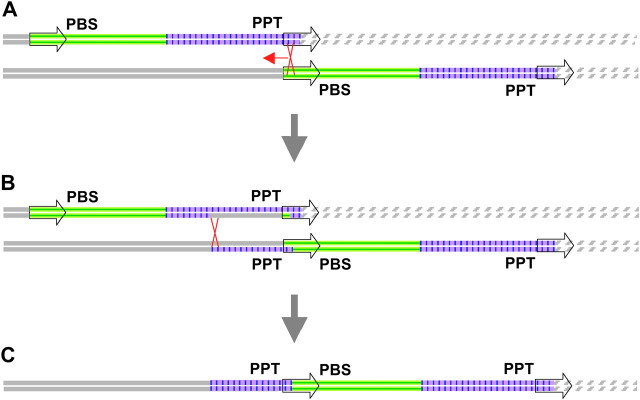
We identified a rearranged LTR retrotransposon, grande_573F08-1, in the Orp1 region of maize. Its unusual property is that a region of 2145 bp directly upstream of the 5′ LTR is very similar (>97% identical) to the sequences upstream of the 3′ LTR. The likely origin of this element structure by unequal conversion is presented in Figure 4. Figure 4C depicts grande_573F08-1 and the sequence flanking it from the region downstream of Orp1. An opie element, likely an insertion subsequent to the events described in Figure 4, is not included. The 13.9-kb grande element is 5′-flanked by 2145 bp sharing 2093 identical base pairs with the 3′ portion of grande_ 573F08-1 immediately upstream of its 3′ LTR (but differing by four indels of 1, 9, 15, and 20 bp). A total of 44 of the mismatches are transitions and 8 are transversions. Both LTRs are 627 bp, of which 615 bp are identical (10 transitions, 2 transversions). This apparent conversion tract of >2145 bp (including an unknown length of sequence in the 5′ LTR) is relatively long, but conversion tracts of >3 kb have been observed in maize (Dooner and Martinez-Ferez 1997; Yandeau-Nelson et al. 2005).
Figure 4.—
Unequal homologous recombination (without exchange of flanking markers) may have replaced the 5′ flanking region of grande_573F08-1 with 2145 bases of its internal sequence. Both strands of unequally paired regions from homeologous chromosomes are shown in A and B. Open arrows denote LTRs. The element is arbitrarily divided into a green, horizontally striped 5′ region and a purple, vertically lined 3′ region. Upstream and downstream flanking DNA are depicted as solid and diagonally striped lines, respectively. (A) Putative initial structure of the grande element prior to recombination. The 3′ LTR of the top homolog unequally pairs with the 5′ LTR of the bottom homolog. The two crossed red lines show the site of initiation of recombination and the red arrow shows the direction of the hypothetical strand transfer. (B) Branch migration progresses beyond the 5′ termini of the LTRs in which the recombination initiated. (C) One of many possible outcomes of this recombination event: the structure present in 573F08. Two landmark sequences associated with LTR-retrotransposon transposition, PBS (primer binding site) and PPT (polypurine tract), are indicated. This figure is not drawn to scale.
DISCUSSION
The comparative genomics approach for gene identification:
In this study, five genes were identified by comparison of colinear regions containing the maize Orp genes and their orthologs in rice and sorghum (Figure 1; Table 3). All of these genes are also present in the Arabidopsis genome (http://www.arabidopsis.org/servlets/sv; Table 1). Except the conserved genes, no other long sequences were shared among these regions, as has been observed for all the orthologous or colinear segments compared among maize, sorghum, and rice (Bennetzen et al. 2005). However, numerous small conserved noncoding sequences have been identified between orthologous genes in multiple plant species, most of which are harbored in intron or promoter domains of genes (Kaplinsky et al. 2002; Guo and Moose 2003; Inada et al. 2003).
TABLE 3 .
Assessments of credibility of gene prediction methods
| Predicted genes based on gene-finding programs and BLAST searches |
||||||
|---|---|---|---|---|---|---|
| Identified geneb |
Unknown sequencec |
|||||
| Region | Transposable elementa |
Shared | Unshared | Shared | Unshared | Total |
| Rice | 28 | 14 | 3 + 2d | 0 | 12 | 59 |
| Sorghum | 2 | 15 | 7 | 0 | 2 | 26 |
| Maize Orp2 | 15 | 4 | 0 | 0 | 1 | 20 |
| Maize Orp1 | 27 | 3 | 1 | 0 | 0 | 31 |
Predicted genes belonging to transposable elements identified in this study or matching that deposited in the public databases.
