Abstract
The glycosylation state of the glycosyl-phosphatidylinositol (GPI) anchored cellular prion protein (PrPC) can influence the formation of the disease form of the protein responsible for the neurodegenerative spongiform encephalopathies. We have investigated the role of membrane topology in the N-glycosylation of PrP by expressing a C-terminal transmembrane anchored form, PrP-CTM, an N-terminal transmembrane anchored form, PrP-NTM, a double-anchored form, PrP-DA, and a truncated form, PrPΔGPI, in human neuroblastoma SH-SY5Y cells. Wild-type PrP, PrP- CTM and PrP-DA were membrane anchored and present on the cell surface as glycosylated forms. In contrast, PrP-NTM, although membrane anchored and localized at the cell surface, was not N-glycosylated. PrPΔGPI was secreted from the cells into the medium in a hydrophilic form that was unglycosylated. The 4-fold slower rate at which PrPΔGPI was trafficked through the cell compared with wild-type PrP was due to the absence of the GPI anchor not the lack of N-glycans. Retention of PrPΔGPI in the endoplasmic reticulum did not lead to its glycosylation. These results indicate that C-terminal membrane anchorage is required for N-glycosylation of PrP.
Keywords: Creutzfeldt–Jakob disease/glycosyl-phosphatidylinositol/oligosaccharyltransferase/prion/scrapie
Introduction
Prion diseases are a group of neurodegenerative diseases that include Creutzfeldt–Jakob disease, Gerstmann– Straussler–Scheinker syndrome, fatal familial insomnia and Kuru in humans, scrapie in sheep and bovine spongiform encephalopathy in cattle. These diseases are caused by a conformational change in the normal cellular isoform of the prion protein (PrPC) to the scrapie isoform (PrPSc) (Prusiner, 1998). Murine PrPC is synthesized as a protein of 254 amino acids, which undergoes a variety of post-translational modifications including glycosylation and glycosyl-phosphatidylinositol (GPI) anchor addition (Stahl et al., 1987). PrPC contains two N-glycosylation sequons at Asn180 and Asn196, both of which can be variably glycosylated, giving rise to unglycosylated, mono-glycosylated and di-glycosylated species (Rudd et al., 1999; Stimson et al., 1999). The ratio of these three glycoforms appears to be characteristic for particular ‘strains’ of PrP (Collinge et al., 1996). Blockade of the glycosylation of PrPC, by either tunicamycin treatment of cells or mutation of the consensus N-glycosylation sites, does not prevent the formation of PrPSc (Taraboulos et al., 1990; Lehmann and Harris, 1997). Indeed, under-glycosylated molecules appear to be preferred substrates in the generation of PrPSc (Kocisko et al., 1994). An inherited form of spongiform encephalopathy has been reported in a Brazilian family with a Thr to Ala substitution at codon 183, which abolishes one of the consensus sites for glycosylation (Nitrini et al., 1997), thus underlining the importance of N-glycosylation to prion biology.
Glycosylation of membrane and secretory proteins involves a series of steps involving a number of oligo- and monosaccharide transferases (Kornfield and Kornfield, 1985; Varki, 1993). This process is initiated in the lumen of the endoplasmic reticulum (ER), concomitant with protein translation and translocation by the transfer of a core N-linked glycosylation unit, Glc3Man9GlcNAc2, onto acceptor Asn residues in the tripeptide sequon Asn-Xaa-Ser/Thr. This initial step is catalysed by the enzyme oligosaccharyltransferase, which is a component of the protein translocation machinery in the ER membrane (Kornfield and Kornfield, 1985). Thus, N-linked glycosylation is considered to be a co-translational event. As the newly glycosylated protein is transported from the ER to the cis-Golgi, trimming and addition of other sugar residues occur. N-linked glycosylation is often essential for the folding, stability, intracellular transport, secretion and function of glycoproteins (Rademacher et al., 1988). The addition of the GPI anchor to the C-terminus of membrane proteins occurs efficiently and rapidly within 1 min of translocation of the polypeptide chain (Bangs et al., 1985). A transamidase enzyme, Gpi8p, is involved in this process, and is closely associated with the translocon apparatus in the membrane of the ER (Benghezal et al., 1996). Thus, addition of the GPI anchor is likely to occur after glycosylation of available Asn residues by oligosaccharyltransferase, and would not be expected to interfere drastically with N-glycosylation of the protein.
We have investigated the role of membrane anchorage and topology in the N-glycosylation of PrP by expressing wild-type murine PrP in human neuroblastoma SH-SY5Y cells, and directly comparing its processing with a form that has a C-terminal transmembrane anchor substituted for the GPI anchor, one that has only an N-terminal transmembrane anchor, one that retains the GPI anchor and has an N-terminal transmembrane anchor, and one that lacks the GPI anchor addition signal. The truncated form of PrP was efficiently secreted from the cells and was unglycosylated, while the N-terminally anchored form was membrane associated and also unglycosylated. In contrast, the C-terminally anchored forms and the double-anchored form were glycosylated. Retention of the secreted form in the ER did not lead to its glycosylation, and the lack of the GPI anchor slowed the rate of trafficking of the protein through the secretory pathway. Thus, C-terminal membrane anchorage of PrP, whether via a GPI moiety or a transmembrane polypeptide, is required for N-glycosylation to occur, and the GPI anchor facilitates forward trafficking through the secretory pathway.
Results
Membrane topology mutants of PrP
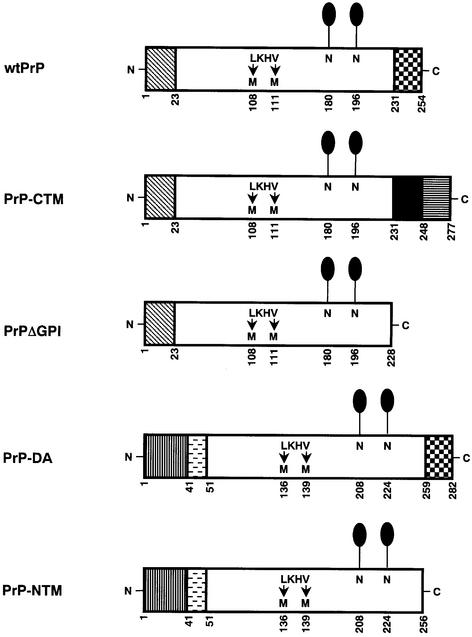
In order to investigate the role of membrane topology in the glycosylation of PrP, alternatively anchored forms and a secreted form of murine PrP were constructed (Figure 1). PrP-CTM has the C-terminal GPI signal peptide replaced with the transmembrane and cytosolic domains from the type I integral membrane glycoprotein, human angiotensin converting enzyme (Soubrier et al., 1988). PrPΔGPI lacks the C-terminal signal sequence required for GPI anchor addition. Both of these constructs retain the cleavable N-terminal signal sequence present in wild-type PrP (wtPrP), whereas in PrP-DA and PrP-NTM this N-terminal signal sequence was replaced with the uncleaved signal sequence/transmembrane and stalk domains from the type II integral membrane glycoprotein, murine aminopeptidase A (Wu et al., 1990). PrP-DA retains the GPI anchor addition sequence at the C-terminus, while PrP-NTM lacks this sequence. All the constructs were epitopically tagged by the substitution of Leu108 and Val111 for methionines, which allows for their recognition by the species-specific antibody 3F4 (Kascsak et al., 1987). Each of the constructs was stably expressed by the human neuroblastoma cell line SH-SY5Y, which lacks detectable levels of human PrP (Perera and Hooper, 1999). Stably expressing cell lines all maintained the ability to take up and release [3H]noradrenaline in a similar manner to untransfected cells (Turner et al., 1994), indicative of their neuronal status.
