Abstract
The interactions between pathogenic bacteria and the host need to be resolved at the molecular level in order to develop novel antiadhesive drugs and vaccines. We have previously identified strepadhesin, a novel glycoprotein-binding activity in Streptococcus pyogenes binding to thyroglobulin, submaxillar mucin, fetuin, and asialofetuin. The activity is known to be regulated by Mga, a regulator of streptococcal virulence factors, and is carried by the surface-associated streptococcal cysteine protease, SpeB. In the present study, we focused on the high strepadhesin activity in an S. pyogenes strain (NZ131rgg) lacking SpeB expression. By extracting surface proteins from the bacteria, a new strepadhesin protein was identified, and mass spectrometric analysis and database search identified it as a putative pullulanase. The gene was cloned, and the recombinant pullulanase (PulA) exhibited pullulanase and starch hydrolyzing activity, as well as strepadhesin activity. Sequencing of the pulA gene revealed an open reading frame with 3,498 bp encoding a protein of 1,165 amino acids with a predicted molecular mass of 129 kDa. PulA exhibited properties typical for a gram-positive surface protein with a putative signal sequence and LPKTGE cell wall anchoring motif and contained the four highly conserved regions common to pullulanases. Mutant bacteria deficient in PulA expression showed diminished strepadhesin activity on bacterial dot blot assay and reduced adherence to thyroglobulin immobilized on microtiter plates. Thus, S. pyogenes strepadhesin activity is carried by a surface-bound pullulanase, which combines glycoprotein-binding and carbohydrate-degrading activities in the same molecule.
In most infectious diseases, the initial event is the adherence of pathogenic organisms to the host. The process is in many cases mediated by the interaction between lectins on the surface of the infectious organism and carbohydrates on the host tissues. The interaction can be blocked by soluble carbohydrates, or analogous structures, recognized by the bacterial lectins or by antibodies binding to the lectins. This approach to prevent and treat infectious diseases, called antiadhesion therapy (5, 29, 30, 52, 59, 60), is a highly promising approach in the fight against pathogens in the era of increasing antibiotic resistance. In order to develop novel antiadhesion drugs, however, we need to first characterize the interactions between bacterial adhesins and host receptors in molecular detail.
Streptococcus pyogenes is a human pathogen that causes a variety of diseases, such as pharyngitis, impetigo, and erysipelas, and also life-threatening infections, necrotizing fasciitis, and toxic shock-like syndrome. The number of S. pyogenes infections, especially the severe ones, has been increasing since the beginning of the last decade (6, 12), and there are several reports concerning the appearance of S. pyogenes strains resistant to antibiotics (37, 51). This emphasizes the need for new preventive methods and therapies against S. pyogenes infections.
Carbohydrate binding has been described in some streptococcal species. In a recent report, Ryan et al. demonstrated that group A streptococcal M protein mediates binding of the bacterium to sialic acid residues of bovine mucin (49). Other adhesion mechanisms involving carbohydrate receptors include the interaction between Streptococcus pneumoniae and a variety of oligosaccharide structures (1, 2, 15, 32). Among well-characterized interactions is the binding of the meningitis-associated Streptococcus suis to Galα1-4Gal and to sialic acid-containing oligosaccharides (20, 21, 35). Streptococcus gordonii expresses the SspB adhesin, which mediates binding to sialic acid-containing salivary glycoproteins (16, 17). Antiadhesion therapy with carbohydrates has proven successful in streptococcal infections. Experiments in an animal model of pneumococcal pneumonia have shown that oligosaccharides can interfere with the establishment and progression of the infection (25).
In previous reports we have studied the mechanism of the binding of S. pyogenes to glycoproteins in order to better understand the molecular interactions between the bacterium and the host. We have characterized a streptococcal “strepadhesin” activity that mediates binding of S. pyogenes to glycoproteins and is present in the majority of the strains studied (23, 24). The activity is controlled by the Mga regulator and is in some strains mediated mainly by the surface-associated virulence factor SpeB. In the present study, we have identified a new protein carrying strepadhesin activity. It was found to be a bacterial surface-associated streptococcal pullulanase, which thus represents a molecule containing both glycoprotein-binding and carbohydrate-degrading activity in the same molecule.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Streptococcus pyogenes clinical isolate A8173 (type M2) was provided by K. Kunnas, National Public Health Institute, Kuopio, Finland. S. pyogenes NZ131 wild-type and the speB and rgg mutant strains were kind gifts from M. Chaussee, National Institutes of Health, Hamilton, Mont. (9, 10). Streptococcal strains were grown on Todd-Hewitt (Difco) plates or media (THY) supplemented with 0.5% yeast extract (Biokar Diagnostics). Escherichia coli was grown in Luria broth. All bacteria were stored at −70°C in growth medium containing 15% glycerol. When appropriate, antibiotics were added to the culture media: for S. pyogenes, erythromycin at 3 μg/ml and kanamycin at 500 μg/ml; for Escherichia coli, kanamycin at 30 μg/ml and chloramphenicol at 34 μg/ml.
Materials.
Bovine thyroglobulin, fetuin, asialofetuin, and submaxillary mucin, horse myoglobin, streptavidin-horseradish peroxide (HRP), the cysteine protease inhibitor trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane (E64), and human placenta laminin were purchased from Sigma. Sequencing-grade modified trypsin was from Promega. NHC-LC biotin was obtained from Pierce, and the biotinylated derivatives were prepared according to the instructions of the manufacturer. Nitrocellulose membrane (0.45 μm [pore size]) was from Schleicher & Schuell. The enhanced chemiluminescence (ECL) Western blotting kit and Hyperfilm were obtained from Amersham. Tran35S-label was from ICN Biomedicals, and Microbeta plates were from Wallac. Bacillus pullulanase was from Fluka, and SpeB antiserum was from Fitzgerald. Mature SpeB was purified from S. pyogenes T19 growth medium and was generously provided by D. Gerlach, Friedrich-Schiller-University, Jena, Germany.
Manipulation of DNA.
Plasmid DNA was prepared from E. coli with the Qiaprep kit (Qiagen). Restriction endonucleases and polymerases were used according to the recommendations of the manufacturers. Chromosomal DNA from S. pyogenes was prepared as described previously (8). S. pyogenes was transformed by electroporation by the method of Simon and Ferretti (54). Sequencing of DNA was performed by using an automated ABI Prism DNA sequencer, and sequences were assembled by using MacDNAsis software.
