Abstract
The genus Salmonella consists of over 2,200 serovars that differ in their host range and ability to cause disease despite their close genetic relatedness. The genetic factors that influence each serovar's level of host adaptation, how they evolved or were acquired, their influence on the evolution of each serovar, and the phylogenic relationships between the serovars are of great interest as they provide insight into the mechanisms behind these differences in host range and disease progression. We have used an Salmonella enterica serovar Typhimurium spotted DNA microarray to perform genomic hybridizations of various serovars and strains of both S. enterica (subspecies I and IIIa) and Salmonella bongori to gain insight into the genetic organization of the serovars. Our results are generally consistent with previously published DNA association and multilocus enzyme electrophoresis data. Our findings also reveal novel information. We observe a more distant relationship of serovar Arizona (subspecies IIIa) from the subspecies I serovars than previously measured. We also observe variability in the Arizona SPI-2 pathogenicity island, indicating that it has evolved in a manner distinct from the other serovars. In addition, we identify shared genetic features of S. enterica serovars Typhi, Paratyphi A, and Sendai that parallel their unique ability to cause enteric fever in humans. Therefore, whereas the taxonomic organization of Salmonella into serogroups provides a good first approximation of genetic relatedness, we show that it does not account for genomic changes that contribute to a serovar's degree of host adaptation.
There are currently over 2,200 serovars in the genus Salmonella. However, classification based upon serotype and other physiological properties has provided limited information regarding the genetic relationship of the serovars and moreover is not sufficient for making disease associations (6, 52). Chromosomal DNA hybridization experiments (13, 30, 31) and multilocus enzyme electrophoresis (MLEE) disclosed that typical serovars share from 85 to 100% of their genetic information and resulted in the categorization of Salmonella into eight subspecies—I, II, IIIa, IIIb, IV, V, VI, and VII—of which subspecies V has been designated Salmonella bongori. Salmonella enterica comprises the remaining subspecies (the members of which we will use the serovar designation alone in the present study) (6, 7, 9, 10, 44, 50, 51). MLEE also confirmed that, whereas there is some justification for using serotype as a measure of relatedness, it is generally not a reliable assessment of phylogenic organization. This observation is indicative of the horizontal exchange of chromosomal genes leading to the emergence of new serovars (6).
More than 60% of all Salmonella strains identified and 99% of the serovars responsible for disease in warm-blooded animals are members of subspecies I. The other Salmonella subspecies, in particular subspecies IIIa (Arizona) and S. bongori, are associated with disease in cold-blooded organisms with Arizona and are occasionally responsible for systemic disease in humans. What is particularly intriguing about subspecies I serovars is that their ability to cause disease in animals encompasses a spectrum of host specificity and disease severity. For example, serovar Typhi causes a systemic disease (typhoid) only in humans and higher primates, whereas serovar Enteritidis produces a self-limiting gastrointestinal disease in many different animals. Serovar Typhimurium causes a gastrointestinal disease in a wide variety of animals and yet is also responsible for a typhoid-like disease in the mouse. In addition, specific isolates have been been found in cases of severe disease in pigeons (42). One of the prevailing questions in Salmonella research today concerns the identification of genetic factors that confer upon these highly related serovars their ability to colonize, and in some cases to cause disease in, a wide variety of animal hosts.
The release of two S. enterica sequences, the imminent completion of six other serovars and strains, and the recent funding to sequence additional serovars and strains (http://www.sanger.ac.uk/Projects/Salmonella/) has initiated a new era of comparative genomics in Salmonella biology (14). This sequence information will provide a valuable resource from which we can begin to dissect the features of Salmonella that are both shared and distinct between serovars and to start exploring how and why differences arose. However, sequencing is still a laborious and expensive technique, making it difficult to obtain answers concerning the genetic composition of serovars, strains, or newly emerged variants of interest in a timely manner. DNA microarray technology provides a useful adjunct to current techniques for the assessment of differences and changes in bacterial genetic content. Indeed, this approach has already been utilized in a variety of bacteria to probe for differences between clinical isolates, vaccine strains, species diversity, and disease endemicity (reviewed in references 22 and 27).
We used a Typhimurium spotted DNA microarray to compare the genomes of a number of Salmonella serovars in order to clarify their genetic relationship and identify features that may serve to profile the serovars and that may correspond to host range and disease. Twenty-four strains of 12 S. enterica serovars and two S. bongori strains were analyzed. We found general agreement with previously published MLEE-based observations but also describe here a more distant relationship of Arizona from subspecies I than was previously determined, as well as identify genetic features that suggest a common origin for the human systemic disease-associated serovars Typhi, Paratyphi A, and Sendai.
MATERIALS AND METHODS
Bacterial strains and preparation of genomic DNA.
Serovars and strains used are detailed in Table 1. Strains from the SARA (7), SARB (9), and SARC (10) reference sets were obtained from the Salmonella Genetic Stock Center, University of Calgary. Serovar Paratyphi A strain 1PA was a kind gift from Bruce Stocker (Stanford University, Stanford, Calif.) and is a wild-type, clinical isolate originally obtained from Peter O'Hanley in Jakarta, Indonesia. SL1344 is an aromatic-independent serovar Typhimurium serovar that is virulent in mice (54). LT2, the sequenced serovar Typhimurium strain, is a common laboratory strain (35, 48). CT18, the serovar Typhi strain recently sequenced, is the multidrug-resistant strain isolated from a young Vietnamese girl (39). TY2 is a common laboratory strain of serovar Typhi. Arizona strains 2323, 2334, 2335, and 5705A were a gift from Donald G. Guiney (University of California, San Diego) and were originally described elsewhere (33). All strains were maintained and grown in Luria-Bertani media, and plates were incubated at 37°C. The identity of the serovars (except for 1PA, 2323, 2334, 2335, and 5705A) was confirmed by standard laboratory agglutination tests by using the Pro-Lab Diagnostics Vision Salmonella Slide Agglutination Antisera (Richmond Hill, Ontario, Canada). Genomic DNA was extracted from overnight cultures by using the CTAB (hexadecyltrimethylammonium bromide) method (2) or Qiagen Genomic-Tips kit according to manufacturer's instructions (Qiagen, Inc.).
TABLE 1.