Fig. 1. Structures of the murine PrP constructs. Murine PrP comprises a 22 amino acid N-terminal signal sequence (diagonally hatched box), two consensus sequences for N-linked glycosylation (N180 and N196) (lollipops) and a 23 amino acid C-terminal GPI anchor addition sequence (chequered box). In PrP-CTM, the C-terminal GPI signal sequence of PrP (231–254) was substituted for the transmembrane (black box) and cytoplasmic (horizontally striped box) domains of human angiotensin converting enzyme. In PrPΔGPI, residues 229–254 of PrP, comprising the C-terminal GPI signal sequence, were deleted. In PrP-NTM and PrP-DA, the signal peptide was replaced with the uncleaved signal sequence/transmembrane domain (vertically striped box) and stalk region (horizontally dashed box) from murine aminopeptidase A. PrP-DA retains the C-terminal GPI addition sequence present in wtPrP, while PrP-NTM lacks this sequence. Numbers beneath the schematics indicate amino acid positions. Wild-type murine PrP and the constructs had methionine residues substituted at the two positions indicated (L108M and V111M), which allowed the proteins to be recognized by the monoclonal antibody 3F4.
PrPΔGPI is hydrophilic and secreted from SH-SY5Y cells
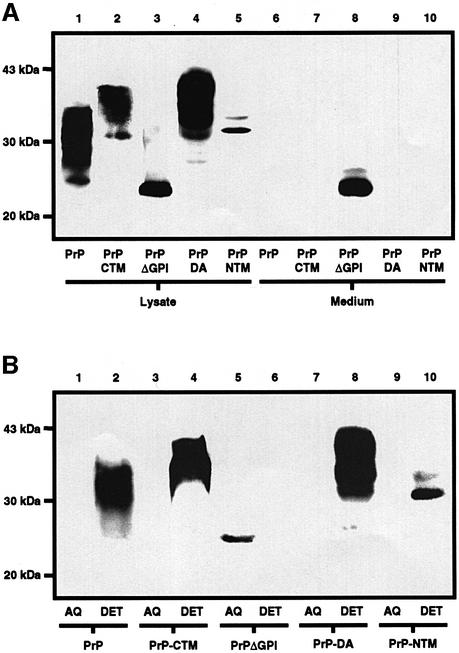
Western blot analysis of the SH-SY5Y cells (Figure 2A) expressing wtPrP revealed a diffuse band of 28–36 kDa in the cell lysate, but no immunoreactive protein was detected in the medium from these cells after 24 h of incubation. Similarly, PrP-CTM and PrP-DA appeared as diffuse bands of 34–43 kDa in the cell lysate but not in the medium, and PrP-NTM appeared as a narrow band of 32 kDa in the cell lysate but not in the medium. In contrast, after 24 h of incubation, PrPΔGPI appeared as a band of 27 kDa in both the cell lysate and the medium, consistent with this protein behaving as a secretory protein. To determine whether the PrP constructs were membrane anchored, the relative hydrophobicity of the proteins was assessed by employing Triton X-114 to partition the cells into detergent and aqueous phases (Figure 2B). wtPrP, PrP-CTM, PrP-DA and PrP-NTM all partitioned completely into the detergent phase, consistent with these constructs possessing hydrophobic membrane-anchoring domains. PrPΔGPI, however, partitioned completely into the aqueous phase, indicating that this protein lacked any mode of membrane anchorage.
Fig. 2. PrPΔGPI is secreted from human neuroblastoma cells and is hydrophilic. (A) SH-SY5Y cells expressing either wtPrP, PrP-CTM, PrPΔGPI, PrP-DA or PrP-NTM were incubated with OptiMEM for 24 h. PrP constructs in the cell lysate and concentrated medium were separated by SDS–PAGE and then western blotted. PrP was detected with antibody 3F4. (B) SH-SY5Y cells expressing either wtPrP, PrP-CTM, PrPΔGPI, PrP-DA or PrP-NTM were lysed in Triton X-114 phase separation buffer and the phases partitioned by centrifugation. PrP constructs present in the aqueous phases and detergent phases were detected with antibody 3F4.
PrP-CTM, PrP-DA and PrP-NTM are all expressed at the cell surface
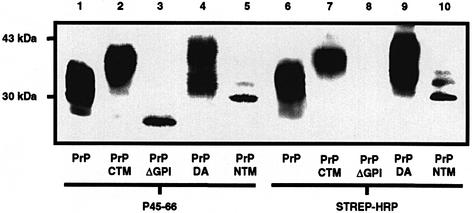
To assess whether the PrP constructs were cell-surface localized, intact cells were treated with the membrane-impermeable reagent biotin-NHS (Figure 3). The biotinylation procedure did not affect the recognition of the constructs by the 3F4 antibody as all the proteins were immunoprecipitated effectively and subsequently detected on the immunoblot with antibody P45-66 (Figure 3, lanes 1–5). That wtPrP, PrP-CTM, PrP-DA and PrP-NTM present in the 3F4 immunoprecipitate had been biotinylated was demonstrated by their binding to streptavidin-conjugated horseradish peroxidase. Thus, these constructs were all present on the surface of the SH-SY5Y cells and their alternative membrane-anchoring domains did not grossly affect the trafficking of PrP to the cell surface. Although PrPΔGPI was present in both the medium and cell lysate samples (Figure 2A), this construct did not bind to the streptavidin-conjugated horseradish peroxidase antibody (Figure 3, lane 8), indicating that the protein was not retained on the cell surface following secretion.
Fig. 3. PrP-CTM, PrP-DA and PrP-NTM are present at the cell surface. SH-SY5Y cells expressing either wtPrP, PrP-CTM, PrPΔGPI, PrP-DA or PrP-NTM were incubated for 30 min with PBS containing biotin sulfo-NHS. PrP constructs were immunoprecipitated from the cell lysates with antibody 3F4, resolved by SDS–PAGE and western blotted. The membranes were incubated with either the P45-66 antibody followed by a secondary peroxidase-conjugated anti-rabbit antibody or peroxidase-conjugated streptavidin.
PrP-DA has both a GPI anchor and a transmembrane anchoring domain
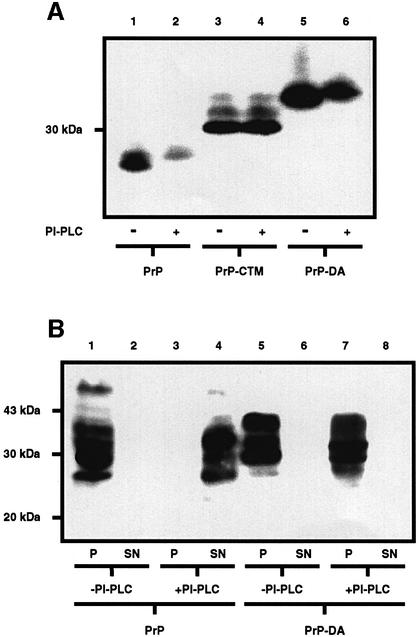
The partitioning of wtPrP, PrP-CTM and PrP-NTM into the detergent phase on phase separation in Triton X-114 (Figure 2B) is consistent with the presence of the GPI anchor, the C-terminal transmembrane anchor or the N-terminal transmembrane anchor, respectively, on these constructs. To confirm the presence of the GPI anchor on PrP-DA, cell lysates were incubated in the absence or presence of bacterial phosphatidylinositol-specific phospholipase C (PI-PLC) and analysed by SDS–PAGE (Figure 4A). PrP, like several other GPI-anchored proteins, undergoes a characteristic shift in mobility on SDS–PAGE, migrating with a higher apparent molecular weight when the GPI anchor has been cleaved by bacterial PI-PLC (Cardoso de Almeida and Turner, 1983; Stahl et al., 1987; Littlewood et al., 1989). In order to visualize this up-shift, the cells were incubated with tunicamycin to prevent the variable N-glycosylation of the proteins complicating the interpretation of the results. Wild-type PrP displayed the characteristic up-shift on the SDS gel following treatment of the cell lysate with bacterial PI-PLC. The mobility of PrP-CTM did not alter on treatment with PI-PLC, as this construct contains only a C-terminal transmembrane anchor. However, like wtPrP, PrP-DA displayed the characteristic up-shift on treatment with the bacterial PI-PLC, indicating the presence of the GPI anchor. To determine whether PrP-DA possesses both a GPI anchor and an N-terminal transmembrane anchor as predicted (Figure 1), membranes were incubated in the presence of bacterial PI-PLC, and the release of PrP into the high speed supernatant examined (Figure 4B). Incubation of membranes from cells expressing wtPrP with PI-PLC resulted in the relocation of the protein from the pellet to the supernatant. In contrast, PrP-DA remained in the pellet fraction after incubation of the membranes with PI-PLC, indicative of the presence of an additional PI-PLC-insensitive anchoring domain. These data are consistent with PrP-DA having both an N-terminal transmembrane anchor and a C-terminal GPI anchor.