Binding assays.
Bacteria grown overnight on THY media were collected by centrifugation, washed twice with phosphate-buffered saline (PBS; 10 mM sodium-phosphate buffer, 0.15 M NaCl [pH 7.4]) and serially diluted (to an optical density at 600 nm [OD600] of 4.0 to 0.125). Aliquots of 1 μl were pipetted onto a gridded nitrocellulose membrane and allowed to dry, and nonspecific binding sites were saturated with 3% bovine serum albumin (BSA)-0.1% Tween 20 in PBS at room temperature for 1 h. The biotinylated ligand was added to the membrane in 1% BSA-0.1% Tween 20 in PBS to a final concentration of 1 μg/ml and then incubated at room temperature for 30 min. The membrane was washed three times with PBS, and 0.1 μg of streptavidin-HRP was added/ml to the membrane in 1% BSA-0.1% Tween 20 in PBS, followed by incubation at room temperature for 30 min. The membrane was washed as described above, and the binding of the ligand was detected by ECL autoradiography. The same protocol was used to detect the binding of glycoproteins to Western blots.
Preparation and analysis of sodium dodecyl sulfate (SDS) lysates of the bacteria.
Bacteria grown and collected as described above were suspended in SDS-PAGE sample buffer, boiled at 95°C for 10 min, and centrifuged. The lysate was analyzed by SDS-PAGE and Western hybridization, and the presence of SpeB on the blots was detected with above protocol by using SpeB specific antiserum (1:1,000) and HRP-conjugated anti-rabbit goat antiserum (1:10,000).
Preparation of NZ131rgg surface extracts and identification of the glycoprotein-binding protein by mass spectrometry.
NZ131rgg bacteria were grown in 50 ml of THY with no agitation overnight at 37°C, collected by centrifugation, washed in PBS, and resuspended in 0.5 ml of the same buffer. The bacterial suspension was digested with SpeB at a concentration of 10 μg/ml for 30 min at 37°C, and the digestion was terminated by the addition of the SpeB inhibitor E64 to a final concentration of 1 μM. The bacteria were pelleted, the supernatant was recovered, and 20 μl of the supernatant was analyzed by SDS-PAGE on a 7.5% polyacrylamide gel and stained with silver. An identical gel was run in parallel, transferred to nitrocellulose membrane, and probed with labeled thyroglobulin as described above.
For identification of the thyroglobulin-binding protein, the protein was cut out from the silver-stained gels and in-gel digested (40, 48, 53). The protein was reduced and alkylated before digestion with trypsin overnight at 37°C. The resulting peptides were extracted by adding 30 to 40 μl of 5% formic acid to the digestion mixture and incubating them at 37°C for 30 min, followed by direct desalting. Desalting was performed with μ-Tips (34) containing Oligo R3-material (PerSeptive Biosystems).
Peptide mass fingerprinting was performed with a Voyager DE PRO matrix-assisted laser desorption ionization-time of flight instrument in the positive ion reflector mode by using α-cyano-4-hydroxycinnamic acid as the matrix. The mass spectra were internally calibrated with autoproteolytic trypsin fragments of 842.50 and 2,211.10 Da. Database searches were performed by using the program MS-Fit (http://prospector.ucsf.edu), and the search criteria were 0.05 Da mass accuracy and a minimum of five matching peptides.
Amplification of the pullulanase gene.
For the PCR amplification of the pullulanase gene (pulA) primers were designed on the basis of the published S. pyogenes M1 genomic sequence (18). Underlined sequences correspond to vector compatible overhangs. Primers 1 (5′-GACGACGACAAGATGAAAAAGAAAGTCAACCAAG-3′) and 2 (5′-GAGGAGAAGCCCGGTCTAATCTTTTTGGCGCTTCAT-3′) were designed to amplify the whole gene. Primers 3 (5′-GACGACGACAAGATGGCTAAAACCATTGTTGGACTA-3′) and 4 (5′-GAGGAGAAGCCCGGTTTACTTTTGGTTTGTTTTAGCTCT-3′) were designed to amplify the sequence between nucleotides 136 and 3396 of the pullulanase gene. This sequence lacks the parts of the gene corresponding to the putative N-terminal signal peptide and the C-terminal membrane-spanning region and cell wall-anchoring motif. PCR was performed by using Vent polymerase and standard conditions in a Perkin-Elmer thermal cycler.
Expression and purification of recombinant pullulanase.
The pullulanase gene was amplified from S. pyogenes NZ131 genomic DNA by using primers 3 and 4. The PCR fragment was treated with T4 DNA polymerase to create the vector-specific compatible overhangs and annealed to pET-30 Ek/LIC expression vector (Novagen). The plasmid was transformed into NovaBlue Singles competent cells (Novagen) and plated on Luria-Bertani (LB) agar plates containing the appropriate antibiotic. Recombinant plasmid was purified from one clone and transformed to the E. coli BL21(DE3)pLysS strain used in the expression of the recombinant protein. Cells were grown in LB medium at 37°C to an OD600 of 0.8, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM, and the cells were grown for an additional 4 h at room temperature. After centrifugation at 3,000 × g for 10 min, the supernatant was discarded and the cell pellet was stored at −70°C. The cells were thawed on ice, and resuspended in 50 mM sodium phosphate buffer (pH 8.0), protease inhibitor cocktail (Boehringer), and Benzonase (Novagen) were added, followed by incubation on ice for 30 min. The cell lysate was separated by centrifugation, and the recombinant pullulanase recovered by affinity purification by using Ni-NTA matrix (Qiagen). The purified protein was stored at 4°C.
Mutagenesis of pulA in NZ131rgg.
