Abstract
Regulation of the GAL genes of Saccharomyces cerevisiae is determined by the interplay of the transcriptional activator Gal4p and the repressor Gal80p, which binds and masks the activation domain of Gal4p under non-inducing conditions. Here we demonstrate that Gal80p dimerizes with high affinity and that this dimerization appears to stabilize the Gal4p–Gal80p interaction and also, indirectly, the Gal4p–DNA interaction in a (Gal4p)2(Gal80p)2DNA complex. In addition, Gal80 dimers transiently interact with each other to form higher order multimers. We provide evidence that adjacent Gal4p binding sites, when correctly spaced, greatly stabilize Gal80p dimer–dimer interactions and that this stabilization results in the complete repression of GAL genes with multiple Gal4p binding sites. In contrast, GAL genes under the control of a single Gal4p binding site do not stabilize Gal80p multimers, resulting in significant and biologically important transcriptional leakage. Cooperative binding experiments indicate that Gal80p dimer–dimer interaction probably does not lead to a stronger Gal4p–Gal80p interaction, but most likely to a more complete shielding of the Gal4p activation domain.
Keywords: GAL4/GAL80/Saccharomyces cerevisiae/transcriptional repression
Introduction
Expression of the galactose catabolic genes of the yeast Saccharomyces cerevisiae is determined by the carbon source due to the action of three regulatory proteins: the transcriptional activator Gal4p, the repressor Gal80p and the signal transducer Gal3p (for reviews see Johnston and Carlson, 1992; Melcher, 1997). In the absence of glucose, the transcriptional activator Gal4p binds constitutively to 17 bp recognition sequences upstream of the GAL genes. While binding of Gal4p to DNA is necessary for transcriptional activation, it is not sufficient. Rather, Gal4p forms a tight poised complex with the repressor Gal80p. The repressor binds to a 28 amino acid region at the very C-terminus of Gal4p, which coincides with Gal4p’s core activation domain and thereby physically blocks interactions of the activation domain with the transcriptional machinery (e.g. Wu et al., 1996; Koh et al., 1998).
Upon induction by galactose, the inhibitory effect of Gal80p is relieved by the action of the signal transducer Gal3p. Gal3p is a homologue of the galactokinase Gal1p, and in the presence of the galactokinase substrates, galactose and ATP, Gal3p physically binds Gal80p. This interaction is believed to cause a conformational change in Gal80p, leading to a release of Gal80p masking of the Gal4p activation domain (Zenke et al., 1996; Yano and Fukasawa, 1997; Platt and Reece, 1998).
There are two types of GAL gene: the catabolic genes GAL1, -2, -7 and -10, which are extremely tightly regulated with essentially no expression in the absence of galactose, and the GAL3, GAL80 and MEL1 genes, which have significant and biologically important basal expression. The physiological role for the two types of uninduced states, which arose from a gene duplication, is particularly well illustrated for the GAL1 and GAL3 genes. The enzymatic and regulatory functions were separated into the catalytically active protein Gal1, whose production needs to be tightly regulated to avoid accumulation of toxic intermediates (Meyer et al., 1991), and into the catalytically inactive regulator Gal3p. Gal3p, as well as the repressor Gal80p, needs to be already expressed at significant levels in the absence of galactose to keep the system both repressed and inducible. Induction of GAL3 and GAL80 is necessary to reach and maintain full induction of the GAL genes and to allow rapid stalling of transcription in the fully induced state. A third example of a GAL gene with critical basal expression is the melibiase (α-galactosidase)-encoding MEL1 gene. Mel1p is a secreted enzyme necessary for extracellular breaking down of melibiose into galactose and glucose. Galactose enters the cell and triggers induction of the GAL genes, including MEL1, via the Gal3p/Gal80p/Gal4p pathway.
Bram et al. (1986) observed that GAL genes with basal expression are driven by a single Gal4p binding site, while the tightly regulated GAL genes are driven by multiple Gal4p sites. Moreover, duplication of an artificial consensus Gal4p binding site in the context of the GAL2 promoter led to decreased uninduced mRNA production, suggesting that binding site architecture modulates the level of basal GAL gene expression. Using a combination of in vivo and quantitative in vitro analyses, we confirmed these observations and investigated the molecular mechanism of this unusual regulation. Our results indicate that correctly spaced Gal4p binding sites stabilize otherwise transient higher order Gal80p multimers and that these multimers are probably necessary for complete shielding of the Gal4p activation domain.
Results
Basal expression of GAL genes and synthetic reporters depends on the number of Gal4p binding sites
Table I recapitulates the basal elements of GAL regulation. The GAL1 promoter is driven by four Gal4p binding sites, and the MEL1 promoter by a single Gal4p binding site. Expression from a GAL1 promoter–lacZ reporter integrated at the chromosomal GAL1 locus is extremely tightly regulated with no detectable basal expression (<0.1%), while basal expression from the endogenous MEL1 (α-galactosidase) gene is readily detectable under non-inducing conditions (1.8% of induced expression).
Table I. Basal expression of GAL genes and synthetic reporters depends on the number of Gal4p binding sites.
| gly/lac (U) | gly/lac + gal (U) | % basal (corrected)a | |
|---|---|---|---|
| A. Natural promoters | |||
| GAL1–lacZ (four sites) | ≤1 | 3000 | <0.1 |
| MEL1 (one site) | 16 | 880 | 1.8 |
| B. Inserted into ΔUAS–CYC1–lacZ reporter pJlb (Finley et al., 1990) | |||
| 1× 17mer MEL1 | 84 | 570 | 12 |
| 2× 17mer MEL1 | 14 | 2300 | 0.2 |
| Four sites of GAL1 | 43 | 5000 | 0.7 |
| Vector alone | 10 | 10 | – |
gly/lac: 3% glycerol, 2% lactic acid; gly/lac + gal: 3% glycerol, 2% lactic acid, 2% galactose.
Integrated Gal1–lacZ and the CYC1–lacZ reporter: units β-galactosidase activity.
MEL1: units α-galactosidase activity.
a[U(gly/lac) – U(gly/lac)vector]:[U(gly/lac + gal) – U(gly/lac + gal)vector]
Standard deviations were ≤20%.
To confirm that basal expression of MEL1 is determined solely by the number of Gal4p binding sites and not by their particular context, one or two Gal4p binding sites from MEL1 were cloned into an upstream activating sequence (UAS)-less CYC1–lacZ reporter. As expected, the reporter gene driven by a single Gal4p binding site has higher basal activity than the one driven by multiple binding sites. Note that the reporter plasmids give high basal activity relative to the endogenous genes and there fore only follow the same trend approximately. The data argue that basal expression is determined by the number of Gal4p binding sites and not by the particular type of Gal4p binding site or by promoter structural context.
Basal expression of Mel1p is Gal4p dependent and due to incomplete repression by Gal80p
If basal expression of MEL1 is indeed independent of promoter context, we would expect it to be completely dependent on Gal4p and not on some other unidentified element. In agreement with a previous report (Post-Beittenmiller et al., 1984), this has been confirmed in Table II. We reasoned that Gal4p-dependent basal expression may be due to incomplete repression by Gal80p. Overexpression of GAL80 may then partially compensate for weakened repression. Indeed, overexpression of GAL80 from a multicopy 2µ plasmid strongly reduced basal MEL1 expression (Table II). Overexpres sion of GAL80 also led to reduced MEL1 expression under inducing conditions, as expected, since induction functions by shifting the equilibrium of the interaction between Gal80p and the Gal4p activation domain (Sil et al., 1999). The decrease in basal Mel1 activity upon moderate overexpression of GAL80 argues that basal expression is at least partially due to incomplete repression by Gal80p.
