Abstract
In Saccharomyces cerevisiae, Cdc13 has been proposed to mediate telomerase recruitment at telomere ends. Stn1, which associates with Cdc13 by the two-hybrid interaction, has been implicated in telomere maintenance. Ten1, a previously uncharacterized protein, was found to associate physically with both Stn1 and Cdc13. A binding defect between Stn1-13 and Ten1 was responsible for the long telomere phenotype of stn1-13 mutant cells. Moreover, rescue of the cdc13-1 mutation by STN1 was much improved when TEN1 was simultaneously overexpressed. Several ten1 mutations were found to confer telomerase-dependent telomere lengthening. Other, temperature-sensitive, mutants of TEN1 arrested at G2/M via activation of the Rad9-dependent DNA damage checkpoint. These ten1 mutant cells were found to accumulate single-stranded DNA in telomeric regions of the chromosomes. We propose that Ten1 is required to regulate telomere length, as well as to prevent lethal damage to telomeric DNA.
Keywords: Cdc13/DNA damage checkpoints/protein–protein interactions/Saccharomyces cerevisiae/telomere length
Introduction
Telomeres, the ends of eukaryotic chromosomes, which are composed of TG-rich sequences elongated by telomerase (Greider, 1996; Lingner and Cech, 1998; Nugent and Lundblad, 1998), are critical for maintaining chromosome stability and genome integrity (Zakian, 1996) and have also been implicated in gene silencing (Lustig, 1998). In the yeast Saccharomyces cerevisiae, mutations in components or regulators of telomerase produce a gradual erosion of chromosomes, which eventually leads to death by senescence (Lundblad and Szostak, 1989; Lundblad and Blackburn, 1993; Lendvay et al., 1996).
Telomeres are capped by proteins that bind to these repeating DNA sequences (Evans and Lundblad, 2000). The S.cerevisiae EST proteins presumably form complexes that regulate telomerase activity and, hence, the length of telomeric tracts (Lendvay et al., 1996; Virta-Pearlman et al., 1996; Evans and Lundblad, 1999; Hughes et al., 2000b), while others, such as Rap1, the SIR and yKU proteins, appear to be involved not only in telomere length control but also in telomeric silencing and DNA repair (Haber, 1999; Evans and Lundblad, 2000). Some telomeric proteins prevent recombinational events that would otherwise occur frequently between repeating telomeric sequences, and also prevent DNA repair enzymes from inappropriately intervening on the double-strand breaks naturally present at the ends of telomeres. In yeast, several proteins have been proposed or suspected to play a role in the protection of telomeric ends, including Cdc13, Stn1 (Garvik et al., 1995; Grandin et al., 1997) and telomerase (Lundblad and Szostak, 1989), but also Rap1 (Krauskopf and Blackburn, 1998; Stavenhagen and Zakian, 1998) and the yKu70/yKu80 and Mre11/Rad50/Xrs2 complexes (Bertuch and Lundblad, 1998).
Cdc13, also known as Est4, together with Est1 and Est3, has been implicated previously as the main regulator of telomerase access to telomeric ends (Garvik et al., 1995; Lendvay et al., 1996; Lin and Zakian, 1996; Nugent et al., 1996; Virta-Pearlman et al., 1996; Evans and Lundblad, 1999; Hughes et al., 2000a; Zhou et al., 2000). In S.cerevisiae, Est2 and Tlc1 represent the reverse transcriptase and RNA template subunits of telomerase, respectively (Singer and Gottschling, 1994; Lendvay et al., 1996; Lingner et al., 1997). Both Cdc13 and Est1 are capable of binding single-stranded telomeric DNA (Lin and Zakian, 1996; Nugent et al., 1996; Virta-Pearlman et al., 1996; Bourns et al., 1998; Hughes et al., 2000b). It is not known yet whether the short single-strand extension of telomeric DNA at the distal end of telomeres acts as an anchoring structure for telomerase (Wellinger et al., 1993; Diede and Gottschling, 1999). Physical association between Est1 and Tlc1, as well as between Est3 and Tlc1, has been demonstrated recently (Hughes et al., 2000a; Zhou et al., 2000). Est1 also associates physically with Cdc13 (Qi and Zakian, 2000). Association of Est1 with Tlc1 does not require Est2, while, in contrast, association of Est3 with Tlc1 does require the presence of Est2 (Hughes et al., 2000a). Finally, immunoprecipitation of Est1 or Est3, but not of Cdc13, resulted in co-precipitation of telomerase activity (Hughes et al., 2000a), thus confirming that Est1 and Est3 are close regulators of telomerase activity, while Cdc13 is more likely to represent a link between telomerase and telomere ends, possibly via specific interactions with Est1 (Evans and Lundblad, 1999; Qi and Zakian, 2000).
