Abstract
Hox genes encoding homeodomain transcriptional regulators are known to specify the body plan of multicellular organisms and are able to induce body plan transformations when misexpressed. These findings led to the hypothesis that duplication events and misexpression of Hox genes during evolution have been necessary for generating the observed morphological diversity found in metazoans. It is known that overexpressing Antennapedia (Antp) in the head induces antenna-to-leg as well as head-to-thorax transformation and eye reduction. At present, little is known about the exact molecular mechanism causing these phenotypes. The aim of this study is to understand the basis of inhibition of eye development. We demonstrate that Antp represses the activity of the eye regulatory cascade. By ectopic expression, we show that Antp antagonizes the activity of the eye selector gene eyeless. Using both in vitro and in vivo experiments, we demonstrate that this inhibitory mechanism involves direct protein–protein interactions between the DNA-binding domains of EY and ANTP, resulting in mutual inhibition.
Keywords: antagonism/eyeless/Hox/interaction/Pax-6
Introduction
The genetic and molecular analysis of the development of different model organisms has yielded a wealth of information about the underlying mechanisms of development. The theme emerging from these studies is that highly conserved genes are involved in the development of animals of strikingly different architecture and embryogenesis. The Hox genes, a subset of the Homeobox gene family, encode transcription factors and are a good example of functional conservation during evolution (Gehring et al., 1994). Hox genes are common to most or all animals, are organized in clusters, and define positional information along the antero–posterior axis.
Homeotic mutations in Drosophila have led to the identification of several ‘master control’ genes. This term, initially introduced by Lewis (1992) for the homeotic genes of the bithorax complex, was illustrated by the genetic construction of four-winged and eight-legged flies. Loss- and gain-of-function in these genes lead to opposite homeotic transformations. For example, in Antennapedia (Antp), recessive loss-of-function mutations are lethal at the embryonic or larval stage and lead to a transformation of the second thoracic segment T2 toward the first thoracic segment T1 (Struhl, 1981; Schneuwly and Gehring, 1982; Abbott and Kaufman, 1986). Dominant gain-of-function mutations lead to a transformation in the opposite direction, i.e. from the anterior head and T1 segments toward T2 (Gehring, 1987). By ubiquitous expression of Antp under the control of a heat shock promoter, Schneuwly et al. (1987) changed the body plan of Drosophila by inducing the formation of middle legs in place of the antennae, second thoracic segment structures on the dorsal head capsule and inhibition of eye development. Similar changes in adult pattern have been observed upon ectopically expressing other Hox proteins.
These transformations resulting from ectopic selector gene expression can be explained by a combinatorial interaction of two or more homeotic genes in order to specify a given body segment. However, the exact molecular mechanisms remain unknown. Recently, additional Drosophila selector genes have been identified that are capable of inducing organogenesis when expressed ectopically. One of the most striking examples is the transcription factor eyeless (ey), a homolog of Pax-6 in vertebrates (reviewed in Callaerts et al., 1997). In mammals, congenital dominant eye diseases known as aniridia (humans) and small eye (mice and rats) are caused by haploinsufficient loss-of-function mutations of Pax-6. Homozygous embryos lack eyes and nostrils completely, have brain and spinal cord malformations, and die prior to birth. In Drosophila, loss-of-function mutations of ey also show eye defects from subtle restructuring to complete loss (Quiring et al., 1994). In gain-of-function experiments, ectopic eyes are formed on the appendages of the fly (Halder et al., 1995). Ectopic expression of Pax-6 homologs from various species is sufficient to induce ectopic eyes in Drosophila, suggesting remarkably conserved mechanisms for eye differentiation (Callaerts et al., 1997; Gehring and Ikeo, 1999).
The Drosophila compound eye develops from the eye–antenna disc, which invaginates from the ectoderm during embryogenesis and grows inside the larva. In the third larval instar, photoreceptor differentiation begins at the posterior margin of the eye disc and spreads anteriorly, led by a depression in the disc known as the morphogenetic furrow. Early determination of the eye primordium requires several nuclear proteins that are likely to act as transcriptional regulators. Like ey, the twin of eyeless (toy) gene encodes a Pax-6 homolog containing a paired and homeo DNA-binding domain (Czerny et al., 1999). Eye gone (eyg) encodes a Pax-like protein (Jun et al., 1998) and sine oculis (so) a homeodomain (HD) protein (Cheyette et al., 1994), while eyes absent (eya) and dachshund (dac) both encode novel nuclear proteins (Bonini et al., 1993; Mardon et al., 1994). Recently, optix, a homeobox gene related to the so gene family, was also shown to play a role in eye development (Toy et al., 1998; Seimiya and Gehring, 2000). Analysis of the expression pattern of these genes combined with a genetic approach in Drosophila has revealed a sequential and hierarchical deployment of these genes during eye development. toy is the first to be expressed and activates ey in the eye primordium (Czerny et al., 1999). eya, so and dac are further downstream and regulated by ey (Halder et al., 1998; Niimi et al., 1999; Zimmerman et al., 2000). eyg and optix are able to induce ectopic eye formation at least in part independently of ey, suggesting that they are involved in a parallel process for eye formation (H.Sun, personal communication; Seimiya and Gehring, 2000).
Despite recent advances in understanding the mechanisms involved in the process of organogenesis, it remains unclear how the selector genes’ activities are controlled and fine-tuned. For example, ey and toy are also expressed in the central nervous system (CNS) and the peripheral nervous system (Czerny et al., 1999), but only a small number of cells comprising the eye primordium will give rise to an eye. Moreover, when ey is misexpressed ubiquitously, ectopic eye development is restricted to specific regions of the disc (Halder et al., 1998; Chen et al., 1999). These findings show that somehow, resident genetic programs in many cells can inhibit Pax-6 function.
The overexpression of various homeobox-containing proteins has been shown to inhibit eye development (Chadwick et al., 1990; Gibson et al., 1990; Zhao et al., 1993; Benassayag et al., 1997; Yao et al., 1999; Curtiss and Mlodzik, 2000). We have investigated the molecular mechanism of dominant eye loss induced by Antp. We find that ectopic ANTP protein induced in the eye is unable to repress ey transcription and translation. Nonetheless, eye development is impaired. Whereas EY is present, the EY target genes so, eya and dac are repressed. These experiments suggest that Antp blocks EY activity. To test this, we ectopically co-expressed ANTP and EY proteins in the same cells of Drosophila imaginal discs, and show that ectopic eye formation (induced by EY) is blocked. Conversely, expression of EY in the antenna disc is able to block the antenna-to-leg transformation induced by ANTP. We show that EY and ANTP interact directly in vitro, via the ANTP HD, and both the paired domain and the HD of EY. In yeast, ANTP inhibits transactivation by EY. Furthermore, in vitro binding of EY to specific DNA target sites is inhibited upon addition of ANTP. These experiments show that homeobox genes can inhibit each other through direct protein–protein interaction. Thus, they support the idea that the relative intracellular level of each protein is crucial for directing the cells into alternative differentiation programs.