Table II. Basal expression of MEL1 is completely dependent on Gal4p and is due to incomplete repression by Gal80p.
| Genotype | Gly/lac (U) | gly/lac + gal (U) |
|---|---|---|
| GAL4 Gal80 | 16 | 880 |
| Δgal4 GAL80 | ≤1 | ≤1 |
| GAL4 mcGAL80 | 1 | 180 |
mcGAL80: multicopy GAL80 plasmid; U: units of α-galactosidase activity.
Standard deviations were ≤20%.
Gal80p and Gal4p form a complex with 2:2 stoichiometry
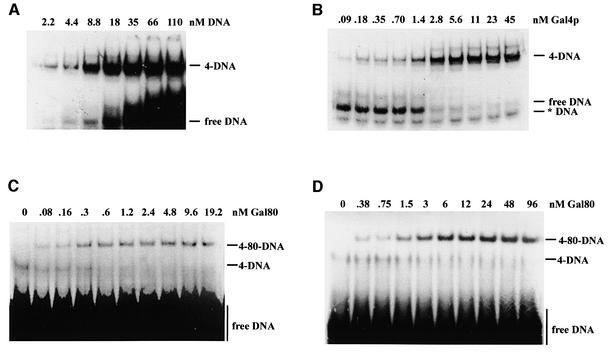
To explore the molecular basis of differential repression, we first determined the kinetic parameters of Gal4p–DNA, Gal4p–Gal80p and Gal80p–Gal80p interactions. We purified from Escherichia coli full-length Gal80p as well as Gal4p(1–147+34), and carefully determined their concentrations. Gal4p(1–147+34) is a truncated version of Gal4p in which the N-terminal 147 amino acids of Gal4p, which include the DNA binding/dimerization domain, were fused to a 34 amino acid peptide (amino acids 841–875) corresponding to the Gal80p interaction/transcriptional activation site. We first determined the DNA binding activity of Gal4p(1–147+34) by titrating a known amount of protein (15 nM) above the dissociation constant (KD) (see below) with increasing amounts of a radiolabelled consensus Gal4p binding site oligonucleotide (Figure 1A). The total DNA binding capacity was 14–15 nM, indicating that Gal4p(1–147+34) was >90% active in specific DNA binding. We determined a KD of 1.3 nM (Figure 1B), in good agreement with published results for both the truncated Gal4p DNA binding domain (Reece and Ptashne, 1993; Vashee et al., 1993) and for full-length Gal4p (Parthun and Jaehning, 1990). However, the affinity of Gal4p(1–147+34) for the same consensus binding site in a longer fragment (108 bp as opposed to a 23 bp oligonucleotide) was ∼10 times higher (see Figure 5), suggesting that Gal4p, similar to many restriction endonucleases (Moreira and Noren, 1995), prefers a more rigid DNA structure for optimal binding, and indicating that Gal4p DNA binding affinity may have previously been underestimated.
Fig. 1. Kinetic determinations of interactions between DNA, Gal4p(1–147+34) and Gal80p. Binding reactions were separated by native PAGE and subjected to autoradiography. Free and bound complexes were quantitated by PhosphorImager densitometry (Molecular Dynamics). DNA: radiolabelled consensus Gal4p binding site oligonucleotide; 4 and Gal4p: Gal4p(1–147+34); 80: Gal80p; *DNA: radiolabelled unannealed oligonucleotide. (A) Quantitative measurement of the DNA binding capacity of Gal4p(1–147+34). Fifteen nanomoles of Gal4p(1–147+34) dimers were incubated with increasing amounts (nM DNA) of the binding site oligonucleotide. The maximum binding capacity Bmax and the apparent dissociation constant Kapp were calculated according to [B] = Bmax × [F]/(Kapp + [F]), with [B] and [F] representing the experimentally determined concentrations of bound and free DNA, respectively. (B) Determination of the Kapp of Gal4p(1–147+34) to the consensus site oligonucleotide. Labelled duplex oligonucleotide (0.4 nM) was incubated with increasing amounts of Gal4p(1–147+34). (C) Determination of the Kapp of Gal80p to a Gal4p(1–147+34)–DNA complex. Gal4p(1–147+34) (90 pM) was incubated with 20 nM radiolabelled oligonucleotide in the presence of increasing amounts of Gal80p. (D) Gal80p stabilizes a Gal4p(1–147+34)–DNA complex in EMSA. Gal4p(1–147+34) at a concentration above the KD (5.7 nM) was incubated with excess DNA and increasing amounts of Gal80p. To accentuate the stabilization, electrophoresis was started after loading of samples, rather than loading reactions onto running gels as for the KD determinations.
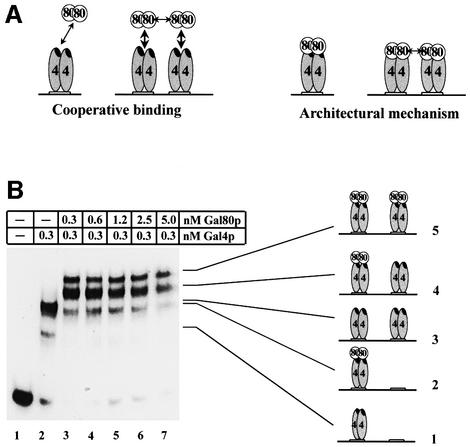
Fig. 5. Gal80p dimers bind non-cooperatively adjacent Gal4p dimers. (A) Cartoon depicting quantitative and qualitative mechanisms on how the Gal80p dimer–dimer interaction could modulate repression. 4: Gal4p; 80: Gal80p; the black oval within each Gal4p monomer represents the activation/Gal80p interaction domain. (B) Gal4p(1– 147+34) (0.3 nM) was incubated with a substoichiometric amount of a 32P-labelled, gel-isolated, 108 bp restriction fragment containing two Gal4p binding sites, spaced 10 bp apart, and with increasing amounts of Gal80p. Reactions were separated by native electrophoresis and subjected to autoradiography. Note that with higher concentrations of Gal80p, total visible labelling decreases, presumably because higher order Gal80p complexes are unstable under native electrophoresis. The five complexes formed are presented as cartoons next to the bands of the autoradiogram.
To determine the affinity of the Gal4p–Gal80p interaction, we first saturated a limiting amount of Gal4p(1–147+34) (0.09 nM) with a large excess of the radiolabelled consensus Gal4p binding site oligonucleotide (20 nM) to drive this very small amount of Gal4p(1–147+34) into a complex with DNA completely. By titrating this complex with increasing amounts of Gal80p, we determined a KD of 0.3 nM (Figure 1C). We determined a binding capacity of >90% for the Gal80p preparation by titrating a saturating amount of Gal4p(1–147+34)–DNA complex (15 nM) with increasing amounts of Gal80p. Binding was stoichiometric, indicating that two Gal80 molecules bind one Gal4p dimer complexed with DNA.