Fig. 4. PrP-DA has both a GPI and a transmembrane anchor. (A) SH-SY5Y cells expressing either wtPrP, PrP-CTM or PrP-DA were incubated in the presence of tunicamycin for 24 h. The resulting cell lysates were incubated for 3 h in the absence or presence of bacterial PI-PLC. PrP constructs were detected with antibody 3F4. (B) Membranes prepared from SH-SY5Y cells expressing either wtPrP or PrP-DA were incubated for 3 h in the absence or presence of bacterial PI-PLC. Released proteins were separated from the membranes by high-speed centrifugation, and PrP in the resulting pellet (P) and supernatant (SN) fractions detected with antibody 3F4.
PrP-NTM and PrPΔGPI are predominantly unglycosylated
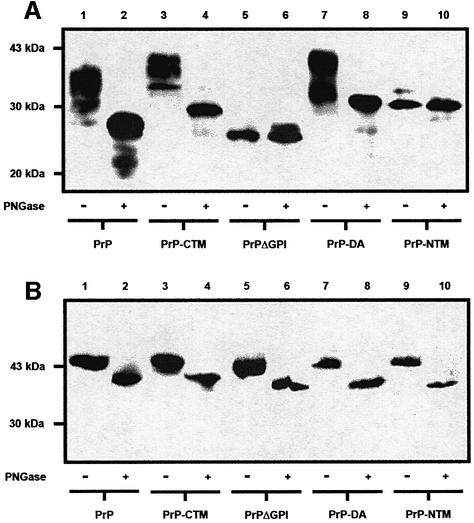
All the murine PrP constructs contain the two potential sites for N-linked glycosylation (Figure 1). In order to assess whether N-linked glycans were attached to the constructs, cell lysates were incubated with peptide N-glycosidase F (PNGase F) (Figure 5A). For the wtPrP, PrP-CTM and PrP-DA constructs, incubation in the presence of PNGase F resulted in a reduction in their molecular weights, indicative of the removal of N-linked glycans from the proteins. In contrast, the molecular weight of PrPΔGPI and PrP-NTM remained unaltered after incubation with PNGase F, indicating that these constructs were not N-glycosylated. A very minor amount of a monoglycosylated form of PrP-NTM was visible on overexposure of the immunoblots. The lack of glycosylation of PrPΔGPI and PrP-NTM was not due (i) to a gross abnormality in the cells’ glycosylation machinery, as another GPI-anchored protein, porcine membrane dipeptidase (Littlewood et al., 1989; White et al., 2000), co-transfected into the various cell lines was fully glycosylated in each case (Figure 5B) nor (ii) to overexpression of these two constructs relative to cells expressing wtPrP, as assessed by comparing the levels of expression of the various PrP constructs relative to endogenous actin (data not shown).
Fig. 5. PrPΔGPI and PrP-NTM are unglycosylated. (A) Cell lysates from SH-SY5Y cells expressing either wtPrP, PrP-CTM, PrPΔGPI, PrP-DA or PrP-NTM were incubated for 16 h in the absence or presence of PNGase F. The PrP constructs in the digests were detected with antibody 3F4. (B) Cells expressing either wtPrP, PrP-CTM, PrPΔGPI, PrP-DA or PrP-NTM were transiently transfected with the cDNA encoding porcine membrane dipeptidase (pEF-MDP) (White et al., 2000). Cells were lysed 48 h post-transfection and incubated with PNGase F as above. The dipeptidase was detected by immuno blotting with an anti-dipeptidase antibody (Littlewood et al., 1989).
Prolonged retention within the cell does not facilitate the glycosylation of PrPΔGPI
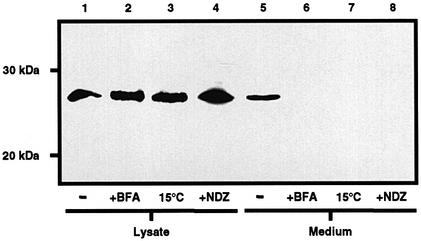
Secretory proteins retained within the ER for prolonged periods tend to become post-translationally overmodified (Chessler and Byers, 1992). In an attempt to promote the glycosylation of PrPΔGPI, the protein was retained within the ER by treatment of the cells either with brefeldin A, which causes a collapse of the Golgi compartment into the ER (Klausner et al., 1992), or with nocodazole, which depolymerizes microtubules, thus preventing vesicular trafficking (Puertollano and Alonso, 1999), or by incubation of the cells at 15°C, which prevents exit from the ER (Figure 6). These three different treatments resulted in the retention of PrPΔGPI within the cell, as indicated by the absence of the protein from the medium. The molecular weight of PrPΔGPI in the cell lysate, however, did not increase after these cellular treatments, indicating that the prolonged retention of PrPΔGPI within the ER had not promoted its glycosylation.
Fig. 6. Retention of PrPΔGPI within the cell does not promote glycosylation. SH-SY5Y cells expressing PrPΔGPI were incubated for 3 h with OptiMEM in the presence of brefeldin A (BFA), at 15°C or in the presence of nocodazole. PrPΔGPI present in the cell lysate and concentrated medium was detected with antibody 3F4.