Results
Antp mainly acts in front of the morphogenetic furrow to inhibit eye development by inducing apoptosis in the eye disc
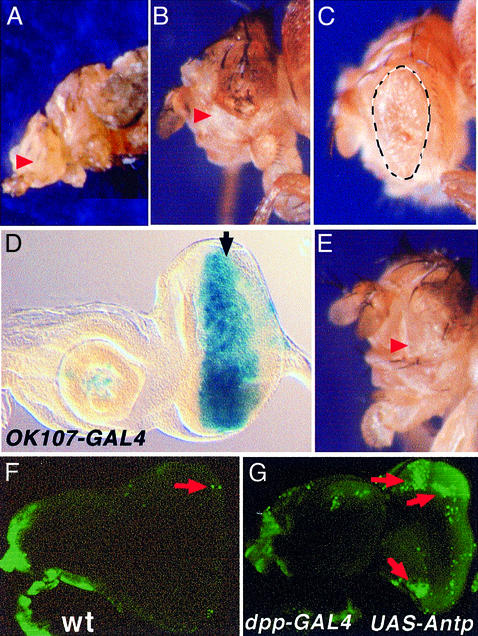
The ectopic expression of several homeotic proteins, including ANTP, has previously been shown to inhibit eye development. These results were obtained using different promoters, including the ubiquitous heat shock promoter (Gibson et al., 1990), the dppblink-GAL4 line (Pai et al., 1998) (Figure 1A) or the ey enhancer-GAL4 (EYE-GAL4) driver (Bello et al., 1998) (Figure 1B). The latter two promoters are expressed during early stages of eye differentiation. The eye-specific enhancer of ey induces gene expression in the eye primordia of the embryo, then maintains expression throughout eye morphogenesis. In contrast to endogenous ey expression, enhancer-driven reporter gene expression in the wild-type eye disc is not down-regulated in the differentiating cells posterior to the morphogenetic furrow but extends throughout the disc (Halder et al., 1998). dppblink expression starts in the undifferentiated cells and is maintained thereafter in the developing photoreceptors (Staehling-Hampton et al., 1994). Similar to results with the ey enhancer, ANTP expression in the eye disc directed by the dppblink-GAL4 driver also induces an eyeless phenotype (Figure 1A and B). However, these results do not clarify the question of whether Antp expression in front of (undifferentiated cells) or behind (photoreceptors) the morphogenetic furrow induces eye loss. Therefore, two additional GAL4 driver lines were employed to direct UAS-Antp expression: the GMR-GAL4 line carrying Glass Multimerized Responsive sites (Ellis et al., 1993) expressed in the differentiated ommatidia posterior to the furrow, and OK107-GAL4 (Connolly et al., 1996) expressed in front of the furrow (Figure 1D). Results show that while expression of Antp in the differentiated cells reduces eye pigmentation, it has only a minor effect on the eye size (Figure 1C). In contrast, expression of Antp in front of the furrow results in an eyeless phenotype (Figure 1E) resembling that obtained with EYE-GAL4 or dppblink-GAL4 (Figure 1A and B). Thus, Antp acts as a repressor of eye development in undifferentiated cells.
Fig. 1. Eye reduction induced by Antp using different drivers. Arrowheads show the lack of eyes (A, B and E), dashed lines delimit the eye size (C). (A) dppblink-Gal4; (B) EYE-GAL4; (C) GMR-GAL4; (E) OK107-GAL4. (D) β-galactosidase expression of OK107-GAL4 in the eye. The morphogenetic furrow is marked by an arrow. (F and G) Acridine orange stainings highlight dead cells (green), (F) wild type and (G) ANTP-expressing disc. Massive cell death is observed in the remaining portion of the disc.
Previous work demonstrated that the eye-loss phenotype associated with the regulatory disruption mutation ey2 was a result of cell death in third instar larvae (Halder et al., 1998). This effect is very similar to those of the so1 and eya1 mutants (Bonini and Fortini, 1999). Thus, we tested whether Antp expression in the eye disc also led to increased apoptosis, assessing cell death by staining with the vital dye acridine orange. Massive cell death was observed in eye discs expressing Antp compared with wild type (Figure 1G versus F). These results clearly show a parallel between the Antp-induced gain-of-function phenotype and that for ey (and so and eya) loss-of-function, supporting the idea that Antp inhibits the eye developmental pathway.
Antp expression in the eye disc disrupts the ey cascade
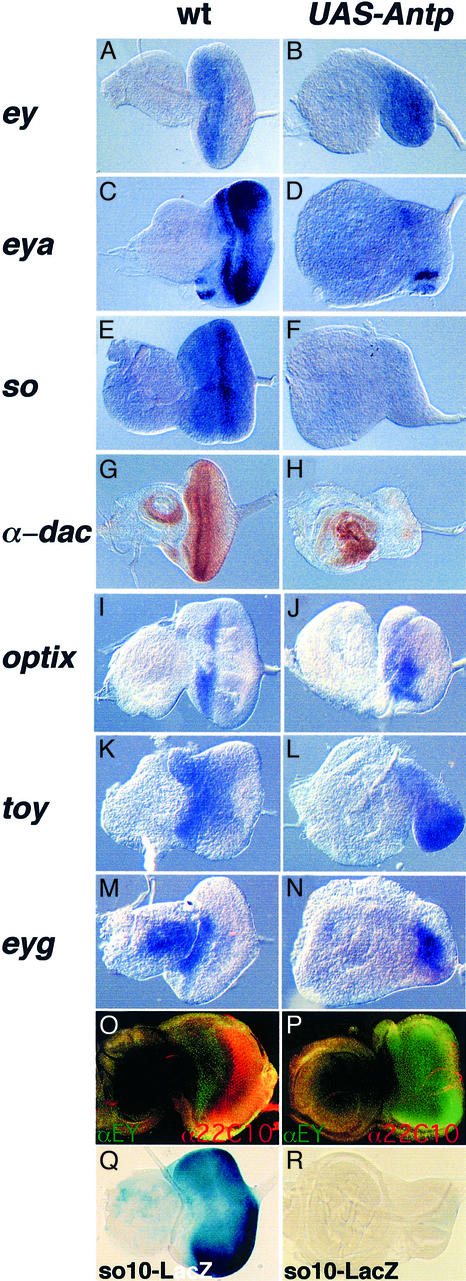
Ectopic Antp expression induces an eyeless phenotype when expressed in front of the morphogenetic furrow. This observation prompted us to analyze the epistatic relationships among the eye determining genes toy, ey, so, eya, dac, eyg and optix. These genes are normally expressed in front of the furrow following a hierarchical pathway. ey, initially defined as the master control gene for eye morphogenesis, induces the expression of so, eya and dac (reviewed in Gehring and Ikeo, 1999; Treisman, 1999). Additional genes involved in the eye development pathway, like optix and eyg (Seimiya and Gehring, 2000; H.Sun, personal communication), may to some extent act in parallel to ey. In order to analyze the effect of Antp on these genes, we performed in situ hybridizations in the eye–antenna disc after expression of Antp. Whereas ey expression is not affected by ANTP (compare Figure 2A and B), the ey target genes eya, so and dac are repressed (Figure 2, compare C and D, E and F, G and H). Interestingly, the expression of optix (Figure 2I and J), toy (Figure 2K and L) or eyg (Figure 2M and N) is not affected. Altogether, these results indicate that Antp expression in eye precursor cells disrupts the ey regulatory cascade and leads to a phenotype resembling those observed in ey2, so1 and eya1 loss-of-function mutants.
Fig. 2. ANTP represses the ey regulatory pathway and blocks photoreceptor differentiation despite the presence of EY. In situ hybridization or immunostaining (G, H, O and P) experiments were performed on eye antenna third instar imaginal discs to study gene expression following Antp expression. (A, C, E, G, I, K, M, O and Q) Wild-type discs; (B, D, F, H, J, L, N, P and R) targeted expression of Antp with dppblink-Gal4. The magnification is 2-fold higher for the ANTP-expressing disc as compared with the wild type. (O and P) Immunostaining experiment using an αEY antibody (in green) and the α22C10 neuronal marker (in red). Note, in wild type (O) the expression of EY is restricted anterior to the furrow. (Q and R) Analysis of the EY responsive element so10 enhancer expression in wild-type and Antp-expressing discs. β-galactosidase staining was performed in parallel in wild-type (Q) as well as in Antp-expressing discs (R).