Strain information and the percentage of SL1344 genes present for each strain
| Strain | SGSC no. | Description | Subspecies | ETa | Designation in this study | No. of arrays | % SL1344 genes present |
|---|---|---|---|---|---|---|---|
| SL1344 | Typhimurium (strain on array) | I | SL1344 | 3 | 100.0 | ||
| SARA 1 | 2182 | Typhimurium | I | Tm1 | Tm1 | 2 | 100.0 |
| LT2 | Sequenced Typhimurium | I | LT2 | 3 | 99.9 | ||
| SARA 6 | 2186 | Typhimurium | I | Tm2 | Tm2 | 2 | 99.5 |
| SARA 53 | 2233 | Paratyphi B | I | Pb3 | Pb3 | 2 | 93.8 |
| SARB 48 | 2505 | Paratyphi C | I | Pc1 | Pc1 | 3 | 93.7 |
| SARB 16 | 2473 | Enteritidis | I | En1 | En1 | 3 | 93.4 |
| SARB 12 | 2469 | Dublin | I | Du1 | Du1 | 3 | 93.0 |
| SARB 4 | 2461 | Choleraesuis | I | Cs1 | Cs1 | 3 | 92.8 |
| SARA 41 | 2221 | Paratyphi B | I | Pb1 | Pb1 | 2 | 92.2 |
| CT18 | Sequenced Typhi | I | CT18 | 3 | 91.5 | ||
| SARB 63 | 2520 | Typhi | I | Tp1 | Tp1 | 3 | 91.1 |
| SARB 51 | 2508 | Pullorum | I | Pu3 | Pu3 | 3 | 91.1 |
| TY2 | Lab Typhi | I | TY2 | 3 | 91.0 | ||
| SARB 64 | 2521 | Typhi | I | Tp2 | Tp2 | 3 | 90.4 |
| 1 PA | Wild type Paratyphi A | I | 1PA | 2 | 89.8 | ||
| SARB 58 | 2515 | Sendai | I | Se1 | Se1 | 2 | 89.7 |
| SARB 21 | 2478 | Gallinarum | I | Ga2 | Ga2 | 3 | 89.7 |
| SARB 42 | 2499 | Paratyphi A | I | Pa1 | Pa1 | 3 | 89.2 |
| SARC 11 | 3100 | S. bongori | V | 1Bg | 3 | 83.7 | |
| SARC 12 | 3103 | S. bongori | V | 2Bg | 3 | 83.4 | |
| SARC 5 | 3061 | Arizona | IIIa | Az | 3 | 77.5 | |
| 2323 | Arizona | IIIa | Az2323 | 1 | 76.4 | ||
| 2335 | Arizona | IIIa | Az2335 | 1 | 76.3 | ||
| 2334 | Arizona | IIIa | Az2334 | 1 | 74.7 | ||
| 5705A | Arizona (clinical isolate) | IIIa | Az5705A | 1 | 73.5 |
ET, electrophoretic type. This refers to the electrophoretic profile of the isolate with regard to a number of metabolic enzymes. A particular serovar can have multiple electrophoretic types depending on the number of alleles that are present for any given enzyme that is assayed.
Construction of the SL1344 microarray.
Our LT2-based array was constructed prior to annotation of the sequence. Open reading frames (ORFs) were selected based on GenBank sequences, ORF prediction was done by using Glimmer (47), and annotation was kindly provided by Robert A. Edwards. An algorithm was written that selected the most 3′ region of each ORF that contained no significant similarity to any other gene (70% identity over 70 nucleotides was considered to be significant) as identified by NCBI BLAST and is available upon request from the author as RedHat Linux executable Perl and batch scripts. Primer pairs were selected from this unique region by using Primer3 (http://www-genome.wi.mit.edu/genome_software/other/primer3.html) (45). Genes that lacked unique regions were subjected to successive iterations with less-stringent cross-hybridization criteria. Primers were synthesized by Illumina, Inc. (San Diego, Calif.) and the Protein and Nucleic Acid Facility at Stanford University. Products were PCR amplified with SL1344 genomic DNA. Amplified products were analyzed on agarose gels and ranged from 70 to 1,496 bp, with 248 bp as the median and 346 bp as the average length. PCR product preparation, polylysine glass slide preparation, printing, and array postprocessing was performed as previously reported (15). The final array consisted of 7,372 spots corresponding to 4,169 ORFs and 414 intergenic regions, with many ORFs represented by multiple spots, either as duplicate or distinct amplicons.
Hybridizations.
Genomic DNA probe preparation was performed essentially as previously described (46), except that 1.5 μg of genomic DNA and one-eighth of one reaction vial of FluoroLink Cy3 or Cy5 monofunctional dye (Amersham) was used per reaction. SL1344 genomic DNA was the reference DNA for all hybridizations and labeled with Cy3, whereas sample DNAs were labeled with Cy5. Multiple hybridizations were performed for the majority of strains analyzed to reduce the effects of variation in array quality. The separate labeling reactions were pooled after each respective Cy dye incorporation step and then again divided into aliquots to minimize inconsistencies in probe generation. The probes were resuspended in 18 μl of Tris-EDTA (TE), 2 μl of 20 mg of yeast tRNA/ml, 4.25 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; 3.4×, final concentration), and 0.75 μl of 10% sodium dodecyl sulfate (0.3%) and then denatured for 2 min at 99°C and centrifuged briefly at 13,800 × g. Probes were hybridized to the array at 50°C for 16 h and washed as described previously (15). Arrays were scanned with a GenePix 4000A scanner (Axon Instruments, Redwood City, Calif.) and processed by using GenePix Pro 3.0. All raw datasets are available from the Stanford Microarray Database (http://genome-www5.stanford.edu/MicroArray/SMD/) (53).
Data analysis.
Normalized data from all 64 hybridizations were filtered for spot quality (Cy3 net mean intensity of ≥350) and downloaded from the Stanford Microarray Database according to their mean log2 Cy5/Cy3 (logRAT2N) ratios for analysis. In addition, spots that gave invalid results for more than 20% of the strains were removed. This allowed for the retrieval of 4,494 spots. The data set was additionally filtered to remove spots whose serovar Typhi CT18 hybridization results were inconsistent with what was expected (see below) and ultimately yielded 4,122 spots corresponding to 3,353 annotated ORFs (73% of the LT2 genome) and 20 intergenic regions. The word “gene” will be used throughout in reference to the ORF that each spot corresponds to unless otherwise specified. The multiple arrays for each serovar or strain were averaged across the datasets. This and all subsequent data analysis were done by using Microsoft Excel and a microarray genomic analysis program called GACK (27a). Briefly, this program is capable of dynamically generating cutoffs for present/absent (conserved/divergent) gene analysis for each array hybridization and functions independently of any normalization process that would otherwise be strongly influenced by differences between the reference strain and serovar of interest. A user manual for the GACK program has been provided (http://falkow.stanford.edu/whatwedo/software).