Gal80p dimerizes with high affinity
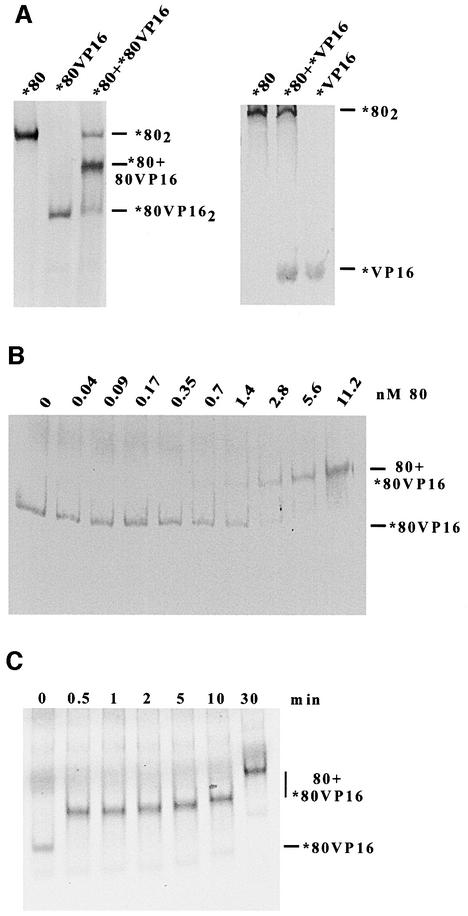
Surprisingly, we found that the repressor Gal80p actually stabilized the binding of Gal4p to DNA (Figure 1D). One mechanism by which Gal80p could affect Gal4p–DNA complex formation would be by stabilization of Gal4p dimers. Gal4p binds DNA exclusively as a dimer (Carey et al., 1989), and it is likely that the combination of Gal4p–Gal4p and Gal4p–DNA interactions is important for a low off-rate. If Gal80p would form stable dimers, then Gal80p dimers could stabilize Gal4p dimers, and thereby Gal4p–DNA complexes. In gel filtration experiments, however, Gal80p appeared to migrate as a monomer (Yun et al., 1991). In order to probe for Gal80p–Gal80p interactions using a more sensitive method, we in vitro translated a radiolabelled fusion protein consisting of Gal80p and the 78 amino acid activation domain of herpes simplex VP16 (Leuther and Johnston, 1992). Due to the high negative net charge of the VP16 activation domain, Gal80pVP16 migrated significantly faster than Gal80p in a native polyacrylamide gel. Co-translated Gal80p and Gal80pVP16 migrate on a native polyacrylamide gel as three distinct complexes (Figure 2A) (Leuther, 1993). One complex corresponded to Gal80p, one to Gal80pVP16, and the more abundant complex to a Gal80p–Gal80pVP16 heteromer. The formation of only one additional complex as well as the 1:2:1 stoichiometry of the complexes indicates that the heteromeric complex is a dimer. This stoichiometry represents the statistical distribution of all equally probable dimerizations (Gal80–Gal80, Gal80–Gal80VP16, Gal80VP16– Gal80 and Gal80VP16–Gal80VP16). We conclude that Gal80p is dimeric at the concentration of the in vitro translated proteins in the reaction and that it can form a heterodimer with Gal80VP16. No additional complex is formed when Gal80 is co-translated with the VP16 activation domain alone (Figure 2A), indicating that Gal80p and VP16 do not interact directly with each other.
Fig. 2. Gal80p dimerizes in vitro. (A) Gal80p (80) and a fusion between Gal80p and the acidic activation domain of herpes simplex virus VP16 (80VP16) were either translated separately or co-translated in vitro in the presence of 35S-labelled methionine. Translations (1 µl, ∼1 ng) in the absence of DNA were separated by native PAGE and subjected to autoradiography. Homodimeric proteins and the heterodimeric complex are indicated on the right. As a control for the specificity of complex formation, co-translated Gal80p and VP16 activation domain alone (VP16) were also tested under the same conditions as above for interaction (right panel). (B) Determination of the Kapp of a Gal80p–Gal80pVP16 complex. Labelled, in vitro translated Gal80pVP16 (*80VP16; ∼50 pM) was incubated with increasing amounts of unlabelled recombinant Gal80p (80). (C) Time course of the dissociation of Gal80pVP16 dimers. Approximately 1 nM labelled, in vitro translated Gal80pVP16 (*80VP16) was incubated with a large molar excess (120 nM) of unlabelled recombinant Gal80p (80). Reactions were incubated for the indicated amount of time before they were loaded onto a running native polyacrylamide gel (note, therefore, the relatively shorter migration of the 80+*80VP16 complex loaded 30 min after the start of electrophoresis). The t0 time point represents the reaction before the addition of unlabelled Gal80p.
We titrated radiolabelled Gal80pVP16 with increasing amounts of unlabelled recombinant Gal80p to determine an apparent dissociation constant (Kapp) of 2 × 10–9 M by native polyacrylamide gel electrophoresis (PAGE; Figure 2B). We also titrated a minimal amount of radiolabelled Gal80p with increasing amounts of unlabelled recombinant Gal80p to follow the transition from a faster migrating monomeric form into a slower migrating dimeric form by native PAGE. Signals were weak and somewhat diffused due to the very low protein concentration and total radioactivity, but we estimate a KD for Gal80p homodimerization of 1–3 × 10–10 M (data not shown). Therefore, Gal80p may homodimerize with ∼10-fold higher affinity than required to form heterodimers with Gal80pVP16 under equilibrium conditions.
To reconcile the apparent instability of Gal80p dimers during gel filtration with the very low KD, we determined the half-life of the dimer (Figure 2C). In vitro translated, 35S-labelled Gal80pVP16 was incubated with a large molar excess of unlabelled recombinant Gal80p and reactions were loaded onto a running native polyacrylamide gel. As seen on the autoradiogram in Figure 2C, even only 0.5 min after addition of unlabelled Gal80p, essentially all Gal80pVP16 migrated to a position expected for the Gal80p–Gal80pVP16 heterodimer. We note that the stability of the Gal80pVP16 dimer does not necessarily have to reflect the stability of Gal80p dimers. Nevertheless, our results together with the migration of Gal80p on a gel filtration chromatogram (Yun et al., 1991) strongly indicate that Gal80p dimerizes with high affinity, but that dimers have an unusually high off-rate when not bound to Gal4p. In contrast, Gal80 dimers are significantly stabilized by Gal4p dimers and have a long half-life when bound to Gal4p–DNA complexes (see below). In summary, a series of strong protein–protein and protein–DNA interactions appears to stabilize all components of a DNA–(Gal4p)2–(Gal80p)2 complex.
Gal80p–Gal80p interaction in vivo
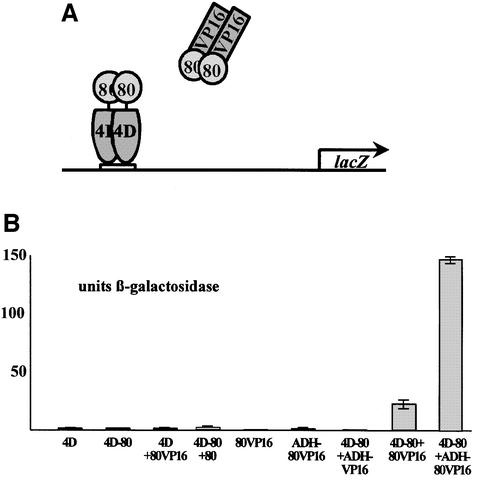
We employed a modified two-hybrid system to probe in vivo for Gal80p self-association. We expressed Gal4p(1–841)–Gal80p in a strain deleted for the wild-type GAL4 and GAL80 genes. In this hybrid, the 34 amino acid activation/Gal80p interaction domain of Gal4p is deleted and replaced by Gal80p directly fused to the truncated protein. Neither Gal4p(1–841)–Gal80p nor Gal80pVP16 alone produced significant β-galactosidase activity from an integrated GAL1 promoter–lacZ reporter (Figure 3A and B). Co-expression of Gal4p(1–841)– Gal80p and Gal80pVP16, however, produced 33 U of β-galactosidase activity, clearly demonstrating that Gal80p also self-associates in vivo. Unlike typical two-hybrid systems, fusion proteins were expressed in single copy from the natural GAL4 and GAL80 promoters, respectively, indicating that the interaction occurs under normal physiological conditions.
