Abstract
Redesign of the bacteriophage 434 Cro repressor was accomplished by using an in vivo genetic screening system to identify new variants that specifically bound previously unrecognized DNA sequences. Site-directed, combinatorial mutagenesis of the 434 Cro helix-turn-helix (HTH) motif generated libraries of new variants which were screened for binding to new target sequences. Multiple mutations of 434 Cro that functionally converted wild-type (wt) 434 Cro DNA binding-sequence specificity to that of a λ bacteriophage-specific repressor were identified. The libraries contained variations within the HTH sequence at only three positions. In vivo and in vitro analysis of several of the identified 434 Cro variants showed that the relatively few changes in the recognition helix of the HTH motif of 434 Cro resulted in specific and tight binding of the target DNA sequences. For the best 434 Cro variant identified, an apparent Kd for λ OR3 of 1 nM was observed. In competition experiments, this Cro variant was observed to be highly selective. We conclude that functional 434 Cro repressor variants with new DNA binding specificities can be generated from wt 434 Cro by mutating just the recognition helix. Important characteristics of the screening system responsible for the successful identifications are discussed. Application of the techniques presented here may allow the identification of DNA binding protein variants that functionally affect DNA regulatory sequences important in disease and industrial and biotechnological processes.
The mechanisms by which DNA binding proteins interact with regulatory DNA sequences are complex (8, 19, 20), and a general code that predicts specific protein-DNA interactions appears to be difficult to attain (5). For DNA binding proteins with helix-turn-helix (HTH) structural motifs (2), such as 434 Cro (16, 22), binding specificity appears to be dependent on side chain contacts of the recognition helix with the DNA and the position of the recognition helix within the major groove (2, 15, 28). Additionally, the conformation of the bound DNA, which is often distorted or bent, and other protein contacts outside of the recognition helix also appear to contribute to binding specificity (11, 12). Because of these complexities, the rational redesign of DNA binding protein specificity, even with relatively simple proteins like 434 Cro, is challenging. The desired DNA specificity redesign of zinc finger DNA binding proteins has been successfully achieved by using phage display technologies (reviewed in reference 21). In a previous report investigating 434 repressor binding specificities, mutation of specific side chains of the recognition helix led in some cases to altered binding specificities (10). Other reports indicated that mutagenesis of the first two residues of the recognition helix of 434 Cro did not lead to proteins that were capable of specific binding to operator mutations (see reference 8). Huang et al. (9) described an in vivo strategy that used negative selection to screen 3 × 106 recognition helix variants of the structurally related 434 repressor protein but failed to identify any uniquely new binding specificities. By using a single chain variant of the 434 repressor dimer and a phenotypic screen of a combinatorial library of recognition helix mutations, Simoncsits et al. (25) more recently showed that variants with specificity changes at one base of the DNA binding sequence could be identified. Here we show that construction of combinatorial mutational libraries at as few as three positions in the recognition helix of 434 Cro and the application of an optimized screening system for transcriptional repression allow the identification of Cro variants that possess a fundamentally new, targeted DNA binding specificity unrelated to that of the parent repressor. Important differences in the screening systems employed here that resulted in successful identifications of new Cro variants are discussed in regard to those methods previously employed.
MATERIALS AND METHODS
Bacterial strains and plasmid vectors used.
The Escherichia coli strains DH5α (29) and JM109 (30) were used for general cloning purposes. Strain DH5α was used for screening and expression of DNA binding proteins. Plasmids pACYC184 (4) and pUC119 (26) were extensively modified for use in the selection procedures (see below). Plasmid pACYC184 was a gift from S. Cohen; pUC119 was obtained from the American Type Culture Collection (Rockville, Md.).
Construction of combinatorial libraries of 434 Cro with mutations in the recognition helix.
Two different types of combinatorial libraries at the N terminus of the recognition helix (positions Q28, Q29, and S30) were constructed by cassette mutagenesis techniques that employed an NNS codon containing oligonucleotides (23) or oligonucleotides synthesized partly using trinucleotide precursors, i.e., the TRIM technique (27). The TRIM technique has several advantages, including the ability to synthesize mutagenic oligonucleotides that have a 1-to-1 ratio of mutations at the DNA level to those at the expressed-protein level. The synthetic HTH-encoding, combinatorial DNA cassettes were cloned in frame into unique restriction sites in plasmid p434cro2. This plasmid, which was used for expression and screening of 434 Cro variants, was constructed from four synthetic oligonucleotides that encoded 434 Cro and was cloned between the HindIII and EcoRI sites of pUC119. The C-terminal remnant of the lacZ′ gene of the pUC vector was removed by restriction digestion and polymerase fill-in reactions. This was found to decrease unwanted α-complementation from the lacZ remnant. In the case of the TRIM libraries, a PCR amplification of the synthetic DNA was performed before cloning into p434cro2. TRIM libraries were designed not to contain proline at the variegated positions. The sizes of the NNS and TRIM libraries were 32,768 and 6,859, respectively. The resulting combinatorial mixtures of mutagenic p434cro2 plasmids were then electroporated into E. coli DH5α. After electroporation, known volumes of cells were plated on appropriate antibiotic-containing media and incubated overnight at 37°C. Colonies were counted, and the total number of viable transformants for the electroporation step was calculated. Results of these control experiments indicated two- to fourfold redundancy of transformants for both libraries. Plasmid DNA was prepared from each library, and aliquots were used to transform DH5α containing an appropriate lacZ′ reporter plasmid (see below). A plasmid containing a 434 cro gene lacking the coding sequence of the HTH structure (residues K14 through G37) was constructed, and the resultant plasmid, p434Δcro, was used in place of full-length Cro-encoding plasmids in negative control experiments. The protein from this plasmid is referred to as CroΔ in the text.