The hybridization data for the three CT18 arrays were downloaded, filtered by using the same parameters indicated above except that a 66% good data filter was applied to ensure at least two datum points for each spot, averaged across the three arrays, and analyzed by using GACK. The percent similarity for the highest high-scoring profile of each amplicon in CT18 was obtained by using WU-BLAST. This percent similarity was compared to the averaged logRAT2N of the hybridization data. Of the 4,712 spots that were retrieved, 85.4% of the data set gave unequivocal results. A total of 687 (14.6% of the data set) had hybridization results contrary to what was expected from the percent similarity analysis. A total of 250 of these were false positives, whereas 160 of were false negatives. This list of 410 spots (8.7% of the data set) was used as an additional filter for the data (see above). Another 277 spots either had percent similarities in the uncertain/slightly divergent range or yielded hybridization results such that they could not be assigned as present or absent and so were not included in the filter.
Genome order analysis was performed by organizing the spots for the entire data set (averaged and processed with GACK) in their genome order according to the LT2 annotation and viewed with TREEVIEW (16). Clustering was performed by using the Pearson correlation, noncentered metric algorithm of XCLUSTER (Gavin Sherlock; http://genome-www.stanford.edu/∼sherlock/cluster.html). Clustered datasets were viewed in TREEVIEW.
Core genes analysis was performed by applying the GACK program to the data set and analyzing the output in Excel to identify spots that were present across all of the serovars and strains. The percent present analysis to analyze the percentage of SL1344 genes that is shared by each serovar was calculated by taking the data set above and determining the number of absent, missing, and present spots for each serovar.
Final datasets have been made available (http://falkow.stanford.edu/whatwedo/supplementarydata/) in a format compatible for viewing with TREEVIEW as noted in the text.
RESULTS AND DISCUSSION
Analysis of the CT18 hybridizations indicates the microarray's level of sensitivity.
The data for the three serovar Typhi CT18 hybridizations were directly compared to expected hybridization results as assessed by the percent identity of each SL1344 amplicon (based on the LT2 sequence) to the CT18 genome sequence. From this analysis, we were able to determine that CT18 genes with ≥93% identity to the amplicon (87% of the data set) would be detected as present/conserved on our array, whereas genes that diverge from more than 77% identity would be assigned as absent/divergent (11% of the data set). Genes that share 92 to 78% identity with the amplicon were classified as uncertain/slightly divergent (2% of the data set). In addition, a percent present analysis was performed on CT18 by using the complete serovar data set (see Materials and Methods and below) and revealed that 91.5% of SL1344 is shared with CT18 (Table 1), a level comparable to the 88% observed in DNA hybridizations involving serovar Typhi strains 643 and LT2 (13), the 89% seen upon direct comparison of the CT18 and LT2 genome sequences (35), and the 90% seen with CT18 hybridized on an LT2 microarray (40).
Genome order analysis reveals discrete regions of variability between serovars and strains.
Salmonella possesses a large amount of horizontally acquired genetic information in the form of prophages and pathogenicity islands (distinguished by their absence in Escherichia coli), within many of which lie genes that have been shown to play a role in pathogenesis (19, 21, 23, 25, 34, 35, 37, 39). It has also been observed that a large proportion of the Typhimurium genome consists of regions of genes with related functions, contributing to the proposal that many horizontally acquired genes are more stably maintained in bacteria when genes of related function are transferred along with them (28). We were therefore interested in seeing whether or not discrete patterns of genes could be identified by arranging our microarray data with respect to the LT2 annotated gene order. This organization of data highlights distinct regions where multiple contiguous genes share the same hybridization pattern regardless of possible translocation or inversion of the region.
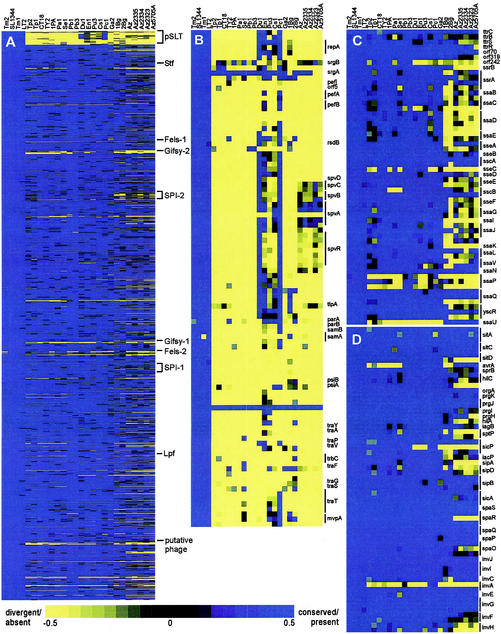
An overview of the Salmonella serovar DNA hybridizations to the SL1344 DNA microarray presented in LT2 gene order is provided in Fig. 1A. It is immediately apparent is that the pSLT Typhimurium virulence plasmid, which is required for systemic disease in the mouse, is predominantly missing from all non-Typhimurium serovars except for serovars Dublin and Paratyphi C (Fig. 1B). It has been demonstrated by Boyd and Hartl (8) by Southern hybridization that the spv gene cluster in pSLT associated with systemic disease is distributed throughout subspecies I and, when present, is always found in a virulence plasmid. In addition, these authors demonstrated that the spv cluster is also found in subspecies II, IIIa, IV, and VIII, but in these instances it is always chromosomally located. The observation that the plasmid and spv region are present in serovars Dublin and Paratyphi C and absent in S. bongori corresponds with what has been previously observed, and comparison of the spv amplicon sequences to the Paratyphi A and Typhi genome sequences confirms that the cluster is absent in these serovars as well (data not shown). However, whereas this previous work examining the same serovar Enteritidis, Pullorum, and Choleraesuis SARB strains as in our study indicates that all possess the spv cluster, our data show that there is some heterogeneity within the cluster, as well as throughout the plasmid, for these serovars. One explanation for the differences between the two data is that, in the Southern hybridizations, a probe against the entire spv cluster was used, whereas our work reflects hybridizations to short, gene-specific amplicons. Interestingly, all serovars appear to be missing or have divergent portions of the virulence plasmid outside of the spv cluster. The array hybridization pattern for Arizona indicates that it possesses the spv locus but is missing the majority of the pSLT plasmid, a finding consistent with previous observations regarding the chromosomal location of spv in this serovar (8). However, our data indicate that there is some variability in the cluster. This is supported by a finding that, although spvRBC of an Arizona clinical isolate are conserved with respect to subspecies I, spvD was absent and spvA possessed a frameshift mutation at the C terminus of the gene that results in a larger protein (8, 33).
FIG.1.
