Abstract
A deletion-insertion mutation in msbB, a gene that encodes a lipid A acyltransferase, was introduced into encapsulated Neisseria meningitidis serogroup B strain NMB and an acapsular mutant of the same strain. These mutants were designated NMBA11K3 and NMBA11K3cap-, respectively. Neither lipooligosaccharide (LOS) nor lipid A could be isolated from NMBA11K3 although a number of techniques were tried, but both were easily extracted from NMBA11K3cap-. Immunoelectron microscopy using monoclonal antibody (MAb) 6B4, which recognizes the terminal Galβ1-4GlcNAc of LOS, demonstrated that NMB, NMBcap-, and NMBA11K3cap- expressed LOS circumferentially, while MAb 6B4 did not bind to the surface of NMBA11K3. However, cytoplasmic staining of NMBA11K3 with MAb 6B4 was a consistent observation. Mass-spectrometric analyses demonstrated that the relative amounts of the lipid A-specific C12:0 3-OH and C14:0 3-OH present in the membrane preparations (MP) from NMBA11K3 were substantially decreased (25- and 23-fold, respectively) compared to the amount in MP from its parent strain, NMB. Western blot analyses of MP from NMBA11K3 demonstrated that the levels of porin in the outer membrane of NMBA11K3 were also substantially decreased. These studies suggest that the lipid A acylation defect in encapsulated NMBA11K3 influences the assembly of the lipid A and consequently the incorporation of porin in the outer membrane.
Neisseria meningitidis is one of the leading causes of bacterial meningitis worldwide (1). Meningococcal disease affects mainly children and young adults. The rapid progression of meningococcal disease makes proper diagnosis and subsequent treatment often vital to the survival of infected individuals. If not properly diagnosed and treated, meningococcal infections can lead to shock and death within a matter of hours (35). Due to the detrimental effects caused by these bacteria, a better understanding of meningococcal pathogenesis may prove valuable in the management of systemic meningococcal disease.
One of the major virulence factors of N. meningitidis is the capsular polysaccharide. N. meningitidis serogroups are based on the capsular polysaccharide. Five serogroups, A, B, C, Y, and W-135, are most often associated with invasive meningococcal strains. Polysaccharide vaccines have been developed for serogroups A, C, Y, and W-135. Additionally, recent work on polysaccharide conjugate vaccines has shown the improved efficacy of these vaccines in infants and young adults (21, 37). Currently a vaccine for serogroup B is not available. The capsular polysaccharide for this serogroup is poorly immunogenic, due to its similarity to human neural adhesion molecules (35).
An additional virulence factor present in N. meningitidis is lipooligosaccharide (LOS). LOS is the principal glycolipid present in the outer membrane of N. meningitidis and is composed of the oligosaccharide chain extensions, the core, and the lipid A. The oligosaccharide chain extensions have been shown to play a role in molecular mimicry (8, 19, 34). The lipid A of N. meningitidis is similar in structure to lipid A from other gram-negative bacteria (11, 12, 28). The lipid A portion of these bacteria is known to be responsible for many of the adverse effects seen with gram-negative bacterial infections (11, 20).
HtrB and MsbB are the acyltransferases responsible for the addition of the secondary acyl substitutions onto the lipid A (3, 5, 16). Unlike Escherichia coli, N. meningitidis does not require the presence of the two 2-keto-3-deoxyoctulosonic acid (Kdo) groups for full lipid A acylation (32). Previous work with htrB and msbB mutants demonstrated that the lipid A portion of their LOS and lipopolysaccharide (LPS) structures were modified (12, 18, 28). These modified forms of LOS and LPS were reduced in their toxicity (9, 16) and in their ability to stimulate cytokine secretion (7, 24). Additionally, the Haemophilus influenzae and Salmonella enterica serovar Typhimurium htrB mutants were reduced in their virulence (9, 16).
Due to the importance of the lipid A structure in pathogenesis we wished to explore the possibility that there is an msbB homologue in N. meningitidis. A gene which showed high similarity to the htrB and msbB genes from E. coli was identified. This gene was cloned, and deletion-insertion mutants were made in N. meningitidis encapsulated strain NMB and N. meningitidis acapsular mutant strain NMBcap-. These mutants were subsequently designated NMBA11K3 and NMBA11K3cap-, respectively. In this study we report our chemical and immunochemical analyses of these mutants.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All bacterial strains and plasmids used in this study are described in Table 1. E. coli was grown in Luria-Bertani medium at 37°C and supplemented with appropriate antibiotics. Encapsulated N. meningitidis serogroup B strain NMB was isolated from the bloodstream of a patient with meningococcal sepsis. An acapsular NMB mutant (PBCC7232-NMB ΔsiaA-D) was a gift from Wyeth-Lederle Vaccines and Pediatrics (West Henrietta, N.Y.). This strain was designated NMBcap- for the studies presented here. N. meningitidis organisms were grown on gonococcal agar (Becton Dickinson, Sparks, Md.) supplemented with 1% IsoVitaleX or on brain heart infusion (BHI) agar (Becton Dickinson) supplemented with 2.5% heat-inactivated fetal calf serum (FCS) at 37°C in 5% CO2. Liquid cultures of N. meningitidis were grown in BHI broth supplemented with 2.5% FCS or in gonococcal broth supplemented with 1% IsoVitaleX at 37°C. Kanamycin-resistant N. meningitidis was grown on supplemented BHI agar plates or in supplemented BHI broth with 50 μg of kanamycin/ml. N. meningitidis organisms grown in the presence of kanamycin were grown without CO2. For growth curve cultures, an overnight bacterial culture was used to inoculate a 3-ml culture to an optical density at 600 nm (OD600) of 0.05. Cultures were grown at 37°C with agitation. Readings were taken once every hour.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype, relevant phenotype, or selection marker | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK−mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| N. meningitidis NMB | Wild type, serogroup B | 26 |
| N. meningitidis NMBcap- | ΔsiaA-siaD, serogroup B | Wyeth-Lederle |
| N. meningitidis NMBA11K3 | Kanamycin resistant, msbB mutant | This study |
| N. meningitidis NMBA11K3cap- | Kanamycin resistant, msbB mutant, ΔsiaA-siaD | This study |
| Plasmids | ||
| pUC19 | Ampicillin, cloning vector | New England Biolabs |
| pCR2.1 | Ampicillin, cloning vector | Invitrogen |
| pNMBA11pUC19 | Ampicillin, msbB PCR product in pUC19 vector | 18 |
| pNMBA11K3 | Ampicillin, kanamycin, insertion-deletion msbB mutant in pUC19 | 18 |
Recombinant DNA and transformation methods.