Since ey transcription is not affected, we next wanted to know whether the EY protein is normally accumulated in the eye disc when Antp is expressed. Immunohisto chemistry experiments were performed using an anti-EY antibody as well as an antibody raised against the 22C10 neuronal marker. As seen in Figure 2P, following Antp expression the EY protein is easily detected throughout the disc; in contrast, neuronal differentiation is impaired (compare Figure 2O and P). Furthermore, immunostainings performed with an anti-ANTP antibody reveal the presence of ANTP in the region where ey is expressed (Figure 3G), indicating that both proteins co-localize.
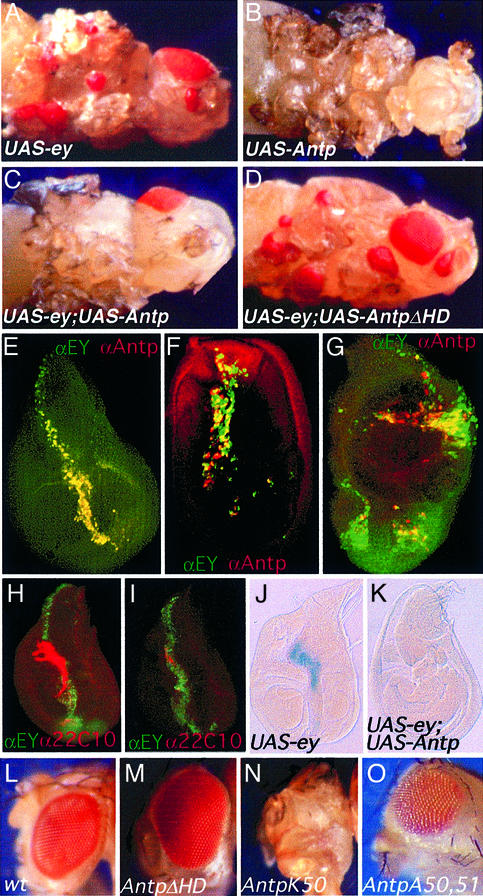
Fig. 3. Ectopic eye induction mediated by ey is inhibited by Antp. Reciprocally, Antp-induced phenotype is inhibited by ey. EY, ANTP and both are expressed using the UAS/GAL4 system with dppblink-Gal4 as driver. (A) Ectopic eyes induced by ey. (B) Antenna-to-leg transformation and eye development inhibition induced by Antp. (C) Lack of ectopic eyes and antenna-to-leg transformation when both proteins are co-expressed. (D) ANTP HD-deleted protein is unable to repress ectopic eye formation. (E–G) EY and ANTP co-localize in the discs, resulting in the inhibition of ectopic eye development. Immunostaining experiments using EY (green) and ANTP (red) antibodies, analyzed by confocal microscopy. Only the merge is presented. (E) Wing disc; (F) leg disc; (G) eye–antenna disc (eye disc at the bottom). (H and I) ANTP-EY co-expression blocks neuronal differentiation and (J and K) so induction. (H) UAS-ey crossed with dppblink-Gal4. Wing disc stained with an EY antibody (green) and the 22C10 neuronal marker (red). (I) Same staining as in (H) but here EY and ANTP are co-expressed. (J and K) β-galactosidase expression of the so enhancer trap line following expression of EY (J) or EY and ANTP (K) in the wing disc. so is not expressed in the wild-type wing disc (not shown). (L–O) Effect on eye development of ANTP-deleted or mutated proteins. Crosses of the UAS constructs indicated in the figure were performed using the driver EYE-GAL4. (L) Wild-type eye; (M) UAS-AntpΔHD construct, HD is deleted; (N) UAS-AntpK50, the Antp DNA-binding specificity is changed to that of Bicoid; (O) UAS-AntpA50,51. Residues involved in DNA contacts have been mutated in order to abolish binding to DNA.
Halder et al. (1998) have demonstrated that so requires ey for its expression in the eye disc. Furthermore, Niimi et al. (1999) recently defined an eye-specific enhancer (so10 fragment) deleted in the so1 mutant, which is directly regulated by ey. This 400 bp element by itself mimics endogenous so expression in the eye disc (compare Figure 2Q with 5A) and constitutes an EY responsive element (Niimi et al., 1999). Therefore, we tested the effect of Antp on so10 fragment expression in order to test whether or not Antp is able to repress this EY responsive element. As seen in Figure 2R, Antp efficiently represses expression directed by the so10 fragment (compare Figure 2Q and R). Taken together, these results strongly support the idea that ANTP blocks EY activity at the protein level.
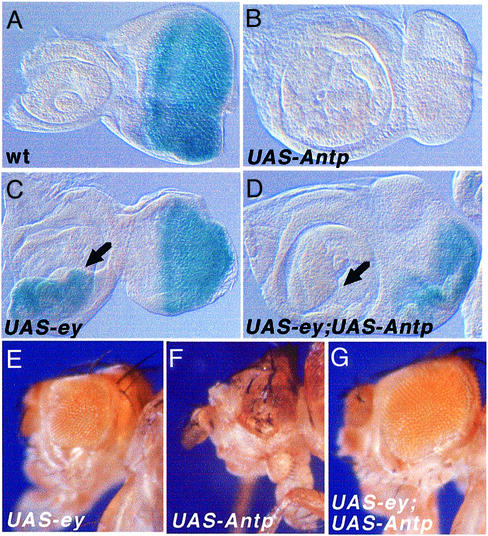
Fig. 5. Competition between EY and ANTP in eye morphogenesis. (A–D) β-galactosidase stainings of the so enhancer trap line following expression of the different constructs indicated. dppblink-Gal4 is the driver. (A) so enhancer trap line expression in wild-type eye disc. (B) Antp represses so in the eye. (C) Arrow: ey induces so in the antenna. (D) ey;Antp co-expression results in so repression in the antenna (arrow) and so derepression in the eye. (E–G) Phenotype of adult flies after expression of the different lines indicated using EYE-GAL4. ey/Antp co-expression resulted in the rescue of Antp-induced eye defects.
Ey and Antp are mutual inhibitors for directing eye and leg development
If the ANTP protein is able to block EY activity, this mechanism should also function in other tissues. Therefore, ectopic eye formation should also be blocked by Antp. To test this prediction, we induced ectopic eyes on wing, antennae and legs using the UAS-GAL4 system (Figure 3A–D). Results show that the ectopic eye formation induced by ey is completely blocked on co-expressing ey and Antp (compare Figure 3A and C). Moreover, the Antp induced antenna-to-leg transformation (Figure 3B) is inhibited by ey (Figure 3C). A series of similar tests employing hs-ey and hs-Antp transgenes, singly or in combination, led to the same conclusions. Furthermore, they revealed a specific requirement for the ANTP HD, since N-terminal deletions of the ANTP protein do not affect its ability to inhibit EY activity, whereas deletion of the HD results in a protein unable to inhibit ey function (data not shown; constructs described in Gibson et al., 1990). Similarly, using the UAS-GAL4 system, the ANTP HD-deleted protein was unable to repress ectopic eye formation (Figure 3D). These results made it necessary to demonstrate that both proteins co-localize in the same cells of the discs. Upon examining protein accumulation by confocal microscopy, we found that both proteins are efficiently co-expressed in these different tissues (Figure 3E, F and G). Furthermore, immunostaining experiments performed using the ey antibody or the neuronal marker 22C10 confirm that, despite the presence of EY in the disc, co-expression of ANTP leads to inhibition of neuronal differentiation (Figure 3H and I).