Stn1 has been shown to interact with Cdc13 by two-hybrid analysis (Grandin et al., 1997). Like Cdc13, Stn1 has been implicated in the physical protection of telomeric ends because stn1-13 mutant cells, like cdc13-1 mutant cells, accumulated single-stranded DNA in telomeric regions of chromosomes when incubated at restrictive growth temperatures (Garvik et al., 1995; Grandin et al., 1997). Moreover, stn1-13 mutant cells have been shown to be deregulated in telomere length control and to exhibit abnormally long telomeres (Grandin et al., 1997). The Cdc13–Stn1 complex may be part of a larger complex comprising additional telomeric proteins and may also be the target of as yet unidentified regulators. We report here the isolation of Ten1, a new protein, which we have found to interact physically with both Stn1 and Cdc13. Analysis of ten1 mutants demonstrates that Ten1 is required both for telomere length regulation and telomere end protection.
Results
Isolation of TEN1
To gain further insight into the function of Stn1 and, hence, of Cdc13 (Grandin et al., 1997), we looked for suppressors of temperature-sensitive (ts) stn1 mutations (see Materials and methods). YLR010 was thus identified as a weak suppressor of the growth defect of stn1-13 and stn1-154 mutant cells (Figure 1A). Due to these genetic interactions and to other properties described below, the product of the YLR010 gene was named Ten1, for protein involved in Telomeric pathways in association with Stn1, number 1. YLR010/TEN1, which potentially codes for a 160 amino acid protein, has never been described before (DDBJ/EMBL/GenBank accession No. AJ296344). The Ten1 protein sequence did not display significant homology to other known sequences in databases or any convincing conserved domains or structural motifs (data not shown). Ten1 can therefore be regarded as a novel protein.
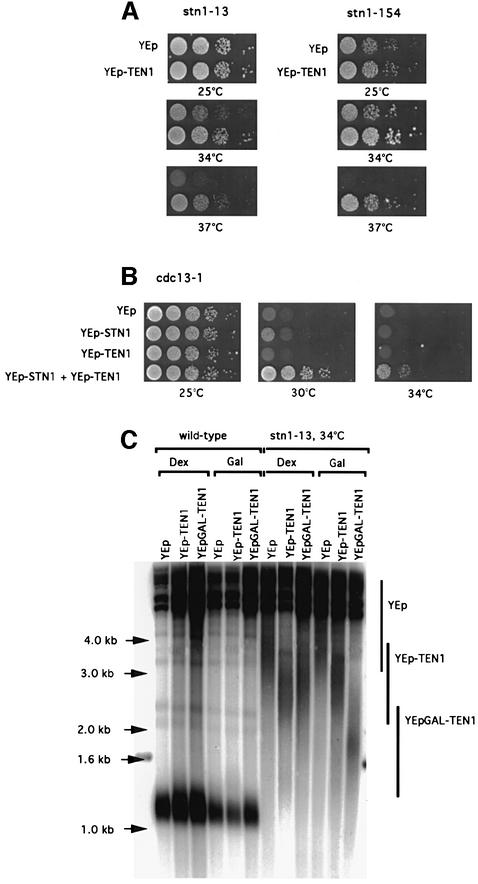
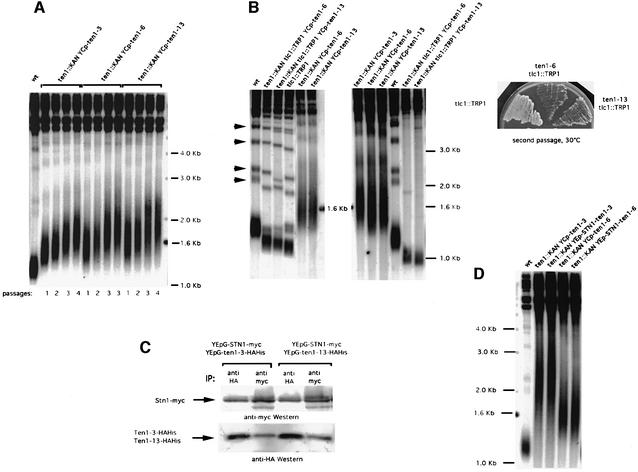
Fig. 1. Suppression of the temperature-sensitive growth and telomere length defects of stn1 and cdc13 mutations following TEN1 overexpression. (A) stn1-13 (left) and stn1::TRP1 YCp111-stn1-154 (right) strains were transformed with a multicopy (episomal, 2µ) plasmid expressing TEN1 under the control of its own promoter (YEp-TEN1) or vector alone (YEp) and 10-fold serial dilutions (from left to right in each row) of transformants grown for 2 days at the indicated temperatures and photographed. (B) Co-overexpression with TEN1 improves the rescue of cdc13-1 by STN1. Both STN1 and TEN1 were overexpressed from a 2µ plasmid under the control of their respective promoter. Temperature-sensitive cdc13-1 cells were transformed with a plasmid expressing STN1 (YEp-STN1), with a plasmid expressing TEN1 (YEp-TEN1), with both plasmids (YEp-STN1 + YEp-TEN1) or with vector alone (YEp), spotted as in (A) and incubated for 2 days at the temperatures indicated. (C) Telomere length in wild-type cells (left) and stn1-13 mutant cells (right) harbouring either YEp-TEN1, YEp-GAL-TEN1 or YEp alone, grown for ∼60 generations either on glucose- (Dex) or galactose-based medium (Gal) to induce expression of the GAL1 promoter. The relevant genotypes are indicated above the lanes. A Y′ 32P-labelled probe was used to detect telomeric sequences (see Materials and methods). Note that the telomere length of stn1-13 cells grown at 34°C was much larger than that at 30 or 32°C (see Figures 5D and 3D, respectively). The existence of such extremely heterogeneous-length telomeres is not unprecedented (see, for instance, rif1 rif2 double mutants; Wotton and Shore, 1997). Vertical bars schematically outline the upper and lower limits of the average telomere lengths in each category of strains considered.
Ten1 cooperates with Stn1 for the accomplishment of Cdc13 function
Overexpression of STN1 allowed cdc13-1 cells to grow at 30°C but not at higher temperatures (Figure 1B), as shown previously (Grandin et al., 1997). On the other hand, overexpression of TEN1 did not complement the loss of function of cdc13-1 cells, even at 30°C (Figure 1B). Strikingly, however, co-overexpression of STN1 and TEN1 could partially rescue the loss of function of Cdc13 at temperatures up to 34–37°C (Figure 1B). Thus, although under these conditions the rescue of cdc13-1 was partial at 37°C, as revealed by the persistence of morphological defects, cells could nevertheless form small colonies (not shown). These observations provide strong genetic arguments to suggest that Ten1 and Stn1 regulate Cdc13 function.
Overexpression of TEN1 reduces telomere length in stn1 mutant cells
stn1 mutant cells exhibit a very dramatic telomere lengthening (Grandin et al., 1997). Overexpression of TEN1 from a multicopy plasmid under the control of its own promoter clearly led to a reduction of telomere lengthening in stn1 mutant cells (Figure 1C). This effect was even more pronounced when TEN1 was overexpressed under the control of the strong, inducible, GAL1 promoter on galactose-based medium (Figure 1C). We noted that overexpression of TEN1 from the YEp-GAL1 plasmid also resulted in a decrease in stn1-associated telomere lengthening even when incubation was on glucose-based medium (Figure 1C; see also below). On the other hand, overexpression of TEN1 had no apparent effect on telomere length in wild-type cells (Figure 1C).
Arrest phenotype of ten1 mutant cells
Sporulation of a ten1::kanMX4/TEN1 diploid strain (see Materials and methods) gave rise to only two out of four viable spores, which were always found to be Kan–. Sporulation of a ten1::kanMX4/TEN1 heterozygous diploid previously transformed with YEp195-TEN1 (URA3) plasmid allowed the recovery of Kan+ spores, which were always also Ura+. Cells derived from such Kan+ Ura+ spores could not grow on 5-fluoro-orotic acid (5-FOA)-containing medium (5-FOA is a drug that counterselects for Ura+ cells). These data indicate that the TEN1 gene is essential for vegetative growth.
