Abstract
Because of the heterogeneity of chromatin, the site of integration of human immunodeficiency virus (HIV) in the genome could have dramatic effects on its transcriptional activity. We have used an HIV-1-derived retroviral vector, in which the green fluorescent protein is under the control of the HIV promoter, to generate by infection 34 Jurkat clonal cell lines each containing a single integration of the HIV-1 vector. In the absence of Tat, a 75-fold difference in expression level between the highest and lowest expressing clones was observed. Basal promoter activity was low in 80% of the clones and moderate to high in the remaining 20% of clones. We found that differences in expression levels are due to the integration site and are not controlled by DNA methylation or histone acetylation. Tat activated transcription in each clone, and an inverse correlation was observed between basal transcriptional activity and inducibility by Tat. These observations demonstrate that the chromatin environment influences basal HIV gene expression and that the HIV Tat protein activates transcription independently of the chromatin environment.
Keywords: acetylation/chromatin/HIV transcription/methylation/trichostatin A
Introduction
After the human immunodeficiency virus (HIV) enters the host cell, the viral RNA is reverse transcribed into DNA and transported to the nucleus in a multiprotein complex called the pre-integration complex. This nucleoprotein structure contains the viral proteins integrase, matrix and Vpr, and host cell proteins (Farnet and Bushman, 1997; Miller et al., 1997), which are involved in nuclear import of the viral genome. Recently, a triple-stranded DNA element located in the center of the HIV genome, the DNA flap, was also found to play a role in HIV-1 DNA nuclear import (Zennou et al., 2000). After entry into the nucleus, the HIV-encoded integrase catalyzes the reaction that integrates the viral DNA into the host genome. While the molecular mechanism of integration has been extensively studied in vitro, little is known about this process in the context of chromatin. Chromatin represents a very heterogeneous environment both structurally and functionally, and there is extensive evidence that the activity of a de novo integrated promoter can be dramatically affected by the site of integration.
Several studies have indicated that retroviruses and other transposons might not integrate at random into the host cell genome. As the most striking example of specific integration, the Ty retrotransposons of yeast integrate close to defined genetic elements: upstream of pol III-transcribed genes for Ty3 (Chalker and Sandmeyer, 1992) and into domains of silent chromatin at the HM loci and telomeres for Ty5 (Zou et al., 1996). This specificity is conferred by a direct interaction between the integrase encoded by the transposon and specific proteins involved in the regulation of transcription by pol III or Sir proteins, respectively (Kirchner et al., 1995; Zhu et al., 1999). While integration appeared non-random for retroviruses of higher species as well (Shih et al., 1988), many studies have failed to define the molecular mechanism of integration site selection. Recent studies on the integration of avian leukosis virus and human T-cell leukemia virus type 1 suggest that integration specificity is determined by local structural features rather than by the accessibility of specific regions (Withers-Ward et al., 1994; Leclercq et al., 2000). A recent study analyzing 61 HIV-1 integration sites did not detect preferential integration near or in transcription units or repetitive elements, as had been previously suggested (Stevens and Griffith, 1994, 1996). This report also found that integration was disfavored in centromeric heterochromatin, a logical consequence of the highly compact and poorly accessible nature of chromatin at these loci (Carteau et al., 1998). In vitro studies have found that integration occurs preferentially in nucleosomal DNA because of the distortion created by DNA wrapping around the histone core (Müller and Varmus, 1994; Pruss et al., 1994). In the case of HIV, the integrase interacts with Ini1/hSNF5, a component of the SWI/SNF ATP-dependent chromatin remodeling complex (Kalpana et al., 1994). Hypothetically, this interaction could direct HIV integration to genomic locations at a subset of genes where the SWI/SNF complex usually resides. Alternatively, the recruitment of this complex to the pre-integration complex could help in remodeling chromatin at the site of integration, thereby facilitating integration (Miller and Bushman, 1995).
Transcription of the HIV provirus is characterized by an early, Tat-independent phase and a late, Tat-dependent phase. In the absence of the viral transactivator Tat, a series of short transcripts are produced due to inefficient elongation by the recruited RNA pol II (Kao et al., 1987). During this phase, the HIV promoter is strictly under the control of the local chromatin environment and cellular transcription factors binding to cis-acting elements in the HIV promoter. However inefficient, this process results in the synthesis of a small fraction of full-length viral transcripts leading to the synthesis of the Tat protein. Tat binds to TAR, a hairpin loop formed at the 5′ termini of the nascent transcripts. Tat specifically recruits the pTEFb complex with its associated CDK9 kinase. CDK9, in turn, phosphorylates the C-terminal domain of pol II, thereby increasing its ability to elongate efficiently on the viral template (recently reviewed in Karn, 1999). This process rapidly leads to the synthesis of more Tat and the establishment of a positive regulatory loop.
We have previously described the chromatin organization of the HIV genome in several latently infected cell lines. Independently of the site of integration, the HIV 5′ long terminal repeat (LTR) is packaged in three unique nucleosomes: nuc-0, -1 and -2. Nuc-1 is positioned immediately downstream of the transcription start and is remodeled upon HIV transcriptional activation by Tat and histone deacetylase (HDAC) inhibitors (Verdin et al., 1993; Van Lint et al., 1996; el Kharroubi et al., 1998). These observations suggest that Nuc-1 plays a repressive role in HIV transcription and that histone acetylation could be involved in overriding this effect. However, since all cell lines examined so far were transcriptionally inactive (latent), further experiments are clearly needed to determine the role of the local genomic chromatin environment in the chromatin organization of the HIV promoter.
To study HIV transcription in its natural context (i.e. integrated in the host genome), we have used an HIV-1-derived retroviral vector to generate a library of Jurkat clones containing single integrations of an HIV-1 minigenome. Given the heterogeneity of the chromatin environment, we anticipate that retrovirus expression will vary depending on the site of integration. However, it is also conceivable that retroviruses have evolved specific mechanisms to select transcriptionally competent integration sites. Few data are available on the effect of distinct integration sites in terms of transcriptional activity or competence of retroviruses. We have used this library to investigate the effect of the genomic integration site on the transcriptional activity of the HIV promoter.
Results
LTR-driven HIV expression is highly heterogeneous in a mixed population
HIV transcription can be divided into two distinct phases. The first phase occurs immediately after integration of the provirus into the cell genome and is dependent on interactions between cellular transcription factors and cis-acting elements located in the HIV promoter region. The second phase of HIV transcription is dependent on the first phase and occurs after significant amounts of the viral transactivator Tat protein have accumulated.
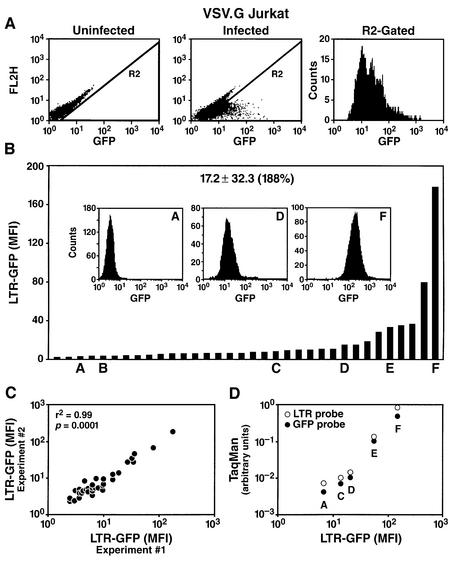
To examine the two phases individually, we first focused on Tat-independent HIV transcription by using an HIV-derived retroviral vector (pRRLGFP-W) to infect the Jurkat lymphoid cell line in vitro. The vector pRRLGFP-W is a minimal non-replicative HIV-1 genome flanked by two LTRs and containing viral cis-acting sequences necessary for packaging and infection (Dull et al., 1998). This construct also contains the cDNA for enhanced green fluorescent protein (GFP) expressed under the control of the HIV promoter. This vector was co-transfected into 293T cells with a packaging construct (pCMV-ΔR8.91) providing all HIV genes required for the production of infective particles (gag, pol, tat, rev) (Dull et al., 1998). An additional plasmid encoding the vesicular stomatitis virus envelope G protein (VSV-G) was also co-transfected to produce pseudotyped viral particles with broad host range and high infectivity (Ory et al., 1996). Viral particles were harvested from the supernatant of transfected cells and used to infect a culture of the lymphocytic cell line Jurkat, with a theoretical multiplicity of infection (m.o.i.) of 0.3 to minimize multiple integration. Flow cytometry analysis of the Jurkat population after infection indicated significant GFP expression in the absence of Tat (Figure 1A). Expression was highly heterogeneous between different cells, with a mean fluorescence intensity (m.f.i.) as low as 5 and as high as 4000 in different cells, a difference in expression levels of three orders of magnitude (Figure 1A). Such heterogeneity was unexpected and could have a profound influence on the rate of viral transcription. A similar heterogeneity of expression was observed after infection of a variety of other cell lines, including HeLa, 293, SW13, SupT1, CEM, A301 and peripheral blood mononuclear cells, indicating that heterogeneity of expression is not influenced by cell-specific factors.