PrPΔGPI is trafficked through the cell slower than wild-type PrP
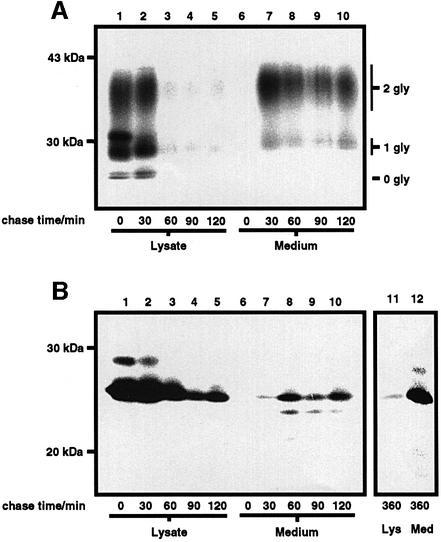
For certain secretory and cell surface glycoproteins, glycosylation is required for their efficient transport to the cell surface (Helenius, 1994), while removal of the GPI moiety from certain GPI-anchored proteins results in an impairment in their rate of transit to the cell surface (Bangs et al., 1997). The rate of transport of PrPΔGPI to the cell surface was compared with that of wtPrP by pulse-labelling (Figure 7). Upon initiation of the chase, radiolabelled wtPrP immunoprecipitated from the cell lysate appeared as three bands corresponding to the un-, mono- and di-glycosylated species. After 30 min of chase, wtPrP had arrived at the cell surface, as demonstrated by the appearance of the protein in the medium following PI-PLC treatment of the cells. After 60 min of chase, the majority of wtPrP was released into the medium by PI-PLC and, thus, was present at the cell surface. From densitometric analysis of the autoradiographs, the transport of wtPrP to the cell surface occurred with a t1/2 of 35 min. The rate of transport of PrPΔGPI to the cell surface was measured as the rate of appearance of PrPΔGPI in the medium (Figure 7B). Upon initiation of the chase, radiolabelled PrPΔGPI was immunoprecipitated from the cell lysate as a major band corresponding to the unglycosylated species and a minor band corresponding to the mono-glycosylated species. PrPΔGPI appeared in the medium after 30 min of chase, after which time the amount of PrPΔGPI in the medium increased with a corresponding decrease in the amount of the protein in the cell lysate. In contrast to wtPrP, the transit to the cell surface of PrPΔGPI was not complete after 120 min of chase. However, after 6 h of chase, the majority of PrPΔGPI was recovered from the medium. From densitometric analysis of the autoradiographs, the transport of PrPΔGPI to the cell surface occurred with a t½ of 135 min. The inefficient export of PrPΔGPI to the cell surface could not be ascribed to a general defect in the secretory pathway of the PrPΔGPI-expressing cells, as wtPrP co-expressed in the same cell line as PrPΔGPI was efficiently transported to the cell surface (data not shown). This impaired rate of transport of PrPΔGPI to the cell surface, in comparison to wtPrP, could be due to the absence of either the N-glycans or the GPI anchor, or both.
Fig. 7. PrPΔGPI traffics to the cell surface slower than wtPrP. SH-SY5Y cells expressing either wtPrP (A) or PrPΔGPI (B) were pulse-labelled for 20 min with [35S]Met/Cys and chased in the presence of cycloheximide and l-Met. Following the chase period, cells expressing wtPrP were incubated at 4°C with OptiMEM containing PI-PLC, lysed, and the medium from the PI-PLC treatment concentrated by methanol precipitation. Cells expressing PrPΔGPI were immediately lysed upon termination of the chase and the chase medium concentrated as above. Radiolabelled PrP constructs present in the cell lysate and concentrated medium were immunoprecipitated with antibody 3F4, resolved by SDS–PAGE, and visualized by autoradio graphy. The unglycosylated (0 gly), monoglycosylated (1 gly) and diglycosylated (2 gly) species of PrP are indicated.
Lack of N-glycans does not impair the forward trafficking of PrP
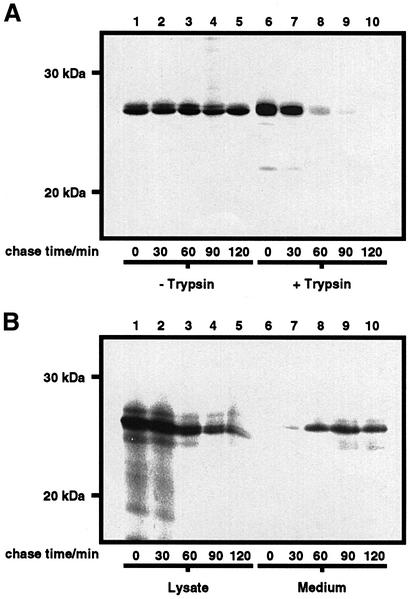
To ascertain the role of N-glycans in the transit of wtPrP to the cell surface, cells were pulse-labelled and chased in the presence of tunicamycin, which inhibits N-linked glycosylation in mammalian cells (Figure 8). The rate of appearance of wtPrP on the cell surface was measured as the rate of disappearance of wtPrP from the cell lysate after treatment of the intact cells with exogenous trypsin. Trypsin hydrolysed all the wtPrP on the cell surface under the conditions of the experiment (data not shown). Bacterial PI-PLC could not be employed as the enzyme was unable to release wtPrP from the cell surface after tunicamycin treatment, in agreement with previous observations (Lehmann and Harris, 1997). Upon initiation of the chase, radiolabelled wtPrP appeared as a single band of 28 kDa, corresponding to the unglycosylated species. In the absence of trypsin treatment, the amount of wtPrP in the cell lysate was constant over the time course of the experiment, indicating that unglycosylated wtPrP was stable and not subject to degradation within the cell. After trypsin treatment, the amount of unglycosylated wtPrP in the cell lysate steadily decreased as the time of chase increased. After 60 min of chase, the majority of wtPrP synthesized during the pulse period was hydrolysed by trypsin and, thus, was present at the cell surface, demonstrating that the transit of wtPrP to the cell surface had not been impaired by the treatment of the cells with tunicamycin. Similarly, the rate of appearance of PrPΔGPI in the medium was not affected by treatment of the cells with tunicamycin (Figure 8B). Consequently, N-glycosylation of PrP was not required for the efficient transport of the protein to the cell surface.
Fig. 8. Tunicamycin does not affect the secretion kinetics of wtPrP and PrPΔGPI. SH-SY5Y cells expressing either wtPrP (A) or PrPΔGPI (B) were pulse-labelled for 20 min with [35S]Met/Cys and chased in the presence of cycloheximide and l-Met. Tunicamycin was present in the starve, pulse and chase medium. Following the chase period, cells expressing wtPrP were either immediately lysed or harvested with trypsin and lysed. Cells expressing PrPΔGPI were immediately lysed upon termination of the chase and the chase medium concentrated by methanol precipitation. Radiolabelled PrP constructs were immuno precipitated with antibody 3F4, resolved by SDS–PAGE, and detected by autoradiography.
Discussion
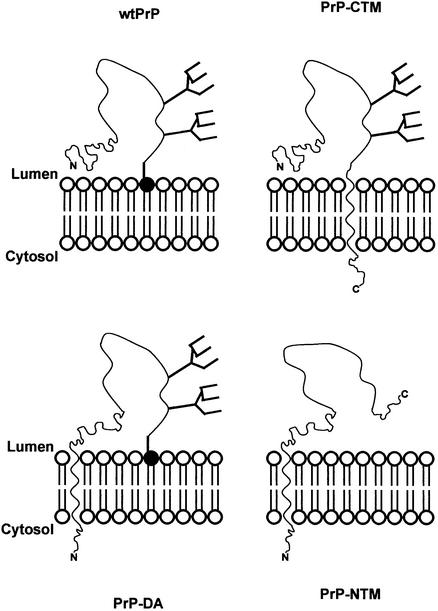
We utilized alternatively anchored forms of murine PrP to investigate the role of membrane topology in the N-glycosylation of the protein (Figure 9). Neither a secreted form of PrP nor one with an N-terminal membrane anchor was N-glycosylated. In contrast, both the GPI-anchored form and one with a C-terminal transmembrane anchor, as well as a form anchored at both ends of the polypeptide chain, were extensively glycosylated. Thus, we show for the first time that not just membrane anchorage, but specifically C-terminal membrane anchorage, whether by a GPI moiety or a transmembrane anchor, affects the efficiency and site occupancy of the N-glycan addition sites of PrP, leading to the conclusion that glycosylation and membrane anchorage are co-operative processes.
Fig. 9. Schematic showing the membrane topology and glycosylation status of the PrP constructs. PrP is depicted as a wavy line with the two N-glycans shown as branched structures. In wtPrP and PrP-DA, the phosphatidylinositol lipid of the GPI anchor is shown by the filled circle. The N- and C-termini of the polypeptide chain are indicated. PrP-NTM is not N-glycosylated, whereas the other three forms are.