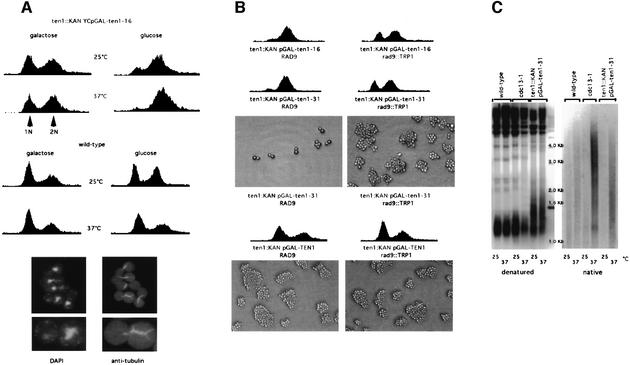
ten1-16 and ten1-31, two ts alleles (see Materials and methods), were found to exhibit a severe growth defect at 37°C. To obtain a tighter arrest, these alleles were recloned into YCp111-GAL1 (LEU2) and re-introduced into a ten1Δ strain. The two resulting strains were found to exhibit wild-type growth and morphology at 25°C when grown on galactose-based medium. However, after transfer to glucose-based medium at 37°C, cells of both strains arrested at G2/M, with a dumb-bell shape, in a very homogeneous manner (Figure 2A). It should be noted that the ten1Δ YCp-GAL1-ten1-16 (and -31) mutant cells could still grow on glucose medium at 25°C, albeit with defects (Figure 2A). This was due to the probable presence of intragenic promoter-like sequences in TEN1, as TEN1 open reading frame (ORF) expressed alone, in the absence of any promoter sequences, could sustain growth of a ten1Δ strain after the plasmid expressing the wild-type TEN1 gene (ORF + natural promoter sequences) had been shuffled out on 5-FOA-containing medium (data not shown). We also noted that the ten1Δ YCp-GAL1-ten1-16 (and -31) strains could grow poorly on galactose medium at 37°C, as the probable result of the leakiness of the mutant alleles under these conditions of overexpression driven by the GAL1 promoter (Figure 2A). Therefore, growth of these ten1 mutants on either galactose medium at 37°C or glucose medium at 25°C represents semi-permissive conditions, as observed on the corresponding fluorescence activated cell sorting (FACS) profiles (Figure 2A), while growth on glucose medium at 37°C represents restrictive conditions, as seen above.
Fig. 2. The arrest conferred by the temperature-sensitive ten1-16 and ten1-31 mutant alleles occurs at G2/M and depends on the Rad9-mediated DNA damage checkpoint. (A) Top: FACS analysis of ten1 arrested cells. Cells disrupted for TEN1 (ten1::kanMX4) and surviving owing to the presence of the temperature-sensitive ten1-16 allele on a single-copy plasmid, under the control of the GAL1 promoter (ten1::KAN YCpGAL-ten1-16), were grown to mid-log phase at 25°C and shifted up to 37°C for 4 h or left at 25°C, either on galactose-based medium, a condition that induced expression of the ten1 allele, or on glucose-based medium, and prepared for FACS analysis, together with wild-type control cells. The ten1 mutant cells arrested in G2/M on glucose medium at 37°C. When grown on glucose medium at 25°C or on galactose medium at 37°C, these mutant strains became enriched in dumb-bell-shaped cells but still continued to grow. ten1-31 cells behaved similarly (not shown). Bottom: cells from the ten1::kanMX4 YCp111-GAL1-ten1-31 strain were grown on galactose-based medium at 25°C, then on glucose-based medium at 37°C (repressive conditions), fixed 4 h later and processed for tubulin immunofluorescence and staining of the DNA with DAPI. Arrested ten1 mutant cells contained a single nucleus located close to the neck joining the mother and daughter. They also exhibited a short tubulin spindle, typical of a G2/M arrest, extending through the nucleus between the two duplicated spindle bodies. (B) Cell cycle profiles of ten1 mutant cells bearing (rad9::TRP1, right) or not (RAD9, left) a disruption of RAD9, a DNA damage checkpoint gene, were fixed for FACS analysis 4 h after transfer to glucose medium at 37°C, a condition that inactivated these temperature-sensitive ten1 alleles. In the presence of the rad9 mutation, both the ten1-16 and ten1-31 mutant cells failed to mark the G2/M arrest exhibited by the RAD9+ ten1 cells. As a consequence, rad9 ten1 double mutants continued to proliferate for a few cell divisions, as attested by their ability to form numerous microcolonies (shown only for ten1-31), a phenotype indicative of a checkpoint defect. In contrast, RAD9+ ten1-31 cells, which are checkpoint proficient, stopped progression through the cell cycle. Meanwhile, ten1 disruptants bearing wild-type TEN1 (ten1::KAN pGAL-TEN1) bearing (rad9::TRP1) or not (RAD9) a disruption of RAD9, and growing on galactose medium, continued to proliferate indefinitely. (C) Non-denaturing Southern hybridization to a Y′ 32P-labelled probe (native, right) revealed the presence of abnormally high levels of single-stranded DNA in the telomeric regions of ten1-31 mutant cells (ten1::TRP1 pGAL-ten1-31) at 37°C. Cells were grown on galactose-based medium at 25°C, then for 4 h on glucose-based medium at 37°C (repressive conditions) or 25°C (permissive conditions) and harvested for preparation of genomic DNA. cdc13-1 mutant cells grown at 37°C were used as a positive control, while cdc13-1 cells grown at the permissive temperature of 25°C and wild-type cells grown at 25 or 37°C served as negative controls. Genomic DNAs from these same strains were run in parallel and processed for hybridization with the same probe under denaturing conditions (denatured, left) to serve as additional controls.