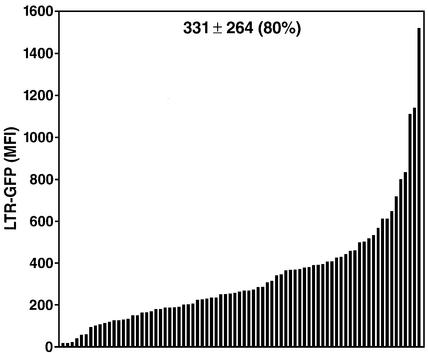
Fig. 1. The site of integration of HIV determines the basal rate of viral transcription. (A) Flow cytometry analysis of a Jurkat culture either uninfected or infected with the HIV-derived vector LTR-GFP (m.o.i. = 0.3). The right panel shows the frequency histogram of R2-gated cells after infection. FL2H is the blank channel used to measure autofluorescence. (B) Basal LTR-GFP expression in 34 individual Jurkat clones. The mean GFP level ± SD is shown. The CV is shown in parentheses and indicates the dispersion of results. Clones A–F are representative clones extensively analyzed below. The frequency histogram for three representative clones (A, D and F) is shown inserted. (C) Flow cytometry analysis of LTR-GFP expression in the collection of clones in two different experiments separated in time by 4 weeks. (D) Relative quantification of LTR-GFP transcription by RT real-time PCR correlates with the flow cytometric quantification of the GFP fluorescence. Five representative clones (A, C, D, E and F) growing exponentially were analyzed by flow cytometry (m.f.i.) and in parallel by real-time RT–PCR with primer pairs corresponding to either the LTR sequence or the GFP open reading frame. Results were normalized to the expression of GAPDH used as endogenous control.
Several viral and cellular factors could explain such heterogeneity in expression levels. These include heterogeneity of promoter activity within a given cell at different stages in its cell cycle (cellular variation), heterogeneity of different cells in terms of their ability to support HIV transcription (clonal variation), a different number of integrated copies in individual cells, the effect of distinct integration sites of the provirus in the cell genome and, finally, the possible role of mutations in viral transcriptional regulatory elements generated during reverse transcription.
Clonal heterogeneity in basal HIV transcription
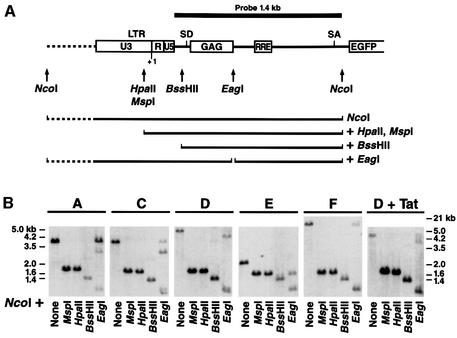
To distinguish between these possibilities, we isolated individual cell clones from this originally infected population. After infection, the culture was serially diluted and individual clones were amplified and characterized by Southern blot analysis to determine the number of integrated copies of the HIV vector. Thirty-four clones were selected by Southern blot analysis based on the presence of a single band, indicative of a single integration site. Southern blot analysis with different enzymes and a single probe demonstrated further that each clone contained a provirus integrated at a different site in the genome (see Figure 7, NcoI digestion for clones A, C, D, E and F). Flow cytometry analysis of GFP showed highly heterogeneous levels among different clones (Figure 1B). Basal activity was low (m.f.i. <15) in 80% of the clones. The remaining clones exhibited much higher expression, such that a 75-fold difference in m.f.i. was measured between the highest and the lowest clones (Figure 1B). To quantify this variability, we calculated the coefficient of variation (CV = 100 × SD/mean) and obtained a value of 188%. Remarkably, the range of expression in different cells of a given clone was significantly narrower (see inserts in Figure 1B; CV = 77%) than in the total population (see R2-Gated in Figure 1A; CV = 194%). This observation indicates that expression levels within a clonal population of cells are restricted and that expression of the integrated HIV promoter is not variegated, in contrast to what was reported for other retroviral vectors (Zentilin et al., 2000).
Fig. 7. The integrated HIV promoter DNA is not methylated in vivo. (A) Schematic representation of the HIV LTR and adjoining sequences in the retroviral vector used to derive our stable Jurkat clones. Restriction sites for the methylation-sensitive endonucleases HpaII, BssHII or EagI and the predicted size of the fragments obtained after a double digestion with NcoI are shown. (B) Genomic DNA purified from six representative clones was assayed for susceptibility to the indicated methylation-sensitive restriction enzyme (HpaII, BssHII or EagI) by Southern blotting. All digestions were further digested with NcoI to define the ends of the hybridization products. Control digestions with NcoI alone or NcoI + MspI (a methylation-insensitive isoschizomer of HpaII) were included.
To determine whether expression levels are constant or fluctuate within a given clone over time, we measured GFP levels in individual clones at two different times separ ated by 4 weeks of continuous culture. This experiment revealed a near perfect correlation between the m.f.i. measured at day 0 and day 30 (Figure 1C). To validate that the fluorescence measured by flow cytometry is an accurate representation of mRNA levels and, therefore, of the transcriptional activity of the HIV promoter, we used reverse transcriptase (RT) semi-quantitative PCR to measure HIV-specific mRNA in individual clones. Total RNA extracted from several clones was reverse transcribed and quantified by real-time PCR (TaqMan; Perkin Elmer) with two different sets of primers specific for the HIV LTR or for the GFP open reading frame. Comparison of the expression levels obtained with LTR- or GFP-specific primers with the values obtained by flow cyto metry (GFP m.f.i.) showed a strong correlation, indicating that GFP measurement is a valid representation of HIV promoter activity in these clones (Figure 1D).
This clonal variation in HIV promoter activity could be explained by a variety of cis- or trans-mechanisms on the HIV promoter. These include local effects at the site of integration of the HIV promoter, the presence of mutations in the integrated vector or clonal differences in the cellular environment necessary to support HIV transcription.
To determine whether mutations had accumulated in the 5′ LTR of the integrated proviruses, we chose 11 clones representative of the complete library (m.f.i. = 3–180), including the seven clones with the highest expression. We used PCR to amplify the 5′ LTR from genomic DNA, cloned the resulting fragment, and sequenced each clone on both strands. No mutations were found in nine of the clones when compared with the sequence of the pRRLGFP-W vector (data not shown). Two additional clones contained some mutations (at position 375 of the 5′ LTR in clone B and at positions 71 and 285 in clone D) of unclear functional consequence. These results indicate that in most cases (9/11) the heterogeneity of expression levels in distinct clones is not due to mutations in the promoter of the integrated construct.
Heterogeneity in HIV expression in different clones is not caused by clonal variation in the cellular environment
Clonal differences in the cellular environment necessary to support HIV transcription could account for differences in basal promoter activity measured in distinct Jurkat clones. To test this possibility, we compared the expression of a transiently transfected LTR-luciferase construct into each clone with the local expression of the integrated LTR-GFP. Assuming that transfection efficiencies are comparable in all clones, we expected transiently transfected templates to give us a picture of the local cellular environment supporting HIV transcription, independently of other confounding variables such as the site of integration.
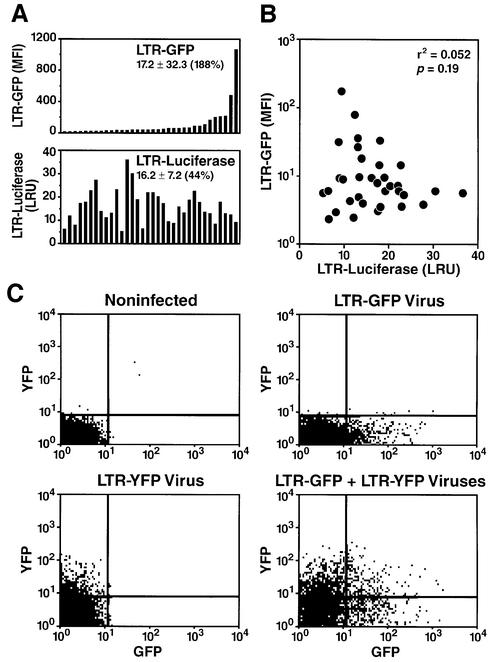
As predicted, distinct clones exhibit significant differences in luciferase activity after transfection of an LTR-luciferase construct (Figure 2A). However, the CV in luciferase activity was significantly lower than that measured for the integrated LTR (44% for luciferase versus 188% for GFP) (Figure 2A). Additionally, no correlation was observed between the activity of the transiently transfected LTR-luciferase construct and the activity of the integrated LTR-GFP template (Figure 2B). These results indicate that the variation in LTR activity measured after integration does not occur as a result of differences in the cellular environment necessary to support HIV transcriptional activity.