For mutagenesis of pulA in NZ131rgg, an internal NsiI-BamHI fragment (nucleotides 297 to 1476) of the pulA gene was cloned into a PstI-BamHI-digested suicide vector pSF151 that does not replicate in streptococci (57). The plasmid was electroporated into E. coli K-12 GM2163 (New England Biolabs), and transformants were selected for kanamycin resistance. One clone was isolated and used for preparing the plasmid pulA-pSF151 for transformation into NZ131rgg. Then, 10 μg of the purified plasmid was electroporated into competent bacteria as described previously (54). Recombinants were selected on THY plates containing 500 μg of kanamycin and 3 μg of erythromycin/ml. The integration of the suicide plasmid in pulA gene was confirmed by PCR with primers specific to pSF151 (5′-TTAGCTCACTCATTAGGCAC-3′) and to 5′ upstream region of pulA gene (5′-GGACACGGTATTAGAGACCAAG-3′). One mutant (NZ131rgg-pulA) was chosen for further analysis.
Binding of radiolabeled streptococci to glycoproteins on microtiter plates.
Bacteria were grown in 5 ml of THY broth containing 0.15 mCi of Tran35S-label at 37°C overnight. The bacteria were concentrated by centrifugation (3,000 × g for 10 min at 4°C) to 1 ml of PBS, and the unbound label was removed by centrifuging the bacteria as described above through 5 ml of 6% BSA in PBS. Microtiter plate wells were coated with 100 μl of the ligand at a concentration of 10 μg/ml at 37°C for 2 h. The wells were rinsed with PBS and saturated with 2% BSA-0.1% Tween 20 in PBS for 1 h at room temperature. The wells were overlaid with 100 μl of the bacteria in 0.1% BSA-0.05% Tween 20 in PBS at a concentration that gave an OD600 of 0.3 corresponding to ca. 0.5 × 108 CFU/ml (determined by plating), incubated for 2 h at 37°C with gentle agitation, and washed three times for 10 min each time with 200 μl of PBS per well. Binding of the bacteria to wells coated with 2% BSA was used as control. Experiments were carried out in eight parallel wells. The bound bacteria were detected by liquid scintillation counter. The results are given as CFU, which were obtained by first determining the counts per minute (cpm)/CFU ratios for each strain, and then dividing the cpm by these ratios.
Enzymatic activity assays.
Pullulanase and amylase activities of the recombinant pullulanase were determined by the dinitrosalisylic acid method (38). The activities were measured at 37°C in a reaction mixture containing 10 mg of the polysaccharide/ml and a suitable aliquot of pullulanase in either 25 mM sodium citrate buffer (pH 5), PBS (pH 7), or 50 mM glycine-NaOH buffer (pH 9). The reducing sugars formed were quantified, after the addition of dinitrosalisylic acid reagent and boiling the sample, by measuring to absorbance of the sample at 550 nm versus sample blanks. Commercially available Bacillus pullulanase was used as a control enzyme.
RESULTS
Strepadhesin activity of SpeB− S. pyogenes strains.
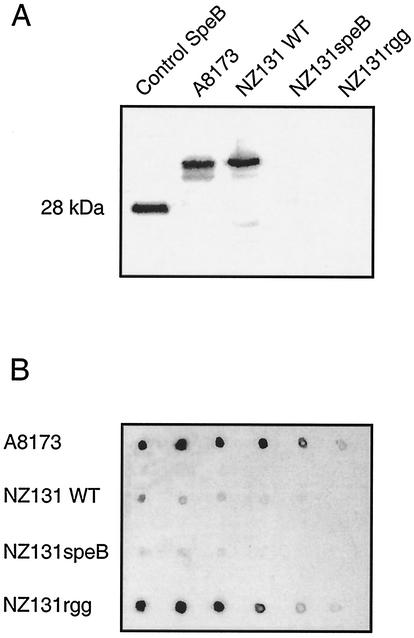
In our previous report we found that the strepadhesin activity is carried by streptococcal virulence factor SpeB (23). S. pyogenes strains that do not express SpeB (NZ131speB and NZ131rgg) were analyzed for their glycoprotein-binding activity compared to the wild-type strains NZ131 and A8173. SDS lysates of the two wild-type strains (A8173 and NZ131) clearly expressed the zymogen form of SpeB, whereas the mutant strains (NZ131speB and NZ131rgg) showed no expression (Fig. 1A). Bacterial dot blot analysis for strepadhesin activity with labeled thyroglobulin demonstrated that both wild-type strains bound it, but the NZ131 wild-type strain bound considerably less than the A8173 wild-type strain (Fig. 1B). The NZ131speB strain with an insertional mutation in the speB gene, expressed as expected weaker thyroglobulin-binding activity compared to the wild-type strain NZ131. Unexpectedly, the NZ131rgg strain with a mutation in the regulatory rgg gene and with no SpeB production bound thyroglobulin with high activity. The thyroglobulin-binding activity of NZ131rgg was similar to that of A8173 in that it was cross-inhibited by fetuin, asialofetuin, and submaxillary mucin and inactivated by trypsin digestion, and thus it represented strepadhesin activity (24) (data not shown). The results suggested that there may be in addition to SpeB another molecule with strepadhesin activity in S. pyogenes.
FIG. 1.
(A) Production of SpeB by A8173 and NZ131 wild-type bacteria and the NZ131speB and NZ131rgg SpeB-defective mutants. Bacteria from overnight cultures were lysed in SDS-PAGE sample buffer, and the samples subjected to SDS-PAGE and transferred to a nitrocellulose membrane, and then the membrane was probed with SpeB antiserum. (B) Strepadhesin activity of wild-type and SpeB-defective mutant streptococci. A membrane containing 1-μl aliquots of serially diluted (OD600 = 4.0 to 0.125) suspensions of A8173 and NZ131 wild-type bacteria and the SpeB-defective NZ131speB and NZ131rgg mutants was incubated with biotinylated thyroglobulin, and the binding was detected with streptavidin-HRP and by ECL autoradiography.
Preparation of NZ131rgg surface extracts and identification of the glycoprotein-binding protein by mass spectrometry and database search.