Genome order analysis of the serovar microarray data. Multiple arrays for each serovar and strain have been averaged, analyzed with GACK, and organized with respect to the LT2 genome order. Each row corresponds to a specific spot on the array, whereas columns represent strains analyzed and are labeled according to the designations in Table 1. The color scheme is located at the bottom of the figure, with the brightest yellow corresponding to spots that are absent/divergent with high certainty, the brightest blue indicating spots that are present/conserved with the greatest certainty, black indicating spots are uncertain or slightly divergent, and gray indicating missing data. (A) The entire data set of 4,122 spots. Indicated are the pSLT virulence plasmid, the SPI-1 and SPI-2 pathogenicity islands, the Stf and Lpf fimbrial operons, and the Fels and Gifsy prophages. Enlargement of the regions corresponding to the pSLT virulence plasmid (B), the SPI-2 pathogenicity island (C), and the SPI-1 pathogenicity island (D) are also shown. Specified are the annotated genes within each region, where vertical bars indicate multiple spots on the array that correspond to the same gene. Not indicated are putative genes, unannotated ORFs, and intergenic regions. This data set is available online (http://falkow.stanford.edu/whatwedo/supplementarydata/, Appendix 1).
The regions of the genome that correspond to prophages and fimbrial clusters are also notably variable across the serovars (http://falkow.stanford.edu/whatwedo/supplementarydata/, Appendices 2 and 3). Previous work had indicated that the Fels prophages were specific to the serovar Typhimurium LT2 strain (1). We show that Fels-1 is absent in all serovars except for the four serovar Typhimurium strains, whereas Fels-2 is absent in the Tm2 strain but present in the other three serovar Typhimurium strains and mostly present in the serovar Enteritidis and Sendai strains and all of the serovar Typhi strains, a finding consistent with what has been seen previously (40). The presence of both Gifsy prophages in all of the Typhimurium serovars is consistent with what had been previously reported (20, 21, 40). The putative phage located at ORFs STM4196 to STM4219 is present in serovars Paratyphi C and Choleraesuis and all of the serovar Typhimurium strain, but is absent in the other serovars. The microarray results for the fimbrial clusters are summarized elsewhere (http://falkow.stanford.edu/whatwedo/supplementarydata/, Appendix 3) and correspond well with the observations made by Porwollik et al. (40) and by the direct comparison of genomes (14).
In addition to these previously characterized regions, there were multiple variable regions throughout the genome that are composed of uncharacterized putative proteins. These regions are potentially of great interest since they represent clusters of genes that may have a serovar-specific association. For example, the region including genes STM4488 to STM4497 houses a putative type II restriction enzyme that is only present in serovar Typhimurium. The region from STM4418 to STM4436 includes sugar transporters, putative endonucleases, and putative cytoplasmic proteins that are present only in serovars Typhimurium and Paratyphi B. STM4258 to STM4264 represent a region of putative genes absent only in Arizona.
Another interesting general observation that can be made from Fig. 1A is the large number of genes missing in the S. bongori and Arizona serovar hybridizations that correlates with the current organization of these two serovars into different subspecies. A major difference between these two serovars (subspecies V and IIIa) and the other serovars (subspecies I) is emphasized in the SPI-2 region (Fig. 1C). We observed here that S. bongori does not hybridize to the majority of the SPI-2 spots, whereas Arizona has a heterogeneous pattern of hybridization, indicating an appreciable degree of sequence variability. This could indicate that the SPI-2 island in Arizona has undergone some modification, possibly consistent with its preferred niche in cold-blooded animals. Previous work looking at the distribution of these pathogenicity islands by using the SARC set demonstrated the absence of SPI-2 in S. bongori but its presence across the other subspecies, including Arizona (36). Since the probe used in the Southern hybridizations spanned the entire SPI-2 region, the sensitivity to detect changes in individual genes is lower than to our ability to probe on a gene-by-gene basis. In contrast, the microarray hybridization pattern for SPI-1 (Fig. 2D ) indicates that it is present across the serovars, a finding consistent with what has been previously shown (36). One SPI-1 gene, avrA, is absent only in the human-specific serovars and S. bongori (Fig. 2D). This gene has been previously shown to encode an effector molecule secreted by the SPI-1 system that bears protein sequence similarity to the Yersinia pseudotuberculosis YopJ protein and the plant pathogen Xanthomonas campestris pv. Vesicatoria-secreted avirulence protein AvrRxv, which is thought to play a role in determining plant host range (26) (see below). With the exception of Sendai (which is associated with enteric fever in humans), the hybridization results show the same distribution for avrA as predicted from Southern hybridization studies (41). Three other SPI-1 genes—sipA, sptP, and sipB—gave microarray hybridization patterns that corresponded with that expected in the SARB set (41).
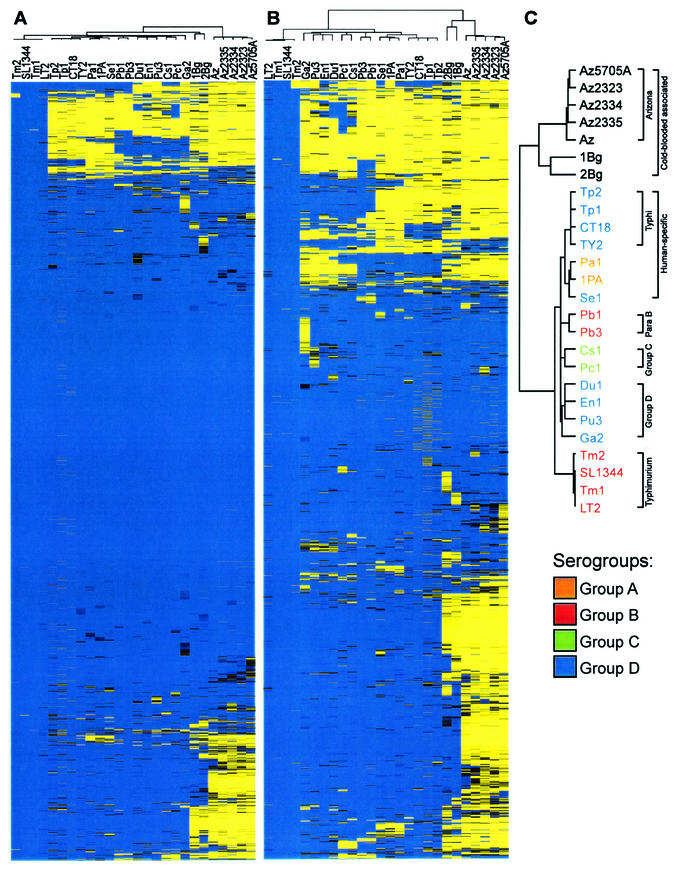
FIG.2.
Hierarchical cluster analysis of the microarray data by XCLUSTER. (A) Clustering of the entire data set by both serovar and gene. Shown at the top is the unrooted tree for the relationship of the serovars. (B) Clustering of the data set is as described in panel A except that the 2,244 core genes have been removed. (C) Enlargement of the tree generated in panel B. Color scheme is as described in Fig. 1. The data set in panel B is available elsewhere (http://falkow.stanford.edu/whatwedo/supplementarydata/, Appendix 5).