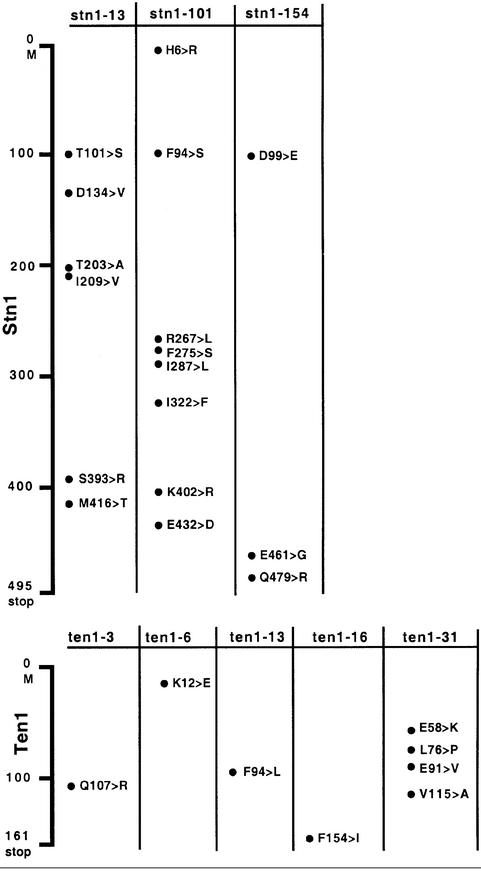
Neither STN1 nor CDC13, when overexpressed from a multi-copy plasmid under the control of their respective promoters, was able to improve growth of the ten1Δ YCp111-GAL1-ten1-16 (and -31) strains on glucose-based medium at temperatures between 25 and 37°C (data not shown). The nature of the mutations present in the ten1-16 and ten1-31 alleles is shown in Table I.
Table I. Sequence analysis of the amino acid changes in the ten1 and stn1 mutant alleles.
The G2/M arrest conferred by the ten1-16 and ten1-31 alleles depends on the DNA damage checkpoint
We next investigated the possibility that the arrest in the ten1-16 and ten1-31 alleles might be due to the activation of DNA damage checkpoints. DNA damage has been shown to activate a set of specific genes, including RAD9, whose main function is to halt the cell cycle in order to allow DNA repair, followed by cell cycle resumption if conditions allow (Weinert and Hartwell, 1988). In contrast to the ten1 RAD9+ mutants, ten1 rad9Δ double mutants no longer arrested after a few cell divisions as large-budded cells, as seen by FACS analysis (Figure 2B). Rather, they continued to divide and formed microcolonies (Figure 2B). The ten1 rad9Δ cells eventually died (data not shown), presumably due to a failure to restrain mitosis in the presence of DNA damage as a consequence of the checkpoint defect (Garvik et al., 1995). Presumably, such cells cannot properly separate their chromosomes and die of mitotic catastrophe, possibly after too many chromosomal aberrations have accumulated. From these experiments, we conclude that ten1-16 and ten1-31 cells arrest at the restrictive temperature due to the presence of a functional DNA damage checkpoint.
cdc13-1 and stn1-13 mutants accumulate single-stranded DNA in telomeric regions of the chromosomes when grown at restrictive temperature (Garvik et al., 1995; Grandin et al., 1997). Hybridization of DNA from ten1-31 cells to a Y′ 32P-labelled probe under native conditions according to the method described by Wellinger et al. (1993) indicated the presence of abnormally high levels of single-stranded DNA at 37°C, but not at 25°C (Figure 2C). Application of the in-gel hybridization method (Dionne and Wellinger, 1996) gave similar results (data not shown). From these experiments, we infer that accumulation of single-stranded DNA at telomeres in ten1-16 and ten1-31 mutants may constitute the DNA damage that causes the activation of the Rad9-dependent DNA damage checkpoint, as explained above.