Fig. 3. Gal80p-modified two-hybrid assay. (A) Outline of the assay. A fusion protein in which Gal80p replaces the activation domain of Gal4p activates transcription in the presence of a Gal80pVP16 hybrid (see text for details). (B) Specific reporter gene activities in different transformants. Constructs were either integrated or transformed on single-copy plasmids. 4D: Gal4p(1–841); 4D-80: fusion between Gal4p(1–841) and Gal80p; 80VP16: fusion between Gal80p and the activation domain of VP16; 80: Gal80p; ADH-80VP16: Gal80pVP16 expressed from the ADH1 promoter; ADH-VP16: His6-tagged VP16 activation domain expressed from the ADH1 promoter. When not indicated otherwise, proteins were expressed from the natural GAL4 and GAL80 promoters.
Gal4p binds DNA exclusively as dimers (Carey et al., 1989). If DNA-bound Gal4p(1–841)–Gal80p represents a stable dimer as well, then the two-hybrid interaction would point to a Gal80p dimer–dimer interaction. In support of this interpretation, overexpression of integrated Gal80pVP16 from an ADH1 promoter directed significantly higher β-galactosidase activity (Figure 3B). This indicates that the intracellular concentration of the fusion proteins expressed from their own promoters is below the KD of the two-hybrid interaction, consistent with a relatively weak affinity rather than the high affinity Gal80p monomer–monomer interaction. Co-expression of the VP16 activation domain alone with Gal4p(1–841)– Gal80p did not yield detectable β-galactosidase activity (Figure 3B).
Gal80p dimers transiently associate with each other
Complete repression by Gal80p on multiple Gal4p sites versus leaky repression on a single site suggest a cooperative repression by Gal80p. Cooperative effects are often mediated by direct protein–protein interactions, such as between a Gal80p dimer assembled on one binding site and a component, most likely Gal80p again, associated with the other binding site. While the two-hybrid interaction pointed to a weak Gal80p dimer–dimer interaction, we could detect no higher order Gal80p complexes stable to native electrophoresis (Figures 1C and D and 2A). Therefore, if Gal80 dimers do associate with each other, the interaction must be relatively transient.
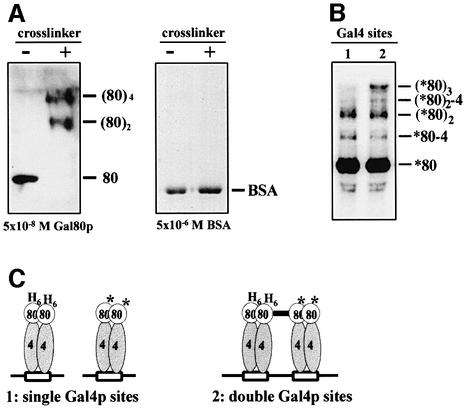
To detect potentially transient Gal80p dimer–dimer interactions physically, we used a novel chemical crosslinking technique (Brown et al., 1995). The reagent employed is a high valency peptide–nickel complex activated by a peracid, because this agent allows efficient and rapid formation of essentially 0 Å crosslinks. A 2 min incubation of 5 × 10–8 M Gal80p with these crosslinking reagents resulted in essentially 100% dimer formation, as detected by SDS–PAGE (Figure 4A). The same result was obtained with Gal80p purified from yeast (data not shown). As a control for the specificity of crosslinking, the reaction was also performed with acetylated bovine serum albumin (BSA), which is strictly monomeric in solution. As shown in the right panel of Figure 4A, incubation of acetylated BSA, even at a 100-fold higher concentration (5 × 10–6 M) than Gal80p (5 × 10–8 M), with the crosslinking reagents did not yield any detectable unspecific complexes. Incubation with a completely different crosslinker, 0.01% glutardialdehyde, led to the formation of dimeric, trimeric and tetrameric Gal80p complexes, and still exclusively monomeric BSA (data not shown).
Fig. 4. Gal80p multimerization is stabilized by adjacent Gal4p binding sites. 80/(80)2/(80)3/(80)4: Gal80p monomer/dimer/trimer/tetramer; 80–4/(80)2-4: Gal80p–Gal4p(1–147+34) heterodimer/heterotrimer; asterisk: radioactively labelled. Compositions of complexes are indicated and are based on molecular weight standards, immunoblots and on control titrations using different concentrations of Gal80p and different crosslinking efficiencies. (A) Gal80p tetramerizes in solution. Gal80p (5 × 10–8 M) or, as a control, BSA (5 × 10–6 M) was incubated for 2 min in the presence or absence of crosslinking reagent (crosslinker). Reactions were separated by SDS–PAGE. Gal80p complexes were visualized by immunoblotting, and BSA by Coomassie Blue staining. (B) Reactions with labelled Gal80p as outlined in (C) were separated by SDS–PAGE and subjected to autoradiography. Asterisks indicate labelled Gal80p and Gal80p–His6Gal80p complexes. Note that with the low crosslinking efficiency in (C), more assembled tetramers are crosslinked to trimers than to tetramers. (C) Schematic outline of the experiment in (B). Gal4p and Gal4p–His6-Gal80p complexes were allowed to assemble on single or double Gal4p binding sites. Reactions were briefly incubated with labelled Gal80p (*80) and crosslinking was initiated. The thick bar indicates a crosslink between His6-80 and *80.
Given the essentially 100% crosslinking efficiency of the peptide–nickel reagent under these conditions, the lack of monomeric Gal80p at a concentration of 5 × 10–8 M is consistent with our KD determination for Gal80p dimerization, which predicts that at this concentration, <1% of Gal80p is monomeric. Almost half of the dimers were covalently connected to tetramers, suggesting that the KD for the Gal80p dimer–dimer interaction is in the range of 5 × 10–8 M. No trimers were detectable, consistent with all higher order complexes arising from covalently crosslinked dimers (Figure 4A). With lower crosslinking efficiency (data not shown and Figure 4B), monomers and trimers were also visible. We conclude that Gal80p dimerizes with high affinity, and in solution it transiently multimerizes with moderate affinity.
Gal80 dimer–dimer interaction is stabilized by adjacent Gal4p binding sites
If locking into a completely repressed form is due to Gal80p dimer–dimer interactions, then this interaction should be weak or transient for a complex assembled on a single Gal4p binding site, but stabilized by the vicinity of two complexes assembled on adjacent Gal4p binding sites. In order to address this important prediction, we performed the experiment outlined in Figure 4C. We constructed and purified two variants of Gal80p, one of which has a His6 tag at its N-terminus (His6-Gal80p) and the other with a five-amino-acid heart muscle kinase (HMK) site for radiolabelling (Blanar and Rutter, 1992) introduced at its C-terminus (Gal80p–HMK). We then incubated Gal4p(1–147+34) dimers with an equimolar amount of consensus Gal4p binding sites and an approximately half-equimolar amount of His6-Gal80p. In one reaction, oligonucleotides with single Gal4p binding sites were used, and in the other reaction, DNA fragments with the same molar amounts of binding sites, but with two adjacent binding sites on each fragment, were used. Complexes were allowed to assemble and assembly reactions were monitored in parallel reactions by shift-westerns. Assembled complexes were then briefly incubated with 32P-labelled Gal80p–HMK and crosslinking was initiated. Reactions were separated by SDS–PAGE and subjected to autoradiography. Figure 4B shows that formation of, and crosslinking to, Gal80p dimers was, as expected, approximately equally efficient in the 1× and 2× binding site reactions. In contrast, efficient crosslinking to trimers and to traces of tetramers was dependent on adjacent Gal4p binding sites, consistent with the prediction that the Gal80p dimer–dimer interaction needs to be stabilized by the close proximity of Gal80p dimers assembled on adjacent Gal4p binding sites. Note that with the low crosslinking efficiency under these conditions, more assembled tetramers were crosslinked to trimers than to tetramers, and only a fraction of complexes were crosslinked to Gal4p(1–147+34) as well.