Construction of lacZ reporter plasmids.
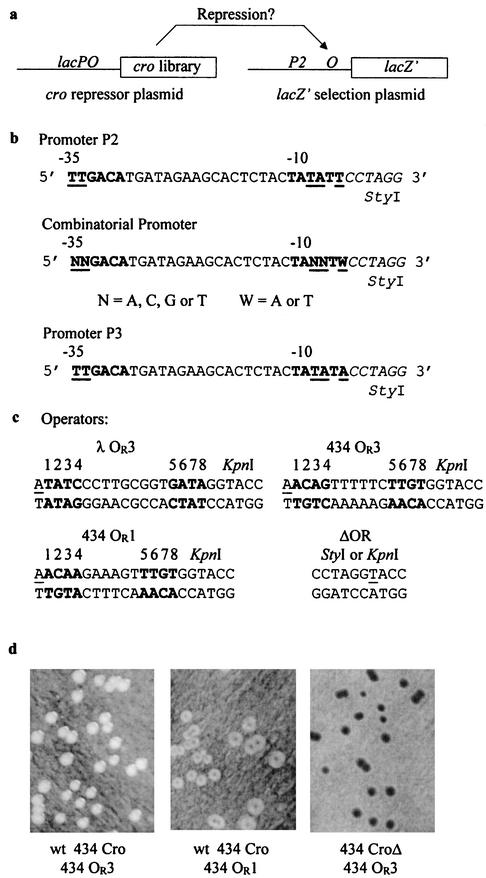
Reporter plasmids for the analysis of DNA binding in in vivo screening experiments by Cro proteins expressed from the repressor plasmids were constructed by using a modified pACYC184 vector that contained either the full-length lacZ+ gene or an α-peptide-encoding lacZ′ gene fragment under transcriptional control of a minimal promoter-operator cassette created from synthetic oligonucleotides (Fig. 1a). The lacZ gene or promoter-operator replaced the 3′ end of the Cmr gene of pACYC184, and a kanamycin resistance gene from transposon Tn903 (18) replaced the Tcr gene. Transcription from the Kanr and lacZ′ genes was designed to occur in opposite orientations, a consideration found to be important for effective screening. The synthetic promoter-operator sequences are given in Fig. 1b and c. Promoter sequences were optimized and balanced with the reporter gene product activity, as described in Results. The target operator sequences used in this study were the natural DNA binding sites OR1 and OR3 from 434 bacteriophage (434 OR1 and 434 OR3, respectively), the OR3 sequence of λ bacteriophage (λ OR3), and a construct lacking any additional DNA between the StyI site at the distal end of the promoter and the KpnI site at the distal end of the operator sequences (ΔOR; see Fig. 1c).
FIG. 1.
Screening for repressor variants. (a) An in vivo combinatorial two-plasmid system for the selection of functional DNA binding variants. The system consists of a repressor plasmid that encodes a variegated cro gene and a compatible reporter plasmid containing a lacZ gene under transcriptional control of a promoter and target operator sequence. LacZ-dependent β-galactosidase activities can be easily visualized on X-Gal-containing medium. (b) Partial sequence of the P2, P3, and combinatorial promoters used. The indicated mutations in the combinatorial promoter were synthesized with the degeneracies at the positions shown. P3 was selected for use with lacZ+ reporters. (c) Target operator sequences of the reporter plasmids used. The operator sequences were cloned between StyI and KpnI sites of the reporter plasmids. The putative +1 position of the promoter is underlined in each of the operators. Pseudopalindromic sequences of the operators are in bold type and are marked with numerals 1 to 8. The construct marked ΔOR shows the fused StyI-KpnI region of pP2ΔOR. (d) Black-and-white photographs of E. coli colonies containing various repressor and reporter plasmid combinations on plates containing X-Gal (and isopropyl-β-d-thiogalactopyranoside [IPTG] to induce 434 Cro transcription and chromosomal lacZΔ 77 15). Left panel, repressor plasmid p434cro2 (wt 434 Cro) and reporter plasmid pP2434OR3 (P2 434 OR3 operator-lacZ′). Middle panel, repressor plasmid p434cro2 (wt 434 Cro) and reporter plasmid pP2434OR1 (P2 434 OR1 operator-lacZ′). Right panel, repressor plasmid p434croΔ (434 Cro with HTH deletion) and reporter plasmid pP2434OR3 (P2 434 OR3 operator-lacZ′). The repression phenotypes of these colonies were designated +++ (white colonies) (left panel), + (white colonies with a light blue middle) (center panel), and −− (dark blue colonies) (right panel). Intermediate phenotypes, designated ++ and +−, had white colonies with less intense or more intense blue middles relative to the + phenotype (center panel).
In vivo and in vitro β-galactosidase assays.
β-Galactosidase activities derived from the full-length lacZ+ gene in the reporter plasmid or from α-complementation derived from the lacZ′ gene of the reporter plasmid and the lacZΔM15 gene of the host DH5α were measured in vivo by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) hydrolysis-oxidation on plates, essentially as described previously (24). The β-galactosidase activities of cell extracts were analyzed in vitro by the spectroscopic determination of hydrolysis of o-nitrophenyl-β-d-galactopyranoside (ONPG) as described by Miller (14). The ratios of repressed to unrepressed β-galactosidase activities (relative repression values) were calculated as described previously (13).