We next tested the effect on subordinate genes taking so as an example, since it is known to be a direct target of EY. Using a so enhancer trap line, we found that expression of Antp blocks so induction, a process normally mediated by EY (compare Figure 3J with K and 5C with D). Taken together, these results parallel those obtained in the eye disc and strongly suggest that the mechanism of EY inhibition is identical under both normal and ectopic conditions.
To further confirm this hypothesis, we targeted expression of the HD-deleted ANTP protein using EYE-GAL4 as driver. These results show that the ANTP protein lacking the HD is unable to repress eye development (Figure 3M). Thus, these results indicate that the same mechanism leads to eye inhibition in the ectopic situation as well as in the normal eye.
In order to test whether the DNA-binding activity of Antp is not required for the inhibition of eye development, we tested an ANTP mutant in which the DNA-binding specificity was changed (Q50K). Interestingly, this mutated protein is still able to repress eye development (Figure 3N). In addition, we performed mutagenesis experiments to convert Q50 and N51, residues shown to be crucial for DNA contacts (reviewed in Gehring et al., 1994), into alanines. This mutant is unable to bind a DNA PS2 probe containing a Hox/Exd/Hth motif, even in the presence of EXD and HTH in the bandshift assay (not shown). This A50,A51 mutant protein is still capable of inhibiting eye development when expressed in the eye disc using a strong EYE-GAL4 line, although with a lower activity than the wild-type ANTP protein (Figures 3O).
EY and ANTP form a protein complex in vitro, mediated by the HD of ANTP and the paired domain and/or HD of EY
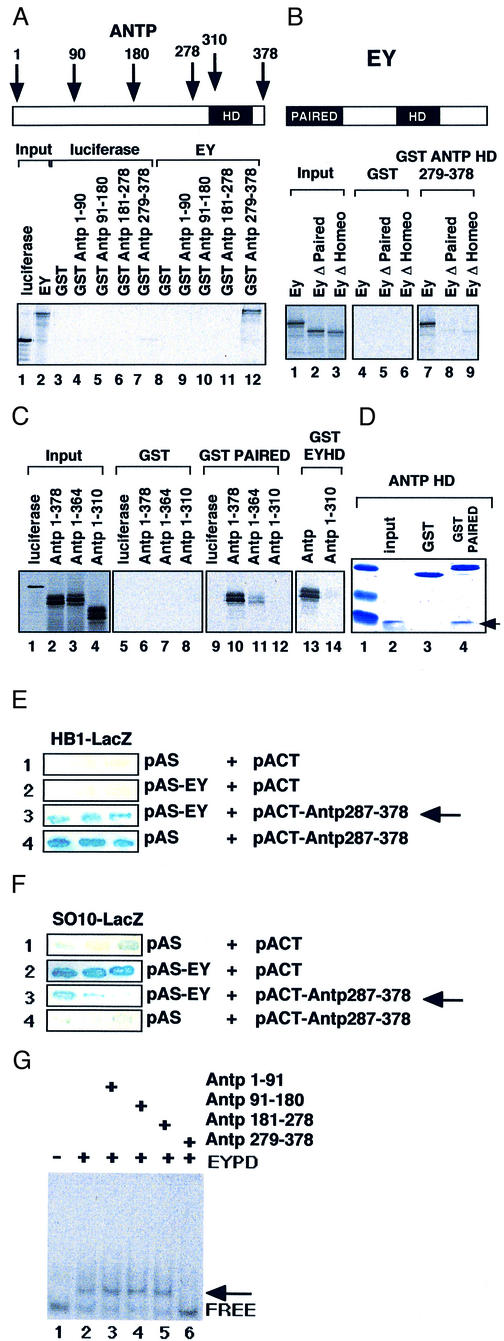
Based on these in vivo results we asked whether the two proteins ANTP and EY might interact directly and thereby inhibit their respective activities. We first examined potential in vitro interactions between ANTP and EY using glutathione S-transferase (GST)–ANTP fusion proteins immobilized on glutathione–Sepharose beads. These were tested for their ability to retain in vitro synthesized 35S-labeled EY protein. Different portions of the ANTP protein were produced and tested separately for their ability to interact with EY. As seen in Figure 4A (lane 12), only the C-terminal portion of ANTP including the HD is able to interact with EY. This interaction is specific, since [35S]luciferase used as a control did not interact with this ANTP fusion protein (lane 7). The specificity of the interaction is further supported by the inability of EY to interact with GST alone (lane 8), and the other fusion proteins tested (lanes 9–11).
Fig. 4. ANTP interacts with EY in vitro and inhibits EY transactivation. (A) GST interaction assays were performed with GST or GST–ANTP constructs and 35S-labeled EY or luciferase as control. Input, 20% of the 35S protein involved in the assay. (B) Effect of PAIRED or HOMEO domain deletion on the binding to ANTP. (C) The EY PAIRED and HOMEO domain interact independently with ANTP. (D) Bacterially expressed purified domains are able to interact. One microgram of ANTP HD (pAop2CS; Müller et al., 1988) incubated with 3 µg of GST or GST–PAIRED was subjected to electrophoresis after interaction with GST beads and stained with Coomassie Blue. The arrow shows the ANTP HD. (E and F) Yeast one-and-a-half hybrid experiment. pAS and pACT: empty vectors. pAS-EY: EY encoding vector. pACT-ANTP 287–378: C-terminus of ANTP encompassing the HD is fused to the Gal4 activation domain. (E) Strain carrying the integrated reporter vector pLacZi containing four multimerized HB1 sites upstream of the minimal promoter of the yeast iso-1-cytochrome C gene. (F) Same as in (E) but instead of HB1 one copy of the so10 enhancer is inserted. Arrow: co-expression of ANTP HD and EY inhibit EY transactivation. (G) Bacterially synthesized ANTP HD inhibits PAIRED DNA binding to the 100 bp so probe (Niimi et al., 1999). His-PAIRED (20 ng) and His-ANTP peptides (1 µg) were co-incubated before adding the so probe (10 ng). The arrow shows the retarded complexes.
To define the regions within EY and ANTP that are required for the interaction of the two proteins, we tested a set of deletion mutants of each protein for their ability to interact in vitro. Structure–function studies of both proteins have delineated specific domains that contribute to their functions as transcription factors as well as their interactions with other proteins. The ANTP HD that mediates DNA binding has also been shown to interact with other HD proteins such as EXD (reviewed in Mann and Affolter, 1998). The EY protein contains two DNA-binding domains, a paired domain and an HD. The paired domain has been shown to interact with different transcription factors (Fitzsimmons et al., 1996; Jun and Desplan, 1996; Bendall et al., 1999). These findings led us to investigate whether these EY paired domains and HDs are involved in the interaction with ANTP. Deleting either of these domains in the EY protein results in a partial loss of the interaction with the ANTP HD (Figure 4B, compare lanes 8 and 9 with 7). Furthermore, either the EY paired domain or the EY HD alone is still able to interact with ANTP (Figure 4C, lanes 10 and 13). These experiments suggest that since each domain is able to interact with ANTP, both domains might cooperate for efficient binding of EY to ANTP (Figure 4B). Moreover, deletion of the C-terminal part of the ANTP protein results in the loss of binding to the paired domain (Figure 4C, lane 12) or HD of EY (Figure 4C, lane 14), confirming that the ANTP HD is essential for the interaction with EY.