In addition, His6-Gal80p with covalently linked proteins was purified on Ni–agarose beads under denaturing conditions. Direct measurement of Cerenkov radiation confirmed that co-purification of radioactivity depended on crosslinking and that significantly more radioactivity co-purified with His6-Gal80p in the 2× than in the 1× binding site reaction (data not shown). Incubation times for radiolabelled Gal80p–HMK with pre-assembled complexes 2–14 min before crosslinking yielded essentially indistinguishable products (data not shown), indicating that Gal80p dimers complexed with DNA-bound Gal4p(1–147+34) dimers are highly stable with little if any exchange during the incubation period.
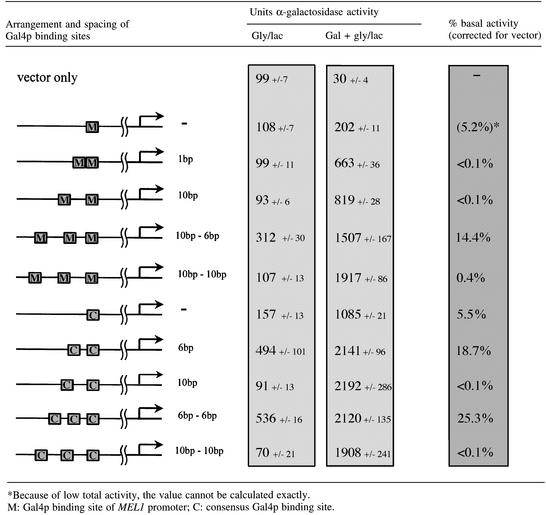
Complete repression depends on the spacing of Gal4p binding sites
Multiple Gal4p binding sites in all constructs used so far were separated 10 bp from each other. We chose this distance to position adjacent Gal4p–Gal80p complexes on the same side of the helix (one helical turn equals ∼10.4 bp in B-form DNA). We noticed that all natural adjacent Gal4p binding sites are either 1, 2 or >47 bp apart, i.e. they are either close enough to contact each other directly on the same side of the DNA or are sufficiently far away from each other to allow looping or bending of the intervening DNA.
Spacing of Gal4p binding sites by one half-helical turn (6 bp) would position Gal80p dimers on opposite sides of a rigid helix, the least favourable position for protein–protein interactions. If complete repression depends on direct binding of adjacent Gal80p dimers, then a 6 bp spacing between Gal4p binding sites would be expected to eliminate or at least reduce dimer–dimer interactions and thereby complete repression. If, on the other hand, complete repression were a consequence of interactions of the transcriptional machinery with either the repressor or the activator, then the positioning of Gal4p binding sites towards each other should have little, if any, effect.
We inserted either strong consensus Gal4p binding sites or the weak Gal4p binding site from the MEL1 promoter into the single copy reporter plasmid pMEL-β (Melcher et al., 2000). pMEL-β contains a UAS-less MEL1 promoter fused to lacZ. Introduced binding sites replace the weak natural Gal4p binding site of the MEL1 promoter, i.e. are in the same context and distance from the transcriptional initiation site as the natural site. As expected and shown in Table III, transcriptional activation as measured by β-galactosidase activity in galactose grown cells does not depend on the spacing of high affinity consensus Gal4p binding sites towards each other. For instance, two consensus Gal4p binding sites spaced 6 bp apart direct 2141 U of β-galactosidase activity, while the same two sites spaced 10 bp apart direct 2192 U. In contrast, complete repression strictly depended on the spacing of adjacent Gal4p binding sites with respect to each other. Binding sites spaced 1 or 10 bp apart had no detectable basal activity (<0.1%), while the same sites separated by 6 bp directed high basal activity (>5%), independent of the type and number of Gal4p binding sites. We conclude that full repression is abolished when adjacent Gal4p binding sites are on opposite sides of a rigid helix, strongly implying that full repression depends on the direct interaction between adjacent repressor complexes.
Table III. Basal expression depends on the spacing of Gal4p binding sites.
Mechanism of differential repression
Gal80p binds non-cooperatively to adjacent Gal4p–DNA complexes. The repressor–repressor interaction could affect the Gal4p–Gal80p interaction either quantitatively (i.e. increases the strength of the Gal4p–Gal80p binding; ‘cooperative binding’ in Figure 5A) or qualitatively (i.e. changes the nature of the Gal4–Gal80 interaction; ‘architectural mechanism’ in Figure 5A). In the first case, a weakened binding could lead to leaky repression by partial dissociation of the complex between Gal80p and the Gal4p activation domain. Although attractive, this model would imply that the Gal80p dimer–dimer interaction has to be kinetically sufficiently stable to allow stabilization of the Gal4p–Gal80p interaction, for which we determined a KD of 3 × 10–10 M. To address this issue experimentally, cooperative binding experiments were performed.
We incubated a radiolabelled restriction fragment with two consensus Gal4p binding sites, separated by 10 bp, with 0.3 nM recombinant Gal4p(1–147+34) and increasing concentrations of Gal80p (Figure 5B). The composition of retarded complexes on a native gel was unambiguously determined by titration with different amounts of Gal4p and Gal80p, and by anti-Gal4p and anti-Gal80p antibody supershifts (data not shown). If the Gal80p dimer–dimer interaction would stabilize the Gal4p–Gal80p interaction, then there should be a fast transition from neither of the two Gal4p dimers being occupied by Gal80p (complex 3 in Figure 5B) to both complexes being fully occupied by Gal80p (complex 5 in Figure 5B). The fraction of complexes with only one Gal4p dimer being occupied by Gal80p (complex 4 in Figure 5B) should then be under-represented. However, we detect a slow transition from the fraction of unoccupied Gal4p dimers to the fraction of completely occupied dimers and a large fraction of complexes in which only one of the two Gal4p dimers is bound by Gal80p (Figure 5B), indicating that under the in vitro conditions used, Gal80p does not bind cooperatively to adjacent Gal4p dimers. This result is consistent with the Gal80 dimer–dimer interaction being transient and much weaker than the Gal4p–Gal80p and the Gal4p–DNA interactions. Note that with higher concentrations of Gal80p, total visible labelling decreases, presumably because higher order Gal80p complexes are unstable during electrophoresis. In summary, under the conditions used we find no evidence for a stabilization of the Gal4p(1–147+34)–Gal80p complex by Gal80p dimer– dimer interaction.