Combinatorial library screening.
Repression of lacZ gene transcription by members of combinatorial libraries of mutations of the 434 Cro gene expressed from pUC-derived repressor plasmids was monitored after transforming DH5α cells containing an appropriate lacZ reporter plasmid. The sequences of the 434 reporter plasmids were optimized in preliminary experiments so that the transcriptional levels of β-galactosidase were repressible by 434 Cro (see Results).
Mobility shift assays.
Mobility shift assays were performed, essentially as described previously (7), by using 5′-32P-end-labeled, double-stranded oligonucleotides as specific probes (40-mers, sequences as described in Results) in 50 mM TrisCl (pH 8.0), 1 mM EDTA, 30 mM KCl, 0.5 mM dithiothreitol, 10% (wt/vol) glycerol, and 0.5 μg of double-stranded poly(dI-dC)/μl. Cellular extracts containing Cro proteins were purified from DH5α strains containing appropriate plasmids, as described previously (3).
Determination of binding constants and Cro protein concentrations in cellular extracts.
Dixon and Webb first reviewed methods for the determination of Kd by the analysis of complex formation between proteins and ligands in 1958 (6). The amounts of Cro DNA binding protein present in the heterogeneous preparations used here were determined in preliminary mobility shift experiments (data not shown). These experiments were performed by titrating fixed amounts of protein extracts with increasing concentrations of 32P-labeled operator DNA. The amounts of free operator and Cro-operator complex at each OR3 concentration were determined by digital integration of the autoradiograms of the mobility shift gels. From the analyses of the resultant binding curves, the concentration of Cro repressor-operator complex at an infinite DNA ligand concentration as well as the Kd for binding was calculated. Because the stoichiometry of binding of DNA to the Cro dimer is known (1 to 1), the concentration of Cro-operator complex at an infinite operator concentration is equal to the concentration of Cro repressor that is active in DNA binding in the respective crude preparations. In these experiments, labeled λ OR3 DNA or 434 OR3 DNA were used with the RCT-Cro variant or wild-type (wt) 434 Cro preparations, respectively. It should be noted that no operator DNA binding was observed in control preparations where the CroΔ plasmid was used instead of the plasmids that encoded wt or variant Cro proteins.
RESULTS
It was of interest in this study to determine if we could create and identify new variants of HTH DNA binding proteins that specifically bound to targeted DNA sequences that were unrelated to those bound by the normal, wt protein. To achieve this goal, we created combinatorial libraries of mutations of the first three residues of the recognition helix of 434 Cro and developed and optimized a phenotypic screening assay to identify new variants with desired binding characteristics.
Combinatorial libraries of mutations of 434 Cro.
We constructed two types of combinatorial libraries at the three N-terminal positions of the recognition helix of 434 Cro (corresponding to Q28, Q29, and S30, numbered as described previously [28]) to test this hypothesis by using NNS mutagenesis (23) or trinucleotide codon mutagenesis (27) techniques. NNS codons encoded all amino acids and one stop codon at positions 28 to 30 (32 codons per variegated position and 32,768 mutations in total), while the TRIM cassette was designed to contain a mixture of codons for all amino acids except proline (19 codons per varied position, with a total of 6,859 mutations). Proline was not employed in order to conserve the α-helical secondary structure of the recognition helix.
In vivo screening of DNA binding by using lacZ-reporter plasmids.
In vivo transcriptional repression was assayed in cells containing both a reporter plasmid with either the full-length lacZ+ gene or the lacZ′ fragment under transcriptional control of a minimal promoter-operator sequence and a compatible repressor plasmid that expressed mutated or wt forms of 434 Cro protein (Fig. 1a). The promoter sequence chosen for use with the lacZ′ reporter (P2 promoter) (Fig. 1b) was based on a minimal promoter described by Benson et al. (1). Target operators (λ OR3, 434 OR1, 434 OR3, or ΔOR) (Fig. 1c) were cloned adjacent to the P2 promoter, and the entire promoter-operator was oriented upstream of the lacZ′ gene of the reporter plasmid. Transformation of E. coli DH5α strains containing these pACYC184-derived reporter plasmids with pUC-derived repressor plasmids that expressed wt or mutated 434 Cro proteins allowed in vivo discrimination of Cro DNA binding by blue-white LacZ repression screening on solid media containing X-Gal. With the P2 promoter-target operator-lacZ′ system, differential repression from 434 OR1 and OR3 promoters was easily discernible (Fig. 1d).
Figure 1d defines the phenotypes we observed for the repression of lacZ′ transcription. When wt 434 Cro was used in combination with the 434 OR3 operator DNA sequence or the 434 OR1 operator sequence, typical repressed LacZ− phenotypes were observed. Repression of 434 OR3 by wt 434 Cro was strongest, as expected (white-colony phenotype) (Fig. 1d, left panel), while repression of 434 OR1 was less extensive, as evidenced by the blue areas within the white colonies (Fig. 1d, middle panel). When a defective form of 434 Cro lacking the HTH structure (CroΔ) was expressed with the reporter plasmid, unrepressed lacZ′ transcription was observed (dark blue colony phenotype) (Fig. 1d, right panel). These phenotypes correlate well with the known characteristics of 434 Cro, binding to its natural operators (8, 10). 434 Cro did not repress λ OR3 DNA in these tests (data not shown).