Because the 35S-labeled proteins used in the binding reactions were synthesized in rabbit reticulocyte lysates, we sought to determine whether the ANTP–EY interaction is direct or dependent on a bridging molecule that might be present in the lysate. Since the complexes were formed using EY paired domain and ANTP HD purified from bacteria (Figure 4D), the two proteins appear to interact directly through their respective DNA-binding regions.
ANTP inhibits EY transactivation in yeast and DNA-binding activity in vitro
Despite considerable efforts, we were unable to confirm the interaction in a two-hybrid assay in yeast. Thus, we hypothesized that the DNA interface might be important to stabilize the interaction. To address this question, we performed ‘one-and-a-half hybrid’ assays that combine elements of the one- and two-hybrid systems. This allows us to test the effect of ANTP on EY-mediated activation and vice versa in yeast. We generated two reporter constructs cloned upstream of the LacZ gene, one carrying the so10 enhancer as an EY responsive element and the second carrying multimeric ANTP binding sites called HB1 (Haerry and Gehring, 1996; Keegan et al., 1997). Furthermore, we generated an ANTP activator by fusing the GAL4 transactivation domain to the HD (pACT-Antp 287–378). This ANTP protein is able to activate HB1 but not the so10 fragment (Figure 4E and F, lane 4). This further indicates that the so10 enhancer is not directly regulated by ANTP. Moreover, EY activates the so10-LacZ reporter but has no effect on HB1-LacZ reporter (Figure 4E and F, lane 2). Interestingly, when both proteins are co-expressed, the activation mediated by one protein is suppressed by the other (Figure 4E and F, lane 3). This result can not be explained by a a squelching effect due to the GAL4 activation domain, since this domain alone does not affect EY activation (Figure 4F, lane 2). In contrast, the presence of the ANTP HD is necessary for repressing EY transactivation. Taken together, these results show that the interaction between the two proteins requires that one of them binds to DNA. We next asked how the interaction between the ANTP and EY proteins might affect DNA binding, using gel retardation assays. A fragment of the so10 enhancer has previously been shown to be bound by EY (Niimi et al., 1999). We used this DNA fragment in a bandshift assay, employing His-tagged EY paired domain and Antp HD. Incubation with the paired domain resulted in a shifted complex (Figure 4G, lane 2), whereas the Antp peptides that failed to interact with EY had no effect on EY binding (Figure 4G, lanes 3–5). Adding the Antp HD (Figure 4G, lane 6) to the paired domain resulted in an inhibition of EY DNA binding. However, we were unable to detect the formation of a predicted ternary complex. We consider that a ternary complex is likely to be formed on the DNA but dissociates during electrophoresis, resulting in an apparent inhibition of binding. Alternatively, the interaction of EY with the ANTP HD may lead to the dissociation of the paired domain from its binding site.
Inhibition of eye development mediated by Antp reflects a balance between the levels of the activator (EY) and the repressor (ANTP)
These findings led to the suggestion that the relative concentration of each protein is important for their activity. Given the developmental roles of these proteins, this mechanism is potentially important for understanding cell fate determination. We therefore investigated this point further using an in vivo assay. We took advantage of the differential expression of ey in the eye and antenna disc and used the UAS-GAL4 system to express more EY protein in the eye disc, exceeding the endogenous level, and to analyze the effect of Antp overexpression under these conditions.
First, using the dppblink-GAL4 driver, we targeted expression of ey, Antp or both in the eye–antenna disc and analyzed their effect on so expression. Figure 5C shows that targeted expression of ey in the antenna disc induces so as expected. Co-expression of ey and Antp in the antenna disc resulted in the repression of so induction (Figure 5D), similar to the results obtained in the leg and wing discs (Figure 3J and K). As a consequence, no ectopic eyes were induced (Figure 3C). In the eye disc with endogenous levels of EY, Antp represses so (Figure 5B). But interestingly, increasing the level of EY by co-expression of ANTP and EY resulted in the partial derepression of so in the eye disc (Figure 5D). This result supports the idea that so expression responds to the respective levels of EY and ANTP, and not simply their presence or absence.
Secondly, using the EYE-GAL4 or dppblink-GAL4 drivers, the inhibition of eye development induced by Antp is relieved when both proteins are co-expressed (compare Figure 3B with C and 5F with G). Overexpression of EY interferes with wild-type eye development (Figure 5E). This confirms the results obtained by Curtiss and Mlodzik (2000), attesting to the importance of a proper level of eye determining genes during eye development.
Thus, taken together with the observation that ANTP inhibits EY activity in yeast, the antagonistic action of ANTP and EY strongly suggests a protein–protein interaction to account for their opposing effect on eye development in vivo.
The inhibition of EY function is not restricted to ANTP
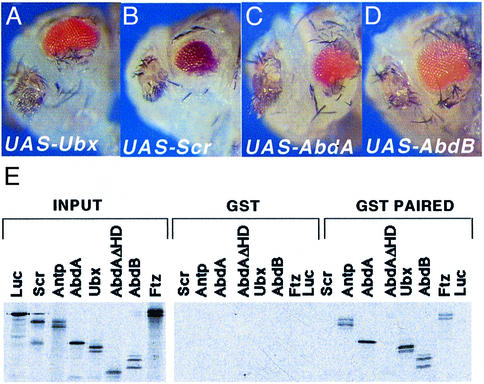
Finally, we addressed the question of whether the ability of Antp to repress eye development and to interact with EY can be extended to other homeotic genes. For this purpose, we targeted expression of Scr, Ubx, abdA and AbdB into the eye disc using dppblink-GAL4. Expression of these different genes also resulted in inhibition of eye development by inducing apoptosis (Figure 6A–D and data not shown). Interestingly, these different proteins are also able to interact with EY in vitro (Figure 6E). Deletion of the ABD-A HD region abrogates binding to EY, suggesting that also for this protein, the HD is required for interaction with EY.
Fig. 6. Different homeotic genes repress eye morphogenesis by interacting with EY. (A–D) The different UAS lines indicated were crossed with dppblink-Gal4. Pictures shown are from dissected pupae. (E) GST interaction assays performed using in vitro reticulocyte lysate synthesis of different homeotic proteins. AbdA ΔHD was generated by internal deletion of the BglII fragment. SCR, which is not detectable here, interacts weakly with EY in other experiments.
Discussion
In this paper, we have studied a molecular mechanism by which homeotic genes repress eye development when they are misexpressed in the eye disc. Analyzing expression patterns of the eye-determining genes ey, so, eya and dac in the eye as well as in ectopically developing eyes, we have demonstrated that Antp blocks ey function. This block leads to increased apoptosis in the eye disc that strongly resembles the phenotype observed in ey, so and eya loss-of-function mutants. Furthermore, we have shown that the ANTP and EY proteins are able to interact directly in vitro. Thus, our findings implicate a functional antagonism through direct protein–protein interaction as a mechanism for regulating selector gene activity. We propose that ANTP and other homeoproteins sequester EY in a protein complex, thereby rendering it unable to activate ey downstream target genes.