Fig. 2. Heterogeneity observed in HIV promoter activity is not secondary to clonal differences. (A) Comparison of expression of a transient LTR-luciferase construct and integrated LTR-GFP in each individual clone. Each clone was tested by flow cytometry to measure GFP expression (upper panel) or transiently transfected by electroporation with an LTR-luciferase construct and an internal transfection control TK-luciferase (Renilla) to correct for transfection efficiency (lower panel). (B) Plotting of luciferase activity (LRU) versus GFP expression (m.f.i.) for each clone demonstrates that there is no correlation between the expression of integrated LTR and transiently transfected LTR. (C) Absence of correlation between LTR-GFP and LTR-YFP integrated in different genomic locations in a single cell. Jurkat cells were infected at an m.o.i. of 0.1 with viral particles containing the retroviral vector LTR-GFP or LTR-YFP, or with both viruses at the same time, and analyzed by flow cytometry 48 h after infection.
To confirm this observation in an independent manner, we constructed a new retroviral vector expressing yellow fluorescent protein (YFP) by substituting GFP by YFP in the retroviral construct pRRLGFP-W. We co-infected Jurkat cells with virus stocks derived from the pRRLGFP-W and the pRRLYFP-W vectors at a low m.o.i. (0.25). After infection, both GFP and YFP levels were measured by flow cytometry. We observed independent expression of GFP and YFP, indicating that distinct levels of HIV expression in distinct cells are not caused by a factor(s) acting in trans (Figure 2C). These experiments collectively show that the heterogeneity observed between clones occurs as a result of different integration sites.
Inverse correlation between Tat transactivation and basal promoter activity
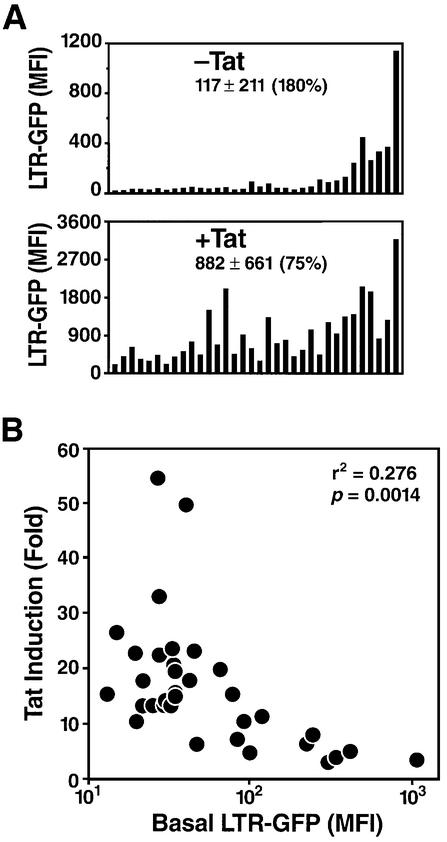
Next, we investigated the second stage of HIV transcription: Tat-dependent transcription. A Tat expression plasmid was transfected into each clone. To identify cells successfully transfected, the Tat-expressing plasmid was co-transfected with a vector containing the cDNA for YFP under the control of a constitutive promoter (cytomegalovirus immediate early promoter). GFP expression was measured in the presence of the Tat plasmid or a control empty vector by flow cytometry after gating on YFP-positive cells. Remarkably, all clones responded to Tat transactivation regardless of the basal rate of HIV transcription (Figure 3A). As had been observed for basal transcription levels, the response of different clones to Tat was heterogeneous, indicating that Tat inducibility depends on the integration site. There was an inverse correlation between HIV basal promoter activity and Tat induction. Clones with high basal levels showed lower induction by Tat (<10-fold), and those with low basal levels showed a higher level of transactivation (>10-fold) (Figure 3B). The differential induction of HIV expression by Tat as a function of basal promoter activity results in a decrease in the CVs of expression after Tat transduction (CV = 75 versus 180% without Tat). These observations suggest that Tat can equalize transcription levels and compensate for variations in expression that occur as a result of distinct integration sites.
Fig. 3. Effect of the HIV promoter integration site on Tat-dependent transcriptional activation. (A) Transcriptional activity of the integrated LTR-GFP construct after transfection of a Tat expression plasmid. Each clone was electroporated with a Tat expression plasmid (pEV280) or the control empty vector pcDNA3.1 (Invitrogen) and a plasmid encoding YFP, pEYFP-C1 (Clontech). LTR-GFP expression was measured in YFP-positive cells (positively transfected) 48 h after transfection. (B) Inverse correlation between HIV basal promoter activity and Tat transactivation. Scatter plot of the Tat induction factor calculated as the ratio of Tat-induced expression versus basal expression against basal LTR-GFP level (m.f.i.). A representative experiment of three is shown.
Expression of an HIV-derived vector containing Tat is also dependent on the integration site
In the experiments described above, we artificially separated the two phases of HIV infection by first infecting with a Tat-defective retroviral construct and then providing the Tat protein via transfection. However, in a normal infection, the two phases are intimately coupled. Indeed, the rate of transcription in the absence of Tat drives the synthesis of the Tat transcripts so that the transition from the first to the second Tat-dependent phase is dependent on the efficacy of the first phase of transcription.
To recapitulate this process in a manner as close to the natural infection as possible, we used another HIV-derived vector (LTR-Tat-IRES-GFP; pEV731), where both the Tat and GFP open reading frames are transcribed by the HIV promoter on a single transcript. To allow for translation of the downstream open reading frame (GFP), an internal ribosome entry site was cloned between the Tat and GFP open reading frames. Jurkat cells were infected at an m.o.i. of 0.1 with viral stocks containing this construct and cloned 6 h after infection. A collection of 79 clones expressing GFP was obtained and analyzed by flow cytometry. Expression of the integrated LTR-Tat–GFP was again highly heterogeneous among clones (CV = 80%) (Figure 4). The fact that the CV at 80% falls between those measured after infection with a Tat-defective construct (CV = 180%) and that measured after introduction of Tat in these clones (CV = 44%), could result from the fact that Tat expression and basal HIV promoter activity are now coupled. While we can not rule out that some degree of heterogeneity is caused by the presence of mutations in Tat/LTR, or by differences in the cellular environment of different clones, it is likely that the site of integration here is, again, the major determinant for distinct expression levels. These data suggest that in a natural HIV infection, the degree of expression of the provirus from cell to cell could also be greatly affected by the site of integration.
Fig. 4. Heterogeneity of HIV expression using a Tat-containing HIV vector after integration. Jurkat cells were infected with viral particles containing the retroviral vector LTR-Tat-IRES-GFP at an m.o.i. of 0.1. Six hours later, individual cells were cloned and a collection of 79 clones was obtained after expansion. LTR-GFP expression measured by flow cytometry is shown for all clones.
Effect of CDK9 kinase inhibition on Tat-independent expression of the HIV promoter
According to our results, ∼20% of HIV integrations exhibit high basal levels of promoter activity in the absence of Tat (Figure 1B), an observation at odds with transient transfection experiments where Tat is essential for efficient HIV transcription. According to current models, Tat activates HIV transcription by recruiting the CDK9 kinase to the elongating polymerase, thereby promoting the assembly of an elongation-competent polymerase complex (reviewed in Karn, 1999). This model assumes that the HIV promoter is deficient in recruiting CDK9 in the absence of Tat. Previous reports documented that basal HIV transcription can also be inhibited by specific CDK9 inhibitors, suggesting that CDK9 may be involved in HIV transcription in the absence of Tat (Mancebo et al., 1997).
We therefore tested the possibility that the differential transcriptional activity of the HIV promoter integrated at different sites in chromatin might occur as a result of a Tat-independent recruitment of CDK9. We measured the effect of a specific inhibitor of CDK9 kinase (DRB) and of a CDK9 dominant-negative mutant (D167N) (Gold et al., 1998) on basal GFP expression and found no difference in susceptibility between clones with low and high basal expression levels (data not shown). These results suggest that recruitment of CDK9 to the HIV promoter does not play a significant role in Tat-independent transcription of the HIV LTR and does not appear to account for the high basal transcription measured at some integration sites.