Importance of balanced promoter-reporter gene strength for in vivo screening.
Unlike the results with the lacZ′ system, no repression in analogous blue-white assays with the P2 promoter and the full-length wt lacZ+ gene were observable; i.e., blue colonies were observed with coexpression of either wt 434 Cro or CroΔ proteins (Table 1). We assayed the β-galactosidase activities of cell extracts in vitro by using the hydrolysis of ONPG to further investigate this lack of observed in vivo repression of the P2 promoter-full-length lacZ+ reporter plasmid by coexpressed 434 Cro. The results of these in vitro β-galactosidase activities are also shown in Table 1. Coexpression of 434 Cro with the P2 promoter-lacZ+ combination with a 434 OR3 target operator resulted in significant repression in the in vitro enzyme assays (compare the results for promoter P2 lacZ+ cro+ with those for P2 lacZ+ croΔ [Table 1]). The relative repression ratio (the quotient of unrepressed β-galactosidase over repressed activity) is a useful comparison value for repression levels (13). The higher the ratio, the stronger the repression of transcription. For the P2 lacZ+ construction, this value was equal to 43. This value can be compared with the much higher value determined by using cultures with the P2-lacZ′ α-complementation system that showed repressed white colony phenotypes in in vivo assays. These results indicated that 434 Cro was binding its target operator and repressing lacZ+ transcription in the P2 lacZ+ constructions, but not to an extent that could be observed in in vivo blue-white assays.
TABLE 1.
β-Galactosidase and relative repression activities of reporter plasmidsa
| Promoter and lacZ and Cro variants | β-Galactosidase specific activity ± SEM | Relative repression | Colony phenotype |
|---|---|---|---|
| P2 lacZ′ cro+ | 0.58 ± 0.13 | 224 | White |
| P2 lacZ′ croΔ | 130 ± 22 | Blue | |
| P3 lacZ+cro+ | 154 ± 25 | 134 | White |
| P3 lacZ+croΔ | 20,634 ± 1,365 | Blue | |
| P2 lacZ+cro+ | 1,274 ± 258 | 43 | Blue |
| P2 lacZ+croΔ | 54,972 ± 4,236 | Blue |
β-Galactosidase activities and relative repression ratios were determined on DH5α cultures with the indicated promoter-434 OR3 operator-lacZ combinations coexpressed with either wt 434 Cro or the repressionally inactive CroΔ HTH deletion mutation. Specific activities of the β-galactosidase are expressed as nmol of ONPG hydrolyzed per min per mg of extract (13). The standard error of the mean is given. Each activity was determined at least four times. Relative repression ratios were calculated as previously described (13) as the quotient of the specific activity of β-galactosidase in cell extracts of unrepressed cultures (reporter plasmid coexpressed with CroΔ protein) over those determined from repressed cultures (reporter plasmid coexpressed with wt 434 Cro protein). The complete absence of repression would result in a relative repression value equal to 1. Colony phenotypes were assayed on agar plates containing IPTG and X-Gal as described in Materials and Methods.
Optimization of the P2-driven wt lacZ+ gene transcription by reducing the promoter strength was accomplished by a combinatorial mutagenesis strategy. Simultaneous mutagenesis of the P2 promoter at the positions in the Pribnow-Schaller box and −35 regions indicated in Fig. 1b was performed by cassette mutagenesis on the P2-434 OR3-lacZ+ reporter plasmid. Subsequent screening in in vivo blue-white colony assays of strains containing the mutated reporter plasmids in the absence of repressor plasmid led to the identification of 10 clones with less intense blue phenotypes relative to the parent P2 promoter-containing strain. In in vitro β-galactosidase assays, 9 of the 10 clones were observed to have less than 1% of the lacZ gene product activity of the unmutated control, while one of the clones had about 75% less activity. Sequencing of this latter plasmid showed that it was mutated in only the last base of the Pribnow Schaller box relative to promoter P2, i.e., TATATT to TATATA. This promoter was named P3. The P3-434OR3-lacZ+ plasmid was assayed with 434 Cro- or CroΔ- expressing repressor plasmids in in vivo blue-white phenotype assays and in vitro enzyme assays. The results of these tests are shown in Table 1. Repression of transcription by using plasmids with the wt lacZ+ driven by the P3 promoter was easily observed in in vivo blue-white assays. In in vitro β-galactosidase assays, a relative repression ratio threefold higher than that determined for the otherwise isogenic P2-promoter construction was observed. These results demonstrate the importance of balancing the strength of gene expression through variation of promoter and reporter gene product activities with the ability to observe repression and therefore target DNA binding. Failure to so balance expression would likely result either in the identification of variants that bind weakly or nonselectively to the target sequence when repression was too easily achieved or in the failure to identify variants with high-affinity, specific target DNA binding characteristics if the total activity of transcription, translation, and enzymatic activities overwhelmed the phenotypic assay. Neither of these outcomes would yield useful binding variants. Because of the balanced expression observed here for the P2 promoter-lacZ′ and the optimized P3-wt lacZ+ constructions, these plasmids were deemed to be effective for screening combinatorial libraries for new binding specificities.
Importance of operator placement for transcriptional repression.