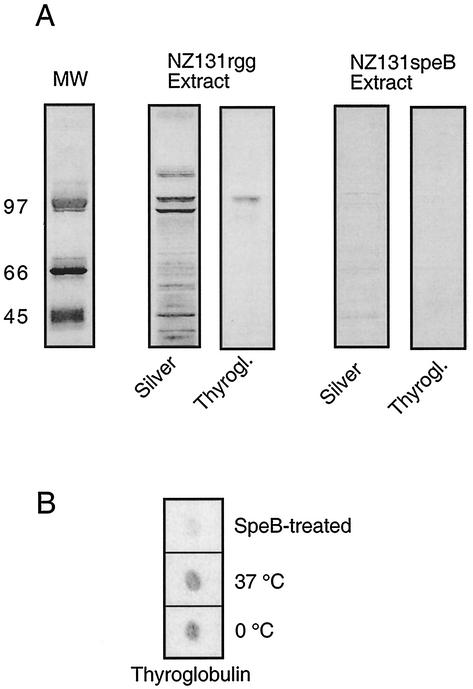
Streptococcal cysteine protease is known to cleave proteins from the bacterial surface (4). In order to extract the new molecule containing strepadhesin activity, NZ131rgg bacteria were digested with SpeB under conditions that allowed partial digestion. By analyzing the NZ131rgg extract by SDS-PAGE and Western blotting we were able to identify a protein fragment that carried thyroglobulin-binding activity (Fig. 2A). This fragment was not present in the extract of the strain NZ131speB, which suggested that the strepadhesin activity carrying fragment was upregulated in the NZ131rgg strain. Complete SpeB digestion abolished the thyroglobulin-binding activity of the treated NZ131rgg bacteria (Fig. 2B). The apparent size of the binding fragment in the NZ131rgg extract was ca. 100 kDa. This fragment was subjected to mass spectrometric analysis, and the obtained peptide mass fingerprint was used to search databases. The result of the search suggested that the protein carrying strepadhesin activity is a putative streptococcal pullulanase.
FIG. 2.
(A) Binding of thyroglobulin to SpeB extracts of the rgg mutant strain NZ131rgg. Bacteria were digested with 10 μg of SpeB/ml at 37°C for 30 min, and the digestion was terminated by adding the SpeB inhibitor E64 to a concentration of 1 μM. The extract was subjected to SDS-PAGE on a 7% gel and then stained with silver. A parallel sample transferred to nitrocellulose membrane was probed with biotinylated thyroglobulin to identify strepadhesin activity. An extract of strain NZ131speB is shown as a control. The molecular weights (MW) in thousands are indicated on the left. (B) Effect of SpeB treatment on the strepadhesin activity of NZ131rgg. Bacteria were either kept on ice (0°C), incubated in PBS at 37°C, or treated with SpeB at a concentration of 100 μg/ml at 37°C for 4 h to totally abolish the strepadhesin activity. The binding of thyroglobulin to 1-μl aliquots of bacterial suspension (OD600 = 4.0) on nitrocellulose membrane was detected as described for Fig. 1B.
Expression and purification of recombinant putative pullulanase.
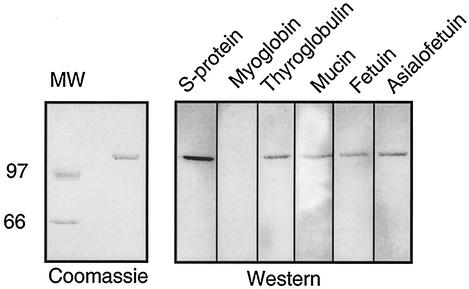
In order to confirm the finding that the putative streptococcal pullulanase carries strepadhesin activity, the pullulanase was recombinantly expressed in E. coli. A PCR fragment lacking the 5′ end of the gene corresponding to the putative N-terminal signal peptide, as well as the 3′ end corresponding to the C-terminal cell wall-anchoring motif and membrane-spanning region, was generated and cloned into the E. coli expression vector pET30 Ek/LIC. The recombinant putative pullulanase was expressed in E. coli BL21(DE3)pLysS as a fusion protein with N-terminal His and S tags and purified by affinity chromatography on Ni-NTA resin. The purified protein had a mobility corresponding to the expected molecular mass of ca. 125 kDa of the recombinant protein (Fig. 3, left panel).
FIG. 3.
Binding of glycoproteins to recombinant putative pullulanase. Recombinant putative pullulanase was subjected to SDS-PAGE on a 7% gel and stained with Coomassie or transferred to nitrocellulose membranes. The membranes were probed with the glycoproteins indicated or with myoglobin (negative control) or S protein (positive control). The molecular weights (MW) in thousands are indicated on the left.
Strepadhesin activity of the recombinant putative pullulanase.
The recombinant putative pullulanase was subjected to SDS-PAGE and blotted to a nitrocellulose membrane. The membrane was probed with biotin-labeled glycoproteins thyroglobulin, submaxillary mucin, fetuin, and asialofetuin to determine whether the recombinant protein exhibited strepadhesin activity. The results demonstrated that the recombinant protein bound these glycoproteins, and thus indicated that the pullulanase carried strepadhesin activity (Fig. 3, right panel).
Generation of a pullulanase-negative double mutant from the NZ131rgg strain.
In order to further investigate the contribution of streptococcal pullulanase to the strepadhesin activity, an isogenic pulA mutant strain was generated from the NZ131rgg strain. An internal 1,180-bp NsiI-BamHI fragment of the pulA gene was cloned into pSF151, a suicide vector incapable of replicating in streptococci, resulting in plasmid pulA-pSF151. The plasmid was electroporated in NZ131rgg, and a mutant, designated NZ131rgg-pulA, was chosen for further experiments (Fig. 4A). Genomic DNA was extracted from the bacteria, and PCR amplification was performed to verify the integration of the plasmid into the pulA gene in NZ131rgg-pulA genome (Fig. 4B). Primers specific for pSF151 plasmid and 5′ upstream region of pulA gene were used in the experiment. A fragment of the expected size (ca. 1.6 kb) could be amplified from NZ131rgg-pulA but not from NZ131rgg, a finding which indicated integration of the suicide plasmid into the pulA gene in NZ131rgg-pulA genome.
FIG. 4.
(A) Strategy for mutagenesis of the streptococcal putative pullulanase gene, pulA. An internal pulA fragment comprising the nucleotides 297 to 1476 was cloned into the suicide plasmid pSF151, generating plasmid pulA-pSF151. The plasmid was electroporated into NZ131rgg, and the transformants were screened for homologous recombination by plating the bacteria on THY-kanamycin-erythromycin agar plates. (B) PCR analysis of the pulA region of the genomic DNA from NZ131rgg and NZ131rgg-pulA. Primers a and b (see Materials and Methods) were used to amplify a 1.6-kb fragment from the NZ131rgg-pulA genome to verify the integration of pulA-pSF151 into the correct location. (C) Binding of thyroglobulin, submaxillary mucin, fetuin, and asialofetuin to dilution series of NZ131rgg and NZ131 rgg-pulA mutant strains.