Analysis of conserved genes identifies a set of Salmonella core genes.
A list of 2,244 spots (54% of the data set) shared by all of the serovars that we have designated as “core” genes is available (http://falkow.stanford.edu/whatwedo/supplementarydata/, Appendix 4). In summary, the list is comprised of a number of genes involved in typical cellular functions such as metabolism, biosynthesis, DNA replication, transcription, translation, ion transport, and cell division. Various structural operons for flagella (fli and flh operons), fimbriae (bcf operon), and lipopolysaccharide synthesis (rfa operon and wzxE) were also conserved. Components of the phs operon involved in hydrogen sulfide production were found to be present across the serovars. Hydrogen sulfide production is a biochemical feature common to all Salmonella strains and is widely employed by clinical laboratories to distinguish it from other Enterobacteriaceae (55), and it has been shown to be a Salmonella-specific operon (40). A number of genes implicated in virulence (slrP, pagP, sopB, the SPI-1 pathogenicity island encoding a type III secretion system required for invasion of epithelial cells) and transcriptional regulatory components (PhoPQ) are shared across the serovars. Whereas the SPI-2 pathogenicity island is purportedly absent from S. bongori and is revealed by our data to be variable in Arizona, some components of the island also appear in the list of core genes (data not shown). The presence or absence of these genes in S. bongori will eventually be verified by the S. bongori sequencing project (http://www.sanger.ac.uk/Projects/Salmonella/).
Measuring serovar relatedness with respect to SL1344 reveals that Arizona is more distantly related than currently thought.
Previous DNA reassociation experiments indicated that subspecies I serovars shared between 85 and 100% of their genetic information with the reference LT2 strain and that Arizona (subspecies IIIa) shared on the order of 70 to 80%, indicating that it is highly related to all of the other salmonellae but is genetically distinct. A third class of “atypical” Salmonella (from subspecies II and IV) had DNA-DNA association ranges that fell between the two classes (13). We were therefore interested in assessing whether or not the microarray data would give us a measure of the relatedness of Salmonella serovars to each other.
Based on an analysis of the microarray data, we estimated the percentage of SL1344 genes contained within each serovar (Table 1). A total of 89 to 100% of SL1344 was shared with the subspecies I serovars and >99% with all other serovar Typhimurium strains. The percentages for serovars Typhi (90.4 to 91.5%), Paratyphi A (89.2 to 89.8%), and Paratyphi B (92.2 to 93.8%) are comparable to genomic comparison values reported for an LT2 microarray and the comparison of annotated sequences (35, 40). A total of 73.5 to 77.5% of the SL1344 genome hybridized with Arizona, with the clinical isolate 5705A sharing the smallest number of genes. These values are consistent with the percent similarities reported in the DNA reassociation experiments (13). Interestingly, SL1344 shares 83% of its genetic information with both S. bongori strains analyzed, placing it in an intermediate range between Arizona and subspecies I. This result indicates that Arizona is the most divergent of the serovars analyzed in the present study, whereas S. bongori assumes a more intermediate degree of difference. A slightly lower percent present value has been previously reported for Arizona relative to S. bongori (83 and 85%, respectively) (35, 40) but not to the same extent as we observe here. MLEE analysis has implicated S. bongori as the most divergent of the Salmonella (10, 44), and the lack of an SPI-2 pathogenicity island indicates that it may have diverged before S. enterica acquired SPI-2 (3). However, the possibility exists that S. bongori originally possessed the island but eliminated it earlier in its evolution due to its restricted niche in cold-blooded animals. Arizona, on the other hand, although considered a pathogen of reptiles, can cause severe disease in humans (24, 57). The island in Arizona may therefore have evolved to maximize its contribution to Arizona's host range. Arizona's genome as a whole may have proceeded down a distinct evolutionary path, leading to its ability to cause disease in such disparate hosts as human and cold-blood animals.
Serogroup and disease-associated relationships are revealed by hierarchical clustering.
We employed a clustering analysis tool to determine how the assayed serovars are associated with one another and to identify patterns in gene composition that drive the associations. The unrooted tree generated from clustering the entire data set by both serovars and genes (Fig. 2A) reveals that Arizona is the most distant serovar relative to subspecies I, followed by the two S. bongori strains, which is consistent with the percent present analysis in Table 1. All of the subspecies I serovars fall under the same major node, whereas all of the serovar Typhi strains and Typhimurium strains form their own subnodes (Fig. 2A). The large blue region in the center of the image illustrates the appreciable proportion of core genes that are shared across the serovars.
Clustering of a data set from which the core genes (54% of the entire data set) were removed produced a different unrooted tree (Fig. 2B and C) without altering the composition of the gene clusters (data not shown). The differences between the serovars were emphasized, and a greater level of resolution with regard to the relationships between the strains and serovars, particular with the subspecies I serovars, was achieved. For example, Tm2 appears to be genetically distinct from the other three strains of serovar Typhimurium. In addition, serovar Gallinarum associated with other group D serovars (Pullorum, Enteritidis, and Dublin), and Arizona and S. bongori emphasized their distinction from the subspecies I serovars by forming a distinct node (Fig. 2C). Analysis of the data by PHYLIP Camin-Sokal parsimony analysis (http://evolution.genetics.washington.edu) generated an unrooted tree that revealed a similar relationship between the serovars and strains (data not shown).
Molecular signatures characterize groups of serovars.
Previous MLEE work focusing on the human enteric fever-associated serovars could not resolve a relationship between them (9, 50). However, we observe the clustering of serovars Typhi, Paratyphi A, and Sendai into a shared node, suggesting shared genetic features may exist for these serovars (Fig. 2C). A cluster of genes absent from S. bongori, Arizona, and the human enteric fever-associated serovars but present in the other warm-blooded disease-associated serovars is shown in Fig. 3. This pattern of absent/divergent genes suggests that a common Salmonella ancestor possessed these genes but that they were lost or diverged specifically in the enteric fever-associated serovars and in Arizona and S. bongori. Alternatively, perhaps the common Salmonella ancestor lacked these genes and the nonenteric fever-associated serovars acquired them, whereas the human enteric fever-associated serovars did not.
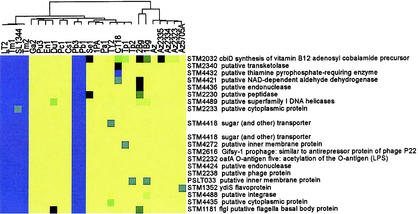
FIG. 3.