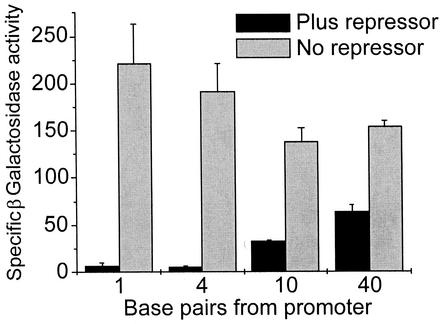
Experiments were conducted to determine the optimal placement of a target operator sequence. When we varied the distance between the promoter and operator sequences of the lacZ′ reporter plasmid by inserting multiples of a self-complementary linker DNA sequence (pCTAGGAGCTC) in the StyI site of the P2-434 OR3-lacZ′ reporter plasmid, we were able to identify clones with 10- or 40-bp inserts between the promoter and OR3 target operator. T4 polymerase fill-in of StyI-restricted P2-434 OR3- lacZ′ reporter plasmid generated a third variant with a 4-bp insertion between promoter and operator. In in vivo blue-white assays, fully repressed white phenotypes were observed only with the original plasmid and the 4-bp insertion. Insertion of 10 and 40 bp between the promoter and operator increased the amount of blue observable in the colonies (light blue [+] and blue-middle [+−] phenotypes, respectively, as described in the legend to Fig. 1). Quantification of repression by in vitro enzyme assays (Fig. 2) confirmed the positional effects on operator placement and showed that there was also a slight context effect of the operators; i.e., β-galactosidase activity in the absence of coexpressed repressor (plus CroΔ) varied by up to ∼25%. These results suggested that we could target new operator sequences for binding by 434 Cro variants by positioning a palindromic target DNA sequence adjacent to the promoter. Those mutated repressors that bind targeted sequences with apparent affinities equivalent to that of the wt 434 Cro binding to 434 OR1 or its high-affinity OR3 site could be selected by picking colonies having in vivo phenotypes equivalent to those shown in the left or center panels of Fig. 1d.
FIG. 2.
Effects on repression as the distance between promoter and target operator were varied. β-Galactosidase activities were measured in cell extracts of strains containing either wt 434 Cro or CroΔ (no repressor) with target 434 OR3 operators separated from the promoter by the indicated number of base pairs. Error bars represent standard errors of the mean.
Alteration of the binding specificity of 434 Cro to the λ OR3 sequence.
Comparison of the sequences of the λ and 434 OR3 operator (Fig. 1b) shows that these operators are dissimilar in almost all characteristics other than their pseudopalindromic nature. Importantly, 434 Cro showed no repression of the λ OR3 operator in in vivo blue-white phenotype assays and only extremely weak repression of the sequence in in vitro ONPG β-galactosidase activity tests. The relative repression value observed in ONPG tests was determined to be 3 ± 0.3, indicative of extremely weak repressional activity of 434 Cro on the λ DNA sequence. Control experiments indicated that repression of the λ OR3 sequence by λ Cro protein was clearly observed in both in vivo and in vitro assays (data not shown), eliminating the possibility that the λ OR3 was unrepressible due to some unforeseen problem.
Screening of combinatorial libraries of 434 Cro for λ OR3 DNA binding.
The NNS and TRIM combinatorial libraries were transformed into E. coli strains containing the P2-lacZ′ reporter plasmid with the λ OR3 operator sequence as the binding target. In initial in vivo screening for repression of the λ OR3 reporter plasmid by Cro variants, 18,000 colonies from the NNS library and 10,000 colonies from the TRIM library were visually examined. Colonies showing any possible repressed phenotype, i.e., +−, +, or ++ (Fig. 1d), were selected. This broad cut was performed so as not to miss any possible repressors in these initial studies. A second screening of these putative 434 Cro variants encoding plasmids for λ operator repression was individually performed for each identified clone to ensure that the phenotypes initially identified were not derived from mutations other than those in the 434 cro gene.
Two clones from the NNS library and 9 clones from the TRIM library were identified that had positive repressor phenotypes with the λ OR3 reporter plasmid in both primary and secondary screening experiments. The 434 cro genes of the 11 variants were sequenced to determine the mutations present. Table 2 shows the phenotypes of the mutant strains carrying the Cro mutations listed in order of the strength of repression on λ OR3. The mutants were named after the deduced amino acids at positions 28 to 30. The mutational combinations found showed amino acid substitutions at positions 28, 29, and 30, as expected. Three of the 11 variants also had mutations downstream of the HTH motif, presumably due to errors induced during library construction.
TABLE 2.
434 Cro variants that bind λ OR3 DNA and their corresponding in vivo blue-white phenotypesa
| Mutations at Cro positions 28 to 30 | Phenotype of the Cro variant with the indicated target operator sequence used in the reporter plasmid
|
|||
|---|---|---|---|---|
| λ OR3 | 434 OR3 | 434 OR1 | ΔOR | |
| RGT | ++ | + | + | +− |
| RCT | + | +− to − | +− to − | +− to − |
| HGC | + | +− | +− | + |
| RKS | + to +− | +− | +− | − |
| LKA | + to +− | +− | +− | − |
| CKT | +− | +++ | +− | − |
| WHT | +− | − | − | − |
| TIQ | +− | +− to − | +− to − | − |
| ILHb | +− | +− to − | +− to − | − |
| IITb | +− | +− to − | +− or − | − |
| ILWb | + | +− | +− | − |
DH5α containing the indicated repressor plasmid and P2-LacZ′ reporter plasmids with the indicated target operator were tested for repression in in vivo blue-white assays. Phenotypes on X-Gal-containing medium are as described in the legend for Fig. 1. The RGT and ILW mutations were identified from NNS library screens. The remainder were identified in screens of libraries.