The strepadhesin activity of NZ131rgg-pulA bacteria was studied on nitrocellulose membrane (Fig. 4C). The binding of thyroglobulin, submaxillary mucin, fetuin, and asialofetuin to the double-mutant bacteria was clearly reduced compared to the parental strain NZ131rgg. This demonstrated that the streptococcal putative pullulanase is a carrier of strepadhesin activity.
Binding of radiolabeled streptococci to glycoproteins on microtiter plates.
From the results of the bacterial dot blot assays above, it is clear that immobilized NZ131 wild-type and mutant strains exhibit differential binding to liquid-phase glycoproteins. In order to study whether the bacteria are able to bind to ligands immobilized on a solid surface, we examined the adhesion of radiolabeled NZ131 wild-type and mutant streptococci to thyroglobulin on microtiter plate wells. The number of NZ131rgg bacteria adhered to wells coated with thyroglobulin was markedly larger than the number of NZ131 wild-type bacteria (Fig. 5). Moreover, the NZ131rgg-pulA strain defective in putative pullulanase adhered less than did NZ131rgg. The binding of the NZ131rgg-pulA bacteria was, however, higher than the binding of the wild-type bacteria, which suggests that there may be additional molecules involved in the binding of the glycoproteins.
FIG. 5.
Binding of radiolabeled streptococci to glycoproteins on microtiter plates. The plates were coated with 10 μg of thyroglobulin, human laminin, or BSA/ml, and the adhesion of radiolabeled NZ131 wild-type and the rgg and rgg-pulA deficient mutant streptococci was detected with a liquid scintillation counter (see Materials and Methods). The results are given as mean values + the standard deviation of eight parallel wells.
In addition, we investigated the binding activities of the bacteria to human laminin and found that the adhesion of the rgg and rgg-pulA mutants was three to four times that of the wild-type strain. However, there was no difference in the binding between the mutants, which suggested that the laminin-binding activity is independent of the putative streptococcal pullulanase.
Sequence of putative pullulanase.
The whole putative pullulanase gene (pulA) was amplified from the genomic DNA by using the Vent polymerase and then subjected to sequencing in order to acquire the pulA nucleotide sequence of the strain under investigation (NZ131). The sequence revealed an open reading frame (ORF) with 3,498 bp encoding a protein of 1,165 amino acids with a predicted molecular mass of 129 kDa (Fig. 6). A putative N-terminal signal sequence with 45 amino acids was predicted by using SignalP V1.1 software (http://www.cbs.dtu.dk/services/SignalP/). At the C terminus, an LPKTGE cell wall-anchoring motif was present, followed by a hydrophobic membrane-spanning region (39). The deduced amino acid sequence contained the four highly conserved regions (I, II, III, and IV) common to pullulanases and forming the catalytic domain. These sequence data have been submitted to the EMBL database under accession number AJ488940.
FIG. 6.
Deduced amino acid sequence of PulA. The putative N-terminal signal sequence with 45 amino acids and the C-terminal cell wall-anchoring motif LPKTGE are underlined. The four conserved regions common to pullulanases are underlined and are indicated by numerals I, II, III, and IV. The amino acids polymorphic between the NZ131, SF370 (18), MGAS8232 (55), MGAS315 (3), and Manfredo strains (http://www.sanger.ac.uk/Projects/S_pyogenes/) are indicated in boldface.
Comparison of the putative pullulanase amino acid sequence with the published S. pyogenes genomic sequences (3, 18, 55) and with the unpublished sequence from S. pyogenes sequencing group at the Sanger Institute (http://www.sanger.ac.uk/Projects/S_pyogenes/) revealed that it is highly conserved with 97% homology among the sequences. The comparison showed that there are 35 amino acids scattered along the sequence showing polymorphism among the five sequences (Fig. 6). They all contained the four identical catalytic domains and the LPKTGE motif. In addition, the catalytic domains were identical to S. pneumoniae pullulanase SpuA and S. agalactiae putative pullulanase catalytic domains. In other bacteria, the pneumococcal pullulanase and the putative pullulanase of S. agalactiae are the closest homologues to S. pyogenes pullulanase, with 52 and 63% homology, respectively (7, 19, 58).
Identification of the putative pullulanase as a genuine pullulanase enzyme.
The pullulanase activity of the recombinant putative pullulanase was measured by using the dinitrosalisylic acid method. An aliquot of the enzyme was incubated with pullulan or starch at different pH values (pH 5, 7, and 9), and the liberated reducing sugars were quantified after addition of the dinitrosalisylic acid reagent and boiling of the sample by determining the absorbance of the sample at 550 nm. The pullulanase showed highest pullulan hydrolyzing activity at neutral pH and a weaker activity at pH 9 (Fig. 7A). At pH 5 the pullulanase was only slightly active against pullulan. In addition to the pullulan hydrolyzing activity, streptococcal pullulanase also showed starch-degrading activity, which was also highest at neutral pH. Therefore, streptococcal pullulanase can be considered as a neutral amylopullulanase. The control enzyme, Bacillus pullulanase, showed highest pullulan-degrading activity at pH 5, in accordance with its nature as an acidic pullulanase.
FIG. 7.
(A) Enzymatic activity of PulA. An aliquot of the recombinant enzyme was incubated in a reaction mixture containing 10 mg of pullulan or starch/ml in either sodium citrate buffer (pH 5), PBS (pH 7), or glycine-NaOH buffer (pH 9) for 10 min at 37°C The hydrolysis of the polysaccharides was quantified by the dinitrosalisylic acid method. Commercially available Bacillus pullulanase (0.03 U) was used as a control enzyme. (B) Enzymatic activity of pullulanase toward glycogen and effect of thyroglobulin and albumin on the pullulan-degrading activity of PulA at pH 7. Recombinant PulA was incubated in a reaction mixture containing either 10 mg of pullulan or glycogen/ml. In the inhibition assays, pullulan was used as the substrate, and thyroglobulin or BSA was added to the reaction at a concentration of 0.1 mg/ml.