Restriction and modifying enzymes were purchased from New England Biolabs (Beverly, Mass.) and Promega (Madison, Wis.). Standard recombinant DNA protocols were performed as previously described (22). Transformation of E. coli with plasmid DNA was done by the CaCl2 method (6). Transformation of N. meningitidis was performed as previously described (27).
DNA isolation.
Plasmid DNA was prepared with the QIAprep Spin Miniprep kit or the QIAprep Midiprep kit, according to manufacturer's instructions (Qiagen Inc., Valencia, Calif.). Chromosomal DNA was isolated with the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.).
DNA sequencing and analysis.
DNA sequencing reactions were performed by using dye terminator cycle sequencing chemistry with AmpliTaq DNA polymerase and the FS enzyme (PE Applied Biosystems, Foster City, Calif.). The reactions were run on and analyzed with an Applied Biosystems model 373A stretch fluorescence automated sequencer at the University of Iowa DNA Facility. All primers were either commercially available or were purchased from either Genosys Corporation (Aldrich, Milwaukee, Wis.) or IDT Technologies (Coralville, Iowa). Sequence analysis was performed using Assembly LIGN, version 1.0 (Oxford Molecular Group Inc., Oxford, United Kingdom), MacVector (Oxford Molecular Group Inc.), and Wisconsin Package, version 10.0 (Genetics Computer Group, Madison, Wis.).
Cloning and mutagenesis of the N. meningitidis msbB gene.
Cloning and mutagenesis of the msbB gene was performed as previously described (18). Briefly, the E. coli htrB gene was used to search the Neisseria gonorrhoeae strain FA1090 sequence at the University of Oklahoma website. The sequence that showed highest homology to the E. coli gene was used to design PCR primers. Since the genomes of N. gonorrhoeae and N. meningitidis are highly homologous, primers gchtrB3 (5′-CAACAGGCGGCGGTGGAACAG-3′) and gchtrB4 (5′-TTCGGCATCCACTCCCCTTTG-3′) were used for amplification of the N. meningitidis strain NMB msbB gene. The 1,443-bp PCR product was cloned with the TA cloning vector pCR2.1 (Invitrogen, Carlsbad, Calif.) and was subsequently subcloned into pUC19. This construct was transformed into E. coli DH5α cells (Invitrogen) and was subsequently designated pNMBA11pUC19. Restriction enzymes BclI and BssHII deleted 138 bp from the msbB gene. A kanamycin resistance cassette was inserted into the modified msbB gene, and the resulting construct was designated pNMBA11K3. The proper construct was confirmed by using PCR and restriction enzyme digests. Plasmid DNA from pNMBA11K3 was used for transformation with N. meningitidis strains NMB and NMBcap-. Transformants were selected for on BHI plates containing kanamycin.
Southern blot and PCR analyses.
Hybridization experiments were carried out according to the manufacturer's protocols. All probes were labeled by either PCR labeling or random labeling with digoxigenin-labeled deoxynucleoside triphosphates (Boehringer Mannheim Corp., Indianapolis, Ind.). Primers gchtrB3 and -4 were used to perform PCRs.
SDS-PAGE of LOS.
LOS was isolated from 6 liters of BHI broth supplemented with 2.5% FCS for strains NMB and NMBcap- and 6 liters of BHI broth supplemented with 2.5% FCS and 50 μg of kanamycin/ml for strain NMBA11K3cap- by using a modified hot-phenol-water preparation (18). Samples were run on a Tris-Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel as previously described by Lesse et al. (13). Silver staining was done according to a previously described protocol (30).
Isolation and SDS-PAGE of MPs.
Overnight 10-ml broth cultures were harvested by centrifugation at ∼3,800 × g for 10 min. The cell pellet was resuspended in membrane preparation (MP) buffer (50 mM Tris-HCl, 150 mM NaCl, 10 mM EDTA, pH 7.4) and warmed at 56°C for 30 min. Then, cultures were cooled to room temperature. Cultures were passed through a syringe by using needles of different gauges to shear the cells. The process was repeated 10 times for each gauge of needle. This procedure was first performed with a 20-gauge needle, then with a 22-gauge needle, and last with a 25-gauge needle. Sheared cells were centrifuged at 16,000 × g for 15 min. The supernatant from this spin was centrifuged at 25,000 × g for 20 min. The supernatant from this spin was centrifuged at 30,000 × g for 20 min. Finally, the supernatant from the previous spin was centrifuged at 100,000 × g for 2 h 15 min. All spins were performed at 20°C. The pellet obtained from the last spin was glass-like and was used as the sample for SDS-PAGE. Samples were separated on a 4 to 20% Tris-Tricine gradient gel (13). Equal quantities of proteins were loaded onto each gel, as determined by spectrophotometric readings. Coomassie blue staining and silver staining were performed according to previously described protocols (17, 30).
Western blot analyses of LOS and MPs.
Western blot analyses were performed as previously described by Towbin et al. (29). MAb 6B4, which recognizes the terminal Galβ1-4GlcNAc moiety of the oligosaccharide chain extension (2), was utilized to detect LOS. The porin antibody 3H1 was a gift from Milan Blake (Baxter Hyland Immuno, Columbia, Md.). The blots were developed by using the Super Signal West Pico chemiluminescent substrate according to the manufacturer's instructions (Pierce, Rockford, Ill.).
SEM and TEM analyses.