Additional mutations in the protein sequences were found outside of the recognition helix downstream of the BstEII site that was used for mutagenesis cassette cloning. These mutations likely arose during construction of the libraries.
In order to gain insight into the ability of the Cro variants to discriminate between several different operator sequences, repressor phenotypes of the 11 Cro variants were examined by using in vivo blue-white assays with each of the λ OR3-, 434 OR3-, 434 OR1-, and ΔOR-containing reporter plasmids (Table 2). One of the Cro variants, encoding amino acids R28C29T30 at the N terminus of its recognition helix (RCT-Cro), strongly repressed the λ OR3 and very weakly repressed or did not repress the 434 OR3, 434 OR1, or ΔOR sequences. This variant showed the highest degree of discrimination of any of the identified variants. It is important to point out that the lack of repression of the ΔOR plasmid by RCT-Cro demonstrated that the RCT variant did indeed bind to the λ OR3 operator and not to other elements of the lacZ′ transcriptional unit. The RGT mutant showed a strong repression of the λ OR3 sequence, but RGT-Cro also repressed the 434 and ΔOR operators and was therefore less discriminatory. Other mutations were weaker and were less discriminatory and are not further discussed here. Neither the ranking of the mutants in Table 2 nor their repressional phenotypes changed between the primary and secondary screens. That the RCT and RGT variants were not identified in the NNS library screens likely reflects the fact that we screened only about 55% of the total candidates in this library.
Quantification of repression of the RCT-Cro variant.
Assays of β-galactosidase-dependent hydrolysis of ONPG by cell extracts of the RCT-Cro variant with the λ OR3 or 434 OR3 reporter plasmids yielded relative repression ratios (± standard errors of the means) of 60.0 ± 7.2 and 16.9 ± 4.2, respectively, based on the results of at least five independent experiments conducted in duplicate. RCT-Cro repressed λ OR3 significantly better than the 434 OR3 operator, indicating that selective repression of the λ sequence was achieved. With wt 434 Cro, we observed very strong repression of the 434 OR3 sequence and strong repression of the 434 OR1 operator, but very weak repression of λ OR3 (relative repression values of 156 ± 11, 84 ± 10, and 3.0 ± 0.3, respectively). Comparison of the extent of repression by wt 434 Cro and the RCT variant shows that the RCT-Cro repressed λ OR3 about as well as the wt 434 Cro repressed 434 OR1. The RCT variant of 434 Cro repressed λ OR3 about fourfold better than it repressed 434 OR3. These results of ONPG hydrolysis demonstrated that the binding specificity of RCT-Cro was reversed with respect to wt 434 Cro.
Binding characteristics of the RCT and wt 434 Cro proteins.
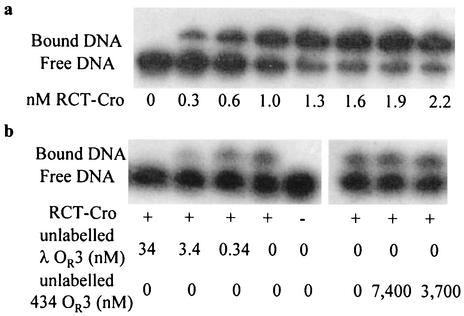
Cellular lysates of E. coli expressing RCT-Cro or wt 434 Cro were analyzed by electrophoretic mobility shift assays to document the binding characteristics more closely. Figure 3a shows a titration of the RCT variant against a fixed concentration of labeled λ OR3 DNA. The apparent Kd for the binding of RCT-Cro to λ OR3 DNA under these conditions was found to be 0.8 nM. In experiments using wt 434 Cro and the 434 OR3 operator, a similar value (1.6 nM) was determined. We were not able to directly observe any binding of wt 434 Cro to λ OR3 or of RCT-Cro to 434 OR3 DNA by using these mobility shift assays. Furthermore, no complex formation between the RCT-Cro variant and the 434 OR3 binding was observed, even at very high RCT-Cro and 434 OR3 concentrations. The limits to detection in these assays were estimated by assuming that ≥10% of the total label in the shifted band could be detected. Under the maximal concentration of Cro protein used, a limit for the Kd could be estimated at ≥1,000 nM.
FIG. 3.
Mobility shift assays of RCT-Cro and wt 434 Cro. (a) Titration of 50 pM 32P-labeled λ OR3 DNA with the indicated concentrations of RCT-Cro. The positions of bound and free DNA are indicated. (b) Competition mobility shift assays with 50 pM 32P-labeled λ OR3 DNA, 0.6 nM RCT-Cro, and the indicated amounts of unlabeled λ OR3 or 434 OR3 DNA.
In further attempts at characterizing binding effects of RCT-Cro to 434 OR3, we performed competition binding mobility shift studies with RCT-Cro by using unlabeled λ and 434 OR3 DNA in the presence of 32P-labeled λ OR3 DNA. Figure 3b shows that formation of the RCT-Cro-labeled λ OR3 complex was significantly decreased by inclusion of fivefold or more unlabeled λ OR3 relative to RCT-Cro in the incubation, while the RCT-Cro-λ OR3 complex formation was not detectably lessened by including a concentration of 434 OR3 more than 11,500 times that of RCT-Cro. In terms of DNA concentrations, the addition of 2,000-fold more 434 OR3 DNA above the amount of unlabeled λ OR3 in the incubation did not decrease observable binding of RCT-Cro to λ DNA. These results indicate that the RCT-Cro variant exhibited a remarkable change in specificity from the 434 OR3 sequence to the λ OR3 sequence. The affinity of the RCT-Cro variant for λ DNA was equivalent to that of wt 434 Cro for its preferential binding sequence, 434 OR3.