All of these genes were found to have homologs in the Arabidopsis whole-genome database.
Genes were predicted only by gene-finding programs.
Two duplicated genes that were predicted to be derived from gene 12-1, a possible ortholog of sorghum gene 12.
In addition to the conserved orthologous genes, we identified 10 more genes in maize, rice, or sorghum that exhibited significant similarity to one or more annotated Arabidopsis genes on the basis of BLASTX searches (Table 1). Because the lineage that gave rise to Arabidopsis has evolved independently from the grass lineage for >150 million years (Wolfe et al. 1989), it is likely that the conserved sequences between Arabidopsis and grasses are genes.
Gene-finding programs such as FGENESH, GENSCAN, and/or Genemark.hmm are useful but imperfect tools for gene identification. These programs predicted 40, 4, 16, and 27 additional genes in the Orp regions of rice and sorghum and the Orp2 and Orp1 regions of maize, respectively, beyond those we consider valid gene candidates (Table 3). Of these predicted genes, 28 (70%), 2 (50%), 15 (94%), and 27 (100%), respectively, were found to have the structure and/or the highest sequence similarity to transposable elements (Table 3). However, 12 predicted genes in rice (11 scattered in the transposon-element-rich area of the Orp segment), 2 predicted genes in sorghum, and 1 predicted gene in maize are unclear in origin, so we did not annotate them as genes. Because these predicted genes have no homologs in Arabidopsis or in any other genome, we think they are rapidly evolving transposable elements or some other nongenic DNA.
Gene content instability in the two maize subgenomes:
Our results are consistent with the hypothesis of a recent tetraploid origin for maize (Swigoňová et al. 2004). Although the two maize segments analyzed in this study share only Orp1 and Orp2 homeologous genes, their comparisons to the orthologous regions of sorghum and rice indicate that they are two homeologous segments. There have been at least two gene deletions near the Orp1 locus. Also, independent insertion of large blocks of retrotransposons in both Orp1 and Orp2 segments have occurred in the last few million years. The maize Orp2 segment remains relatively “intact,” with no gene deletion detected in this region. This result parallels the recent finding by Langham et al. (2004). By comparing the maize lg2 region and its homeologous lrs1 region, Langham et al. (2004) found that a cluster of four predicted genes 3′ to the lrs1 locus have been deleted, leading to “zero retention” of duplicated factors, excluding the lg2/lrs1 gene pair. In contrast, >40% of the total genes from each homeologous region were found to have been deleted by several separate deletion events in the maize adh1 region and its homeologue, indicating that both regions have been equally unstable compared to their orthologs in sorghum and rice (Ilic et al. 2003). However, at least one copy of all orthologous genes appears to be conserved between the two homeologous regions in all cases investigated, suggesting that natural selection has acted against loss of all copies of any of these genes.
Timing of gene loss in the Orp region of maize:
We cannot precisely determine the times of gene deletion or insertion events in the Orp1 region of maize, although our comparative data indicate that they took place after the divergence of maize and sorghum. The extensive deletion of genes and low-copy-number sequences appears to be a common feature of genomes with polyploid origins, such as Arabidopsis (Arabidopsis Genome Initiative 2000) and maize (Ahn et al. 1993; Song et al. 2002; Ilic et al. 2003). The elimination of low-copy-number sequences has also been detected in newly formed polyploids (Song et al. 1995; Feldman et al. 1997; Ozkan et al. 2001), indicating that genome changes often happen in the first few generations in response to the formation of a polyploid. Gene deletion and transposon accumulation have also been seen to differentiate haplotypes in the allelic regions of different maize inbreds (Fu and Dooner 2002; Song and Messing 2003).
Genic rearrangements: Deletion, insertion, and/or translocation?
Comparison of the orthologous regions of the rice and sorghum genomes reveals numerous small genic rearrangements. Apparent insertions of genes 1, 4, and 6 were detected in sorghum compared to rice and maize. No gene deletion or insertion was found in the two gene-clustered regions that are separated by a cluster of transposable elements in rice. This observation parallels observations in adh (Tikhonov et al. 1999; Ilic et al. 2003), sh2/a1 (Chen et al. 1997; Li and Gill 2002), and php200725 (Song et al. 2002) orthologous regions, indicating that rice has a relatively stable gene content and order compared with maize, sorghum, or wheat.