For microscopy studies, all bacterial strains were grown in BHI broth to an OD600 of 0.8 to 1.0. Samples for transmission electron microscopy (TEM) analysis were placed in an equal volume of 4% paraformaldehyde (final concentration, 2%). Samples for scanning electron microscopy (SEM) analysis were settled onto silicon wafers and fixed in 2% paraformaldehyde. Cells for immuno-TEM were dehydrated by using a standard graded-ethanol series, followed by embedment in London Resin White (Ted Pella Inc., Redding, Calif.). For resolution of cell membrane morphology by TEM, bacterial cells were treated with osmium according to standard protocols and then dehydrated through graded ethanols and embedded in Epon acrylic resin. Thin sections of embedded cells were mounted on nickel grids. The Epon-embedded sections were stained with 5% uranyl acetate and lead citrate for contrast. Samples for SEM and immuno-TEM were treated with neuraminidase (1 U/ml; Oxford GlycoScience, Novato, Calif.) for 2 h at 37°C prior to labeling with MAb 6B4. Following overnight incubation with the primary antibody, specimens were incubated with goat anti-mouse IgM conjugated to gold beads, either 12-nm gold bead-conjugate (Jackson ImmunoResearch, West Grove, Pa.) for TEM or 25-nm gold bead-conjugate for SEM (EMS, Ft. Washington, Pa.). The TEM samples were finally counter-stained with 5% uranyl acetate. Immunolabeled samples for SEM were incubated in 2.5% glutaraldehyde to cross-link the antibodies and then processed by using a standard graded-ethanol series. These samples were carbon coated before being viewed on an S-4000 scanning electron microscope (Hitachi, Mountain View, Calif.). TEM samples were viewed with a Hitachi H-7000 transmission electron microscope. All samples were viewed with microscopes located in the Central Microscopy Research Facility at the University of Iowa.
GC/MS analysis of membrane fatty acids.
The MPs from strains NMB, NMBA11K3, NMBcap-, and NMBA11K3cap- were treated with 0.5 ml of 10% (wt/wt) BF3-methanol (Supelco, Bellefonte, Pa.) and heated at 100°C for 6 h. Samples were allowed to cool to room temperature and then were treated with 0.5 ml of saturated NaCl solution, followed by 0.5 ml of high-performance liquid chromatography grade hexanes (Aldrich, St. Louis, Mo.). After the samples were vortexed and centrifuged, the organic layers were removed and transferred to clean vessels. The aqueous layers were then extracted a second time with 0.5 ml of hexanes. The combined organic layers were evaporated to dryness under a stream of nitrogen and later redissolved in hexanes for gas chromatography/mass spectrometry (GC/MS) analysis. Samples were analyzed with a Hewlett-Packard 5890 gas chromatograph interfaced with a VG70SE mass spectrometer. The gas chromatograph was equipped with an on-column injector (J & W Scientific, Folsom, Calif.), and samples were separated on a 30-m by 0.25-mm BPX70 column with a 0.25-μm film thickness (SGE, Inc., Austin, Tex.). The initial oven temperature, 100°C, was held for 5 min, and then data acquisition was started and the samples were eluted by using a temperature gradient from 100 to 220°C at 4°C/min. The carrier gas was helium at ∼6 lb/in2. Relative peak areas were measured from the total ion chromatograms for each run and normalized to the C16:0 component.
RESULTS
Cloning and mutagenesis of the N. meningitidis msbB gene.
The N. meningitidis msbB gene was amplified by PCR and cloned. This gene has been previously shown to be able to complement for both the temperature sensitivity and the LPS phenotype of an E. coli htrB mutant (18). A deletion-insertion mutation was generated in the N. meningitidis msbB gene and the resulting construct was designated pNMBA11K3.
Transformations of pNMBA11K3 into N. meningitidis strains NMB and NMBcap- were performed as previously described (27). Selection for transformants was done on plates containing kanamycin. Southern blot analyses and PCR demonstrated that the proper mutations had been incorporated into the N. meningitidis NMB and NMBcap- genomic DNA (data not shown). The resulting transformants were designated NMBA11K3 and NMBA11K3cap-, respectively. These mutations should not have a polar effect on downstream genes. First, a kanamycin cassette, which has previously been shown to produce nonpolar mutations (15), was utilized to construct the pNMBA11K3 mutant. Second, the msbB gene is not part of an operon. We sequenced over 200 bp of DNA downstream from the msbB gene, and no open reading frames were found. In addition, the annotated sequence from N. meningitidis strain MC58 indicates that the closest gene is almost 300 bp downstream from the msbB gene, and it is transcribed in the opposite orientation. Proper capsule expression phenotypes for the four different strains were confirmed by using MAb 2-2-B, a serogroup B-specific capsular MAb, which was a gift from Wendell Zollinger (Walter Reed Army Institute of Research, Silver Spring, Md.) (data not shown).
Comparison of growth rates of N. meningitidis strains NMB, NMBcap-, NMBA11K3, and NMBA11K3cap-.
To determine whether the mutation in the msbB gene had any effect on the growth rate of the bacteria, growth curves were determined. These growth curves demonstrated that there was no difference in the growth rates of strains NMB, NMBcap-, and NMBA11K3cap- (data not shown). However, there was a reduction in the growth rate of strain NMBA11K3 compared with those of the other strains. In the first 8 h of growth, NMBA11K3 reached 50% of the density of the other strains, and after 24 h growth was reduced by approximately one-third. The growth curves were performed three times, and the results of all three experiments were consistent.
Characterization of the LOS by SDS-PAGE and Western blot analysis.