DISCUSSION
In this study we identified 434 Cro variants that are able to specifically bind a previously unrecognized DNA target sequence. We used relatively small combinatorial libraries of the recognition helix of a HTH binding protein and an optimized screening system to reach this goal. The results of this study contradict previous reports, where no identification of such binding specificity changes was obtained or only relatively minor specificity changes involving single positions in the DNA were identified (8, 9). The reasons why others may not have been able to identify new HTH protein variants with significantly different binding specificities may be related to two phenomena. The first possibility is that the systems used by others to screen or select such new variants may not have been optimized in terms of the strength of gene expression versus the ability of a biologically functional DNA binding protein to repress the phenotype. Insensitivity in in vivo assays due to unbalanced reporter gene activity relative to repressional ability of the DNA binding protein was demonstrated in this report in the results of promoter strength experiments shown in Table 1. Even though repression was observed with the highly sensitive in vitro enzymatic assays, no in vivo repressor phenotypes were found for the strong promoter-strong reporter combination. It is possible that other systems that were not optimized in this respect might fail to identify HTH variants with new binding specificities. For those methods dependent on the repression of an activity detrimental to the survival of the cell (negative selection schemes), the use of large libraries, for example, those with >106 members, may also select for random mutations that inactivate the selection processes. In unpublished work with a variety of such systems, we have found that the high rate of false positives in these types of selections are a major impediment to the utility of such negative selection systems. The lack of negative genetic pressure in phage display systems used to identify protein variants that bind new DNA sequences may support this latter point (see reference 17 for a HTH protein application and reference 21 for a review for zinc fingers).
It is apparent from the results presented here that the best λ DNA binding 434 Cro variant identified, RCT-Cro, was not selected simply because it had a changed binding affinity for all DNA and an increase in nondiscriminatory binding, i.e., an increase in nonspecific high-affinity binding characteristics. On the contrary, RCT-Cro bound λ OR3 DNA with an affinity equal to that with which its parent protein bound to its preferred 434 DNA sequence, and it was selective in this binding, discriminating well between λ and 434 OR3 sequences. Such selective binding by the RCT-Cro variant was found in in vivo assays (Table 2), in vitro cell extract assays (described in the text), as well as in mobility shift competition assays (Fig. 3). The in vitro repression data derived from the ONPG hydrolysis assays using RCT-Cro and the data from in vivo phenotype assays indicated that the RCT Cro repressed the λ target about as well as wt 434 Cro represses the 434 OR1 operator. In summary, selective binding and functional repression of the new, desired target DNA was observed with the RCT Cro, albeit with some lowered overall selectivity in some assays compared with wt 434 Cro.
The observations of the much higher selectivity for the RCT Cro variant from the results of the competition mobility shifts (Fig. 3) compared to the results of in vivo phenotype assays and the β-galactosidase activity tests should be commented on. The differences might reflect the sensitivity of inhibiting enzymatic reactions that are likely running at near-maximal velocities for the conditions present in the latter two tests, while the competition mobility shift assay results might have a much higher threshold for visualizing differences in banding intensities. It is also very likely that the RCT sequence itself plays a deciding role in the differences observed. It is quite possible that the cysteine residue of the RCT variant undergoes some oxidation during the in vivo phenotype and ONPG hydrolysis tests. The oxidation of the RCT cysteine would be favored because of its proximal location to the arginine residue, where the deprotonation of the sulfhydryl might be enhanced. This, in turn, would favor the formation of disulfide bonds or a negative thiolate, which would be less receptive to DNA binding. The electrophoretic mobility shift assays would be much less prone to this problem because of the inclusion of dithiothreitol in the medium. In addition, the methods used for determining the amounts of active repressor present in cell lysates would not detect inactivated (oxidized) repressor. The oxidation would therefore only lower the specific activity of the protein in the in vivo and ONPG hydrolysis tests and would thereby account for the observed differences with the results of electrophoretic mobility shift assays.
None of the HTH proteins, including the wt 434 Cro, 434 repressor, λ Cro, and λ repressor, encodes the amino acid sequence RCT in their respective recognition helices. The prediction of R, C, and T for mutations at positions 28 to 30 in 434 Cro in a hypothetical mutagenesis experiment based on a rational design scheme of protein-DNA interactions for Cro would not have been likely prior to this experimental report. The results demonstrate the power of combinatorial approaches that follow biological activities such as transcriptional repression in attempts at discovering new interactions between DNA and protein that are responsible for specificity. It will be of interest in the future to gain higher-resolution structural information about the interactions between the λ OR3 DNA sequence and the HTH structures of the RCT Cro variant, perhaps through nuclear magnetic resonance or crystallographic studies.
The results presented in this report that show that combinatorial mutagenesis of just three residues of the recognition helix of 434 Cro can be successfully used with an optimized screening system to identify new protein variants that bind previously unrecognized targeted DNA sequences with nanomolar affinity and high selectivity suggest that HTH proteins can be used to generate proteins with new DNA binding specificities.