Interestingly, all of the noncolinear genes present in the Orp region of sorghum and/or maize were found to have very similar copy numbers in both rice and Arabidopsis, indicating copy-number conservation for >150 million years of independent evolution (Wolfe et al. 1989). For four noncolinear genes, only single homologs were detected in both rice and Arabidopsis. If one assumes that these single-copy homologs are orthologous to the corresponding genes identified in sorghum, then it is clear that synteny or colinearity is not a perfect indicator of orthology. The relocations of these genes may have occurred in the rice and/or sorghum lineages. Alternatively, these four genes may be paralogous to the corresponding genes detected in rice because deletions removed the actual orthologs. Hence, on the basis of current data it is impossible to say whether these genes were deleted, inserted, or relocated in the rice and sorghum genomes.
All identified genes in the japonica Orp region were found to have homologs in the homologous region of indica rice. Because most assembled shotgun sequences from the indica genome are relatively small, we did not obtain the complete Orp region of indica and thus cannot compare order or orientation of these sequence fragments. It is also not clear whether any genes are uniquely present in the indica region. However, complete japonica and indica sequences of the php200725 region show complete conservation of gene order in both subspecies (Song et al. 2002). Furthermore, previous comparison of ∼1.1 Mb of orthologous regions between indica and japonica has demonstrated a lack of gene acquisition or loss from either indica or japonica (Ma and Bennetzen 2004), supporting the previous observation that the rice genome exhibits relatively stable gene content in contrast to the maize genome (Song et al. 2002; Ilic et al. 2003).
A hotspot for gene rearrangement and the insertion of LTR retrotransposons in rice:
We found a large LTR-retrotransposon-rich segment in the rice genome that contains few genes, and all of the genes within this retrotransposon block were either duplicates or noncolinear inserts relative to sorghum. Our data indicate that this rice region has expanded rapidly by insertion of LTR retrotransposons in the past 2 MY, with most insertions in the few hundred thousand years since the divergence of indica and japonica ancestors. The ancient insertions (>1 MY old) in this region indicate that it has been a hotspot for transposon accumulation for a long time, while the recent insertions suggest that this insertion affinity is still present.
In our dating of relatively intact LTR retrotransposons in the rice genome, we found that the average age is ∼1.3 MY (Ma et al. 2004), while in the retrotransposon block of the rice Orp region, the average age of all datable LTR retrotransposons is ∼0.7 MY. Moreover, we demonstrated a minimum of eight new transposon insertions within the japonica region since the divergence from a common ancestor with indica, adding at least 53 kb of new DNA to a target region of 190 kb. This is about a fourfold higher frequency of insertion than that observed for 1 Mb of chromosome 4 DNA from our earlier indica and japonica comparison (Ma and Bennetzen 2004).
A high percentage of repetitive DNA was observed in the centromeric region of rice chromosome 8. In this region, LTR retrotransposons account for at least 50% of the DNA (Wu et al. 2004), and >80% of these elements were amplified before the divergence of indica and japonica (J. Ma and J. L. Bennetzen, unpublished observations). In contrast to the centromeric region, 55% of the transposon-rich segment of the rice Orp region is composed of LTR retrotransposons, and about half of them were amplified after the divergence of these two subspecies (Figure 2). This high rate of transposon insertion, plus the presence of a nontandem gene triplication and several noncolinear truncated genes in the Orp region, suggests that this block is a hotspot for several different kinds of genome rearrangement. It will be interesting to see if other retrotransposon blocks exhibit this type of exceptional instability when other comparative studies are performed.
An intraelement retrotransposon conversion event:
While maize retrotransposons are frequently intact at their termini, including the presence of short, flanking host-site duplications, there is no shortage of more tattered elements present in any maize BAC sequence. Unequal recombination is frequently invoked to explain the presence of solo LTRs (Devos et al. 2002; Ma et al. 2004). Here we suggest that this phenomenon may explain a larger group of rearrangements simply by positing that an unequal recombination event initiating inside LTRs might migrate outside a terminus of these LTRs. While Figure 4 depicts a recombination event with symmetric exchange of strands, nonsymmetric events should also occur. These would yield the same outcome. Repair of heteroduplex DNA will also play a role in these sorts of recombination events and this could result in more complex rearrangements than depicted if the repair was noncontinuous over the recombination tract.
One other model could be proposed to explain the structure that we found. Two grande elements (most likely proximate to one another) on the same chromosome in the same orientation could recombine unequally to create a double element, sharing an LTR. But a second event would be required to explain the deletion of the 5′-end of the 5′-element. This second model is also unlikely because the duplicated region resulting from this mechanism would likely have a greater percentage of mismatched bases over the duplication than the 3% that is observed. Comparison of a grande element from the 22-kD α-zein gene family (Song et al. 2001) and this grande element yields a 15% mismatch frequency over aligned bases. Rarely are retrotransposons (even from the same family) >90% similar over >2-kb regions.