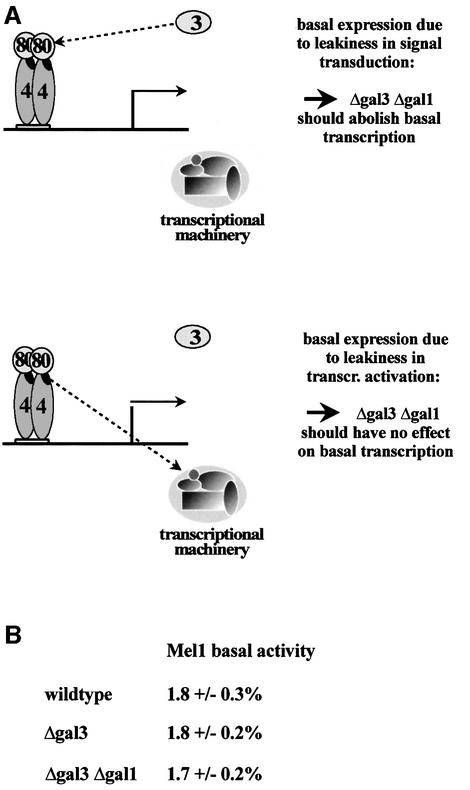
Incomplete repression is not due to leakiness of the signal transduction pathway. We can envisage two qualitative (architectural) models by which Gal80p multimerization on adjacent Gal4p binding sites could abolish basal transcription (Figure 6A). In one model, the signal transducer Gal3p would still have a low residual affinity for Gal80p in the absence of galactose, sufficient to allow some low level expression from genes regulated by a single Gal4p binding site. Locking of Gal80p dimers into multimers might then occlude accessibility for Gal3p. In the second model, signal transduction to Gal80p would be unaffected, but rather Gal4p’s activation domain would not be completely masked by dimeric Gal80p. Formation of Gal80p multimeric complexes would then completely block accessibility of the activation domain and hence fully prevent the activation domain from making protein– protein contacts with the transcriptional machinery (Figure 6A). To distinguish between these models we completely disrupted the signal transduction pathway by deleting both the GAL3 and GAL1 genes. Mel1p expression under non-inducing conditions was unaffected by the lack of Gal1p and Gal3p (Figure 6B). We conclude that basal expression is not a consequence of a leaky signal transduction pathway, but of a process subsequent to Gal3p’s activation of Gal80p.
Fig. 6. Basal expression is not affected by disruption of the GAL3 GAL1 signal transduction pathway. (A) Experimental outline to distinguish between two models for MEL1 basal expression. 4: Gal4p; 80: Gal80p; 3: Gal3p. (B) Percentage basal Mel1p activities (relative to galactose-grown, wild-type cells) in strains that are isogenic except for GAL1 and GAL3.
Discussion
The GAL genes of S.cerevisiae fall into two classes. One group is completely repressed under non-inducing conditions and characterized by multiple Gal4p binding sites. Genes of the other class have biologically important basal expression and single Gal4p binding sites. We demonstrated that basal expression can be strongly reduced by moderate overexpression of the repressor Gal80p. An increase in the concentration of Gal80p can therefore partially overcome the need for adjacent Gal4p–Gal80p complexes for complete repression. This observation suggested a simple model. Adjacent Gal4p binding sites could stabilize weak or transient interactions between Gal80 molecules. Gal80p dimer–dimer interaction could in turn stabilize the Gal4p–Gal80p interaction or could prevent residual interactions of the complex with the transcription apparatus or with components of the signal transduction pathway. This model makes several specific predictions and we used a combination of in vivo and quantitative in vitro assays to test them.
First, Gal80 molecules on adjacent binding sites must be capable of interacting with each other. We established that the unit of Gal80p binding to a Gal4p–DNA complex is a homodimer. We could then trap interactions between Gal80p dimers by chemical crosslinking. Secondly, this interaction must be weak or transient without the proximity arising from adjacent Gal4p binding sites. Moreover, an increase in the concentration of Gal80p should drive dimer–dimer interaction in solution or on a single Gal4p binding site. In a series of in vitro experiments, we demonstrated that the Gal80p dimer–dimer interaction is weak, with a KD that appears to be at least two orders of magnitude higher than the monomer–monomer interaction, that it is not stable to native electrophoresis, and that it is kinetically too transient to promote cooperative binding of Gal80p dimers on adjacent Gal4p–DNA complexes. In addition, overexpression of the Gal80pVP16 hybrid resulted in strongly increased reporter gene activity (Figure 3), as predicted if the expression level from integrated GAL80VP16 under the control of the wild-type GAL80 promoter is so low that only a fraction of Gal80p hybrids engage in dimer–dimer interactions on a single Gal4p binding site. Thirdly, assembly of Gal4p–Gal80p complexes on adjacent DNA sites must stabilize the inherently transient interaction between Gal80p dimers. We confirmed this prediction by using a 0 Å crosslinker in two assembly reactions that differed only in having the same concentration of Gal4p binding sites above the KD either as isolated sites or as neighbouring sites. Finally, the ability or inability of adjacent Gal80p dimers to interact with each other should correlate with complete or incomplete repression. We used a DNA spacing assay to probe for the importance of protein–protein interactions. Complexes assembled right next to each other or one helical turn apart are on the same side of the DNA. In this orientation they can interact with each other, which correlates with complete repression of transcription. Complexes assembled a half-helical turn from each other are on opposite sides of a rigid helix. This orientation was incompatible with complete repression, providing strong evidence for an architectural mechanism that links full repression with a direct interaction between the assembled complexes (Table III).
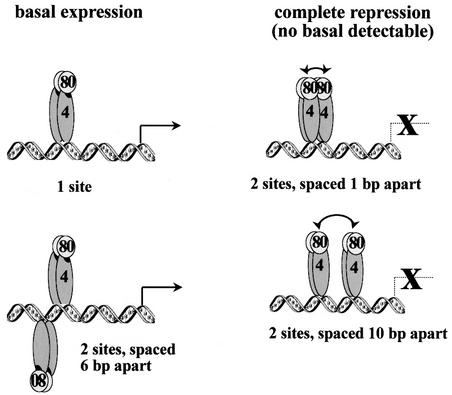
How could a stabilized Gal80p dimer–dimer interaction increase repression? The simplest explanation would be that it does so by stabilizing the Gal4p–Gal80p interaction and thereby the masking of the Gal4p activation domain. However, cooperative binding assays indicated that Gal80p dimer–dimer interactions may be kinetically too labile to increase the strength of the Gal4p–Gal80p interaction significantly. We therefore favour a mechanism in which Gal80p multimerization reduces residual accessibility of the Gal4p–Gal80p complexes either to the signal transducer or to the transcriptional machinery. Since basal expression was independent of a functional Gal3p signal transduction pathway, we believe that it most probably results from partial access of the activation domain to the transcriptional machinery (Figure 7).
Fig. 7. Model for differential repression in the yeast GAL system. On a single Gal4p binding site or on two binding sites on opposite sides of the DNA helix, Gal80p dimers do not interact with each other and only partially mask the Gal4p activation domain. On binding sites on the same side of the DNA helix (1 and 10 bp spacing), the Gal80p dimer–dimer interaction is stabilized, causing complete shielding of the activation domain.
Our analysis of the mechanism of differential repression clearly points to the potential of weak interactions that are difficult to detect by standard biochemical methods, yet can have profound regulatory consequences. In order to understand the interactions between the regulatory factors, it was important to put them into a quantitative framework. We have shown that Gal80p dimerizes in solution with high affinity and that Gal4p forms a heterotetramer with Gal80p on DNA. We have found that in this complex, the half-life of the Gal80p dimer is greatly extended relative to that of the free dimer and that, in turn, Gal80p stabilizes the interaction between Gal4p and DNA. While our KD determination for Gal4p bound to a consensus site oligonucleotide agreed well with that of several other laboratories, our KD for the Gal4p–Gal80p interaction is more than one order of magnitude lower than that determined by Lue et al. (1987; 5 × 10–9 M) using an electrophoretic mobility shift assay (EMSA), and two orders of magnitude lower than that determined by Ptashne and colleagues using a BIACORE microchip (Wu et al., 1996). The apparent difficulty in determining this binding constant probably reflects the complicated linkage of equilibria between Gal4p and DNA, Gal4p and Gal80p, and Gal80p and Gal80p. In particular, we needed to DNA-drive the lower affinity interaction of Gal4p to its consensus site oligonucleotide at a Gal4p concentration that is below the KD of its higher affinity interaction with Gal80p. We therefore believe that the lowest value, determined at a Gal4p–DNA concentration below the 0.3 nM KD of the Gal4p–Gal80p interaction, is probably the correct one. With complexes assembled on a consensus site restriction fragment, rather than a minimal oligonucleotide, all pairwise interactions within the DNA-bound complexes have KDs in the subnanomolar range and are stabilized further by neighbouring interactions. These strong interactions stabilize transient dimer–dimer interactions by positioning dimers in suitable close proximity.