A cluster pattern showing genes that are absent in the serovars associated with human enteric fever and cold-blooded animals but present in the other warm-blooded disease-associated serovars. This cluster was pulled out from the larger image shown in Fig. 2B. Indicated is the annotation gene number (STM) and annotated gene information. The color scheme is as described in Fig. 1. Intergenic regions are not shown. Putative genes are indicated by the term “put.”
Within the human-specific node, the serovar Typhi strains fall into a separate grouping, whereas the two Paratyphi A (serogroup A) strains and Sendai (serogroup D) form another group (Fig. 2C). This organization of the serovar Typhi strains is consistent with the highly clonal nature of this serovar (44). The close association of serovars Paratyphi A and Sendai has also been previously noted (9, 50). Additional evidence supporting a close relationship between these two serovars arises from their serotypes. For example, in addition to sharing the same phase-1 flagellar antigen, serovar Paratyphi A also possesses the genetic information for the same phase-2 antigen as serovar Sendai, although it is not expressed unless under selective pressure (Bruce A. Stocker, unpublished data). Both share the same minor O antigens and differ only in the major serogroup-determining antigen. However, both group A and group D serovars proceed through the same intermediate during O-antigen synthesis, at which point conversion of a paratose sugar residue to a tyvelose residue by the product of the rfbE locus generates the group D O antigen (43). It is possible that serovar Paratyphi A, in addition to preferentially not expressing the phase-2 flagellar antigen, either does not possess or does not express rfbE. Our data for these two serovars substantiates a close genetic relationship, since they share the same profile for present and absent genes with the exception of 166 genes (4% of 4,124 total in a separate Paratyphi A and Sendai specific analysis [data not shown]).
Figure 4 is a node of genes specific to group B serovars, which includes Typhimurium and Paratyphi B. Contained within this group B signature is the gene for OafA responsible for the O-antigen acetylation step that generates the group B serotype antigen. Interestingly, even though these serovars share a strong group B pattern, Paratyphi B (which causes a typhoid-like disease in humans) forms a node in the tree distinct from Typhimurium (Fig. 2C). In contrast, MLEE analysis indicated that serovars Typhimurium and Paratyphi B are relatively similar (9, 50). There may be additional disease-associated genetic features that drive the separation of Typhimurium and Paratyphi B (Fig. 4). Nevertheless, the existence of a specific signature for this serogroup indicates that shared genetic factors do not necessarily correlate with disease phenotype.
FIG. 4.
(A) A cluster indicating genes that are conserved only in group B serovars. This cluster was pulled out from the larger image shown in Fig. 2B. Indicated is the annotation information from the LT2 sequence including the gene number (STM number) and the gene name if available. Not indicated are intergenic regions. The color scheme is as described in Fig. 1.
The organization of the two serogroup C serovars into a single node (Fig. 2C) illustrates a second situation where clustering is not as strongly influenced by a disease correlation. Serovar Paratyphi C is associated with human typhoid-like disease, whereas serovar Choleraesuis, which is occasionally associated with serious human disease, is generally considered a pathogen of swine. This association between Paratyphi C and Choleraesuis has been previously observed and was attributed to the two serovars sharing an ancestor, with Choleraesuis subsequently gaining the Vi antigen during the course of its evolution (50). The organization of serovars Enteritidis, Dublin, Gallinarum, and Pullorum into the same node is another example of organization according to serogroup irrespective of disease correlation. The association between serovars Enteritidis and Dublin has been previously observed by MLEE (9). In addition, it has also been demonstrated by sequence analysis of various loci that serovars Gallinarum and Pullorum (both are nonmotile serovars associated with disease in fowl) are highly related and share a recent common lineage (32). This organization may be driven by a gene cluster unique to these serogroup D (non-Typhi) serovars.
The presence of a cluster of absent genes unique to serovars associated with human-specific enteric fever (Fig. 3) indicates that there may have been a convergent evolution of the serovars into their disease niche or that they originated from a common ancestral serovar. This cluster includes genes from the Lpf fimbrial operon previously shown to be absent in serovars Typhi, Paratyphi A, and Sendai (••Appendix 3••) (56), the sodC-1 gene encoding a periplasmic superoxide dismutase (located within the Gifsy-2 prophage region), and the avrA gene mentioned above. Lpf has been implicated in mediating attachment of serovar Typhimurium to the Peyer's patches in the mouse intestine, a critical early step in the disease process (4, 5). SodC-1 is one of three periplasmic superoxide dismutases found in Salmonella and one of two horizontally acquired from lysogenic bacteriophages (21). The periplasmic location of the Sod enzymes positions them for the neutralization of the toxic effects of exogenous superoxide produced by the host, a feature that is critical for persistence in macrophages and pathogenesis in mice (11, 18, 49). Mutation of either sodC-1 or sodC-2 in serovar Typhimurium has been shown to attenuate the bacteria during mouse infection while mutation of both genes leads to an even more severely attenuated phenotype (17). Interestingly, although mutation of either sodC-1 or sodC-2 in serovar Choleraesuis leads to attenuation in mice, mutation of both does not result in a more severe phenotype (49). While all Salmonella serovars possess sodC-2, the distribution of sodC-1 has been shown to be restricted to a smaller set of serovars, including isolates of the serovars Dublin, Enteritidis, Gallinarum, Pullorum, and Paratyphi B (17), a finding in agreement with our observations and its location in a prophage. Taking into account the important role that this enzyme plays in resistance to the host superoxide response, it has been proposed that there may be selective pressure for the horizontal acquisition of additional Sod genes (49). AvrA, as mentioned above, possesses homology to the Y. pseudotuberculosis-secreted effector YopJ and the plant pathogen X. campestris-secreted protein AvrRxv. YopJ has been shown to interfere with the host immune response by modulating the mitogen-activated protein kinase and NF-κB signaling pathways to prevent the release of proinflammatory cytokines and block the antiapoptotic pathway (38). AvrRxv is a member of a family of plant-secreted “avirulence” proteins whose presence elicits a hypersensitivity response from the plant that limits the spread of disease and thereby restrict host range (29). Recent work has demonstrated a potential role for serovar Typhimurium AvrA in modulating virulence in vertebrates by interfering with proinflammatory NF-κB activation (12).
Concluding remarks.
There are several features of microarray-based analysis that make it a particularly attractive complement to the MLEE method for the study of strain and serovar relationships. The MLEE method is limited by the number of enzymes that can be assayed, whereas the microarray allows us to probe most of the genome of a strain of interest. In addition, MLEE has been determined to have a limited ability to accurately assess some relationships due to the presence of different alleles giving the same pattern of mobility (52). Also, by being restricted to alleles of metabolic enzymes, the contribution of horizontally acquired genes and other larger changes to the evolution of a particular genome are overlooked.