Thyroglobulin did not inhibit the enzymatic activity of pullulanase (Fig. 7B). Instead, there was a slight increase in the activity, but a similar increase caused by albumin suggested that this was due to an unspecific stabilizing effect of the added proteins. This demonstrates that thyroglobulin does not interfere with the enzymatic activity of pullulanase and thus suggests that the glycoprotein-binding and the enzymatic activities are independent of each other. In addition, pullulanase did not cleave glycogen, nor was pullulan-degrading activity inhibited by this polysaccharide (Fig. 7B). The recombinant protein lost pullulanase activity upon freezing and thawing and was therefore stored at 4°C, at which temperature it remained active for months.
DISCUSSION
In previous studies we identified a novel glycoprotein-binding activity in S. pyogenes binding to thyroglobulin, submaxillary mucin, fetuin, and asialofetuin (23, 24). The activity, called strepadhesin activity, was found to be present in the majority of strains studied representing different M types and various infection foci. The strepadhesin activity of the M2 strain A8173 was demonstrated to be regulated by the virulence factor regulator Mga. In subsequent experiments, we were able to identify a molecule carrying strepadhesin activity to be a bacterial-surface-associated streptococcal cysteine protease, SpeB. We describe here the identification of another strepadhesin activity-carrying molecule, streptococcal pullulanase.
The occurrence of more than one molecule displaying similar biological activity is not unusual. As an example of this, there are several adhesins described in various S. pyogenes strains mediating adhesion to fibronectin. These include among others protein F (SfbI) (22, 56), protein F2 (26), fibronectin-binding protein FBP54 (14), SfbII/serum opacity factor (31, 45), 28-kDa fibronectin-binding protein (13), and PFBP (47). Therefore, it is not unexpected that S. pyogenes strepadhesin activity is mediated by more than just one molecule, which may vary among strains. The receptor structure for strepadhesin in host tissues and the binding specificity of the activity in terms of the molecular details is yet unknown. The apparently broad binding specificity toward different glycoproteins is, however, not unusual among bacterial adhesins. For example, antigen I and II adhesins of oral streptococci mediate binding to a wide variety of molecules, such as salivary agglutinin, collagen, laminin, and fibronectin, and also to other microorganisms of the oral cavity without any apparent specificity toward a single molecular determinant (28, 36, 43, 44, 50).
The increased amount of streptococcal pullulanase on the surface of the Rgg-deficient strain NZ131rgg and the increased strepadhesin activity could be due to at least two cellular mechanisms. An obvious explanation would be that in this strain the lack of secretion of SpeB, which is known to cleave proteins from the bacterial surface, results in decreased shedding of pullulanase and thereby in increased strepadhesin activity. However, in this case one would expect increased strepadhesin activity also in the speB mutant strain NZ131speB, but this is not the case. In addition, the bacterial surface protein extract prepared from NZ131speB had no strepadhesin reactivity in Western blots, neither were there any pullulanase-sized fragments, as judged by silver staining of SDS-PAGE gels (Fig. 2A). Another possibility is that the increase in surface-associated pullulanase is due to an increase in the expression of the protein. In addition to the positive influence on speB expression, Rgg is known to either up- or downregulate the expression of several streptococcal genes (11). Therefore, the most plausible explanation for the increased strepadhesin activity in NZ131rgg strain is that pullulanase is upregulated in the absence of Rgg activity.
The biological role of an enzyme-degrading pullulan, a polysaccharide consisting mainly of maltotriose units joined through α1-6 linkages, on the surface of a S. pyogenes is not clear. Pullulanase may hydrolyze potential carbon sources to be used as energy by the bacteria, but since pullulan is a carbohydrate not encountered by S. pyogenes in its natural habitat, this seems unlikely. However, the streptococcal pullulanase carries also starch-degrading activity, which could be used by the bacterium during the colonization of the nasopharyngeal epithelium. Based on the structural similarities between pullulan and glycogen, where the glucose units are similarly linked by α1-4 bonds with branches formed by α1-6 bonds, one could assume that streptococcal pullulanase could have a role in the glycogen-degradation. However, we did not observe any cleavage of glycogen by pullulanase (Fig. 7B). Another possibility could be that glycogen has pullulanase-inhibiting activity, but we did not detect any such activity either.
The surface association of enzymes that are normally cytosolic or extracellular or that have no known function in the bacterial metabolism has been reported increasingly during the last few years. In an earlier study we described the surface localization of streptococcal cysteine protease, an enzyme normally secreted outside the bacterium, mediating streptococcal adherence to glycoproteins (23). The bacterial cytosolic enzymes GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and α-enolase have been found as bacterial surface constituents (41, 42). GAPDH, also called streptococcal surface dehydrogenase, has been shown to bind fibronectin, lysozyme, and the cytoskeletal proteins myosin and actin and may therefore have a role in the colonization or subsequent steps of infection (41). The surface α-enolase was shown to exhibit strong plasminogen-binding activity (42). We show here that S. pyogenes expresses on its surface an enzyme with no obvious enzymatic role in the streptococcal life cycle but which supports the adhesion of the pathogen to solid surfaces coated with a ligand molecule.
The presence in S. pyogenes genome of an ORF with homology to known pullulanases has been reported, but the identity of the putative enzyme as a genuine pullulanase or other properties of the protein encoded by this ORF was not addressed (27). Reid et al. (46), by searching streptococcal genomes for putative surface proteins based on the presence of the LPXTG motif or on the homology of the sequences with known virulence-associated proteins, identified putative streptococcal pullulanase as a possible surface protein. These authors expressed fragments of such proteins and found antibodies to a fragment of the pullulanase in sera of healthy and S. pyogenes-infected subjects. We have expressed the whole surface-exposed region of streptococcal pullulanase and found antibodies specific to pullulanase in the sera of patients who have had S. pyogenes infections (unpublished result). These findings indicate that streptococcal pullulanase is expressed also in vivo. These results, in addition to the close sequence homology among pullulanases from different S. pyogenes strains, also suggest streptococcal pullulanase as a vaccine candidate.