The nature of the C-terminal membrane anchor of PrP does not affect the N-glycosylation of the protein. In PrP-CTM, the GPI anchor addition signal was replaced with the transmembrane and cytosolic domains from the type I integral membrane glycoprotein, angiotensin converting enzyme (Soubrier et al., 1988) (Figure 9). This construct was anchored at the surface of the SH-SY5Y cells in a highly glycosylated form, consistent with previous studies in which the GPI anchor was replaced with a transmembrane anchor (Taraboulos et al., 1995; Kaneko et al., 1997). However, membrane anchorage per se does not lead to glycosylation of PrP, as clearly demonstrated by the lack of glycosylation of PrP-NTM. This N-terminally anchored form of the protein (Figure 9) was targeted to the cell surface, indicating that tethering of the protein via its opposite end does not lead to gross mis-folding and intracellular retention; however, it was predominantly unglycosylated. An amber mutation at codon 145 of the PrP gene results in the inherited Gerstmann–Straussler–Scheinker prion disease (Ghetti et al., 1996). Recently, it has been reported that a significant proportion of the truncated PrP145 produced by this mutation retained the N-terminal signal peptide and was degraded rapidly by the proteasome (Zanusso et al., 1999). Although PrP-NTM retains the N-terminal signal peptide/transmembrane domain, incubation of the SH-SY5Y cells with a proteasome inhibitor did not increase its level of expression (data not shown), indicating that it is probably the lack of the C-terminal half of PrP, rather than retention of the signal peptide, that is responsible for the rapid clearance of PrP145.
We also generated a construct PrP-DA, which retained the GPI anchor signal sequence and had an uncleaved signal peptide/transmembrane domain at the N-terminus. This form of the protein was modified with a GPI anchor and had an additional N-terminal membrane anchoring domain (Figure 9). Somewhat surprisingly, this construct was trafficked to the cell surface and extensively glycosylated as for wtPrP, indicating that tethering of the protein at both ends does not grossly affect its biosynthesis. This may reflect the fact that the N-terminus of wtPrP is relatively close to the membrane surface, such that the presence of an additional transmembrane domain at this end of the protein does not restrict its folding. Alternatively, it may reflect the fact that the N-terminus is highly flexible, as observed in the recent NMR structural determinations of recombinant full-length PrP (Donne et al., 1997; Riek et al., 1997), and can therefore accommodate being restricted by membrane attachment. To our knowledge, a similar double-anchored form of another GPI-anchored protein has not been reported, although the type II integral membrane glycoprotein neprilysin was converted into a double-anchored form by the addition of a GPI anchor attachment signal at the C-terminus (Howell et al., 1994).
The truncated form of PrP lacking the GPI signal sequence, PrPΔGPI, although expressed in, and secreted from, human SH-SY5Y cells, was almost exclusively unglycosylated. This is in marked contrast to numerous reports that both membrane anchored and secreted proteins are efficiently N-glycosylated, and that disruption of the anchoring of a normally membrane-attached protein does not lead usually to abrogation of N-linked glycosylation. For example, expression of truncated forms of the GPI-anchored decay accelerating factor, or placental alkaline phosphatase that terminated at or near the normal site of GPI anchor attachment, resulted in the secretion of soluble forms of the proteins that were still N-glycosylated to a similar extent as the GPI-anchored forms (Brown et al., 1989; Caras et al., 1989). Furthermore, in leukocytes from patients with paroxysmal nocturnal haemoglobinuria, in which GPI anchor biosynthesis is defective, the normally GPI-anchored urokinase-type plasminogen activator receptor is secreted in a soluble, N-glycosylated form (Ploug et al., 1992).
Although it has been observed previously that deletion of the GPI anchor attachment signal from PrP results in the synthesis of an under-glycosylated form of the protein, in most cases the relative amount of the unglycosylated form of the protein secreted from the cell was not reported, and no direct correlation was made between membrane anchorage and glycosylation. A truncated form of murine PrP expressed in ScN2a neuroblastoma cells appeared predominantly as an unglycosylated form, from which the authors concluded that the lack of the GPI anchor either prevents the addition and processing of the N-linked glycans within the ER and Golgi or renders these carbohydrates labile (Rogers et al., 1993). However, no indication was given as to whether the truncated protein was secreted. A truncated form of syrian hamster PrP expressed in NIH 3T3 cells appeared as an unglycosylated form, although a significant amount of a mono-glycosylated form was also apparent, but again the relative amounts of retained and secreted protein were not reported (Kocisko et al., 1994). The majority of immunoreactive protein expressed from a truncated form of syrian hamster PrP in Chinese hamster ovary (CHO) cells appeared in the cell lysates, as opposed to the medium, with a molecular weight lower than that of wild-type PrP, which the authors explained as due either to a lack of glycosylation or to degradation (Blochberger et al., 1997). In contrast, during the course of the present study, it was reported that a truncated form of murine PrP was efficiently secreted from CHO cells as both unglycosylated and mono-glycosylated forms (Brimacombe et al., 1999). This phenomenon does not appear to be restricted to PrP. Recently it was reported that a truncated form of the GPI-anchored Thy-1, when expressed in CHO cells, was not as extensively glycosylated as the membrane-bound form (Devasahayam et al., 1999).
Although PrPΔGPI was clearly being secreted from the SH-SY5Y cells, pulse–chase analysis revealed that it was transported ∼4-fold less efficiently than wtPrP to the cell surface. Thus, despite a slower rate of movement through the secretory pathway, PrPΔGPI was under-glycosylated as compared with wtPrP. As there was no observable difference in the transport of wtPrP in the presence or absence of tunicamycin, we attribute this decrease in the rate of transport of PrPΔGPI to the lack of the GPI anchor rather than the lack of N-glycans. Previously, from studies in trypanosomes, it has been proposed that the GPI anchor facilitates forward trafficking through the secretory pathway by promoting the association of the protein with particular secretory vesicles (McDowell et al., 1998). In transformed procyclic trypanosomes, the absence of the GPI anchor from the variant surface glycoprotein caused a 5-fold reduction in the rate of transport of the protein through the secretory pathway as compared with the GPI-anchored protein. We show for the first time that the GPI anchor on a mammalian protein also appears to act as a positive forward trafficking signal.
The utilization of N-linked glycosylation sequons within a protein is dependent upon their accessibility to the glycosylation enzymes of the ER during their biosynthesis (Allen et al., 1995). Potential N-glycosylation sites, which are located within the hydrophobic core of the native protein, may be occluded from glycosylating enzymes at an early stage of the folding pathway and never glycosylated. Consequently, the lack of utilization of the N-glycosylation sites within PrP-NTM and PrPΔGPI could be due to a difference in the conformation of the protein as compared with that of wtPrP. However, PrP-NTM and PrPΔGPI cannot be grossly misfolded as the proteins were not retained within the ER (Hammond and Helenius, 1995). Another possibility for the apparent lack of glycosylation of PrP-NTM and PrPΔGPI is that the Asn residues are initially modified, but then subsequently deglycosylated as the protein traffics through the secretory pathway. To investigate these possibilities we examined whether retention of PrPΔGPI within the ER by disruption of the secretory pathway would lead to an increase in the glycosylation of the protein. Although treatment of the SH-SY5Y cells with either brefeldin A or nocodazole, or incubation of the cells at 15°C, prevented the secretion of PrPΔGPI, there was no evidence for increased glycosylation of the protein upon its prolonged retention in the ER. Thus, it would appear that the consensus sequons for N-glycosylation in PrP-NTM and PrPΔGPI are not being utilized by oligosaccharyltransferase, even when the proteins were retained within the ER.