There are also several limitations with spotted DNA microarrays. Single-nucleotide polymorphisms are not currently detectable with spotted DNA arrays; for instance, the limit of resolution for conserved genes in the microarray used for these studies (based on a CT18 analysis) is ∼93% identity. Consequently, caution needs to be employed when applying the term “divergent” to microarray comparison studies since this cutoff is higher than what is currently used in direct sequence comparisons. As a result of this limitation, small genetic changes during serovar evolution and minor gene differences between serovars may not be detected. These restrictions could be addressed by using other independent approaches to verify observations. For example, phylogenic observations made with the microarray could be substantiated and complemented by MLEE or sequence comparison. Microarrays are also incapable of identifying regions present in the serovar of interest but absent from the strain or serovar from which the array was constructed. For example, a comprehensive analysis of clinical isolates of serovar Typhi and correlating them to disease outcome may not be feasible with a serovar Typhimurium array since Typhi possesses genetic elements important in disease (such as multidrug resistance genes and the Vi capsule) that are absent in Typhimurium. Finally, genomic comparisons with microarrays reveal nothing about the effect of gene expression on a serovar or strain's ability to cause disease or persist in a particular environment. One potentially powerful study to address this would be to synchronize the growth of strains of interest and perform RNA expression comparisons at defined time points or under specific growth conditions. Nevertheless, microarrays provide a powerful, high-capacity means to characterize serovars and strains and clearly complement other techniques.
While this study was in preparation, Porwollik et al. described genomic comparisons of Salmonella serovars performed by using a serovar Typhimurium LT2 array (40). We have found that the results of that study and ours are both highly comparable and complementary. We had chosen to incorporate serovars into our analysis that were primarily representative of those in subspecies I associated with disease in humans and animals and had included Arizona and S. bongori as references for their occasional disease association and phylogenic distance, respectively. Porwollik et al. selected serovars that represent all of the subspecies of both S. enterica and S. bongori and incorporated genome information from other genera in order to trace the history of gene acquisition and loss that may have contributed to the emergence of Salmonella and its the widely diverse serovars. One major difference in observation between our analyses is the location of Arizona relative to both S. bongori and subspecies I. This difference can most likely be attributed to the respective designs of the arrays. Whereas Porwollik et al. constructed their array to include entire annotated ORFs and used LT2 as the source of genomic DNA, our group built an SL1344 genomic DNA array to include shorter amplicons corresponding to the region of each ORF that was the most unique and therefore less likely to cross-hybridize. Since the two arrays are inherently different in their design, some dissimilarity would be expected in the results.
We have shown here that a Typhimurium microarray is a useful tool for the genomic comparison of Salmonella serovars. With a few exceptions, the observations made here correspond well with those made in MLEE analysis and DNA association experiments, lending credibility to the use of this tool in inferring phylogenic relatedness. The observation that Arizona is more distant from the subspecies I serovars than currently thought is where our results have significantly deviated from previous studies. In general, although we have observed some correlation between the descriptive taxonomy of the serovars and their genetic relatedness, there are additional genetic loci that influence the clustering of some of the serovars together in a manner that reflects their disease associations. An example of this is the clustering of human enteric fever-associated serovars Typhi, Paratyphi A, and Sendai that we observed despite their distribution in two distinct serogroups. The identification of genes that drive the clustering of these serovars may provide information regarding the role of these genes in determining host range and disease phenotype. Thus, while serogrouping may provide a good first approximation of the genetic relationship between the serovars, it is not necessarily predictive of the phylogenic organization of Salmonella (6), nor does it account for the horizontal exchange of genetic information that could alter the serotype of a Salmonella or influence its evolution into a particular disease or host affiliation. Application of microarray-based genomic comparison could therefore serve as a tool to clarify, if not modify, the way in which we understand the organization of Salmonella.
Acknowledgments
We gratefully acknowledge Dolores Cunanan, Denise Monack, Igor Brodsky, and Inna Bilis for their contribution to the construction of the microarray; Robert A. Edwards for the annotation of the LT2 genome; and Craig Cummings for assistance with primer design. In addition, we thank David Botstein, Timothy McDaniel, Karen Guillemin, and Nina Salama for assistance and advice during the planning stages of the array. We also thank Elizabeth A. Joyce and Erin C. Gaynor for critical reading of the manuscript and Bruce A. D. Stocker for being an invaluable resource.
This work was supported by the Ellison Medical Foundation, grant AI26195 from the National Institutes of Health, and Digestive Disease Center grant DK56339. K.C. is a National Science Foundation Graduate Research Fellow. C.C.K. is supported by a Howard Hughes Medical Institute Predoctoral Fellowship and a Stanford Graduate Fellowship. S.B. and G.D. are supported by The Wellcome Trust and the Biotechnology and Biological Sciences Research Council. C.S.D. is supported by a fellowship from the American Cancer Society.
REFERENCES
- 1.Affolter, M., C. Parent-Vaugeois, and A. Anderson. 1983. Curing and induction of the Fels 1 and Fels 2 prophages in the Ames mutagen tester strains of Salmonella typhimurium. Mutat. Res. 110:243-262. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M. 1995. Preparation of genomic DNA from bacteria, p. 2.11-2.22. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology, 3rd ed. John Wiley & Sons, Inc., New York, N.Y.