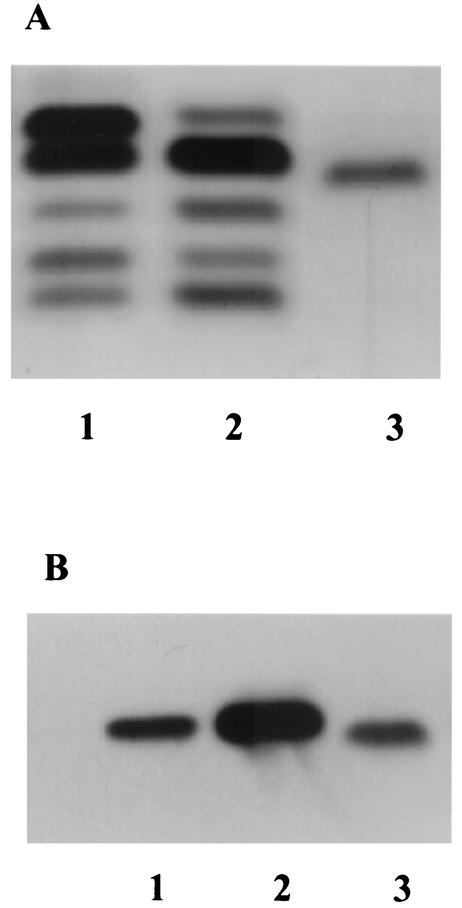
Silver staining showed that the NMBA11K3cap- LOS migrated through the gel slightly faster than the NMB and NMBcap- LOS (Fig. 1A) and that the NMBA11K3cap- LOS stained brown instead of black. This staining pattern was consistent with previous reports of LPS and LOS isolated from htrB and msbB mutants (12, 18, 23). Western blot analysis using LOS isolated from NMB, NMBcap-, and NMBA11K3cap- was performed. The blot showed that MAb 6B4 bound to all of the LOS samples tested (Fig. 1B). These results indicated that the oligosaccharide portion of the NMBA11K3cap- LOS was intact. LOS could not be purified from NMBA11K3 by using phenol-water, proteinase K, and petroleum ether-phenol extraction methods. Subsequent MPs from NMBA11K3 failed to reveal the presence of LOS bands.
FIG. 1.
Characterization of NMBA11K3cap- LOS by SDS-PAGE and Western blot analyses. (A) Silver staining analysis of an SDS-PAGE gel. Lane 1, NMB LOS; lane 2, NMBcap- LOS; lane 3, NMBA11K3cap- LOS. The sialylated LOS (top band) is absent from the NMBcap- LOS because the sialylation genes were deleted in this strain. A different glycoform of LOS is visible in the NMBcap- LOS sample where the sialylated LOS band would normally migrate. (B) Western blot analysis with MAb 6B4. Lane 1, NMB LOS; lane 2, NMBcap- LOS; lane 3, NMBA11K3cap- LOS.
TEM analyses of MAb 6B4-immunolabeled NMB, NMBA11K3, NMBcap-, and NMBA11K3cap-.
Figure 2 shows micrographs representative of each of the samples. Immunoelectron micrographs of NMB, NMBcap-, and NMBA11K3cap- (Fig. 2A, C, and D, respectively) show the typical diplococcus structure of N. meningitidis. These micrographs also show structurally intact membranes and an electron-dense cytoplasm. The NMBA11K3 sample (Fig. 2B) shows bacteria that still have the coccoid shape but that are somewhat larger than the bacteria from the NMB, NMB cap-, and NMBA11K3cap- samples. Additionally, there appear to be patches, instead of an even distribution, of electron-dense material in the cytoplasm. Immunoelectron micrographs of NMB, NMBcap-, and NMBA11K3cap- (Fig. 2A, C, and D, respectively) show the meningococci were labeled circumferentially with MAb 6B4. The immunoelectron micrograph of NMBA11K3 (Fig. 2B) shows the meningococci were labeled with MAb 6B4 predominately in the cytoplasm of the bacteria, with essentially no MAb 6B4 label present on the outer membrane. Higher-power electron microscopy revealed that both NMB and NMBA11K3 had evidence of bilamellar outer membranes (Fig. 2E and F, respectively).
FIG. 2.
Examination of MAb 6B4-immunolabeled N. meningitidis using TEM analyses. (A) NMB; (B) NMBA11K3; (C) NMBcap-; (D) NMBA11K3cap-; (E and F) Epon-embedded sections showing the structure of the bacterial cell membranes of NMB (E) and NMBA11K3 (F). Scale bars, 1 μm (A to D) and 100 nm (E and F).
SEM analyses of MAb 6B4-immunolabeled NMB, NMBA11K3, NMBcap-, and NMBA11K3cap-.
Figure 3 shows micrographs representative of data collected from each of the samples. All strains showed the typical diplococcus shape when viewed by SEM. NMB, NMBcap-, and NMBA11K3cap- (Fig. 3A, C, and D, respectively) showed surface labeling with MAb 6B4. MAb 6B4 did not label the surface of NMBA11K3 (Fig. 3B).
FIG. 3.
SEM analyses of MAb 6B4-immunolabeled N. meningitidis. (A) NMB; (B) NMBA11K3; (C) NMBcap-; (D) NMBA11K3cap-. Scale bars, 100 nm.
SDS-PAGE and Western blot analyses of MPs from NMB, NMBcap-, NMBA11K3, and NMBA11K3cap-.