Our data imply that with PrP, glycosylation and membrane anchorage are co-operative processes. This is a somewhat surprising finding given that the two N-glycosylation sequons in PrP will become accessible to oligosaccharyltransferase after the N-terminal signal peptide, and before the C-terminus of the polypeptide, have been translated and translocated through the ER membrane. One possibility is that the anchorless construct is initially glycosylated and then the lack of membrane anchorage causes it to be deglycosylated. However, this seems unlikely as retention of the protein within the ER did not lead to increased glycosylation, and numerous other anchorless proteins are fully glycosylated. Alternatively, as all the enzymes involved in glycan processing that have been characterized to date are membrane bound, membrane anchorage may affect the access of these enzymes to the protein. However, neither of these possibilities would explain why the N-terminally anchored PrP-NTM is unglycosylated. Another possibility is that N-glycosylation occurs after the entire polypeptide chain has been translocated, such that C-terminal rather than N-terminal membrane anchorage somehow influences whether oligosaccharyltransferase recognizes and modifies the N-linked glycan sequons. As the glycosylation state of PrP is known to influence its conversion into the infectious PrPSc isoform (Taraboulos et al., 1990; Kocisko et al., 1994; Lehmann and Harris, 1997; Nitrini et al., 1997), the present study indicates that membrane topology may influence this process. Furthermore, it has recently been reported that certain inherited PrP mutations appear to cause neurodegeneration in the absence of PrPSc, working instead by favoured synthesis of a transmembrane form of PrP (Hegde et al., 1999).
Materials and methods
Generation of PrP constructs
The coding sequence of murine PrP containing a 3F4 epitope tag was excised from the PrP:pBC12/CMV plasmid (a gift from Dr D.A.Harris, Washington University, MO) using HindIII and BamHI, and inserted into the BamHI site of pIRESneo (Clontech). DNA encoding PrP-CTM was constructed by PCR using the following primers: primer 1, 5′-ATAAGAATGCGGGCCGCATGGCGAACCTTGGCTAC-3′; primer 2, 5′-CAGGAAGAGCAGCAGCCAGGATCTTCTCCCGTCGTA-3′; primer 3, 5′-CGGGATCCTCAGGAGTGTCTCAGCTC-3′. Primers 1 and 2 were used to generate from the PrP:pBC12/CMV plasmid a primary PCR product comprising a 5′ NotI site followed by a sequence encoding residues 1–230 of PrP fused to a sequence encoding the first six residues of the transmembrane domain of angiotensin converting enzyme. The primary PCR product and primer 3 were used to generate from the hACE/pECE plasmid (a gift from Prof. P.Corvol, INSERM, Paris, France) a secondary PCR product comprising a 5′ NotI site followed by a sequence encoding residues 1–230 of PrP fused with a sequence encoding the transmembrane and cytoplasmic domains of angiotensin converting enzyme (Wei et al., 1991) and terminating with a 3′ BamHI site. The secondary PCR product was cleaved with NotI and BamHI, and inserted into the respective sites of pIRESneo. DNA encoding PrP-DA was constructed using the following primers: primer 4, 5′-CAG GACACAACCCCAGCTAAAAAGCGGCCAAAGCCT-3′; primer 5, 5′-CGGGATCCTCATCCCACGATCAGGAA-3′; primer 6, 5′-ATA AGAATGCGGCCGCATGAACTTTGCAGAGGAA-3′. Primers 4 and 5 were used to generate from the PrP:pBC12/CMV plasmid a primary PCR product comprising a 3′ BamHI site followed by a sequence encoding residues 23–230 of PrP fused to a sequence encoding the first six residues of the stalk region of murine aminopeptidase A (Wu et al., 1990). The primary PCR product and primer 6 were used to generate from the APA(Cys43Ser)/pUC19 plasmid (a gift from Prof. M.Cooper, University of Alabama at Birmingham, AL) a secondary PCR product comprising a 5′ NotI site followed by a sequence encoding the uncleaved signal anchor and stalk domains of aminopeptidase A fused with the sequence encoding residues 23–230 of PrP and terminating with a 3′ BamHI site. PrPΔGPI and PrP-NTM were constructed from PrP and PrP-DA, respectively, by mutating the codon for R229 in PrP and R257 in PrP-DA to a stop codon by site-directed mutagenesis using the following primers (mutant bases are indicated in bold): sense primer, 5′-CGA CGGAGATGATCCAGCAGCACC-3′; antisense primer, 5′-GGTGCT GCTGGATCATCTCCCGTCG-3′.
Cell culture, transfection and lysis
SH-SY5Y cells were maintained at 37°C in Dulbecco’s modified Eagle’s medium with Glutamax-I, sodium pyruvate, 4.5 mg/ml glucose and pyridoxine supplemented with 10% (v/v) fetal calf serum (FCS) in a humidified atmosphere of 5% CO2/95% air. Cells at mid-confluency were harvested with trypsin and resuspended in complete medium at a concentration of ∼1 × 106 cells/ml. An 800 µl aliquot of cell suspension was placed in a 4 mm electroporation cuvette and incubated for 1 min with 30 µg of linearized DNA prior to the pulse. Cells were pulsed at 1650 µF/250 V using the Easy-Ject electroporator (Flowgen, Lichfield, UK) and immediately transferred to fresh complete medium. Selection for antibiotic resistance was imposed 48 h after electroporation by incubating the cells with complete medium containing 0.5 mg/ml G-418. G-418-resistant colonies were harvested with trypsin and pooled. Each stably expressing cell line represented a pooled population of G-418-resistant colonies (<50 colonies/cell line) that expressed their target protein at levels sufficient for detection by western blotting and pulse–chase analyses.
Cells at confluency were rinsed twice with phosphate-buffered saline (PBS; 1.5 mM KH2PO4, 2.7 mM Na2HPO4, 150 mM NaCl pH 7.4), scraped into the same buffer and harvested by centrifugation for 3 min at 500 g. The cells were resuspended in lysis buffer [10 mM Tris–HCl pH 7.8, 0.5% (w/v) sodium deoxycholate, 0.5% (v/v) NP-40, 100 mM NaCl, 10 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride] and incubated for 30 min at room temperature. The lysates were clarified by centrifugation for 1 min at 13 000 g. Medium samples were concentrated by the addition of 5 vols of methanol and incubated for 30 min at –20°C. The precipitate was harvested by centrifugation for 5 min at 13 000 g, dried and resuspended in lysis buffer.
Immunoprecipitation
Cell lysates and medium samples were made 1% (w/v) with respect to N-lauroylsarcosine and incubated for 30 min with 0.5% (w/v) protein A–Sepharose. The protein A–Sepharose was pelleted by centrifugation for 1 min at 13 000 g and the supernatant incubated overnight at 4°C with 0.1% (v/v) 3F4 antibody (obtained from Dr R.Kascsak, Institute for Basic Developmental Research, Staten Island, NY). Protein A–Sepharose was added to 0.5% (w/v) and incubation continued for 1 h. The immunocomplexes were pelleted by centrifugation at 13 000 g for 1 min and washed three times with 10 mM Tris–HCl pH 7.8, 1% (w/v) N-lauroylsarcosine, 100 mM NaCl.