Acknowledgments
We thank Katrien M. Devos and Zuzana Swigoňová for useful discussions and two anonymous reviewers for their valuable comments. This work was supported by the National Science Foundation Plant Genome Program (grant no. 9975618).
References
- Ahn, S., and S. D. Tanksley, 1993. Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. USA 90 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S., J. A. Anderson, M. E. Sorrells and S. D. Tanksley, 1993. Homoeologous relationships of rice, wheat and maize chromosomes. Mol. Gen. Genet. 241 483–490. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and J. Ma, 2003. The genetic collinearity of rice and other cereals on the basis of genomic sequence analysis. Curr. Opin. Plant Biol. 6 128–133. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., C. Coleman, R. Liu, J. Ma and W. Ramakrishna, 2004. Consistent over-estimation of gene number in complex plant genomes. Curr. Opin. Plant Biol. 7 732–736. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., J. Ma and K. M. Devos, 2005. Mechanisms of recent genome size variation in flowering plants. Ann. Bot. 95 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman, R., N. Rostoks, D. Kudrna, A. Kilian, F. Han et al., 2002. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 99 9328–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, S., B. Keller and C. Feuillet, 2003. A large rearrangement involving genes and low-copy DNA interrupts the microcollinearity between rice and barley at the Rph7 locus. Genetics 164 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., P. J. SanMiguel, A. C. de Oliveira, S. S. Woo, H. Zhang et al., 1997. Microcollinearity in sh2-homologous regions of the maize, rice, and sorghum genomes. Proc. Natl. Acad. Sci. USA 94 3431–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, G. L., M. D. McMullen, C. Baysdorfer, T. Musket, D. Grant et al., 1999. A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequence tagged sites (ESTs) in a 1736-locus map. Genetics 152 1137–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K. M., J. K. Brown and J. L. Bennetzen, 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., and I. M. Martinez-Ferez, 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky, J., W. Ramakrishna, P. J. SanMiguel, C. S. Busso, L. Yan et al., 2001. Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol. 125 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using PHRED: II. Error probabilities. Genome Res. 8 186–194. [PubMed] [Google Scholar]
- Feldman, M., B. Liu, G. Segal, S. Abbo, A. A. Levy et al., 1997. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q., Y. Zhang, P. Hao, S. Wang, G. Fu et al., 2002. Sequence and analysis of rice chromosome 4. Nature 420 316–320. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., and B. Keller, 1999. High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl. Acad. Sci. USA 96 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., S. Travella, N. Stein, L. Albar, A. Nublat et al., 2003. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. USA 100 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., and H. K. Dooner, 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M. D., and K. M. Devos, 1998. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and J. F. Doebley, 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S. A., D. Ricke, T.-H. Lan, G. Presting, R. Wang et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100. [DOI] [PubMed] [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. CONSED: a graphical tool for sequence finishing. Genome Res. 8 195–202. [DOI] [PubMed] [Google Scholar]
- Guo, H., and S. P. Moose, 2003. Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjaris, T., D. Weber and S. Wright, 1988. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, K., J. P. SanMiguel and J. L. Bennetzen, 2003. A complex history of rearrangement in an orthologous region of the maize, sorghum and rice genomes. Proc. Natl. Acad. Sci. USA 100 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada, D. C., A. Bashir, C. Lee, B. C. Thomas and C. Ko, 2003. Conserved noncoding sequences in the grasses. Genome Res. 13 2030–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinsky, N. J., D. M. Braun, J. Penterman, S. A. Goff and M. Freeling, 2002. Utility and distribution of conserved noncoding sequences in the grasses. Proc. Natl. Acad. Sci. USA 99 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E. A., 2001. Evolutionary history of the grasses. Plant Physiol. 125 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J., J. Ma, Z. Swigonova, W. Ramakrishna, E. Linton et al., 2004. Gene loss and movement in the maize genome. Genome Res. 14 1924–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langham, R. J., J. Walsh, M. Dunn, C. Ko, S. A. Goff et al., 2004. Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., and B. S. Gill, 2002. The colinearity of the Sh2/A1 orthologous region in rice, sorghum and maize is interrupted and accompanied by genome expansion in the Triticeae. Genetics 160 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2004. Recent rapid growth and divergence of the rice nuclear genome. Proc. Natl. Acad. Sci. USA 101 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., K. M. Devos and J. L. Bennetzen, 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1930. A cytological demonstration of the location of an interchange between two non-homologous chromosomes of Zea mays. Proc. Natl. Acad. Sci. USA 16 791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan, H., A. A. Levy and M. Feldman, 2001. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A. H., J. E. Bowers, M. D. Burow, X. Draye, C. G. Elsik et al., 2000. Comparative genomics of plant chromosomes. Plant Cell 12 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., J. Dubcovsky, Y.-J. Park, C. Busso, J. Emberton et al., 2002. a Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., J. Emberton, P. SanMiguel, M. Ogden, V. Llaca et al., 2002. b Comparative sequence analysis of the sorghum Rph region and the maize Rp1 resistance gene complex. Plant Physiol. 130 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M. M., 1951. Duplicated genes in maize. Am. Nat. 85 105–110. [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium, 2003 In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300: 1566–1569. [DOI] [PubMed]
- SanMiguel, P., A. Tikhonov, Y. K. Jin, N. Motchoulskaia, D. Zakharov et al., 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., B. S. Gaut, A. Tikhonov, Y. Nakajima and J. L. Bennetzen, 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20 43–45. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P. J., W. Ramakrishna, J. L. Bennetzen, C. S. Busso and J. Dubcovsky, 2002. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A(m). Funct. Integr. Genomics 2 70–80. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., T. Matsumoto, K. Yamamoto, K. Sakata, T. Baba et al., 2002. The genome sequence and structure of rice chromosome 1. Nature 420 312–316. [DOI] [PubMed] [Google Scholar]
- Song, K., P. Lu, K. Tang and T. C. Osborn, 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2002. Contiguous genomic DNA sequence comprising the 19-kD zein gene family from maize. Plant Physiol. 130 1626–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2003. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., V. Llaca, E. Linton and J. Messing, 2001. Sequence, regulation, and evolution of the maize 22-kD alpha zein gene family. Genome Res. 11 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., V. Llaca and J. Messing, 2002. Mosaic organization of the orthologous sequences in grass genomes. Genome Res. 12 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigoňová, Z., J. Lai, J. Ma, W. Ramakrishna, V. Llaca et al., 2004. Close split of maize and sorghum genome progenitors. Genome Res. 14 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigoňová, Z., J. L. Bennetzen and J. Messing, 2005. Structure and evolution of the r/b chromosomal regions in rice, maize and sorghum. Genetics 169 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova, T. A., and T. L. Madden, 1999. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174 247–250. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A. P., P. J. SanMiguel, Y. Nakajima, N. M. Gorenstein, J. L. Bennetzen et al., 1999. Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw, C. A., W. B. Barbazuk, G. Pertea, A. P. Chan, F. Cheung et al., 2003. Enrichment of gene-coding sequences in maize by genome filtration. Science 302 2118–2120. [DOI] [PubMed] [Google Scholar]
- Wicker, T., N. Yahiaoui, R. Guyot, E. Schlagenhauf, Z. D. Liu et al., 2003. Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15 1186–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H., M. Gouy, Y. W. Yang, P. M. Sharp and W. H. Li, 1989. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, A. D., C. A. Moehlenkamp, G. H. Perrot, M. G. Neuffer and K. C. Cone, 1992. The maize auxotrophic mutant orange pericarp is defective in duplicate genes for tryptophan synthase beta. Plant Cell 4 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., H. Yamagata, M. Hayashi-Tsugane, S. Hijishita, M. Fujisawa et al., 2004. Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahiaoui, N., P. Srichumpa, R. Dudler and B. Keller, 2004. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 37 528–538. [DOI] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, G. Tranquilli, M. Helguera, T. Fahima et al., 2003. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, A. Blechl, G. Tranquilli, W. Ramakrishna et al., 2004. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandeau-Nelson, M. D., Q. Zhou, H. Yao, X. Xu, B. J. Nikolau et al., 2005. MuDR Transposase increases the frequency of meiotic crossovers in the vicinity of a Mu insertion in the maize a1 gene. Genetics 169 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim, Y. S., G. L. Davis, N. A. Duru, T. A. Musket, E. W. Linton et al., 2002. Characterization of three maize bacterial artificial chromosome libraries toward anchoring of the physical map to the genetic map using high-density bacterial artificial chromosome filter hybridization. Plant Physiol. 130 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., S. N. Hu, J. Wang, K. W. Gane, S. G. Li et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. [DOI] [PubMed] [Google Scholar]
- Zhao, W. M., J. Wang, X. He, X. Huang, Y. Jiao et al., 2004. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomes. Nucleic Acids Res. 32 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]