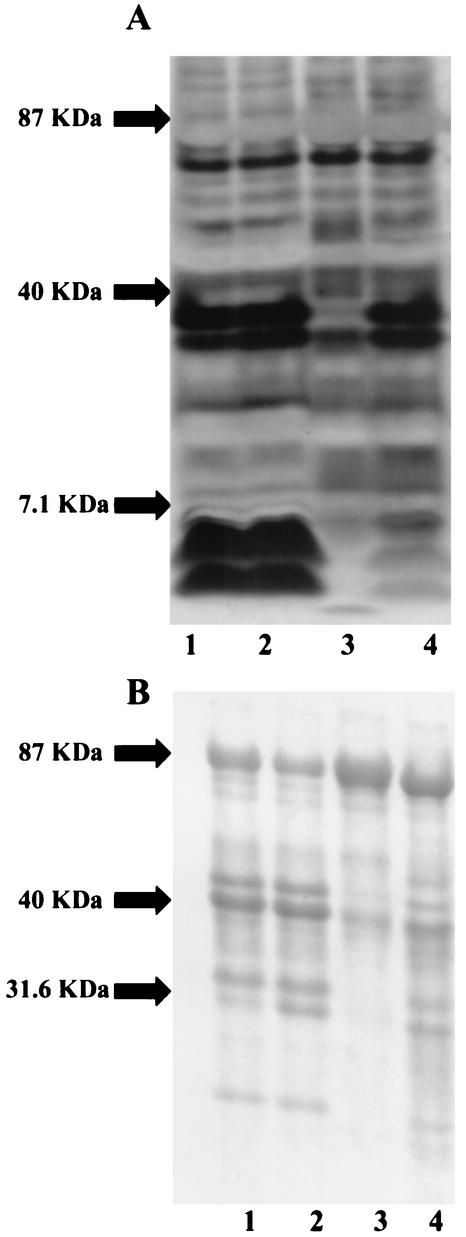
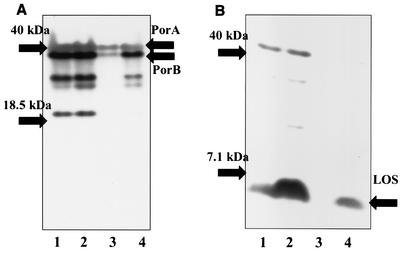
Both silver staining and Coomassie blue staining demonstrated that there were differences in the components of the MP from NMBA11K3 compared with those from NMB, NMBcap-, and NMBA11K3cap- (Fig. 4). Silver-staining analysis showed that there was no detectable LOS present in the NMBA11K3 sample (Fig. 4A). Western blot analysis utilizing MAb 6B4 confirmed the absence of a full-length LOS structure in the MP from NMBA11K3 (Fig. 5B). Examination of the MP also suggested that there were decreases in the levels of at least two other components of the outer membrane, those with molecular masses of ∼42 and ∼35 kDa, in NMBA11K3 compared with levels in its parent strain, NMB. Western blot analyses determined that the proteins with altered levels were PorA and PorB, respectively (Fig. 5A). The expected molecular masses of these proteins are approximately 42 kDa for PorA and 35.7 kDa for PorB.
FIG. 4.
Silver staining (A) and Coomassie blue staining (B) analyses of MPs from N. meningitidis strains NMB (lane 1), NMBcap- (lane 2), NMBA11K3 (lane 3), and NMBA11K3cap- (lane 4).
FIG. 5.
Western blot analyses of MPs from N. meningitidis strains NMB (lanes 1), NMBcap- (lanes 2), NMBA11K3 (lanes 3), and NMBA11K3cap- (lanes 4). (A) MAb 3H1 for porin; (B) MAb 6B4 for LOS. Expected molecular masses: PorA, ∼42 kDa; PorB, ∼35 kDa; LOS, ∼5 kDa.
MS analyses of MP fatty acids from NMB, NMBcap-, NMBA11K3, and NMBA11K3cap-.
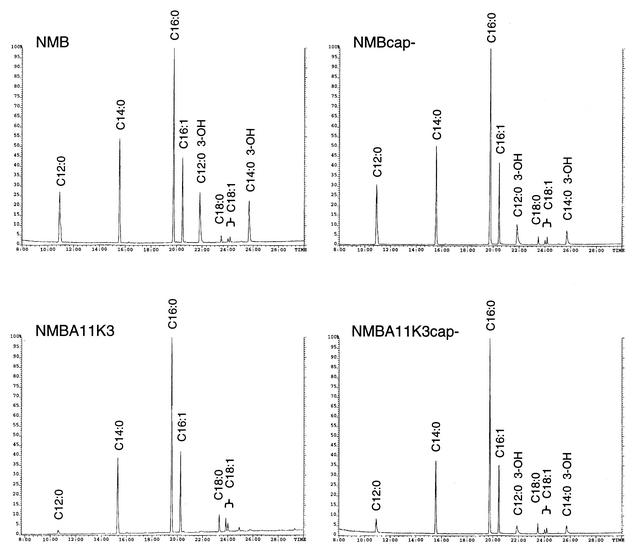
Fatty acid methyl esters were prepared from the MPs and were subsequently analyzed by GC/MS (Fig. 6 and Table 2). All of the expected fatty acids were present in each sample; however, the relative abundances of the fatty acids detected from the samples varied. For both the encapsulated and acapsular strains, the hydroxylated fatty acids derived exclusively from lipid A (C12:0 3-OH and C14:0 3-OH) had lower relative abundances in the msbB mutants than in the parental strains. This phenomenon was most dramatic for the encapsulated strains, where the levels of C12:0 3-OH and C14:0 3-OH were 25- and 23-fold higher, respectively, in the NMB sample than in the NMBA11K3 sample. The NMBcap- sample had C12:0 3-OH and C14:0 3-OH levels that were both approximately twofold higher than those of the NMBA11K3cap- sample. Additionally, the C12:0 fatty acid, which is also found in lipid A, was recovered in lower relative abundance in the msbB mutants than in their respective parental strains (Table 2). Compared to the lipid A fatty acid ratios in the NMBcap- strain, the relative amounts of C12:0, C12:0 3-OH, and C14:0 3-OH detected in the NMBA11K3cap- strain suggest the loss of a single C12:0 fatty acid from the lipid A structure in the msbB mutant. This observation is consistent with molecular mass measurements of the intact lipid A from NMB, NMBcap-, and NMBA11K3cap- obtained by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS, which showed a shift to lower mass (∼182 Da, corresponding to the loss of a lauric acid residue) for the msbB mutant (data not shown). No LOS could be isolated from the NMBA11K3 mutant strain; therefore, MALDI-TOF analysis of the intact lipid A was not possible. These data demonstrate that the relative abundance of the lipid A fatty acids present in the MPs from the msbB mutants is dramatically reduced compared with their relative abundance in the parental strains. However, this reduction is significantly more pronounced in the encapsulated msbB mutant (NMBA11K3) than in the acapsular msbB mutant (NMBA11K3cap-). These data suggest that the amounts of lipid A, and hence LOS, expressed in the outer membranes of NMBA11K3 and NMBA11K3cap- are significantly reduced compared to the amounts expressed in the outer membranes of their parent strains. Additionally, the relative amounts of C16:0 in the MPs from the msbB mutants were higher than those in the MPs from the wild-type strains. Consistent with these findings, a previous study by Steeghs et al. also found that the relative amounts of short-chain fatty acids increased in a LOS-deficient N. meningitidis lpxA mutant (25).