Activator–repressor systems that involve activation domain masking are not a pecularity of the yeast GAL system, but rather have been found in a number of biologically important systems, including the binding of the tumour suppressor RB to the activation domains of E2F (Nevins, 1992) and E1A (Trouche and Kouzarides, 1996), and the masking of the p53 activation domain by MDM2 (Momand et al., 1992; Kussie et al., 1996). Therefore, the type of differential repression characterized here may apply to other systems as well.
Finally, we would like to point out that while MEL1 basal expression is abolished in the absence of Gal4p, other GAL genes may be regulated by additional elements (see, for instance, Sakurai et al., 1994). While basal expression can be achieved either by only a single Gal4p binding site or by introducing additional regulators, complete repression of GAL genes can only be achieved by correctly spaced multiple Gal4p binding sites.
Materials and methods
Strains, enzyme assays and genetic techniques
Yeast strains used were 21R (GAL4 GAL80 MEL1 ura3-52 leu2-3,112 ade1) (Johnston and Hopper, 1982) for testing expression driven from Gal4p binding sites (Tables I–III), and YJ0Z (Δgal4 Δgal80 MEL1 ura3-52 leu2-3,112 his3 ade2-101 trp1 with an integrated GAL1–lacZ reporter) (Leuther and Johnston, 1992) for two-hybrid experiments and disruption of the signal transduction pathway. Derivatives of YJ0Z were described previously (Leuther and Johnston, 1992; Ding and Johnston, 1997). GAL4 integrants were constructed by transforming linearized YIp351-GAL4 into YJ0Z80 and YJ0ZΔgal3. GAL1 was disrupted by using pYES-TRP1-GAL1-3′ untranslated region (Ding and Johnston, 1997). Single-site integration and disruptions were confirmed by PCR. α- and β-galactosidase assays were performed as described previously (Melcher et al., 2000).
Protein purification and labelling
Gal4p(1–147+34), Gal80p and Gal80p–HMK were purified as glutathione S-transferase (GST) fusion proteins using standard protocols. GST tags were removed with TEV protease as described (Melcher, 2000). His6-Gal80p was expressed from plasmid pPROEX-Gal80 (U.Kuchibhotla and S.A.Johnston, unpublished) and purified by Ni-chelate chromatography. Gal80p–HMK was radioactively labelled with [γ-32P]ATP and 10 U of HMK (Sigma) for 70 min at room temperature in buffer as described previously (Blanar and Rutter, 1992).
Determination of protein concentrations
Concentrations of the purified proteins were determined by Bradford assay, by SDS–PAGE/Coomassie Blue titration with BSA standards and by absorbance at 280 nm with εGal4p(1-147+34) = 17 700 cm2/mol (2W, 2F, 4Y) and εGal80p = 41 570 cm2/mol (3W, 20F, 22Y). Results of all three assays agreed well with each other.
Gel retardation assays (EMSA) and DNA-free gel shifts
For EMSA with a single Gal4p binding site, oligonucleotides 1 and 2 (see Table IV, available supplementary data at The EMBO Journal Online) were annealed to each other and end-labelled with [γ-32P]ATP and T4 kinase. Oligonucleotides 3 and 4, containing two completely palindromic consensus Gal4p binding sites, annealed almost exclusively intramolecularly. We therefore first cloned the mixture with a fraction of intermolecularly annealed oligonucleotides into the polylinker of YEp351. Only intermolecularly annealed oligonucleotides had correct ends for cloning. We then isolated a 160 bp 2× binding site fragment by PCR with standard M13 primers. The PCR fragment was partially digested with XbaI and PstI, and end-labelled with Klenow polymerase and [α-32P]dATP. Restriction fragments together with end-labelled pUC19/Sau3A standards were separated on a 12% polyacrylamide gel and visualized by autoradiography. The band corresponding to the 108 bp fragment was excised, and the DNA isolated by the crush and soak method (Sambrook et al., 1989).
Binding reactions (15 µl) were performed with the indicated concentration of proteins in buffer A(50) [25 mM Tris–HCl pH 7.5, 50 mM KCl, 5 mM MgCl2, 0.125 mM EDTA, 10% glycerol, 1 mM dithiothreitol (DTT)]. Reactions also contained 1 mg/ml unprogrammed rabbit reticulocyte lysate proteins (except for Figure 1D) and 1–2 µg of sheared salmon sperm DNA. For antibody supershifts, reactions were incubated with 1 µl of purified IgGs. Protein–DNA complexes were resolved by running 6% polyacrylamide gels in 0.5× TBE (12.5 mM Tris, 95 mM glycine, 0.5 mM EDTA) at 10 V/cm and 4°C.
For DNA-free gel shifts, Gal80p and Gal80pVP16 were in vitro translated with [35S]methionine. Indicated amounts of translated and recombinant proteins were incubated in 15 µl reactions in buffer A(50) for 15 min at room temperature (Figure 2A and B) or for the indicated amount of time (Figure 2C) before complexes were resolved as above. Gels were fixed for 30 min in 40% methanol/10% acetic acid prior to drying and autoradiography.
Immunoblots and shift-westerns
SDS or native polyacrylamide gels were blotted to polyvinylidene difluoride (PVDF) membranes. Shift-westerns were briefly immersed in 100% methanol, air-dried, and subjected to autoradiography prior to re-wetting and blocking. Blots were incubated with polyclonal rabbit anti-Gal80p serum (a kind gift from T.Fukasawa) or anti-Gal4p(729–875) serum (a kind gift from K.K.Leuther) at 1:10 000 dilutions. Secondary antibodies were horseradish peroxidase conjugated (Bio-Rad) for use with the chemiluminescence kit from Dupont.
Crosslinking
GlyGlyHis-Ni-mediated crosslinking was described by Brown et al. (1995). All crossslinking reactions were performed in buffer A(50). Reactions were allowed to equilibrate before incubation with 100 µM freshly Ni-complexed (or as control, uncomplexed) GlyGlyHis tripeptide and 100 µM magnesium monoperoxyphthalic acid (MMPP) for 2 min (crosslinking in solution) or 90 s (crosslinking of assembled complexes) at room temperature. Acetylated BSA was purchased from Promega.
For the assembly reaction, 3.1 pmol of Gal4p(1–147+34) dimers and 1.5 pmol of His6-Gal80p dimers were incubated in 80 µl of buffer A(50), with either 3 pmol of the annealed consensus site oligonucleotide or 1.5 pmol of the 163 bp PCR fragment with two adjacent consensus Gal4p binding sites (3 pmol sites in total) for 20 min at room temperature. Expected molar ratios and quantative binding were confirmed by titrating all components of the reaction in parallel shift-westerns using anti-Gal80p and anti-Gal4p antibodies for detection. 32P-labelled Gal80p–HMK (0.3 pmol) was incubated with pre-assembled complexes for 3 min prior to crosslinking. Aliquots (10 µl) per reaction were quenched by the addition of SDS sample buffer, separated by SDS–PAGE, blotted to PVDF membranes, and subjected to autoradiography. Aliquots (40 µl) of each reaction, together with 40 µl of ‘no crosslinking’ controls were quenched by the addition of 600 µl of denaturing wash buffer (6 M guanidium hydrochloride, 20 mM Tris pH 7.9, 500 mM NaCl, 20 mM imidazole) and 10 µl of Ni-charged Sepharose (Qiagen) for denaturing Ni-affinity chromatography as described (Fancy et al., 1996). Ni-bound complexes were washed as described previously (Fancy et al., 1996) and Cerenkov radiation of retained radioactivity determined by scintillation counting.