Synergistic activation of the HIV promoter by Tat, deacetylase inhibitors and phorbol esters
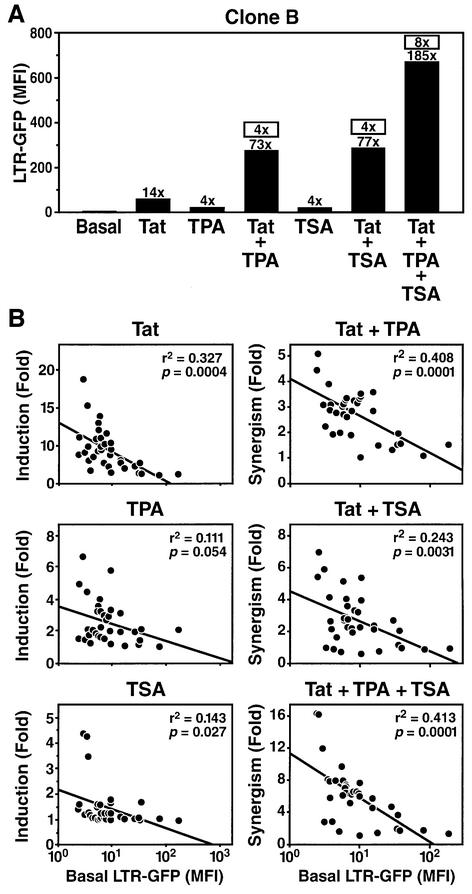
To determine how the integration site of the provirus modulates the transcriptional activation of the HIV LTR in response to cellular signals, we used phorbol esters (tetradecanoyl phorbol acetate, TPA) and an HDAC inhibitor (trichostatin A, TSA). These agents were selected because of their ability to activate HIV expression via distinct pathways (Verdin et al., 1993; Van Lint et al., 1996). Phorbol esters mimic T-cell mitogen activation and activate the NF-κB pathway, a transcription factor involved in HIV transcription initiation. HDAC inhibitors activate HIV transcription in latently HIV-infected cell lines or in transiently transfected HIV LTR reporter constructs, presumably by causing a relative hyperacetylation of histones. Each clone of our library was treated with 400 nM TSA or 10 nM TPA, alone or in combination with extracellular Tat protein (recombinant Escherichia coli purified His6-Tat), and GFP expression was measured by flow cytometry. Preliminary experiments conducted in a representative clone showed a modest 4-fold activation by TPA or TSA (Figure 5A). Tat activated HIV expression 14-fold after incubation of the target cells with the recombinant protein. Interestingly, a marked synergistic activation of HIV expression was observed when Tat was combined with TPA (73-fold activation), with TSA (77-fold activation) or with both TPA and TSA (185-fold activation).
Fig. 5. Synergistic activation of the integrated HIV promoter in response to Tat, TPA and TSA. (A) Each Jurkat clone was treated with recombinant Tat (25 µg/ml), TPA (10 nM) and TSA (400 nM) for 24 h, and LTR activity was measured by flow cytometry. Fold induction versus control untreated samples are shown immediately above each bar. Synergism factors are shown above each bar (boxed) for the combinations Tat + TPA, Tat + TSA and Tat + TPA + TSA. They are calculated using the formula: induction by A + B/induction by A + induction by B. (B) Scatter plots of induction factors (defined as GFP m.f.i. in response to agents relative to untreated samples) for Tat, TPA or TSA against basal GFP levels in each clone. Scatter plots of the synergism factors (defined above) for the combinations Tat + TPA, Tat + TSA or Tat + TPA + TSA versus GFP m.f.i. for each clone.
To determine whether the response to these activating signals is conditioned by the basal rate of HIV transcription, we extended this analysis to all our Jurkat clones. The response of individual clones to Tat, TPA and TSA varied widely (Figure 5B). In response to exogenous Tat, GFP expression was induced up to 19-fold, and an inverse correlation was observed between the Tat response and basal GFP expression. GFP induction in response to TPA varied from 2- to 6-fold, and an inverse correlation, weaker than for Tat, was also observed between induction and basal expression. Most clones did not respond to TSA treatment alone; however, three clones showed a 4-fold response. The synergism detected in clone B between Tat and TPA was observed in most clones and varied from 1 to 5. Here again, an inverse correlation was observed between basal GFP expression and Tat + TPA synergistic response. Similar results were obtained in response to Tat + TSA or in response to the three agents together (Figure 5B).
Low basal HIV expression is not caused by histone deacetylation or DNA methylation
Histone deacetylation and DNA methylation both play a significant role in the silencing of virally transduced genes (Hoeben et al., 1991; Chen et al., 1997). Silencing occurs progressively after integration and can be reversed by HDAC inhibitors (TSA) or DNA methylation inhibitors, such as 5-azacytidine (5-azaC) or 5-aza-2-deoxycytidine (5-azadC).
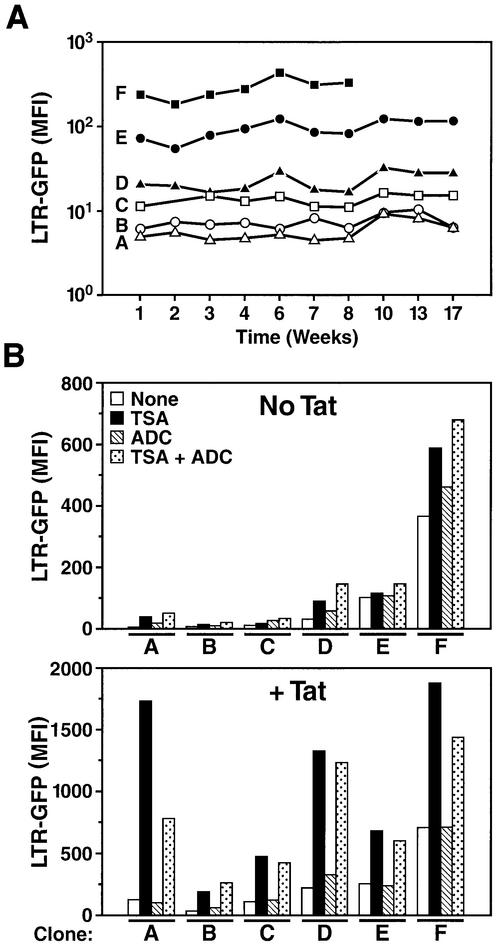
To determine whether long-term silencing occurs in our stably transduced clones, we measured basal expression of the integrated LTR-GFP construct for up to 17 weeks in six representative clones exhibiting the full range of basal expression levels (Figure 6A). We observed no change in expression over a 17-week period, indicating that, once established, the level of expression of a clone is maintained over extended periods of time. However, we reasoned that silencing of the HIV promoter by DNA methylation or histone deacetylation could have occurred immediately after integration during the 3-week interval separating the infection and our first analysis after clones had been expanded. Such silencing, occurring in the majority of integration sites, would have resulted in the picture observed (Figure 1B) and should be reversed by treatment with TSA, 5-azadC or both, as reported in other systems (Groudine et al., 1981; Chen et al., 1997).
Fig. 6. The HIV promoter is stably expressed over time and unresponsive to HDAC inhibitors or DNA methylation inhibitors. (A) Basal activity of the integrated LTR-GFP construct was measured weekly by flow cytometry over a 17-week period in six representative clones (A–F). The basal LTR activity of these individual clones is shown in Figure 1B. (B) The same six clones were treated with TSA (400 nM), 5-azadC (5 µM) or both agents, in the absence or presence of recombinant Tat (12.5 µg/ml). On day 1, 5-azadC was added to an exponentially growing culture; on day 2, Tat, TSA and a second aliquot of 5-azadC were added; on day 3, the experiment was completed and LTR-GFP expression analyzed by flow cytometry.
To test this hypothesis, we treated each clone at weekly intervals either with 5 µM 5-azadC for 48 h or 400 nM TSA for 24 h, or with a combination of both drugs, and measured GFP expression. We observed that 5-azadC had a negligible effect on LTR-GFP expression, whereas TSA modestly induced expression of the LTR between 1.6- and 4-fold, depending on the clone (Figure 6B). No synergy was observed between TSA and 5-azadC. In contrast, we observed that an HIV LTR integrated after transient transfection into Jurkat cells (clone 1G5 provided by the AIDS Research and Reference Reagent Program) was activated by the same concentrations and the same stock of 5-azadC (C.Callebaut and E.Verdin, unpublished observations). Tat synergized with TSA, as shown earlier, but not with 5-azadC (Figure 6B). These results indicate that DNA methylation is not responsible for silencing of the HIV promoter upon integration, at least during the 5-month period that we studied.
We have also studied the extent of DNA methylation affecting the integrated LTR region. Genomic DNA was extracted from representative clones and digested with endonucleases sensitive to the methylation state of DNA (HpaII, BssHII and EagI). These endonucleases are unable to digest DNA when cytosines are methylated. Digested DNA was analyzed by Southern hybridization to quantify the extent of the digestion. We observed that restriction sites in the integrated vector are digested by each of these endonucleases, indicating that the integrated HIV LTR DNA is not significantly methylated (Figure 7). These results are in agreement with our observations described above that HIV expression is not modified by 5-azaC.
Remodeling of nuc-1 correlates with basal transcriptional activity
We have reported that a single nucleosome (nuc-1) located immediately downstream of the HIV transcription start is remodeled upon transcriptional activation (Verdin et al., 1993; Van Lint et al., 1996). This remodeling is unaffected by α-amanitin, an inhibitor of RNA pol II elongation, indicating that it does not occur simply as a result of transit of the pol II in this region of the HIV genome. We proposed that the position of this nucleosome immediately after the transcription start site could pose a unique elongation barrier for the polymerase and that remodeling of the nucleosome could play a significant role in the transcriptional activation of the HIV promoter.