It is at present not understood, why a carbohydrate-degrading enzyme such as pullulanase occurs at the streptococcal surface. Enzymatic roles of S. pyogenes pullulanase other than hydrolysis of potential carbon sources are not known. It might cleave some (as-yet-uncharacterized) host carbohydrates, thereby contributing to penetration of the tissues. To this end, we have previously found in rat tissues a developmentally regulated 1-4-linked glucose polymer, the identity or possible glycogen nature of which could not be determined (33). Glucose is also a common finding in the monosaccharide analysis of tissue-derived glycoproteins, although it is usually regarded as an external contaminant. Modification of host glycogonjugates by a carbohydrate-degrading enzyme may lead to removal of binding sites or to the uncovering of more internal binding sites of the bacteria. Pullulanase could also exert its activity toward carbohydrates on the bacterial surface of itself or of other bacteria.
In summary, we described here the identification of streptococcal pullulanase, a novel cell surface-associated molecule that carries strepadhesin activity in addition to polysaccharidase activity. Although several carbohydrate-degrading roles can be envisioned for pullulanase, the enzymatic function of the protein remains obscure in terms of in vivo substrate specificity. Therefore, from the viewpoint of the evolution of streptococcal virulence, it is possible that S. pyogenes has acquired the gene coding for pullulanase not because of the enzymatic activity but because of the advantage gained through the strepadhesin activity it encodes.
Acknowledgments
This work was supported by grants from the Sigrid Jusélius Foundation and the Academy of Finland.
We thank Dieter Gerlach for the SpeB enzyme, Kyllikki Kunnas, and Michael Chaussee for providing the bacterial strains and Terttu Jompero for technical assistance.
Editor: A. D. O'Brien
REFERENCES
- 1.Andersson, B., J. Dahmen, T. Frejd, H. Leffler, G. Magnusson, G. Noori, and C. Svanborg-Eden. 1983. Identification of an active disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells. J. Exp. Med. 158:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthelson, R., A. Mobasseri, D. Zopf, and P. Simon. 1998. Adherence of Streptococcus pneumoniae to respiratory epithelial cells is inhibited by sialylated oligosaccharides. Infect. Immun. 66:1439-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berge, A., and L. Björck. 1995. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem. 270:9862-9867. [DOI] [PubMed] [Google Scholar]
- 5.Beuth, J., B. Stoffel, and G. Pulverer. 1996. Inhibition of bacterial adhesion and infections by lectin blocking. Adv. Exp. Med. Biol. 408:51-56. [DOI] [PubMed] [Google Scholar]
- 6.Bochicchio, G. V., M. Joshi, M. Joshi, S. Henry, and T. Scalea. 2001. Group A streptococcus (GAS) soft-tissue infections: a lethal organism on the rise. Am. Surg. 67:1089-1092. [PubMed] [Google Scholar]
- 7.Bongaerts, R. J., H. P. Heinz, U. Hadding, and G. Zysk. 2000. Antigenicity, expression, and molecular characterization of surface-located pullulanase of Streptococcus pneumoniae. Infect. Immun. 68:7141-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 9.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaussee, M. S., D. Gerlach, C. E. Yu, and J. J. Ferretti. 1993. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect. Immun. 61:3719-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chelsom, J., A. Halstensen, T. Haga, and E. A. Hoiby. 1994. Necrotising fasciitis due to group A streptococci in western Norway: incidence and clinical features. Lancet 344:1111-1115. [DOI] [PubMed] [Google Scholar]
- 13.Courtney, H. S., D. L. Hasty, J. B. Dale, and T. P. Poirier. 1992. A 28-kilodalton fibronectin-binding protein of group A streptococci. Curr. Microbiol. 25:245-250. [DOI] [PubMed] [Google Scholar]
- 14.Courtney, H. S., Y. Li, J. B. Dale, and D. L. Hasty. 1994. Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect. Immun. 62:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cundell, D. R., and E. I. Tuomanen. 1994. Receptor specificity of adherence of Streptococcus pneumoniae to human type-II pneumocytes and vascular endothelial cells in vitro. Microb. Pathog. 17:361-374. [DOI] [PubMed] [Google Scholar]
- 16.Demuth, D. R., C. A. Davis, A. M. Corner, R. J. Lamont, P. S. Leboy, and D. Malamud. 1988. Cloning and expression of a Streptococcus sanguis surface antigen that interacts with a human salivary agglutinin. Infect. Immun. 56:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demuth, D. R., E. E. Golub, and D. Malamud. 1990. Streptococcal-host interactions. Structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein. J. Biol. Chem. 265:7120-7126. [PubMed] [Google Scholar]
- 18.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 20.Haataja, S., K. Tikkanen, J. Liukkonen, C. Francois-Gerard, and J. Finne. 1993. Characterization of a novel bacterial adhesion specificity of Streptococcus suis recognizing blood group P receptor oligosaccharides. J. Biol. Chem. 268:4311-4317. [PubMed] [Google Scholar]
- 21.Haataja, S., K. Tikkanen, U. Nilsson, G. Magnusson, K. A. Karlsson, and J. Finne. 1994. Oligosaccharide-receptor interaction of the Galα1-4Gal binding adhesin of Streptococcus suis. Combining site architecture and characterization of two variant adhesin specificities. J. Biol. Chem. 269:27466-27472. [PubMed] [Google Scholar]
- 22.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hytönen, J., S. Haataja, D. Gerlach, A. Podbielski, and J. Finne. 2001. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol. Microbiol. 39:512-519. [DOI] [PubMed] [Google Scholar]
- 24.Hytönen, J., S. Haataja, P. Isomäki, and J. Finne. 2000. Identification of a novel glycoprotein-binding activity in Streptococcus pyogenes regulated by the mga gene. Microbiology 146:31-39. [DOI] [PubMed] [Google Scholar]
- 25.Idänpään-Heikkilä, I., P. M. Simon, D. Zopf, T. Vullo, P. Cahill, K. Sokol, and E. Tuomanen. 1997. Oligosaccharides interfere with the establishment and progression of experimental pneumococcal pneumonia. J. Infect. Dis. 176:704-712. [DOI] [PubMed] [Google Scholar]