Plasmid constructions and oligonucleotides
For detailed information see the Supplementary data, available at The EMBO Journal Online.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
Most of the experimental work was performed while K.M. was a postdoctoral fellow, and H.E.X. a graduate student, in the laboratory of S.A.Johnston. We thank T.Fukasawa for antibodies against Gal80p and T.Kodadek for purified Gal80p, as well as U.Kuchibhotla and W.Ding for plasmids indicated in the supplementary data. We especially acknowledge K.Leuther for developing the Gal80p–Gal80pVP16 interaction assay. Finally, we thank S.A.Johnston, T.Kodadek, W.V.Ding, L.Nover, K.Breunig, J.Klein, S.Russel and W.Zheng for helpful discussions, and K.-D.Entian for continuous support. This work was supported by grants to K.M. from the Deutsche Forschungsgemeinschaft (Me 1573 and SFB 474) and to Stephen A.Johnston from the National Institutes of Health (GM-40700).
REFERENCES
- Blanar M.A. and Rutter,W.J. (1992) Interaction cloning: identification of a helix–loop–helix zipper protein that interacts with c-Fos. Science, 256, 1014–1018. [DOI] [PubMed] [Google Scholar]
- Bram R.J., Lue,N.F. and Kornberg,R.D. (1986) A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J., 5, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.C., Yang,S.-H. and Kodadek,T. (1995) Highly specific oxidative cross-linking of proteins mediated by a nickel–peptide complex. Biochemistry, 34, 4733–4739. [DOI] [PubMed] [Google Scholar]
- Carey M., Kakidani,H., Leatherwood,J., Mostashari,F. and Ptashne,M. (1989) An amino-terminal fragment of GAL4 binds DNA as a dimer. J. Mol. Biol., 209, 423–432. [DOI] [PubMed] [Google Scholar]
- Ding W.V. and Johnston,S.A. (1997) The DNA binding and activation domains of Gal4p are sufficient for conveying its regulatory signals. Mol. Cell. Biol., 17, 2538–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy D.A., Melcher,K., Johnston,S.A. and Kodadek,T. (1996) New chemistry for the study of multiprotein complexes: the six-histidine tag as receptor for a protein crosslinking reagent. Chem. Biol., 3, 551–559. [DOI] [PubMed] [Google Scholar]
- Finley R.J., Chen,S., Ma,J., Byrne,P. and West,R.J. (1990) Opposing regulatory functions of positive and negative elements in UASG control transcription of the yeast GAL genes. Mol. Cell. Biol., 10, 5663–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. (1987) A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev., 51, 458–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.A. and Hopper,J.E. (1982) Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc. Natl Acad. Sci. USA, 79, 6971–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S.S., Ansari,A.Z., Ptashne,M. and Young,R.A. (1998) An activator target in the RNA polymerase II holoenzyme. Mol. Cell, 1, 895–904. [DOI] [PubMed] [Google Scholar]
- Kussie P.H., Gorina,S., Marechal,V., Elenbaas,B., Moreau,J., Levine, A.J. and Pavletich,N.P. (1996) Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science, 274, 948–953. [DOI] [PubMed] [Google Scholar]
- Leuther K.K. (1993) Genetic, biochemical, and biophysical analysis of the transcriptional activator GAL4 and its interaction with the negative regulator GAL80. PhD thesis, University of Texas Southwestern Medical Center, Dallas, TX.
- Leuther K.K. and Johnston,S.A. (1992) Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science, 256, 1333–1335. [DOI] [PubMed] [Google Scholar]
- Lue N.F., Chasman,D.I., Buchman,A.R. and Kornberg,R.D. (1987) Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol. Cell. Biol., 7, 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K. (1997) Galactose metabolism in Saccharomyces cerevisiae: a paradigm for eukaryotic gene regulation. In Zimmermann,F.K. and Entian,K.-D. (eds), Yeast Sugar Metabolism. Technomic Publishing Inc., Lancaster, PA, pp. 235–269.
- Melcher K. (2000) A modular set of prokaryotic and eukaryotic expression vectors. Anal. Biochem., 277, 109–120. [DOI] [PubMed] [Google Scholar]
- Melcher K., Sharma,B., Ding,W.V. and Nolden,M. (2000) Zero background yeast reporter plasmids. Gene, 247, 53–61. [DOI] [PubMed] [Google Scholar]
- Meyer J., Walker-Jonah,A. and Hollenberg,C.P. (1991) Galactokinase encoded by GAL1 is a bifunctional protein required for the induction of the GAL genes in Kluyveromyces lactis and able to suppress the gal3 phenotype in Saccharomyces cerevisae. Mol. Cell. Biol., 11, 5454–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J., Zambetti,G.P., Olson,D.C., George,D. and Levine,A.J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell, 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- Moreira R.F. and Noren,C.J. (1995) Minimum duplex requirements for restriction enzyme cleavage near the termini of linear DNA fragments. Biotechniques, 19, 56–59. [PubMed] [Google Scholar]
- Nevins J.R. (1992) E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science, 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Parthun M.R. and Jaehning,J.A. (1990) Purification and characterization of the yeast transcriptional activator GAL4. J. Biol. Chem., 265, 209–213. [PubMed] [Google Scholar]
- Platt A. and Reece,R.J. (1998) The yeast galactose genetic switch is mediated by the formation of a Gal4p–Gal80p–Gal3p complex. EMBO J., 17, 4086–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller M.A., Hamilton,R.W. and Hopper,J.E. (1984) Regulation of basal and induced levels of the MEL1 transcript in Saccharomyces cerevisiae. Mol. Cell. Biol., 4, 1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece R.J. and Ptashne,M. (1993) Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science, 261, 909–911. [DOI] [PubMed] [Google Scholar]
- Sakurai H., Ohishi,T. and Fukasawa,T. (1994) Two alternative pathways of transcription initiation in the yeast negative regulatory gene GAL80. Mol. Cell. Biol., 14, 6819–6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sil A.K., Alam,S., Xin,P., Ma,L., Morgan,M., Lebo,C.M., Woods,M.P. and Hopper,J.E. (1999) The Gal3p–Gal80p–Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p–Gal4p complex in response to galactose and ATP. Mol. Cell. Biol., 19, 7828–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche D. and Kouzarides,T. (1996) E2F1 and E1A (12S) have a homologous activation domain regulated by RB and CBP. Proc. Natl Acad. Sci. USA, 93, 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S., Xu,H., Johnston,S.A. and Kodadek,T. (1993) How do Zn2 Cys6 proteins distinguish between similar upstream activation sites? Comparison of the DNA-binding specificity of the GAL4 protein in vitro and in vivo. J. Biol. Chem., 268, 24699–24706. [PubMed] [Google Scholar]
- Wu Y., Reece,R.J. and Ptashne,M. (1996) Quantitation of putative activator-target affinities predicts transcriptional activating potentials. EMBO J., 15, 3951–3963. [PMC free article] [PubMed] [Google Scholar]
- Yano K. and Fukasawa,T. (1997) Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 1721–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.J., Hiraoka,Y., Nishizawa,M., Takio,K., Titani,K., Nogi,Y. and Fukasawa,T. (1991) Purification and characterization of the yeast negative regulatory protein GAL80. J. Biol. Chem., 266, 693–697. [PubMed] [Google Scholar]
- Zenke F.T., Engels,R., Vollenbroich,V., Meyer,J., Hollenberg,C.P. and Breunig,K.D. (1996) Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science, 272, 1662–1665. [DOI] [PubMed] [Google Scholar]