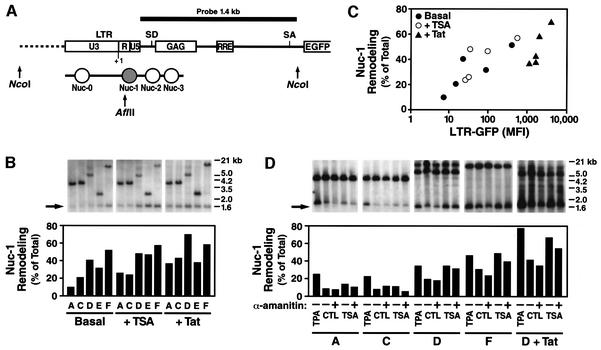
To confirm this model, we have investigated the status of nuc-1 in the integrated HIV promoter present in our clones. To assay for nuc-1 remodeling, we measured the ability of a restriction endonuclease cutting within nuc-1 DNA (AflII) to cut genomic DNA in the context of intact nuclei (see scheme in Figure 8A). Purified nuclei were incubated with the restriction endonuclease, DNA was purified, digested with NcoI and analyzed by Southern blotting using indirect end labeling as previously described (Verdin et al., 1993). In this assay, nuc-1 remodeling leads to increased accessibility of AflII to its recognition site within nuc-1, and thereby increases the intensity of the 1.5 kb AflII–NcoI fragment. Analysis of five Jurkat clones with differing basal transcriptional activity using this assay showed that nuc-1 remodeling correlated with basal transcriptional activity (Figure 8B and C). Clones with low basal transcriptional activity exhibited low accessibility to AflII, indicating that nuc-1 is present and hindering access for the endonuclease. Clones with high basal levels showed increased AflII digestion, indicating increased remodeling. Similar results were obtained with HinfI, another restriction endonuclease cutting within nuc-1 DNA (data not shown). Next, the effect of Tat and TSA on nuc-1 remodeling was examined. In all clones, Tat activity resulted in nuc-1 remodeling, as previously reported (el Kharroubi et al., 1998). Interestingly, TSA treatment resulted in nuc-1 remodeling to the same extent as Tat, while the activation of transcription was significantly greater in response to Tat (Figure 8B and C). Similarly, TPA also induced extensive remodeling of nuc-1 (Figure 8D). As previously reported (Van Lint et al., 1996), blocking of transcriptional elongation with α-amanitin (10 µg/ml for 5 h) did not dramatically change the remodeling of nuc-1 that occurs under basal conditions or in response to TSA (Figure 8D). These observations are consistent with our model that nuc-1 remodeling occurs as a cause rather than a consequence of increased transcriptional elongation.
Fig. 8. Remodeling of nuc-1 correlates with basal and activated promoter activity. (A) Diagram of the LTR promoter in the integrated HIV-derived vector (modified from Verdin et al., 1993). The positions of nucleosomes are indicated with respect to the HIV promoter, including nuc-1, which is remodeled upon HIV transcriptional activation. Positions of restriction sites for AflII and NcoI and the probe used in indirect end labeling are indicated. (B) AflII accessibility of the HIV promoter in five clones under basal conditions and in response to 400 nM TSA (4 h) or Tat. Purified nuclei were digested with AflII, DNA extracted and analyzed by indirect end labeling after NcoI digestion. The larger band in each clone corresponds to the NcoI–NcoI fragment and the shorter band corresponds to the AflII–NcoI fragment. The extent of nuc-1 remodeling was quantified by measuring the ratio of the intensity of the AflII-digested band and the total intensity of the two bands (undigested + digested). (C) The accessibility of nuc-1 to AflII correlates with the transcriptional activity in each clone. (D) Endonuclease accessibility after α-amanitin treatment. The same experimental conditions as in Figure 8B were used, but α-amanitin (10 µg/ml) was added 1 h before TSA addition and 5 h before analyzing nuc-1 accessibility. The response to 10 nM TPA is shown.
Discussion
Our current understanding of HIV transcriptional regulation has relied exclusively on analyses performed on cell populations. Since most cell lines are thought to provide a homogeneous cellular environment, it is generally assumed that predictions can be made on what happens in individual cells based on the analysis of the population. However, the HIV genome can integrate at multiple sites in chromatin and, given the heterogeneity of chromatin in terms of its transcriptional potential, this could dramatically alter the rate of transcription.
To begin addressing this problem, we have generated a library of Jurkat-derived cellular clones containing a single integration of an HIV-1 minigenome. We observed that HIV-1 transcription is highly heterogeneous at the clonal level. In contrast to what has been observed in population-based assays, where basal promoter activity is low and promoter activation strictly dependent on Tat, we found that transcription is significant in 20% of clones in the absence of Tat. The remaining 80% of clones showed significantly lower basal expression levels. In the presence of Tat, an inverse correlation between basal promoter activity and Tat response was noted. Using a variety of approaches, we demonstrate that the site of integration of the provirus into chromatin is responsible for these variations in basal promoter activity and Tat response.
By using a retrovirus vector that does not encode Tat, we artificially separated the two phases of HIV transcription into an early, Tat-independent phase and a late, Tat-dependent phase, which could be examined independently. Our results demonstrate that the integration site of the provirus has a profound effect on the early Tat-independent phase. We predict that in sites where basal transcription is strong, Tat will be produced early and the transition to the late phase will occur rapidly. In other sites, the low efficiency of elongation in the absence of Tat will cause a relative delay in transition to the late phase. To an extreme, if basal transcription is very low, Tat will not accumulate and the provirus could remain in a non-productive state equivalent to latency.
To examine the late, Tat-dependent phase of HIV transcription, we introduced the Tat protein into each clone, either in the form of a recombinant protein or in the form of an expression vector. We observed that Tat enhances transcription in all clones and that an inverse correlation existed between basal level and the transcriptional activation promoted by Tat. As a result, Tat-mediated transcription in all clones was more homogeneous than that in the absence of Tat. However, even in the presence of Tat, significant differences in transcription rates were observed, indicating that the site of integration plays a significant role in HIV production. In a final experiment, we used a retroviral construct expressing both GFP and the Tat protein to mimic a natural HIV-1 infection where the early and late phases of transcription are coupled. Again, we observed a great degree of heterogeneity in viral transcription among different clones.
The heterogeneity in HIV promoter basal activity is dependent on the local chromatin environment. While the unintegrated HIV promoter is generally inactive, we observed that 20% of clones exhibited very significant transcriptional activity. These observations are consistent with a model in which the default state of the HIV promoter is to be transcriptionally inactive. Experiments with TSA and 5-azadC provide evidence that the level of histone acetylation or DNA methylation of the HIV promoter is not responsible for low level expression observed in most clones. It is likely that negative elongation factors (Garber and Jones, 1999) and the assembly of a poorly processive RNA polymerase at the level of the HIV promoter contribute to low transcription rates in the absence of Tat.
Interestingly, we have observed that the basal level of expression of our clones is stable over a 4-month period, indicating that no silencing occurs. This observation is to be contrasted with previous reports describing the silencing of retroviruses and other extraneously introduced genetic elements in mammalian cells (Bestor, 2000). Silencing of integrated viruses occurs via de novo DNA methylation (Groudine et al., 1981; Hoeben et al., 1991; Lorincz et al., 2000; Zentilin et al., 2000) or via a repressive histone acetylation code (Chen et al., 1997; Chen and Townes, 2000; Pannell et al., 2000). In some cases, the extent of methylation-associated silencing has been reported to be dependent on the chromosomal position (Hoeben et al., 1991) and to show a variegated expression pattern (Zentilin et al., 2000). Cloning of chromatin insulators into retroviral vectors protects them from this chromosomal position effect (Emery et al., 2000; Rivella et al., 2000). Previous studies proposed that the HIV-1 promoter was inactivated by CpG methylation; however, HIV LTR was introduced via transfection in this particular study (Bednarik et al., 1990). In the current study, we have found no evidence of DNA methylation of the HIV promoter in any of the clones and no stimulation of transcription after treatment with 5-azadC, alone or in combination with TSA. In addition, we observed minimal stimulation of viral transcription in response to TSA and no variegation of expression. These observations collectively support the possibility that unique sites are selected by the virus integration machinery for integration in chromatin.
In 20% of clones, however, the chromatin environment at the site of integration provides factors that contribute to increased basal activity. It has been shown that cloning of a cellular enhancer upstream of the HIV LTR stimulated HIV promoter activity in a Tat-independent manner by increasing the elongation capacity of the RNA polymerase (West and Karn, 1999). This experiment suggests that by inserting its genome close to unique cellular transcriptional regulatory elements, the HIV promoter can function independently of Tat.