The discovery of additional new DNA-protein interactions through the identification of mutations that are not readily predictable, such as the RCT-Cro variant, should enhance our knowledge of protein-nucleic acid interactions and may allow the identification of DNA binding protein variants that functionally affect DNA regulatory sequences important in disease and industrial and biotechnological processes.
Acknowledgments
We acknowledge B. Virnakäs for supplying the TRIM oligonucleotide and Colette Kananura, Oliver Haase, and Jochen Baβler (Univ. Kaiserslautern) for technical help. The authors also thank S. Cohen (Stanford) for providing plasmid pACYC184 and C. Wolberger (Johns Hopkins) for helpful discussions.
K.F. was supported by a graduate stipend from the Landesgraduiertenförderungsgesetz Rheinland-Pfalz.
REFERENCES
- 1.Benson, N., P. Sugiono, S. Bass, L. Medelman, and P. Youderian. 1986. General selection for specific DNA-binding activities. Genetics 114:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan, R. G., and B. W. Matthews. 1989. The helix-turn-helix DNA binding motif. J. Biol. Chem. 264:1903-1906. [PubMed] [Google Scholar]
- 3.Bruice, T. W., J. G. Wise, D. S. E. Rosser, and D. S. Sigman. 1991. Conversion of λ phage cro into an operator-specific nuclease. J. Am. Chem. Soc. 113:5446-5447.
- 4.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 135:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo, Y., and A. Klug. 1997. Physical basis of a protein-DNA recognition code. Curr. Opin. Struct. Biol. 7:117-125. [DOI] [PubMed] [Google Scholar]
- 6.Dixon, M., and E. C. Webb. 1979. Enzymes, 3rd ed., p. 120. Academic Press, New York, N.Y.
- 7.Fried, M. G., and D. M. Crothers. 1981. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 9:6505-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison, S. C., and A. K. Aggarwal. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 59:933-969. [DOI] [PubMed] [Google Scholar]
- 9.Huang, L., T. Sera, and P. G. Schultz. 1994. A permutational approach toward protein-DNA recognition. Proc. Natl. Acad. Sci. USA 91:3969-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koudelka, G. B., and C.-Y. Lam. 1993. Differential recognition of OR1 and OR3 by bacteriophage 434 repressor and Cro. J. Biol. Chem. 268:23812-23817. [PubMed] [Google Scholar]
- 11.Koudelka, G. B. 1991. Bending of synthetic bacteriophage 434 operators by bacteriophage 434 proteins. Nucleic Acids Res. 19:4115-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koudelka, G. B., S. C. Harrison, and M. Ptashne. 1987. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. Nature 326:886-889. [DOI] [PubMed] [Google Scholar]
- 13.Lehming, N., J. Sartorious, B. Kisters-Woike, B. von Wilcken-Bergmann, and B. Müller-Hill. 1990. Mutant lac repressors with new specificities hint at rules for protein-DNA recognition. EMBO J. 9:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. H. 1972. Experiments in molecular genetics, 1st ed., p. 352. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Mondragón, A., and S. C. Harrison. 1991. The phage 434 Cro/OR1 complex at 2.5 Å resolution. J. Mol. Biol. 219:321-334. [DOI] [PubMed] [Google Scholar]
- 16.Mondragón, A., C. Wolberger, and S. C. Harrison. 1989. Structure of phage 434 Cro protein at 2.35 A resolution. J. Mol. Biol. 205:179-188. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson, M. T., M. C. Mossing, and M. Widersten. 2000. Functional expression and affinity selection of single-chain Cro by phage display: isolation of novel DNA-binding proteins. Protein Eng. 13:519-526. [DOI] [PubMed] [Google Scholar]
- 18.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 19.Pabo, C. O., and R. T. Sauer. 1984. Protein-DNA recognition. Annu. Rev. Biochem. 53:293-321. [DOI] [PubMed] [Google Scholar]
- 20.Pabo, C. O., and R. T. Sauer. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 61:1053-1095. [DOI] [PubMed] [Google Scholar]
- 21.Pabo, C. O., E. Peisach, and R. A. Grant. 2001. Design and selection of novel cys2his2 zinc finger proteins. Annu. Rev. Biochem. 70:313-340. [DOI] [PubMed] [Google Scholar]
- 22.Padmanabhan, S., M. A. Jimenez, C. Gonzalez, J. M. Sanz, G. Gimenez-Gallego, and M. Rico. 1997. Three-dimensional solution structure and stability of phage 434 Cro protein. Biochemistry 36:6424-6436. [DOI] [PubMed] [Google Scholar]
- 23.Reidhaar-Olson, J. F., and R. T. Sauer. 1988. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science 241:53-57. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Simoncsits, A., M.-L. Tjörnhammer, S. Wang, and S. Pongor. 1999. Single-chain 434 repressors with altered DNA-binding specificities. Genetica 106:85-92. [DOI] [PubMed] [Google Scholar]
- 26.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 27.Virnekäs, B., G. Liming, A. Plückhun, K. C. Schneider, G. Wellnhofer, and S. E. Moroney. 1994. Trinucleotide phosphoramidites: ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res. 22:5600-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wohlberger, C., Y. C. Dong, M. Ptashne, and S. C. Harrison. 1988. Structure of a phage 434 Cro/DNA complex. Nature 335:789-795. [DOI] [PubMed] [Google Scholar]
- 29.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]