- 3.Baumler, A. J., R. M. Tsolis, T. A. Ficht, and L. G. Adams. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler, A. J., R. M. Tsolis, and F. Heffron. 1996. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. USA 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, W. M. Frerichs, I. K. Wachsmuth, K. Ferris, A. C. McWhorter, J. G. Wells, A. Cravioto, et al. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltran, P., S. A. Plock, N. H. Smith, T. S. Whittam, D. C. Old, and R. K. Selander. 1991. Reference collection of strains of the Salmonella typhimurium complex from natural populations. J. Gen. Microbiol. 137(Pt. 3):601-606. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, E. F., and D. L. Hartl. 1998. Salmonella virulence plasmid: modular acquisition of the spv virulence region by an F-plasmid in Salmonella enterica subspecies I and insertion into the chromosome of subspecies II, IIIa, IV and VII isolates. Genetics 149:1183-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, E. F., F. S. Wang, P. Beltran, S. A. Plock, K. Nelson, and R. K. Selander. 1993. Salmonella reference collection B (SARB): strains of 37 serovars of subspecies I. J. Gen. Microbiol. 139(Pt. 6):1125-1132. [DOI] [PubMed] [Google Scholar]
- 10.Boyd, E. F., F. S. Wang, T. S. Whittam, and R. K. Selander. 1996. Molecular genetic relationships of the salmonellae. Appl. Environ. Microbiol. 62:804-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canvin, J., P. R. Langford, K. E. Wilks, and J. S. Kroll. 1996. Identification of sodC encoding periplasmic [Cu,Zn]-superoxide dismutase in Salmonella. FEMS Microbiol. Lett. 136:215-220. [DOI] [PubMed] [Google Scholar]
- 12.Collier-Hyams, L. S., H. Zeng, J. Sun, A. D. Tomlinson, Z. Q. Bao, H. Chen, J. L. Madara, K. Orth, and A. S. Neish. 2002. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-κB pathway. J. Immunol. 169:2846-2850. [DOI] [PubMed] [Google Scholar]
- 13.Crosa, J. H., D. J. Brenner, W. H. Ewing, and S. Falkow. 1973. Molecular relationships among the salmonellae. J. Bacteriol. 115:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 15.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 16.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Baumler, U. Ochsner, T. Testerman, S. Bearson, J. C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu,Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrant, J. L., A. Sansone, J. R. Canvin, M. J. Pallen, P. R. Langford, T. S. Wallis, G. Dougan, and J. S. Kroll. 1997. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol. Microbiol. 25:785-796. [DOI] [PubMed] [Google Scholar]
- 19.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueroa-Bossi, N., E. Coissac, P. Netter, and L. Bossi. 1997. Unsuspected prophage-like elements in Salmonella typhimurium. Mol. Microbiol. 25:161-173. [DOI] [PubMed] [Google Scholar]
- 21.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald, J. R., and J. M. Musser. 2001. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 9:547-553. [DOI] [PubMed] [Google Scholar]
- 23.Groisman, E. A., and H. Ochman. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791-794. [DOI] [PubMed] [Google Scholar]
- 24.Hall, M. L., and B. Rowe. 1992. Salmonella arizonae in the United Kingdom from 1966 to 1990. Epidemiol. Infect. 108:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 26.Hardt, W. D., and J. E. Galan. 1997. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc. Natl. Acad. Sci. USA 94:9887-9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joyce, E. A., K. Chan, N. Salama, and S. Falkow. 2002. Redefining bacterial populations: a post-genomic reformation. Nat. Rev. Genet. 3:462-473. [DOI] [PubMed] [Google Scholar]
- 27a.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:RESEARCH0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leach, J. E., and F. F. White. 1996. Bacterial avirulence genes. Annu. Rev. Phytopathol. 64:156-179. [DOI] [PubMed] [Google Scholar]
- 30.Le Minor, L., M. Y. Popoff, B. Laurent, and D. Hermant. 1986. Characterization of a 7th subspecies of Salmonella: S. choleraesuis subsp. indica subsp. nov. Ann. Inst. Pasteur Microbiol. 137B:211-217. [PubMed] [Google Scholar]
- 31.Le Minor, L., M. Veron, and M. Popoff. 1982. The taxonomy of Salmonella. Ann. Microbiol. 133:223-243. [PubMed] [Google Scholar]
- 32.Li, J., N. H. Smith, K. Nelson, P. B. Crichton, D. C. Old, T. S. Whittam, and R. K. Selander. 1993. Evolutionary origin and radiation of the avian-adapted non-motile salmonellae. J. Med. Microbiol. 38:129-139. [DOI] [PubMed] [Google Scholar]
- 33.Libby, S. J., M. Lesnick, P. Hasegawa, M. Kurth, C. Belcher, J. Fierer, and D. G. Guiney. 2002. Characterization of the spv locus in Salmonella enterica serovar Arizona. Infect. Immun. 70:3290-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus, S. L., J. H. Brumell, C. G. Pfeifer, and B. B. Finlay. 2000. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2:145-156. [DOI] [PubMed] [Google Scholar]
- 35.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 36.Ochman, H., and E. A. Groisman. 1996. Distribution of pathogenicity islands in Salmonella spp. Infect. Immun. 64:5410-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 38.Orth, K. 2002. Function of the Yersinia effector YopJ. Curr. Opin. Microbiol. 5:38-43. [DOI] [PubMed] [Google Scholar]
- 39.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 40.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prager, R., S. Mirold, E. Tietze, U. Strutz, B. Knuppel, W. Rabsch, W. D. Hardt, and H. Tschape. 2000. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical isolates of Salmonella enterica. Int. J. Med. Microbiol. 290:605-617. [DOI] [PubMed] [Google Scholar]
- 42.Rabsch, W., H. L. Andrews, R. A. Kingsley, R. Prager, H. Tschape, L. G. Adams, and A. J. Baumler. 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect. Immun. 70:2249-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raetz, C. R. H. 1996. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles, p. 1035-1063. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 44.Reeves, M. W., G. M. Evins, A. A. Heiba, B. D. Plikaytis, and J. J. Farmer III. 1989. Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J. Clin. Microbiol. 27:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 46.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanderson, K. E., A. Hessel, and B. A. D. Stocker. 1996. Strains of Salmonella typhimurium and other Salmonella species used in genetic analysis, p. 2496-2503. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 49.Sansone, A., P. R. Watson, T. S. Wallis, P. R. Langford, and J. S. Kroll. 2002. The role of two periplasmic copper- and zinc-cofactored superoxide dismutases in the virulence of Salmonella choleraesuis. Microbiology 148:719-726. [DOI] [PubMed] [Google Scholar]
- 50.Selander, R. K., P. Beltran, N. H. Smith, R. Helmuth, F. A. Rubin, D. J. Kopecko, K. Ferris, B. D. Tall, A. Cravioto, and J. M. Musser. 1990. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect. Immun. 58:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selander, R. K., L. J., and K. Nelson. 1996. Evolutionary genetics of Salmonella enterica, p. 2691-2707. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C.
- 53.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford microarray database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, B. P., M. Reina-Guerra, S. K. Hoiseth, B. A. Stocker, F. Habasha, E. Johnson, and F. Merritt. 1984. Aromatic-dependent Salmonella typhimurium as modified live vaccines for calves. Am. J. Vet. Res. 45:59-66. [PubMed] [Google Scholar]
- 55.Sulkin, S. E., and J. C. Willett. 1940. A triple sugar-ferrous sulfate medium for use in identification of enteric organisms. J. Lab. Clin. Med. 25:649-653. [Google Scholar]
- 56.Townsend, S. M., N. E. Kramer, R. Edwards, S. Baker, N. Hamlin, M. Simmonds, K. Stevens, S. Maloy, J. Parkhill, G. Dougan, and A. J. Baumler. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 69:2894-2901. [DOI] [PMC free article] [PubMed]
- 57.Weiss, S. H., M. J. Blaser, F. P. Paleologo, R. E. Black, A. C. McWhorter, M. A. Asbury, G. P. Carter, R. A. Feldman, and D. J. Brenner. 1986. Occurrence and distribution of serotypes of the Arizona subgroup of Salmonella strains in the United States from 1967 to 1976. J. Clin. Microbiol. 23:1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]