Tat increases the ability of the polymerase assembled by the HIV-1 promoter to elongate efficiently by recruiting CDK9, a kinase specific for the C-terminal domain (CTD) of RNA pol II. The inverse correlation between basal LTR-GFP expression levels and Tat response suggested that the local environment at the site of integration might activate transcription by a mechanism similar to that used by Tat. According to this model, cellular transcriptional elements would increase transcription elongation by recruiting CDK9 or another transcriptional elongation factor to the HIV promoter independently of Tat. Tat recruitment, therefore, would favor elongation to a degree inversely proportional to the elongation defect present in the absence of Tat. To test whether CDK9 plays a role in the basal rate of HIV expression, we measured the effect of CDK9 inhibitors and a mutant form of CDK9 with dominant-negative activity on basal GFP expression. Our results show that the activity of the HIV promoter is inhibited by CDK9 inhibition to the same degree in clones with high basal transcription as in those with low basal expression. We conclude that high basal HIV-1 transcription at some integration sites results from the recruitment of a highly processive RNA polymerase complex, in a Tat- and CDK9-independent manner. These observations are in agreement with a previous report that enhancer elements can promote HIV elongation in a CDK9-independent manner (West and Karn, 1999).
We previously reported that phorbol esters (TPA) and HDAC inhibitors (TSA) can activate HIV transcription in latently infected cell lines (Verdin et al., 1993; Van Lint et al., 1996). We also reported that HIV genomes integrated in these latently infected cell lines (ACH2 and U1 cell lines) carried mutations in either Tat or TAR (Emiliani et al., 1996, 1998). In this context, we were unable previously to determine whether the activation of latent viruses by TSA or TPA was due to an effect on the LTR or on the Tat protein, or both. In the experiments described here, we demonstrate that the LTR is poorly responsive to TPA or TSA in the absence of Tat. In contrast, most clones were strongly induced by both TPA and TSA in the presence of Tat and a strong synergy was demonstrated with Tat. Synergism between Tat and TPA or TSA could be due to a direct effect of either TSA or TPA on Tat or a Tat cofactor. In the case of TPA, synergy between Tat and protein kinase C, the target of action of TPA, has previously been reported (Jakobovits et al., 1990). In the case of TSA, it has recently been reported that acetylation of Tat by p300 is important for its transcriptional activity (Kiernan et al., 1999; Ott et al., 1999). In this context, TSA could promote the hyperacetylation of Tat, leading to increased Tat activity, or could lead to increased transcription reinitiation as previously demonstrated in vitro on chromatinized templates (Sheridan et al., 1997).
We reported previously that a single nucleosome (nuc-1) is located immediately downstream of the transcription start site of the HIV promoter under latency conditions. Nuc-1 is remodeled upon activation of the HIV promoter in response to Tat, phorbol esters and HDAC inhibitors (Verdin et al., 1993; Van Lint et al., 1996; el Kharroubi et al., 1998). Because of the previously reported inhibitory effect of chromatin on transcriptional elongation and the known elongation block in this region of the HIV promoter, nuc-1 was seen as a repressive element for HIV transcription, and its remodeling as a prerequisite for viral expression. We have now extended this chromatin analysis to the HIV promoter integrated at different sites. We have found that the extent of nuc-1 remodeling correlates closely with the basal transcriptional level. Since nuc-1 remodeling is largely insensitive to α-amanitin, an inhibitor of transcriptional elongation, we conclude that nuc-1 is a crucial determinant of the transcriptional activity of the HIV promoter and that its remodeling does not occur simply as a consequence of transcriptional activation. According to this model, differences in transcription rates observed between different clones are due to the local chromatin environment at the site of integration, which dictates the degree of nuc-1 remodeling. The site of integration could modulate nuc-1 remodeling either via changes in histone acetylation at the site of integration or via the recruitment of specific chromatin remodeling complexes. Both possibilities are currently being addressed experimentally. However, our experiments also allow us to conclude that nuc-1 remodeling, although necessary for full transcriptional activation, is not sufficient. We indeed observed significant nuc-1 remodeling in response to treatment with TSA or TPA alone, while either agent only activated transcription marginally. These observations are consistent with a model in which activation of the HIV promoter is dependent on the concerted action of factors acting at the level of transcription initiation and elongation.
Remarkably, the HIV promoter can be activated by Tat in all clones that we studied, indicating that Tat can activate transcription independently of the integration site. This observation is different from what has been observed after random integration of the HIV promoter by stable transfection protocols. In such experiments, it is generally observed that up to 50% of clones are silent and can not be reactivated by Tat (S.Emiliani, C.Van Lint and E.Verdin, unpublished observations). This discrepancy could indicate that delivery of the HIV-1 genome via the infection process leads to integration of the provirus at sites in chromatin that are fully competent for transcription, and suggests that some degree of site selection occurs during the process of virus-mediated entry. Evidence has been presented that retroviruses integrate non-randomly in genomic DNA (Shih et al., 1988; Withers-Ward et al., 1994; Leclercq et al., 2000) and that HIV integration is disfavored in heterochromatin (Carteau et al., 1998). The Saccharomyces cerevisiae retrotransposon Ty3 is uniquely targeted to rRNA genes via the interaction of the retrotransposon integrase with the polymerase complex transcribing these genes (Chalker and Sandmeyer, 1992; Kirchner et al., 1995). The HIV integrase interacts with Ini1/BAF57, a subunit of the mammalian SWI/SNF complex, and could target the HIV promoter to the subset of cellular genes that are usually under the control of the SWI/SNF complex (Kalpana et al., 1994; Miller and Bushman, 1995; Holstege et al., 1998). Such targeting could account for the universal response of all integrated HIV proviruses to Tat activation. As an alternative explanation, it is also possible that Tat can activate transcription independently of the integration site. In the context of such a model, the role of Tat is to counterbalance the role of chromatin in the transcriptional activity of the integrated HIV promoter.
In summary, our observations indicate significant clonal variation in HIV expression dependent on the integration site, resistance to DNA methylation and a universal response to transcriptional activation by Tat. These experiments indicate either that the HIV promoter is targeted to unique regions of chromatin or that once integrated, the provirus creates a minilocus that becomes resistant to the surrounding chromatin environment. Further studies will focus on characterizing the factors present at distinct integration sites and their influence on transcription. These studies should increase our understanding of HIV transcription in its natural context and the modulating role of chromatin on this process.
Materials and methods
Cell culture and transfection
Jurkat cell line was grown in RPMI 1640 medium (Mediatech Cellgro) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 µg/ml streptomycin and 2 mM l-glutamine at 37°C under a 95% air/5% CO2 atmosphere. HeLa and 293T cells were grown under the same conditions on Dulbecco’s modified Eagle’s medium (Mediatech Cellgro).
293T cells were routinely transfected with calcium phosphate. Jurkat cells (107 cells/0.4 ml serum-free medium) were electroporated on 0.4 cm gap cuvettes at 250 V and 950 mA (Biorad Gene Pulser II). Plasmid DNA was purified with the Qiagen Plasmid Maxi kit, followed by phenol extraction and ethanol precipitation.
Viral production and infection
For the production of viral particles containing an HIV-derived vector, 5 × 106 293T cells were transfected with plasmids (provided by D.Trono, University of Geneva) pRRLGFP-W (10 µg), pCMV-R8.91 (6.5 µg) and pMD.G (3.5 µg) in 10 cm dishes. After 16 h, medium was replaced, and supernatants containing viral particles were harvested 24 h later. Viral particles were quantified with the HIV-1 p24 ELISA assay (NEN). p24 (150 ng, corresponding to ∼6 × 104 particles) was used to infect 2 × 105 Jurkat cells. After 48 h, the efficiency of the infection process was monitored by FACS analysis in the presence of 10 nM TPA to induce the expression of the integrated LTR-GFP vector. Approximately 30% of the cells were expressing GFP levels above the autofluorescence background. The infected culture, non-TPA treated, was serially diluted and plated on 96-well plates. Individual clones were obtained after 3 weeks.
To produce viral particles containing a retroviral vector with the YFP reporter gene under the control of the HIV promoter, the GFP gene from pRRLGFP-W was substituted for the YFP gene obtained from plasmid pEYFP-C1 (Clontech) by standard cloning procedures (pEV735).
Plasmid pEV731 is an HIV-based vector derived from the pHR′ series (provided by D.Trono), where Tat101 (corresponding to the two-exon form of the HIV-1 Tat gene) and GFP are under the control of the HIV-1 LTR by using an internal ribosome entry site (LTR-Tat-IRES-EGFP). The IRES-EGFP fragment was obtained from plasmid pIRES2-EGFP (Clontech). Plasmid pEV695 is a pHR′-derived vector containing Tat72 (corresponding to the first exon of the HIV-1 Tat gene) transcribed from the HIV promoter (LTR-Tat).