Schneuwly et al. (1987) have demonstrated by overexpressing Antp in the head, that a single gene is able to induce a change in the body plan. In this case, these modifications resulted in head-to-thorax transformation. The question remains as to how homeotic proteins operate at the cellular and molecular levels to ensure and to coordinate segmental development. Based on their dissection of the ANTP protein, Gibson et al. (1990) proposed that just a small portion of the protein, containing the evolutionarily conserved HD, is sufficient to define functional specificity. Interestingly, we found that the same domain is necessary for the interaction with EY.
Considerable effort has been expended in order to establish the molecular mechanisms for the antenna-to-leg transformation. It has been shown that several different homeotic genes, including Antp but also Scr and Ultrabithorax, are able to reprogram the antenna into a leg structure. These leg-inducing homeotic genes have been proposed to antagonize the apparent antenna determining gene hth, thereby preventing the nuclear localization of EXD required for antenna formation (Casares and Mann, 1998). It has recently been shown that several HOM-C genes share a common capacity to repress transcription of the hth gene (Yao et al., 1999). Since the mechanism for eye inhibition reported here is different, it is of interest to note that both mechanisms occur through inhibition of resident head genes (hth for the antenna and ey for the eye).
The results presented here corroborate previous observations suggesting that this inhibition is not restricted to Antp, since the HOM-C genes Scr, Ubx, abdA, AbdB (this study), pb (Benassayag et al., 1997), hth and exd (Pai et al., 1998; Yao et al., 1999) are able to repress eye development. Moreover, the observation of Gibson et al. (1990) that Scr is less efficient than Antp in inhibiting eye development in vivo nicely parallels our finding that SCR also shows less effective binding to EY when compared with ANTP. Since the HDs of SCR and ANTP mainly differ in their N-terminal arms, these observations imply that the N-terminal arm is critically involved in the interaction with EY. Apart from ANTP, ABD-A (this study) and PB (D.Cribbs, personal communication) likewise interact with EY through their HD. On the other hand, TOY interacts with ANTP (not shown). Moreover, an inhibition through physical association has been proposed between En-1 and Pax-6 in quail during eye development (Plaza et al., 1997), and between Pax-3 and Msx-1 for muscle development in chicken (Bendall et al., 1999), again suggesting that similar regulatory mechanisms occur in other species and might be diversely employed to control organ development. These observations suggest that the phenotypic and molecular inhibition described here might constitute a general mechanism for controlling EY activity.
This suggestion has numerous potential ramifications, given the complex nature of the developmental pathways involved. Both homeotic and eye selector functions are influenced, whether directly or indirectly, by other transcription factors and by impinging signaling pathways. For example, the ectopic eye formation induced by some loss-of-function clones of hth or exd suggests that these genes restrict eye development within the adult head region (Gonzalez-Crespo and Morata, 1995; Pai et al., 1998). However, the mechanism by which this induction occurs is not yet known. One interesting possibility involves the regulation of signaling pathways implicated in normal eye development. The graded distribution of DPP protein resulting from spatially localized gene expression provides a crucial piece of information toward the appendage or organ structure; this information is in turn utilized by selector genes to mobilize groups of cells into a differentiation pathway. Indeed, dpp function is required for eye development and for the activation of the EY target so (Chen et al., 1999; Treisman, 1999; Curtiss and Mlodzik 2000). Conversely, functional SO is required to activate dpp (Chen et al., 1999; Treisman, 1999; Curtiss and Mlodzik 2000). This interdependence may provide an elegant means for progressively differentiating the eye via feedback loops. In such a situation, an accurate measure of the relative levels of gene products as implied in this study and in Curtiss and Mlodzik (2000) may have a particular importance for the correct outcome.
Other recently described examples of functional repression of transcription factors involve direct interaction. It has been shown that PU1, an ets gene family transcription factor, blocks erythrocyte differentiation by binding to and thereby inhibiting the activity of the transcription factor GATA1 (Rekhtman et al., 1999).
A Hox protein may possess both activating and repressing functions in vivo, depending on the target gene. However, the mechanisms permitting such a conversion are not well understood. Intermolecular interaction between DNA-binding domains is one commonly encountered strategy to generate functional diversity. Most of the examples available show cooperative interaction (Lassar et al., 1991; Luisi et al., 1991; Ellenberger et al., 1992; Wilson and Desplan, 1995). Interestingly, Zappavigna et al. (1994) described a mutual antagonism for HOX proteins through direct interaction. A variation of this strategy is to associate HOX proteins with co-factors such as EXD and HTH, thus facilitating the diversification of regulatory roles on different genes (Mann and Affolter, 1998).
Prominent developmental selector roles for ey and Antp affect two distinct regions of the head and thorax, respectively. However, the apparent specificity of the molecular interactions described above lead us to predict that ey and Antp might be co-expressed in vivo during normal development. Evolutionarily conserved expression and function of Hox and Pax-6 genes in the CNS may offer one relevant situation for measuring the relative quantities of HOX and PAX proteins. In support of this idea, preliminary experiments indicate a co-expression of these two genes in the nervous system during Drosophila embryogenesis.
The finding of specific protein–protein interactions between the HD and the paired domain also has other important evolutionary implications. One of the most striking features of these two domains is their strong evolutionary conservation. This high degree of sequence conservation can be explained to some extent by evolutionary constraints imposed by their specific binding to numerous DNA target sites. The recent demonstration that some HDs also bind to specific RNA sequences (Rivera-Pomar et al., 1996) adds an additional potential constraint. Our demonstration that HD and paired domain are not only DNA-binding domains, but also specific protein–protein interaction domains, confers another evolutionary constraint to the Hox and Pax genes. This may provide an additional explanation for their evolutionary conservation, and offers novel possibilities for functional diversification.
Materials and methods
Fly strains
Flies were reared on standard medium at 25°C. Lines used were: so-LacZ (Cheyette et al., 1994), so10-LacZ (Niimi et al., 1999), GMR-Gal4 (Ellis et al., 1993), dppblink-Gal4 (Staehling-Hampton et al., 1994), UAS-ey (Halder et al., 1995), EYE-Gal4 and hs-ey (Halder et al., 1998), UAS-Antp and UAS-AntpΔHD (Bello et al., 1998), and hs-Antp constructs H45, G2, G8, G10 and G11 (Gibson et al., 1990). UAS-AntpK50 is a generous gift of J.Botas. OK107 (Connolly et al., 1996) was made available by P.Callaerts, who found expression in the eye disc.
Specific genotypes generated for this publication were: (i) UAS-ey/UAS-ey;UAS-Antp/UAS-Antp; (ii) so10-LacZ/so10-LacZ;UAS-Antp/TM6B, Tb, Hu; (iii) so10-LacZ/so10-LacZ;dppblink-Gal4/TM6B, Tb, Hu;spapol/spapol; (iv) so-LacZ/CyO;dppblink-Gal4/TM6B, Tb, Hu; and (v) UAS-ey/UAS-ey;UAS-AntpΔHD/UAS-AntpΔHD. Transgenic lines (pUAST-Antp50A,51A) were generated as described by Niimi et al. (1999).
Histology
Since crosses of EYE-GAL4 and UAS-Antp resulted in ‘headless’ larvae lacking an eye disc, analysis of gene expression was performed with dppblink-Gal4. In situ hybridizations were performed using digoxigenin-labeled probes according to Halder et al. (1998). Antibody stainings were performed according to Halder et al. (1998) at the following dilutions: rat α-ey, 1/600; mouse α-Antp, 1/500; mouse α-22C10, 1/20; mouse α-Dac, 1/100. Apoptosis experiments were carried out according to Halder et al. (1998). β-galactosidase expression was detected as described in Niimi et al. (1999). Correct amounts of EY or ANTP proteins were monitored by western blotting of discs. We ensured that creating the double UAS lines did not affect protein expression and that the ANTP HD-deleted protein was produced efficiently.