SDS–PAGE and western blot analysis
Samples containing ∼10 µg of total protein were mixed with an equal volume of SDS dissociation buffer [125 mM Tris–HCl pH 6.8, 2% (w/v) SDS, 20% (v/v) glycerol, 100 mM dithiothreitol, 0.002% (w/v) bromophenol blue] and boiled for 5 min. Immunoprecipitates were resuspended in SDS dissociation buffer and boiled as above. Proteins were resolved by electrophoresis through 15% polyacrylamide gels. For western blot analysis, resolved proteins were transferred to a Hybond-P poly(vinylidene) difluoride membrane (Amersham, Little Chalfont, UK). The membrane was blocked by incubation for 1 h with PBS containing 0.1% (v/v) Tween-20 and 5% (w/v) dried milk powder. Incubations with primary and peroxidase-conjugated secondary antibodies were performed for 1 h in the same buffer at dilutions of 1/5000 and 1/2000, respectively. Incubation with peroxidase-conjugated streptavidin was performed for 1 h at a dilution of 1/1000 in PBS containing 0.1% (v/v) Tween-20. Bound peroxidase conjugates were visualized using an enhanced chemiluminescence detection system (Amersham).
Surface biotinylation and treatment with brefeldin A and nocodazole
Cells at confluency were incubated for 30 min at 4°C with PBS containing 0.5 mg/ml biotin sulfo-NHS, rinsed three times with PBS containing 50 mM glycine and lysed in lysis buffer. PrP constructs present in the cell lysate were immunoprecipitated with the 3F4 antibody and resolved by SDS–PAGE prior to western blot analysis. Cells at confluency were rinsed twice with PBS and incubated for 3 h at 37°C with OptiMEM containing either 10 µg/ml brefeldin A or 20 µM nocodazole. Cells were lysed in lysis buffer and the medium concentrated by methanol precipitation.
Triton X-114 phase separation and PI-PLC and PNGase F incubations
Cells at confluency were lysed in 10 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% (v/v) pre-condensed Triton X-114 and the lysate was successively incubated for 10 min at 4°C and then 37°C. The aqueous and detergent phases were separated by centrifugation for 3 min at 3000 g. The volume of the detergent phase was made equal to that of the aqueous phase with 10 mM Tris–HCl pH 7.4 and 150 mM NaCl. For cleavage of the GPI anchor with PI-PLC, cell lysates or membranes were incubated for 3 h at 37°C with 1 U/ml Bacillus thuringiensis PI-PLC (a gift from Dr M.G.Low, Columbia University, NY), followed by centrifugation at 100 000 g for 90 min as required. For enzymic deglycosylation, samples of cell lysate were made 20 mM with respect to sodium phosphate pH 7.6, 50 mM with respect to EDTA, 5% (w/v) with respect to SDS and 5% (v/v) with respect to β-mercaptoethanol. Samples were boiled for 5 min, diluted 5-fold with 1.25% (v/v) Triton X-100 and incubated for 16 h at 37°C with 1 U peptide N-glycosidase F (PNGase F).
Pulse–chase analysis
Cells at confluency were starved of methionine and cysteine by incubation for 45 min at 37°C with Met/Cys-free modified Eagle’s medium supplemented with 0.3 mg/ml glutamine (starve medium). Cells were labelled by incubation for 20 min at 37°C with starve medium containing 50 µCi of [35S]Met/Cys per 25 cm2 flask, rinsed twice with PBS, and incubated for various chase times with OptiMEM containing 5 mM cycloheximide and 5 mM l-Met. Where indicated, tunicamycin was present in the starve, pulse and chase medium at a concentration of 5 µg/ml. Following chase periods performed in the absence of tunicamycin, cells expressing PrP were rinsed twice with PBS, and incubated for 30 min at 4°C with OptiMEM containing 0.2 U/ml B.thuringiensis PI-PLC. Cells were lysed in lysis buffer and the medium from the PI-PLC treatment was concentrated by methanol precipitation. Following chase periods performed in the presence of tunicamycin, cells expressing PrP were incubated for 5 min with 0.5 mg/ml trypsin, 0.5 mM EDTA and 150 mM NaCl. Cells were washed once with FCS, twice with PBS and lysed in lysis buffer containing 0.5 mg/ml soybean trypsin inhibitor. Cells expressing PrPΔGPI were immediately lysed upon termination of the chase, and the chase medium concentrated by methanol precipitation. PrP constructs present in the cell lysate and concentrated medium samples were immunoprecipitated with the 3F4 antibody and resolved by 15% SDS–PAGE. Gels were fixed in 5% (v/v) 2-propanol/10% (v/v) acetic acid, dried, and subjected to autoradiography using Kodak Biomax MR film.
Acknowledgments
Acknowledgements
We thank Dr P.F.T.Vaughan (University of Leeds) for performing the noradrenaline uptake and release experiments, and Dr D.A.Harris (Washington University) for provision of the 3F4 epitope-tagged murine PrP cDNA. This work was supported by a Strategic Project Grant from the Medical Research Council of Great Britain. F.Z. was in receipt of an Overseas Research Scholarship and a University of Leeds Tetley and Lupton Scholarship.
REFERENCES
- Allen S., Naim,H.Y. and Bulleid,N.J. (1995) Intracellular folding of tissue-type plasminogen activator. Effects of disulfide bond formation on N-linked glycosylation and secretion. J. Biol. Chem., 270, 4797–4804. [DOI] [PubMed] [Google Scholar]
- Bangs J.D., Hereld,D., Krakow,J.L., Hart,G.W. and Englund,P.T. (1985) Rapid processing of the carboxyl terminus of a trypanosome variant surface glycoprotein. Proc. Natl Acad. Sci. USA, 82, 3207–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs J.D., Ransom,D.M., McDowell,M.A. and Brouch,E.M. (1997) Expression of bloodstream variant surface glycoproteins in procyclic stage Trypanosoma brucei: role of GPI anchors in secretion. EMBO J., 16, 4285–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M., Benachour,A., Rusconi,S., Aebi,M. and Conzelmann,A. (1996) Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J., 15, 6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Blochberger T.C., Cooper,C., Peretz,D., Tatzelt,J., Griffith,O.H., Baldwin,M.A. and Prusiner,S.B. (1997) Prion protein expression in Chinese hamster ovary cells using a glutamine synthetase selection and amplification system. Protein Eng., 10, 1465–1473. [DOI] [PubMed] [Google Scholar]
- Brimacombe D.B., Bennett,A.D., Wusteman,F.S., Gill,A.C., Dann,J.C. and Bostock,C.J. (1999) Characterisation and polyanion-binding properties of purified recombinant prion protein. Biochem. J., 342, 605–613. [PMC free article] [PubMed] [Google Scholar]
- Brown D.A., Crise,B. and Rose,J.K. (1989) Mechanism of membrane anchoring affects polarised expression of two proteins in MDCK cells. Science, 245, 1499–1501. [DOI] [PubMed] [Google Scholar]
- Caras I.W., Weddell,G.N. and Williams,S.R. (1989) Analysis of the signal for attachment of a glycophospholipid membrane anchor. J. Cell Biol., 108, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso de Almeida M.L. and Turner,M.J. (1983) The membrane form of variant surface glycoproteins of Trypanosoma brucei. Nature, 302, 349–352. [DOI] [PubMed] [Google Scholar]
- Chessler S.D. and Byers,P.H. (1992) Defective folding and stable association with protein disulfide isomerase/prolyl hydroxylase of type I procollagen with a deletion in the pro α2(I) chain that preserves the Gly-X-Y repeat pattern. J. Biol. Chem., 267, 7751–7757. [PubMed] [Google Scholar]
- Collinge J., Sidle,K.C.L., Meads,J., Ironside,J. and Hill,A.F. (1996) Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature, 383, 685–690. [DOI] [PubMed] [Google Scholar]