Flow cytometry analysis
Cells were washed in phosphate-buffered saline (PBS) and resuspended in PBS containing 1% paraformaldehyde. GFP fluorescence was measured by a FACScan machine (Becton Dickinson). A two-parameter analysis to distinguish GFP-derived fluorescence from background fluorescence was used: GFP was measured in FL1 and cellular autofluorescence was monitored in FL2. Electronic compensation was applied during analysis. For the dual detection of GFP and YFP fluorescence, a FACSVantage (Becton Dickinson) was used with bandpass filters of 510/10 and 550/30, respectively. Results shown throughout the manuscript correspond to representative data of experiments repeated at least three times. Cell sorting was carried out with the FACSVantage.
Luciferase assays
The Dual-Luciferase Reporter Assay system (Promega) was used for luciferase activity measurement. To correct for transfection efficiency, plasmid pRL-TK encoding the Renilla luciferase gene under the control of the TK promoter was co-transfected and detected as described above.
Southern blotting and DNA methylation analysis
Genomic DNA from Jurkat-derived cells was extracted with the DNeasy Tissue kit (Qiagen). DNA was digested with EcoRI and run on a 0.7% agarose gel in Tris–acetate buffer. Agarose gels were incubated for 20 min in denaturation solution (0.5 N NaOH, 1.5 M NaCl) and 20 min in neutralization solution (0.5 M Tris–HCl pH 7.5, 3 M NaCl). Transfer was carried out by vacuum blotting in 20× SSC (3 M NaCl, 0.3 M sodium citrate pH 7) for 1 h with a VacuGene XL (Pharmacia Biotech) to nylon positively charged membranes (Roche). DNA was cross-linked to the membrane by exposure to UV light and prehybridized for 1–2 h at 42°C in the Ultrahyb hybridization solution (Ambion). A digoxigenin-labeled probe corresponding to a 1.4 kb fragment internal to the pRRLGFP-W vector was generated with the PCR DIG Probe Synthesis kit (Roche) and primers EV1048 (5′-GTGGCGCCCGAACAGGGACC-3′) and EV1049 (antisense, 5′-CCGTCGAGATCCGTTCACTA-3′). Hybridization was carried out overnight at 42°C. Membranes were washed at 42°C for 2× 10 min in 2× SSC/0.1% SDS, and 2× 30 min in 0.1× SSC/0.1% SDS, followed by a 65°C wash for 30 min in 0.2× SSC/0.1% SDS. Detection of the digoxigenin label was carried out with the DIG Detection kit and the chemiluminiscent reagent CSPD (Roche). Alternatively, a radioactive probe was generated from the 1.4 kb PCR fragment (primers EV1048 and EV1049) with the Multiprime DNA labeling system (Amersham) and [α-32P]dCTP (3000 Ci/mmol), purified on a Sephadex G-50 Nick column (Pharmacia Biotech), and allowed to hybridize to the membrane overnight at 42°C. Autoradiographic exposures were carried out for 1–5 days at –70°C, or the signal was detected directly on an imaging analyzer (Fujix BAS1000).
To quantitate LTR methylation, genomic DNA was digested with HpaII, NarI, BssHII or EagI. As a control, MspI, a methylation-insensitive isoschizomer of HpaII, was also used. DNA was digested further to completion with NcoI, and Southern blotting was performed as described above.
Purification of recombinant Tat protein and treatments
Recombinant His6-tagged Tat protein was produced as previously reported (Ott et al., 1997). Purified protein was aliquoted and lyophilized. Aliquots were kept under vacuum at 4°C to prevent Tat oxidation, and Tat was resuspended in serum-free cell culture media (Opti-Mem I, Gibco-BRL) immediately before use at 250 µg/ml. Protein determinations were carried out with the Bio-Rad Protein Assay and with bovine serum albumin (BSA) as a standard.
In a typical experiment, 1 ml of an exponentially growing culture (106 cells) was centrifuged, and the cell pellet resuspended in 0.1 ml of reconstituted Tat (25 µg). If required, TPA (10 nM) or TSA (400 nM) was added, and cells were incubated for 5 h at 37°C under a 95% air/5% CO2 atmosphere. Next, cell culture medium (10% serum-supplemented RPMI 1640) was added up to 1 ml, containing TPA or TSA if required, and incubation continued overnight; the final Tat protein concentration was 25 µg/ml.
PCR amplification and sequencing of LTR
The 5′ LTR from selected clones was amplified from genomic DNA with primers EV976 (5′-GCTAATTCACTCCCAACGAAGAC-3′; 5′ end of LTR) and EV997 (antisense, 5′-TCGCTTTCAGGTCCCTGTTCG-3′; immediately downstream of the 5′ LTR) and Pfu DNA polymerase (Stratagene). The reaction was run with the following program: (a) 45 s at 94°C; (b) 30 cycles of 45 s at 94°C, 45 s at 58°C and 1.5 min at 72°C; and (c) 10 min at 72°C. The amplified 663 bp product was purified from ethidium bromide-containing 1% agarose gel with the GenClean Spin kit (Bio101) and cloned in the pCR-Blunt vector provided in the Zero Blunt PCR Cloning kit (Invitrogen). Two recombinant clones containing the expected DNA insert were sequenced for each original LTR-GFP integration clone, both with primers M13 Forward and M13 Reverse. For the sequencing reaction, 0.5 µg of plasmid DNA and 3.2 pmol of primer were mixed with 8 µl of Big Dye d-Rhodamine Terminator Ready Reaction mix (Perkin Elmer Applied Biosystems) in a 20 µl total volume. The reaction was run with the PCR program: (a) 1.5 min at 96°C; and (b) 25 cycles of 30 s at 96°C, 30 s at 50°C and 4 min at 60°C. The reaction products were purified on a Centri-Sep spin column (Princeton Separations) and sequenced.
RNA analysis
HIV-1-specific transcripts were detected by RT semi-quantitative real-time PCR. Total RNA was extracted from exponentially growing cells with the Trizol reagent (Gibco-BRL) following the manufacturer’s instructions. RNA concentration was determined by spectrophotometry at 260 nm. RNA was treated with RQ1 RNase-free DNase (Promega). Treated RNA (2.5 µg) was used for cDNA synthesis with the SuperScript First-Strand Synthesis system for RT–PCR (Gibco-BRL) with random hexanucleotide primers. After the reaction was terminated, RNA was removed by treatment with RNase H.
For real-time PCR, one-tenth of the cDNA was amplified with 200 nM each of the specific primers and 100 nM specific TaqMan probe, and with the TaqMan Universal PCR Master mix (Perkin Elmer). The reaction was run for 40 cycles (15 s at 95°C plus 1 min at 60°C) in an AbiPrism 7700 Sequence Detector (Perkin Elmer). Two sets of specific primers plus FAM-labeled probe were used to detect the integrated HIV-derived vector (LTR-EGFP) specific transcripts. One corresponding to the R-U5 junction of the LTR [amplicon 77 bp; primers EV1143 (5′-GCTAACTAGGGAACCCACTGCTT-3′) and EV1144 (antisense, 5′-ACAACAGACGGGCACACACTAC-3′); probe EV1145 (5′-6 FAM-AGCCTCAATAAAGCTTGCCTTGAGTGCTTC-TAMRA–3′)], the other corresponding to the EGFP gene [amplicon 75 bp; primers EV1252 (5′-GGAGCGCACCATCTTCTTCA-3′) and EV1253 (antisense, 5′-AGGGTGTCGCCCTCGAA-3′); probe EV1254 (5′-6 FAM-CTACAAGACCCGCGCCGAGGTG-TAMRA-3′)]. Primers were from Genset; the labeled probe from Operon. A GAPDH-specific set of primers and JOE-labeled probe (TaqMan GAPDH Control reagents; Perkin Elmer) were used as an endogenous control to standardize the amount of sample RNA added to the reactions. Periodically, products of amplification were checked on 2% agarose gels for purity. In parallel, real-time amplifications were carried out from samples of DNase-treated RNA not submitted to RT to check that no contaminant DNA was present. For the relative quantitation of HIV expression among clones, we have used the relative standard curve method.
The effect of α-amanitin treatment on global transcription in intact cells was evaluated by incorporation of [3H]uridine into mRNA after pulse labeling. Exponentially growing cells (2 × 106 cells) were or were not treated with α-amanitin (10 µg/ml) for 4 h and then labeled with [3H]uridine (10 µCi/ml; 40 Ci/mmol; Amersham Pharmacia) for 90 min. mRNA was extracted with the QuickPrep Micro mRNA Purification kit (Amersham Pharmacia). Radioactivity incorporated into the mRNA eluted from the oligo(dT)-cellulose column was measured by scintillation counting and expressed as c.p.m./106 cells. α-amanitin reduced mRNA synthesis to 39% in comparison with untreated cells.