- 26.Jaffe, J., S. Natanson-Yaron, M. G. Caparon, and E. Hanski. 1996. Protein F2, a novel fibronectin-binding protein from Streptococcus pyogenes, possesses two binding domains. Mol. Microbiol. 21:373-384. [DOI] [PubMed] [Google Scholar]
- 27.Janulczyk, R., and M. Rasmussen. 2001. Improved pattern for genome-based screening identifies novel cell wall-attached proteins in gram-positive bacteria. Infect. Immun. 69:4019-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkinson, H. F., and D. R. Demuth. 1997. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol. Microbiol. 23:183-190. [DOI] [PubMed] [Google Scholar]
- 29.Karlsson, K. A. 1995. Microbial recognition of target-cell glycoconjugates. Curr. Opin. Struct. Biol. 5:622-635. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson, K. A. 1998. Meaning and therapeutic potential of microbial recognition of host glycoconjugates. Mol. Microbiol. 29:1-11. [DOI] [PubMed] [Google Scholar]
- 31.Kreikemeyer, B., S. R. Talay, and G. S. Chhatwal. 1995. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Mol. Microbiol. 17:137-145. [DOI] [PubMed] [Google Scholar]
- 32.Krivan, H. C., D. D. Roberts, and V. Ginsburg. 1988. Many pulmonary pathogenic bacteria bind specifically to the carbohydrate sequence GalNAc β1-4Gal found in some glycolipids. Proc. Natl. Acad. Sci. USA 85:6157-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krusius, T., J. Finne, J. Kärkkäinen, and J. Järnefelt. 1974. Neutral and acidic glycopeptides in adult and developing rat brain. Biochim. Biophys. Acta 365:80-92. [DOI] [PubMed] [Google Scholar]
- 34.Kussmann, M., E. Nordhoff, H. Rahbek-Nielsen, S. Haebel, M. Rossel-Larssen, L. Jakobsen, J. Gobom, E. Mirgorodskaya, A. Kroll-Kristensen, L. Palm, and P. Roepstroff. 1997. Matrix-assisted laser desorption/ionization mass spectrometry sample preparation techniques designed for various peptide and protein analytes. J. Mass Spectrom. 32:593-601. [Google Scholar]
- 35.Liukkonen, J., S. Haataja, K. Tikkanen, S. Kelm, and J. Finne. 1992. Identification of N-acetylneuraminyl α2→3 poly-N-acetyllactosamine glycans as the receptors of sialic acid-binding Streptococcus suis strains. J. Biol. Chem. 267:21105-21111. [PubMed] [Google Scholar]
- 36.Love, R. M., M. D. McMillan, and H. F. Jenkinson. 1997. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect. Immun. 65:5157-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin, J. M., M. Green, K. A. Barbadora, and E. R. Wald. 2002. Erythromycin-resistant group A streptococci in schoolchildren in Pittsburgh. N. Engl. J. Med. 346:1200-1206. [DOI] [PubMed] [Google Scholar]
- 38.Miller, G. L., R. Blum, W. E. Glennon, and A. L. Burton. 1960. Measurement of carboxymethylcellulase activity. Anal. Biochem. 1:127-132. [Google Scholar]
- 39.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 40.Nyman, T. A., S. Matikainen, T. Sareneva, I. Julkunen, and N. Kalkkinen. 2000. Proteome analysis reveals ubiquitin-conjugating enzymes to be a new family of interferon-alpha-regulated genes. Eur. J. Biochem. 267:4011-4019. [DOI] [PubMed] [Google Scholar]
- 41.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pancholi, V., and V. A. Fischetti. 1998. α-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 43.Petersen, F. C., S. Assev, H. C. van der Mei, H. J. Busscher, and A. A. Scheie. 2002. Functional variation of the antigen I/II surface protein in Streptococcus mutans and Streptococcus intermedius. Infect. Immun. 70:249-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petersen, F. C., S. Pasco, J. Ogier, J. P. Klein, S. Assev, and A. A. Scheie. 2001. Expression and functional properties of the Streptococcus intermedius surface protein antigen I/II. Infect. Immun. 69:4647-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rakonjac, J. V., J. C. Robbins, and V. A. Fischetti. 1995. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect. Immun. 63:622-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reid, S. D., N. M. Green, J. K. Buss, B. Lei, and J. M. Musser. 2001. Multilocus analysis of extracellular putative virulence proteins made by group A streptococcus: population genetics, human serologic response, and gene transcription. Proc. Natl. Acad. Sci. USA 98:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rocha, C. L., and V. A. Fischetti. 1999. Identification and characterization of a novel fibronectin-binding protein on the surface of group A streptococci. Infect. Immun. 67:2720-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 49.Ryan, P. A., V. Pancholi, and V. A. Fischetti. 2001. Group A streptococci bind to mucin and human pharyngeal cells through sialic acid-containing receptors. Infect. Immun. 69:7402-7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sciotti, M. A., I. Yamodo, J. P. Klein, and J. A. Ogier. 1997. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct binding domains. FEMS Microbiol. Lett. 153:439-445. [DOI] [PubMed] [Google Scholar]
- 51.Seppälä, H., T. Klaukka, J. Vuopio-Varkila, A. Muotiala, H. Helenius, K. Lager, P. Huovinen, et al. 1997. The effect of changes in the consumption of macrolide antibiotics on erythromycin resistance in group A streptococci in Finland. N. Engl. J. Med. 337:441-446. [DOI] [PubMed] [Google Scholar]
- 52.Sharon, N., and I. Ofek. 2000. Safe as mother's milk: carbohydrates as future anti-adhesion drugs for bacterial diseases. Glycoconj. J. 17:659-664. [DOI] [PubMed] [Google Scholar]
- 53.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 54.Simon, D., and J. J. Ferretti. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 66:219-224. [DOI] [PubMed] [Google Scholar]
- 55.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talay, S. R., P. Valentin-Weigand, K. N. Timmis, and G. S. Chhatwal. 1994. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol. Microbiol. 13:531-539. [DOI] [PubMed] [Google Scholar]
- 57.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 58.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zopf, D., and S. Roth. 1996. Oligosaccharide anti-infective agents. Lancet 347:1017-1021. [DOI] [PubMed] [Google Scholar]
- 60.Zopf, D., P. Simon, R. Barthelson, D. Cundell, I. Idänpään-Heikkilä, and E. Tuomanen. 1996. Development of anti-adhesion carbohydrate drugs for clinical use. Adv. Exp. Med. Biol. 408:35-38. [DOI] [PubMed] [Google Scholar]