FIG. 6.
GC/MS chromatograms of fatty acid methyl esters derived from membrane preparations of the designated N. meningitidis strains. Relevant peaks are labeled in each chromatogram. The y axes show the relative peak intensities of the summed ion abundances.
TABLE 2.
Fatty acid analysis of outer membranes from designated strainsa
| Fatty acid | Relative abundance (%) in:
|
|||
|---|---|---|---|---|
| NMB | NMBA11K3 | NMBcap- | NMBA11K3cap- | |
| C12:0 | 42.1 | 3.2 | 43.2 | 10.0 |
| C14:0 | 63.4 | 42.5 | 51.3 | 40.8 |
| C16:0 | 100.0 | 100.0 | 100.0 | 100.0 |
| C16:19 | 40.9 | 36.9 | 34.4 | 34.8 |
| C18:0 | 3.1 | 7.5 | 3.1 | 4.6 |
| C18:19trans | 1.9 | 5.9 | 1.7 | 1.7 |
| C18:19cis | 2.9 | 3.9 | 3.3 | 2.7 |
| C12:0 3-OH | 37.4 | 1.5 | 18.0 | 8.4 |
| C14:0 3-OH | 30.1 | 1.3 | 10.8 | 6.5 |
Samples were normalized to the C16:0 component.
DISCUSSION
Previous studies involving htrB mutants from E. coli, H. influenzae, and S. enterica serovar Typhimurium have demonstrated that they exhibit a number of phenotypes (10, 12, 16, 28). E. coli, H. influenzae, and S. enterica serovar Typhimurium htrB mutants were all shown to be initially sensitive to temperatures above 32°C (10, 12, 28). However, work from our laboratory and others has demonstrated that N. gonorrhoeae and E. coli msbB mutants are not temperature sensitive (18, 24). In agreement with these studies, both NMBA11K3 and NMBA11K3cap- were able to grow on solid medium at 37°C. Growth curves demonstrated that NMBA11K3cap- was able to grow at the same rate as NMB and NMBcap-. However, NMBA11K3 had a slower growth rate and was unable to reach the same OD, after 56 h of growth, as the other three strains. Since the msbB mutants were able to grow at 37°C and since NMBA11K3cap- was able to grow at the same rate as NMB and NMBcap-, it seems unlikely that the lag in the growth of NMBA11K3 is due to temperature sensitivity.
An additional characteristic of htrB and msbB mutants is modification of the LPS and LOS structures. Studies performed with S. enterica serovar Typhimurium and H. influenzae htrB mutants demonstrated that the lipid A structures of both were modified (12, 28). The lipid A from the H. influenzae htrB mutant was determined to be approximately 90% tetraacyl and 10% pentaacyl instead of the normal hexaacyl structure. Additionally, studies of an N. gonorrhoeae msbB mutant demonstrated that the lipid A was pentaacyl instead of hexaacyl (18). In the study presented here, we were able to isolate and begin to characterize LOS from NMBA11K3cap-. The increase in migration rate and the change in the staining pattern of the NMBA11K3cap- LOS were consistent with data that have been previously reported for msbB and htrB mutants (12, 18, 23). Additionally, the binding of MAb 6B4, which recognizes the terminal two sugars of the oligosaccharide, to NMBA11K3cap- LOS indicates that the oligosaccharide region of the LOS is intact. Similar to findings with the N. gonorrhoeae msbB mutant (18), MS analysis of the NMBA11K3cap- lipid A demonstrated that it is missing one lauric acid (C12:0) substitution and thus may have a pentaacyl rather than a hexaacyl structure. These results are consistent with a previous study of an N. meningitidis strain H44/76 msbB mutant (33).
We were unable to isolate LOS from NMBA11K3 despite using a variety of standard methods. Immuno-SEM using MAb 6B4 demonstrated a lack of surface labeling of NMBA11K3, while NMB, NMBcap-, and NMBA11K3cap- all showed surface binding of the antibody. Immuno-TEM micrographs showed that, unlike what was found for NMB, NMBcap-, and NMBA11K3cap-, there was no MAb 6B4 binding to the outer membrane of NMBA11K3. However, binding of MAb 6B4 was visible in the cytoplasm of NMBA11K3. Since we could not isolate LOS from this mutant, we speculate that this labeling represents MAb 6B4 binding to the Galβ1-4GlcNAc of the oligosaccharide chain extensions still attached to its carrier, undecaprenol phosphate. MAb 6B4 has been previously shown to bind to lacto-N-neotetraose-ceramide in human erythrocytes (14). That study, by Mandrell et al., demonstrated that MAb 6B4 is able to recognize its epitope on a lipid carrier other than lipid A. Recent work in our laboratory with a meningococcal bacA mutant suggests that N. meningitidis LOS is assembled similarly to LPS (D. Post, A. Zaleski, E. Johansen, B. Gibson, and M. Apicella, unpublished data). Based on these studies and the current model for LPS assembly (36), it appears that the oligosaccharide chain extensions are assembled on the carrier lipid undecaprenol phosphate. Then, the oligosaccharide chain extensions and the lipid A-core region are transported to the periplasm by separate mechanisms, where they are subsequently ligated and transported to the outer membrane. The failure to isolate the lipid A-core region alone suggests that there is a defect in the assembly of the lipid A-core complex in NMBA11K3. Interestingly, recent findings by Tzeng et al. demonstrated that two N. meningitidis strain NMB mutants, defective in Kdo biosynthesis and Kdo transfer to the lipid A, expressed an LOS that consisted only of lipid A (31, 32). These results demonstrate that the lipid A can be assembled and transported to the bacterial surface in the absence of Kdo.