Heat shock experiments
Virgin females homozygous for hs-Antp constructs (Gibson et al., 1990) were crossed with males homozygous for hs-ey (Halder et al., 1998). Control crosses for expressing Antp or ey separately were performed by crossing these homozygous fly lines to the recipient lines yw or ry506, respectively. Eggs laid during 2–4 h periods were collected and incubated at 25°C. Heat shock cycles were applied according to Halder et al. (1998) except that the first heat shock was applied at 74 ± 2 h after egg laying to ensure proper Antp transformation (Gibson et al., 1990). After hatching, adult escapers were counted and analyzed. Results given are based on at least 50 flies.
Pull-down experiments and gel shift assays
GST fusion proteins (pGEX4T3) were produced and purified according to the manufacturer’s specifications (Pharmacia). Reticulocyte lysate proteins were produced using the TNT reticulocyte lysate synthesis kit (Promega). Pull-down experiments were carried out essentially as described in Plaza et al. (1997). Equal amounts of GST fusion protein were monitored by Coomassie Blue staining.
Gel shift assays were performed with the 100 bp so fragment essentially as described in Niimi et al. (1999). His fusion proteins (pQE30-EY paired and ANTP) were produced according to the manufacturer’s specification (Qiagen) and finally diluted in the binding reaction to give 10 mM Tris pH 7.5, 75 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 10% glycerol, 25 mM imidazole and 1 µg/ml poly[(dI)–(dC)].
One-hybrid experiment
One-hybrid experiments were performed using the MATCHMAKER one-hybrid system (Clontech) according to the manufacturer’s specifications. The NcoI–SmaI fragment of ey cDNA was subcloned in-frame with the Gal4DB in the pAS vector. We made sure that the fusion had no effect on EY activation. The Antp (amino acids 287–378) HD-containing fragment was fused to the Gal4 transactivation domain in the pACT vector. pAS and pACT are from Durfee et al. (1993).
Cloning procedure and plasmids
Point mutations and fusion proteins generated were constructed using standard PCR amplification procedures. Cloning sites were created at both ends of the amplification product to ensure the correct orientation. After cloning, each construct was verified by sequencing. A detailed description of the primers used and cloning procedure will be given upon request. In vitro translation experiments were performed using full-length cDNA cloned in pBSKII+. Subclones were: EcoRI Antp cDNA and EcoRI abdA cDNA into pBSKII at EcoRI site; BamHI Ubx cDNA into pBSKII at BamHI site. The Scr and AbdB constructs (p728) were generous gifts from M.Berry and F.Karch, respectively. The pBSKII-ey plasmid was from Quiring et al. (1994). Truncations of the Antp cDNA were performed by linearizing the plasmid, which was further used to synthesize truncated proteins in vitro using the TNT system. The Antp A50,51 mutant was created by PCR, replacing the XbaI–BamHI fragment with the mutated one in pBSKII Antp. This mutated cDNA was further cloned into the pUAST vector using EcoRI–BglII sites.
Acknowledgments
Acknowledgements
We acknowledge F.Karch for providing the abdA, Ubx and AbdB clones, H.Sun and D.Brower for ANTP antibodies, U.Walldorf for EY antibodies, G.Mardon for Dac antibodies, and S.Benzer for mAb22C10. This work was supported by the Kantons of Basel-Stadt and Basel-Land, and a grant from the Swiss National Science Foundation. S.P. was supported by the Centre National de la Recherche Scientifique and by an EMBO fellowship.
Note added in proof
A similar inhibitory mechanism involving the HD of Hox protein has recently been published by Kataoka et al. (2001) and Abramovich et al. (2000). Because of space limitation we were unable to cite all the references related to the work, for which we apologize.
Abramovich,C., Shen,W.F., Pineault,N., Imren,S., Montpetit,B., Largman,C. and Humphries,R.K. (2000) Functional cloning and characterization of a novel nonhomeodomain protein that inhibits the binding of PBX1–HOX complexes to DNA. J. Biol. Chem., 275, 26172--26177.
Kataoka,K., Yoshitomo-Nakagawa,K., Shioda,S. and Nishizawa,M. (2001) A set of Hox proteins interact with the Maf oncoprotein to inhibit its DNA binding, transactivation, and transforming activities. J. Biol. Chem., 276, 819–826.
REFERENCES
- Abbott M.K. and Kaufman,T.C. (1986) The relationship between the functional complexity and the molecular organization of the Antennapedia locus of Drosophila melanogaster. Genetics, 114, 919–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B., Resendez-Perez,D. and Gehring,W.J. (1998) Spatial and temporal targeting of gene expression in Drosophila by means of a tetracycline-dependent transactivator system. Development, 125, 2193–2202. [DOI] [PubMed] [Google Scholar]
- Benassayag C., Seroude,L., Boube,M., Erard,M. and Cribbs,D.L. (1997) A homeodomain point mutation of the Drosophila proboscipedia protein provokes eye loss independently of homeotic function. Mech. Dev., 63, 187–198. [DOI] [PubMed] [Google Scholar]
- Bendall A.J., Ding,J., Hu,G., Shen,M.M. and Abate-Shen,C. (1999) Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development, 126, 4965–4976. [DOI] [PubMed] [Google Scholar]
- Bonini N.M. and Fortini,M.E. (1999) Surviving Drosophila eye development: integrating cell death with differentiation during formation of a neural structure. BioEssays, 21, 991–1003. [DOI] [PubMed] [Google Scholar]
- Bonini N.M., Leiserson,W.M. and Benzer,S. (1993) The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell, 72, 379–395. [DOI] [PubMed] [Google Scholar]
- Callaerts P., Halder,G. and Gehring,W.J. (1997) PAX-6 in development and evolution. Annu. Rev. Neurosci., 20, 483–532. [DOI] [PubMed] [Google Scholar]
- Casares F. and Mann,R.S. (1998) Control of antennal versus leg development in Drosophila. Nature, 392, 723–726. [DOI] [PubMed] [Google Scholar]
- Chadwick R., Jones,B., Jack,T. and McGinnis,W. (1990) Ectopic expression from the Deformed gene triggers a dominant defect in Drosophila adult head development. Dev. Biol., 141, 130–140. [DOI] [PubMed] [Google Scholar]
- Chen R., Halder,G., Zhang,Z. and Mardon,G. (1999) Signaling by the TGF-β homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development, 126, 935–943. [DOI] [PubMed] [Google Scholar]
- Cheyette B.N., Green,P.J., Martin,K., Garren,H., Hartenstein,V. and Zipursky,S.L. (1994) The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron, 12, 977–996. [DOI] [PubMed] [Google Scholar]
- Connolly J.B., Roberts,I.J., Armstrong,J.D., Kaiser,K., Forte,M., Tully,T. and O’Kane,C.J. (1996) Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science, 274, 2104–2107. [DOI] [PubMed] [Google Scholar]
- Curtiss J. and Mlodzik,M. (2000) Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development, 127, 1325–1336. [DOI] [PubMed] [Google Scholar]