Accessibility of nucleosomal DNA to restriction enzymes
Exponentially growing cells were harvested by centrifugation and washed with ice-cold PBS. Subsequent steps were performed on ice with precooled buffers. Cells were resuspended at 25 × 106 cells/ml in buffer A (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.3 M sucrose) and incubated on ice for 10 min. An equal volume of buffer A/0.2% NP-40 was added, and cells were incubated for a further 10 min. Nuclei were pelleted at 230 g for 10 min, resuspended at 108 nuclei/ml in buffer B (10 mM Tris pH 7.9, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, 100 µg/ml BSA, 0.1 mM phenylmethylsulfonyl fluoride) and digested for 20 min with AflII (1 U/µl) at 37°C. Digestion reactions were placed on ice and genomic DNA was purified with the DNeasy Tissue kit (Qiagen). The same amounts of DNA from each sample were digested to completion with NcoI, and Southern blotting was performed to detect the extent of AflII cleavage by indirect end labeling. As described above, a 1.4 kb probe internal to the pRRLGFP-W vector was used.
Acknowledgments
Acknowledgements
We thank John C.W.Carroll for graphics, Heather Gravois for manuscript preparation, and Stephen Ordway and Gary Howard for editorial assistance. The HIV-derived retroviral vectors pRRLGFP-W and the pHR′ series were kindly provided by D.Trono and R.Zufferey, and plasmids pFLAG-CMV2-hPITALRE (D167N) by A.P.Rice. We thank Christian Callebaut and Jen-Kuei Wang for plasmids and Veronique Kiermer for helpful discussions. We also thank the Gladstone Institute Flow Cytometry core for technical assistance. This work was supported in part by a Public Health Service grant from the NIH (GM 51671-05A2). A.J. was a recipient of fellowships from the Secretaria de Estado de Universidades, Investigacion y Desarrollo (Fulbright/Ministry of Education & Culture, Spain) and the Human Frontier Science Program Organization. P.D. was a recipient of fellowships from the Belgian National Fund for Scientific Research and the Belgian American Educational Foundation.
References
- Bednarik D.P., Cook,J.A. and Pitha,P.M. (1990) Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J., 9, 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T.H. (2000) Gene silencing as a threat to the success of gene therapy. J. Clin. Invest., 105, 409–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carteau S., Hoffmann,C. and Bushman,F. (1998) Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol., 72, 4005–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D.L. and Sandmeyer,S.B. (1992) Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev., 6, 117–128. [DOI] [PubMed] [Google Scholar]
- Chen W.Y. and Townes,T.M. (2000) Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc. Natl Acad. Sci. USA, 97, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.Y., Bailey,E.C., McCune,S.L., Dong,J.Y. and Townes,T.M. (1997) Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc. Natl Acad. Sci. USA, 94, 5798–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T., Zufferey,R., Kelly,M., Mandel,R.J., Nguyen,M., Trono,D. and Naldini,L. (1998) A third-generation lentivirus vector with a conditional packaging system. J. Virol., 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Kharroubi A., Piras,G., Zensen,R. and Martin,M.A. (1998) Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol. Cell. Biol., 18, 2535–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery D.W., Yannaki,E., Tubb,J. and Stamatoyannopoulos,G. (2000) A chromatin insulator protects retrovirus vectors from chromosomal position effects. Proc. Natl Acad. Sci. USA, 97, 9150–9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S., Van Lint,C., Fischle,W., Paras,P.,Jr, Ott,M., Brady,J. and Verdin,E. (1996) A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl Acad. Sci. USA, 93, 6377–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S., Fischle,W., Ott,M., Van Lint,C., Amella,C.A. and Verdin,E. (1998) Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol., 72, 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnet C.M. and Bushman,F.D. (1997) HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell, 88, 483–492. [DOI] [PubMed] [Google Scholar]
- Garber M.E. and Jones,K.A. (1999) HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol., 11, 460–465. [DOI] [PubMed] [Google Scholar]
- Gold M.O., Yang,X., Herrmann,C.H. and Rice,A.P. (1998) PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J. Virol., 72, 4448–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Eisenman,R. and Weintraub,H. (1981) Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature, 292, 311–317. [DOI] [PubMed] [Google Scholar]
- Hoeben R.C., Migchielsen,A.A., van der Jagt,R.C., van Ormondt,H. and van der Eb,A.J. (1991) Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J. Virol., 65, 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Jakobovits A., Rosenthal,A. and Capon,D.J. (1990) Trans-activation of HIV-1 LTR-directed gene expression by tat requires protein kinase C. EMBO J., 9, 1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalpana G.V., Marmon,S., Wang,W., Crabtree,G.R. and Goff,S.P. (1994) Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science, 266, 2002–2006. [DOI] [PubMed] [Google Scholar]
- Kao S.Y., Calman,A.F., Luciw,P.A. and Peterlin,B.M. (1987) Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature, 330, 489–493. [DOI] [PubMed] [Google Scholar]
- Karn J. (1999) Tackling Tat. J. Mol. Biol., 293, 235–254. [DOI] [PubMed] [Google Scholar]
- Kiernan R.E. et al. (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J., 18, 6106–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J., Connolly,C.M. and Sandmeyer,S.B. (1995) Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retrovirus-like element. Science, 267, 1488–1491. [DOI] [PubMed] [Google Scholar]
- Leclercq I., Mortreux,F., Cavrois,M., Leroy,A., Gessain,A., Wain-Hobson,S. and Wattel,E. (2000) Host sequences flanking the human T-cell leukemia virus type 1 provirus in vivo. J. Virol., 74, 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz M.C., Schübeler,D., Goeke,S.C., Walters,M., Groudine,M. and Martin,D.I. (2000) Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional re pression. Mol. Cell. Biol., 20, 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo H.S. et al. (1997) P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev., 11, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.D. and Bushman,F.D. (1995) HIV integration. Ini1 for integration? Curr. Biol., 5, 368–370. [DOI] [PubMed] [Google Scholar]
- Miller M.D., Farnet,C.M. and Bushman,F.D. (1997) Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol., 71, 5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.P. and Varmus,H.E. (1994) DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J., 13, 4704–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory D.S., Neugeboren,B.A. and Mulligan,R.C. (1996) A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl Acad. Sci. USA, 93, 11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M., Emiliani,S., Van Lint,C., Herbein,G., Lovett,J., Chirmule,N., McCloskey,T., Pahwa,S. and Verdin,E. (1997) Immune hyper activation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science, 275, 1481–1485. [DOI] [PubMed] [Google Scholar]
- Ott M., Schnölzer,M., Garnica,J., Fischle,W., Emiliani,S., Rackwitz,H.R. and Verdin,E. (1999) Acetylation of the HIV-1 Tat protein by p300 is important for its transcriptional activity. Curr. Biol., 9, 1489–1492. [DOI] [PubMed] [Google Scholar]
- Pannell D. et al. (2000) Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J., 19, 5884–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss D., Bushman,F.D. and Wolffe,A.P. (1994) Human immuno deficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl Acad. Sci. USA, 91, 5913–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivella S., Callegari,J.A., May,C., Tan,C.W. and Sadelain,M. (2000) The cHS4 insulator increases the probability of retroviral expression at random chromosomal integration sites. J. Virol., 74, 4679–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan P.L., Mayall,T.P., Verdin,E. and Jones,K.A. (1997) Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev., 11, 3327–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C.C., Stoye,J.P. and Coffin,J.M. (1988) Highly preferred targets for retrovirus integration. Cell, 53, 531–537. [DOI] [PubMed] [Google Scholar]
- Stevens S.W. and Griffith,J.D. (1994) Human immunodeficiency virus type 1 may preferentially integrate into chromatin occupied by L1Hs repetitive elements. Proc. Natl Acad. Sci. USA, 91, 5557–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens S.W. and Griffith,J.D. (1996) Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J. Virol., 70, 6459–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C., Emiliani,S., Ott,M. and Verdin,E. (1996) Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J., 15, 1112–1120. [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Paras,P.,Jr and Van Lint,C. (1993) Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J., 12, 3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M.J. and Karn,J. (1999) Stimulation of Tat-associated kinase-independent transcriptional elongation from the human immunodeficiency virus type-1 long terminal repeat by a cellular enhancer. EMBO J., 18, 1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers-Ward E.S., Kitamura,Y., Barnes,J.P. and Coffin,J.M. (1994) Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev., 8, 1473–1487. [DOI] [PubMed] [Google Scholar]
- Zennou V., Petit,C., Guetard,D., Nerhbass,U., Montagnier,L. and Charneau,P. (2000) HIV-1 genome nuclear import is mediated by a central DNA flap. Cell, 101, 173–185. [DOI] [PubMed] [Google Scholar]
- Zentilin L., Qin,G., Tafuro,S., Dinauer,M.C., Baum,C. and Giacca,M. (2000) Variegation of retroviral vector gene expression in myeloid cells. Gene Ther., 7, 153–166. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Zou,S., Wright,D.A. and Voytas,D.F. (1999) Tagging chromatin with retrotransposons: target specificity of the Saccharomyces Ty5 retrotransposon changes with the chromosomal localization of Sir3p and Sir4p. Genes Dev., 13, 2738–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S., Ke,N., Kim,J.M. and Voytas,D.F. (1996) The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev., 10, 634–645. [DOI] [PubMed] [Google Scholar]