MS analyses of outer membranes isolated from NMBA11K3 demonstrated that the amounts of the lipid A-specific C12:0 3-OH and C14:0 3-OH present in these samples were 25- and 23-fold, respectively, less than the amount present in the MP from NMB. These data taken together with the microscopy data suggest that the amount of LOS expressed in the outer membranes of NMBA11K3 is significantly reduced compared to the amount expressed in the outer membranes of its parent strain. MP from NMBA11K3cap- showed 2- and 1.7-fold decreases in the amounts of C12:0 3-OH and C14:0 3-OH, respectively, present in these samples compared to the amounts present in MP from NMBcap-. Additionally, the relative amounts of C16:0 in the MPs from the msbB mutants were higher than those in the MPs from their respective wild-type strains. These data suggest that both NMBA11K3 and NMBA11K3cap- are defective in their abilities to assemble their LOS and that the presence of the capsular polysaccharide makes the defect in LOS assembly and subsequent transport more pronounced. These results also suggest that NMBA11K3 may place other lipids, most likely C16:0, the lipid that anchors the capsule to the outer membrane, in their outer membranes to compensate for the loss of the lipid A portion of the LOS. These mutants may preferentially express the lipidated capsule on their surfaces when the lipid A is altered. However, if the capsule components are absent in the msbB mutant, the meningococcus may be able to incorporate the modified LOS structure into the outer membrane. Interestingly, Steeghs et al. determined that the presence of the capsular polysaccharide is essential for the viability of the LOS-deficient N. meningitidis lpxA mutant H44/76(pHBK30) (25), further suggesting that the C16:0 lipid may have a role in stabilizing the outer membranes of LOS-depleted mutants.
Western blot analyses demonstrated that the levels of porin expressed in the outer membrane of NMBA11K3 were also altered. Since porin is known to closely associate with LOS (4), it was not surprising to see a decrease in the levels of PorA and PorB in the outer membranes. SDS-PAGE analyses of MP from NMBA11K3 demonstrated that a number of other proteins expressed in the outer membrane were present at levels similar to those in the NMB MP. An explanation for the decrease in the levels of porin and LOS could be that they are transported to the outer membrane as a complex. The modification in the lipid A structure may have decreased the efficiency of LOS assembly; therefore, the transport and subsequent surface expression of the whole complex are altered.
A decrease in LOS and LPS expression on the bacterial surface has not been previously reported for any htrB or msbB mutant. Previous studies of an N. meningitidis msbB mutant by van der Ley et al. did not report any changes in the amount of LOS or porin expressed on the bacterial surface (33). One explanation for this difference may be the use of different strains of N. meningitidis in our respective studies.
Further study is required to more clearly determine the components involved in the maintenance of the outer membrane structure of NMBA11K3. This strain may prove to be an important tool for further elucidating the mechanisms of LOS assembly and transport in pathogenic Neisseria. Additionally, since NMBA11K3 is not greatly impaired in its growth rate and since the surface expression of LOS is markedly reduced, this strain may prove useful in the future development of meningococcal vaccines.
Acknowledgments
The University of Iowa DNA facility is supported in part by the Diabetes Endocrinology Research Center with National Institutes of Health grant DK25295 and by the College of Medicine. Research in M. A. Apicella's laboratory was supported by AI45728 and AI44642. D. M. B. Post's work was in part supported by NIH training grant T32A107511. Research in B. W. Gibson's laboratory was supported by AI44642 and by the UCSF mass spectrometry facility, which is partially supported by NCRR 06164.
Editor: J. T. Barbieri
REFERENCES
- 1.Apicella, M. A. 2000. Neisseria meningitidis, p. 2228-2241. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed., vol. 2. Churchill Livingstone, Philadelphia, Pa.
- 2.Apicella, M. A., R. E. Mandrell, M. Shero, M. E. Wilson, J. M. Griffiss, G. F. Brooks, C. Lammel, J. F. Breen, and P. A. Rice. 1990. Modification of sialic acid of Neisseria gonorrhoeae lipooligosaccharide epitope expression in human urethral exudates: an immunoelectron microscopic analysis. J. Infect. Dis. 162:506-512. [DOI] [PubMed] [Google Scholar]
- 3.Brozek, K. A., and C. R. H. Raetz. 1990. Biosynthesis of lipid A in Escherichia coli. Acyl carrier protein-dependent incorporation of laurate and myristate. J. Biol. Chem. 265:15410-15417. [PubMed] [Google Scholar]
- 4.Buehler, L. K., S. Kusumoto, H. Zhang, and J. P. Rosenbusch. 1991. Plasticity of Escherichia coli porin channels. J. Biol. Chem. 266:24446-24450. [PubMed] [Google Scholar]
- 5.Clementz, T., Z. Zhou, and C. R. H. Raetz. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. J. Biol. Chem. 272:10353-10360. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 7.Harvey, H. A., D. M. B. Post, and M. A. Apicella. 2002. Immortalization of human urethral epithelial cells: a model for the study of the pathogenesis of and the inflammatory cytokine response to Neisseria gonorrhoeae infection. Infect. Immun. 70:5808-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey, H. A., M. P. Jennings, C. A. Campbell, R. Williams, and M. A. Apicella. 2001. Receptor-mediated endocytosis of Neisseria gonorrhoeae into primary human urethral epithelial cells: the role of the asialoglycoprotein receptor. Mol. Microbiol. 42:659-672. [DOI] [PubMed] [Google Scholar]