- Czerny T., Halder,G., Kloter,U., Souabni,A., Gehring,W.J. and Busslinger,M. (1999) twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell, 3, 297–307. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer,K., Chen,P.L., Yeh,S.H., Yang,Y., Kilburn,A.E., Lee,W.H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Ellenberger T.E., Brandl,C.J., Struhl,K. and Harrison,S.C. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein–DNA complex. Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Ellis M.C., O’Neill,E.M. and Rubin,G.M. (1993) Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development, 119, 855–865. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons D., Hodsdon,W., Wheat,W., Maira,S.M., Wasylyk,B. and Hagman,J. (1996) Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev., 10, 2198–2211. [DOI] [PubMed] [Google Scholar]
- Gehring W.J. (1987) Homeo boxes in the study of development. Science, 236, 1245–1252. [DOI] [PubMed] [Google Scholar]
- Gehring W.J. and Ikeo,K. (1999) Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet., 15, 371–377. [DOI] [PubMed] [Google Scholar]
- Gehring W.J., Affolter,M. and Burglin,T. (1994) Homeodomain proteins. Annu. Rev. Biochem., 63, 487–526. [DOI] [PubMed] [Google Scholar]
- Gibson G., Schier,A., LeMotte,P. and Gehring,W.J. (1990) The specificities of Sex combs reduced and Antennapedia are defined by a distinct portion of each protein that includes the homeodomain. Cell, 62, 1087–1103. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Crespo S. and Morata,G. (1995) Control of Drosophila adult pattern by extradenticle. Development, 121, 2117–2125. [DOI] [PubMed] [Google Scholar]
- Haerry T.E. and Gehring,W.J. (1996) Intron of the mouse Hoxa-7 gene contains conserved homeodomain binding sites that can function as an enhancer element in Drosophila. Proc. Natl Acad. Sci. USA, 93, 13884–13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G., Callaerts,P. and Gehring,W.J. (1995) Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science, 267, 1788–1792. [DOI] [PubMed] [Google Scholar]
- Halder G., Callaerts,P., Flister,S., Walldorf,U., Kloter,U. and Gehring,W.J. (1998) Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development, 125, 2181–2191. [DOI] [PubMed] [Google Scholar]
- Jun S. and Desplan,C. (1996) Cooperative interactions between paired domain and homeodomain. Development, 122, 2639–2650. [DOI] [PubMed] [Google Scholar]
- Jun S., Wallen,R.V., Goriely,A., Kalionis,B. and Desplan,C. (1998) Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc. Natl Acad. Sci. USA, 95, 13720–13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan L.P., Haerry,T.E., Crotty,D.A., Packer,A.I., Wolgemuth,D.J. and Gehring,W.J. (1997) A sequence conserved in vertebrate Hox gene introns functions as an enhancer regulated by posterior homeotic genes in Drosophila imaginal discs. Mech. Dev., 63, 145–157. [DOI] [PubMed] [Google Scholar]
- Lassar A.B., Davis,R.L., Wright,W.E., Kadesch,T., Murre,C., Voronova,A., Baltimore,D. and Weintraub,H. (1991) Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell, 66, 305–315. [DOI] [PubMed] [Google Scholar]
- Lewis E.B. (1992) Clusters of master control genes regulate the development of higher organisms. J. Am. Med. Assoc., 267, 1524–1531. [PubMed] [Google Scholar]
- Luisi B.F., Xu,W.X., Otwinowski,Z., Freedman,L.P., Yamamoto,K.R. and Sigler,P.B. (1991) Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature, 352, 497–505. [DOI] [PubMed] [Google Scholar]
- Mann R.S. and Affolter,M. (1998) Hox proteins meet more partners. Curr. Opin. Genet. Dev., 8, 423–429. [DOI] [PubMed] [Google Scholar]
- Mardon G., Solomon,N.M. and Rubin,G.M. (1994) dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development, 120, 3473–3486. [DOI] [PubMed] [Google Scholar]
- Müller M., Affolter,M., Leupin,W., Otting,G., Wuthrich,K. and Gehring,W.J. (1988) Isolation and sequence-specific DNA binding of the Antennapedia homeodomain. EMBO J., 7, 4299–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T., Seimiya,M., Kloter,U., Flister,S. and Gehring,W.J. (1999) Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development, 126, 2253–2260. [DOI] [PubMed] [Google Scholar]
- Pai C.Y., Kuo,T.S., Jaw,T.J., Kurant,E., Chen,C.T., Bessarab,D.A., Salzberg,A. and Sun,Y.H. (1998) The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev., 12, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza S., Langlois,M.C., Turque,N., LeCornet,S., Bailly,M., Begue,A., Quatannens,B., Dozier,C. and Saule,S. (1997) The homeobox-containing Engrailed (En-1) product down-regulates the expression of Pax-6 through a DNA binding-independent mechanism. Cell Growth Differ., 8, 1115–1125. [PubMed] [Google Scholar]
- Quiring R., Walldorf,U., Kloter,U. and Gehring,W.J. (1994) Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science, 265, 785–789. [DOI] [PubMed] [Google Scholar]
- Rekhtman N., Radparvar,F., Evans,T. and Skoultchi,A.I. (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev., 13, 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Pomar R., Niessing,D., Schmidt-Ott,U., Gehring,W.J. and Jackle,H. (1996) RNA binding and translational suppression by bicoid. Nature, 379, 746–749. [DOI] [PubMed] [Google Scholar]
- Schneuwly S. and Gehring,W.J. (1982) Homeotic transformation of thorax into head: developmental analysis of a new Antennapedia allele in Drosophila melanogaster. Dev. Biol., 108, 377–386. [Google Scholar]
- Schneuwly S., Klemenz,R. and Gehring,W.J. (1987) Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature, 325, 816–818. [DOI] [PubMed] [Google Scholar]
- Seimiya M. and Gehring,W.J. (2000) The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless independent mechanism. Development, 127, 1879–1886. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K., Jackson,P.D., Clark,M.J., Brand,A.H. and Hoffmann,F.M. (1994) Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ., 5, 585–593. [PubMed] [Google Scholar]
- Struhl G. (1981) A homoeotic mutation transforming leg to antenna in Drosophila. Nature, 292, 635–638. [DOI] [PubMed] [Google Scholar]
- Toy J., Yang,J.M., Leppert,G.S. and Sundin,O.H. (1998) The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc. Natl Acad. Sci. USA, 95, 10643–10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman J.E. (1999) A conserved blueprint for the eye? BioEssays, 21, 843–850. [DOI] [PubMed] [Google Scholar]
- Wilson D.S. and Desplan,C. (1995) Homeodomain proteins. Cooperating to be different. Curr. Biol., 5, 32–34. [DOI] [PubMed] [Google Scholar]
- Yao L.C., Liaw,G.J., Pai,C.Y. and Sun,Y.H. (1999) A common mechanism for antenna-to-leg transformation in Drosophila: suppression of homothorax transcription by four HOM-C genes. Dev. Biol., 211, 268–276. [DOI] [PubMed] [Google Scholar]
- Zappavigna V., Sartori,D. and Mavilio,F. (1994) Specificity of HOX protein function depends on DNA–protein and protein–protein interactions, both mediated by the homeo domain. Genes Dev., 8, 732–744. [DOI] [PubMed] [Google Scholar]
- Zhao J.J., Lazzarini,R.A. and Pick,L. (1993) The mouse Hox-1.3 gene is functionally equivalent to the Drosophila Sex Combs reduced gene. Genes Dev., 7, 343–354. [DOI] [PubMed] [Google Scholar]
- Zimmerman J.E., Bui,Q.T., Liu,H. and Bonini,N.M. (2000) Molecular genetic analysis of Drosophila eyes absent mutants reveals an eye enhancer element. Genetics, 154, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]