- Devasahayam M., Catalino,P.D., Rudd,P.M., Dwek,R.A. and Barclay,A.N. (1999) The glycan processing and site occupancy of recombinant Thy-1 is markedly affected by the presence of a glycosylphosphatidylinositol anchor. Glycobiology, 9, 1381–1387. [DOI] [PubMed] [Google Scholar]
- Donne D.G., Viles,J.H., Groth,D., Mehlhorn,I., James,T.L., Cohen,F.E., Prusiner,S.B., Wright,P.E. and Dyson,H.J. (1997) Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl Acad. Sci. USA, 94, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti B. et al. (1996) Vascular variant of prion protein cerebral amyloidosis with τ-positive neurofibrillary tangles: the phenotype of the stop codon 145 mutation in PRNP. Proc. Natl Acad. Sci. USA, 93, 744–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C. and Helenius,A. (1995) Quality control in the secretory pathway. Curr. Opin. Cell Biol., 7, 523–529. [DOI] [PubMed] [Google Scholar]
- Hegde R.S., Tremblay,P., Groth,D., DeArmond,S.J., Prusiner,S.B. and Lingappa,V.R. (1999) Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature, 402, 822–826. [DOI] [PubMed] [Google Scholar]
- Helenius A. (1994) How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol. Biol. Cell, 5, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell S., Lanctot,C., Boileau,G. and Crine,P. (1994) A cleavable N-terminal signal peptide is not a prerequisite for the biosynthesis of glycosylphosphatidylinositol-anchored proteins. J. Biol. Chem., 269, 16993–16996. [PubMed] [Google Scholar]
- Kaneko K., Vey,M., Scott,M., Pilkuhn,S., Cohen,F.E. and Prusiner,S.B. (1997) COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scapie isoform. Proc. Natl Acad. Sci. USA, 94, 2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kascsak R.J., Rubinstein,R., Merz,P.A., Tonna-DeMasi,M., Fersko,R., Carp,R.I., Wisniewski,H.M. and Diringer,H. (1987) Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol., 61, 3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner R.D., Donaldson,J.G. and Lippincott-Schwartz,J. (1992) Brefeldin A—insights into the control of membrane traffic and organelle structure. J. Cell Biol., 116, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocisko D.A., Come,J.H., Priola,S.A., Chesebro,B., Raymond,G.J., Lansbury,P.T. and Caughey,B. (1994) Cell-free formation of protease-resistant prion protein. Nature, 370, 471–474. [DOI] [PubMed] [Google Scholar]
- Kornfield R. and Kornfield,S. (1985) Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem., 54, 631–664. [DOI] [PubMed] [Google Scholar]
- Lehmann S. and Harris,D.A. (1997) Blockade of glycosylation promotes acquisition of scrapie-like properties by the prion protein in cultured cells. J. Biol. Chem., 272, 21479–21487. [DOI] [PubMed] [Google Scholar]
- Littlewood G.M., Hooper,N.M. and Turner,A.J. (1989) Ectoenzymes of the kidney microvillar membrane. Affinity purification, characterization and localization of the phospholipase C-solubilized form of renal dipeptidase. Biochem. J., 257, 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell M.A., Ransom,D.M. and Bangs,J.D. (1998) Glycosyl phosphatidylinositol-dependent secretory transport in Trypanosoma brucei. Biochem. J., 335, 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitrini R. et al. (1997) Familial spongiform encephalopathy associated with a novel prion protein gene mutation. Ann. Neurol., 42, 138–146. [DOI] [PubMed] [Google Scholar]
- Perera W.S.S. and Hooper,N.M. (1999) Proteolytic fragmentation of the murine prion protein: role of Tyr-128 and His-177. FEBS Lett., 463, 273–276. [DOI] [PubMed] [Google Scholar]
- Ploug M., Eriksen,J., Plesner,T., Hansen,N.E. and Dano,K. (1992) A soluble form of the glycolipid-anchored receptor for urokinase-type plasminogen activator is secreted from peripheral blood leukocytes from patients with paroxysmal nocturnal haemoglobinuria. Eur. J. Biochem., 208, 397–404. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. (1998) Prions. Proc. Natl Acad. Sci. USA, 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R. and Alonso,M.A. (1999) MAL, an integral element of the apical sorting machinery, is an itinerant protein that cycles between the trans-Golgi network and the plasma membrane. Mol. Biol. Cell, 10, 3435–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher J.W., Parekh,R.B. and Dwek,R.A. (1988) Glycobiology. Annu. Rev. Biochem., 57, 785–838. [DOI] [PubMed] [Google Scholar]
- Riek R., Hornemann,S., Wider,G., Glockshuber,R. and Wuthrich,K. (1997) NMR characterisation of the full length recombinant murie prion protein, mPrP(23–231). FEBS Lett., 413, 282–288. [DOI] [PubMed] [Google Scholar]
- Rogers M., Yehiely,F., Scott,M. and Prusiner,S.B. (1993) Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc. Natl Acad. Sci. USA, 90, 3182–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd P.M. et al. (1999) Glycosylation differences between the normal and pathogenic prion protein isoforms. Proc. Natl Acad. Sci. USA, 96, 13044–13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubrier F., Alhenc-Gelas,F., Hubert,C., Allegrini,J., John,M., Tregear,G. and Corvol,P. (1988) Two putative active centers in human angiotensin I-converting enzyme revealed by molecular cloning. Proc. Natl Acad. Sci. USA, 85, 9386–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl N., Borchelt,D.R., Hsiao,K. and Prusiner,S.B. (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell, 51, 229–240. [DOI] [PubMed] [Google Scholar]
- Stimson E., Hope,J., Chong,A. and Burlingame,A.L. (1999) Site-specific characterization of the N-linked glycans of murine prion protein by high-performance liquid chromatography/electrospray mass spectrometry and exoglycosidase digestions. Biochemistry, 38, 4885–4895. [DOI] [PubMed] [Google Scholar]
- Taraboulos A., Rogers,M., Borchelt,D.R., McKinley,M.P., Scott,M., Serban,D. and Prusiner,S.B. (1990) Acquisition of protease resistance by prion proteins in scrapie-infected cells does not require asparagine-linked glycosylation. Proc. Natl Acad. Sci. USA, 87, 8262–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Scott,M., Semenov,A., Avraham,D., Laszlo,L. and Prusiner,S.B. (1995) Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J. Cell Biol., 129, 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N.A., Rumsby,M.G., Walker,J.H., McMorris,F.A., Ball,S.G. and Vaughan,P.F. (1994) A role for protein kinase C subtypes α and ε in phorbol-ester-enhanced K(+)- and carbachol-evoked noradrenaline release from the human neuroblastoma SH-SY5Y. Biochem. J., 297, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology, 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Alhenc-Gelas,F., Corvol,P. and Clauser,E. (1991) The two homologous domains of human angiotensin I converting enzyme are both catalytically active. J. Biol. Chem., 266, 9002–9008. [PubMed] [Google Scholar]
- White I.J., Souabni,A. and Hooper,N.M. (2000) Comparison of the glycosyl-phosphatidylinositol cleavage/attachment site between mammalian cells and parasitic protozoa. J. Cell Sci., 113, 721–727. [DOI] [PubMed] [Google Scholar]
- Wu Q., Lahti,J.M., Air,G.M., Burrows,P.D. and Cooper,M.D. (1990) Molecular cloning of the murine BP-1/6C3 antigen: a member of the zinc-dependent metallopeptidase family. Proc. Natl Acad. Sci. USA, 87, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanusso G., Petersen,R.B., Jin,T., Jing,Y., Kanoush,R., Ferrari,S., Gambetti,P. and Singh,N. (1999) Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J. Biol. Chem., 274, 23396–23404. [DOI] [PubMed] [Google Scholar]