- 9.Jones, B. D., W. A. Nichols, B. W. Gibson, M. G. Sunshine, and M. A. Apicella. 1997. Study of the role of the htrB gene in Salmonella typhimurium virulence. Infect. Immun. 65:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karow, M., O. Fayet, A. Cegielska, T. Ziegelhoffer, and C. Georgopoulos. 1991. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J. Bacteriol. 173:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulshin, V. A., U. Zahringer, B. Lindner, C. E. Frasch, C. M. Tsai, B. A. Dmitriev, and E. T. Rietschel. 1992. Structural characterization of the lipid A component of pathogenic Neisseria meningitidis. J. Bacteriol. 174:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, N., M. Sunshine, J. Engstrom, B. Gibson, and M. A. Apicella. 1995. Mutation of the htrB locus of Haemophilus influenzae nontypeable strain 2019 is associated with modifications of lipid A and phosphorylation of the lipooligosaccharide. J. Biol. Chem. 270:27151-27159. [PubMed] [Google Scholar]
- 13.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine sodium dodecyl sulfate polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 14.Mandrell, R. E., J. M. Griffiss, and B. A. Macher. 1988. Lipooligosaccharides (LOS) of Neisseria gonorrhoeae and Neisseria meningitidis have components that are immunochemically similar to precursors of human blood group antigens. Carbohydrate sequence specificity of the mouse monoclonal antibodies that recognize crossreacting antigens on LOS and human erythrocytes. J. Exp. Med. 168:107-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menard, R., P. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols, W. A., C. R. H. Raetz, T. Clementz, A. L. Smith, J. A. Hanson, M. R. Ketterer, M. Sunshine, and M. A., Apicella. 1997. htrB of Haemophilus influenzae: determination of biochemical activity and effects on virulence and lipooligosaccharide toxicity. J. Endotoxin Res. 4:163-172. [Google Scholar]
- 17.Pederson, S., S. V. Reeh, J. Parker, R. J. Watson, J. D. Friesen, and N. P. Fiil. 1976. Analysis of the proteins synthesized in ultraviolet light-irradiated Escherichia coli following infection with the bacteriophages λdrifd 18 and λdfus-3. Mol. Gen. Genet. 144:339-343. [DOI] [PubMed] [Google Scholar]
- 18.Post, D. M. B., N. J. Phillips, J. Q. Shao, D. D. Entz, B. W. Gibson, and M. A. Apicella. 2002. Intracellular survival of Neisseria gonorrhoeae in male urethral epithelial cells: importance of a hexaacyl lipid A. Infect. Immun. 70:909-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preston, A., R. E. Mandrell, B. W. Gibson, and M. A. Apicella. 1996. The lipooligosaccharide of pathogenic gram-negative bacteria. Crit. Rev. Microbiol. 22:139-180. [DOI] [PubMed] [Google Scholar]
- 20.Raetz, C. R. H. 1993. Bacterial endotoxins: extraordinary lipids that activate eucaryotic signal transduction. J. Bacteriol. 175:5745-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richmond, P., R. Borrow, J. Findlow, S. Martin, C. Thornton, K. Cartwright, and E. Miller. 2001. Evaluation of de-O-acetylated meningococcal C polysaccharide-tetanus toxoid conjugate vaccine in infancy: reactogenicity, immunogenicity, immunologic priming, and bactericidal activity against O-acetylated and de-O-acetylated serogroup C strains. Infect. Immun. 69:2378-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Schnaitman, C., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somerville, J. E., Jr., L. Cassiano, B. Bainbridge, M. D. Cunningham, and R. P. Darveau. 1996. A novel Escherichia coli lipid A mutant that produces an anti-inflammatory lipopolysaccharide. J. Clin. Investig. 97:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steeghs, L., H. de Cock, E. Evers, B. Zomer, J. Tommassen, and P. van der Ley. 2001. Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. EMBO J. 20:6937-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens, D. S., C. F. McAllister, D. Zhou, F. K. Lee, and M. A. Apicella. 1994. Tn916-generated, lipooligosaccharide mutants of Neisseria meningitidis and Neisseria gonorrhoeae. Infect. Immun. 62:2947-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens, D. S., J. S. Swartley, S. Kathariou, and S. A. Morse. 1991. Insertion of Tn916 in Neisseria meningitidis resulting in loss of group B capsular polysaccharide. Infect. Immun. 59:4097-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunshine, M., B. W. Gibson, J. J. Engstrom, W. A. Nichols, B. D. Jones, and M. A. Apicella. 1997. Mutation of the htrB gene in a virulent Salmonella typhimurium strain by intergenic transduction: strain construction and phenotypic characterization. J. Bacteriol. 179:5521-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 31.Tzeng, Y. L., A. Datta, C. Strole, V. S. K. Kolli, M. R. Birck, W. P. Taylor, R. W. Carlson, R. W. Woodard, and D. S. Stephens. 2002. KpsF is the arabinose-5-phosphate isomerase required for 3-deoxy-d-manno-octulosonic acid biosynthesis and for both lipooligosaccharide assembly and capsular polysaccharide expression in Neisseria meningitidis. J. Biol. Chem. 277:24103-24113. [DOI] [PubMed] [Google Scholar]
- 32.Tzeng, Y. L., A. Datta, V. K. Kolli, R. W. Carlson, and D. S. Stephens. 2002. Endotoxin of Neisseria meningitidis composed only of intact lipid A: inactivation of the meningococcal 3-deoxy-d-manno-octulosonic acid transferase. Infect. Immun. 184:2379-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Ley, P., L. Steeghs, H. J. Hamstra, J. ten Hove, B. Zomer, and L. van Alphen. 2001. Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect. Immun. 69:5981-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel, U., and M. Frosch. 1999. Mechanisms of neisserial serum resistance. Mol. Microbiol. 32:1133-1139. [DOI] [PubMed] [Google Scholar]
- 35.West, D., K. Reddin, M. Matheson, R. Heath, S. Funnell, M. Hudson, A. Robinson, and A. Gorringe. 2001. Recombinant Neisseria meningitidis transferrin binding protein A protects against experimental meningococcal infection. Infect. Immun. 69:1561-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitfield, C. 1995. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 3:178-185. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, Q., R. Lakshman, R. Burkinshaw, S. Choo, J. Everard, S. Akhtar, and A. Finn. 2001. Primary and booster mucosal immune responses to meningococcal group A and C conjugate and polysaccharide vaccines administered to university students in the United Kingdom. Infect. Immun. 69:4337-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]