Ten1 binds Stn1 in vivo
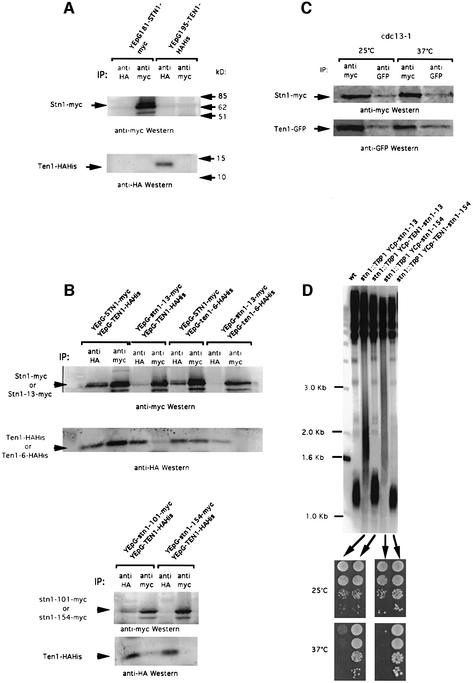
To verify the possibility that Ten1 and Stn1 might form a complex together, an HA-His-tagged version of TEN1 and a myc-tagged version of STN1 were constructed, which allowed controlled expression by the inducible GAL1 promoter (Figure 3A). Both constructs were fully functional because their expression alone could complement a deletion of the corresponding gene. In cells expressing both constructs simultaneously, immunoprecipitation with anti-myc antibody revealed the presence of Ten1–HAHis on anti-HA western blots. Conversely, immunoprecipitation with anti-HA antibody allowed Stn1–myc to be visualized on anti-myc westerns (Figure 3B). The physical association between Ten1 and Stn1 was confirmed by two-hybrid analysis (see below).
Fig. 3. Ten1 associates with Stn1 in vivo, an interaction that is defective in stn1 mutants. (A) Control experiments were performed to reveal the position of Stn1–myc and Ten1–HAHis on western blots. Extracts from wild-type cells overexpressing STN1–myc alone under GAL1 promoter control (lanes 1 and 2 in both top and bottom panels) or TEN1–HAHis alone under GAL1 promoter control (lanes 3 and 4 in both top and bottom panels), at 30°C, were subjected to immunoprecipitation with monoclonal anti-HA antibody (IP: anti-HA, lanes 1 and 3 in both panels) or monoclonal anti-myc antibody (IP: anti-myc, lanes 2 and 4 in both panels). The band migrating below Stn1–myc in this and subsequent blots presumably represents an Stn1 degradation product. (B) Co-immunoprecipitation of Stn1–myc and Ten1–HAHis. Protein extracts were prepared from cells of the indicated genotype. All experiments were performed at 30°C. (C) Stn1 and Ten1 still co-immunoprecipitate in cdc13-1 mutant cells co-expressing YEp181-GAL1-STN1–myc and YEp195-GAL1-TEN1–GFP and shifted up for 4 h at the restrictive temperature of 37°C; same methods and nomenclature as in (A) and (B). (D) Suppression of stn1-associated telomere elongation following expression of fusions between wild-type Ten1 and mutant Stn1 proteins. An stn1::TRP1 haploid strain kept alive on galactose owing to the presence of YCp33-GAL1-STN1 was transformed with either one of the indicated centromeric plasmids. The plasmid containing wild-type STN1 was then shuffled out following counterselection on 5-FOA medium. The resulting strains were then grown for ∼60 generations at 32°C and the length of their telomeres measured using a Y′ probe. Expression of either stn1-13 or stn1-154 alone produced dramatic telomere elongation (lanes 2 and 4, respectively) compared with wild-type cells (wt, lane 1). In contrast, expression of the Ten1–Stn1-13 (lane 3) or Ten1–Stn1-154 (lane 5) fusions totally suppressed the stn1-induced telomere elongation. In addition, the Ten1–Stn1 hybrid proteins rescued not only the inviability due to the stn1 disruption, but also the temperature-sensitive growth defect conferred by the Stn1-13 or Stn1-154 mutant protein. The four patches of cells in each row represent 10-fold dilutions of the same culture.
Since Stn1 has been shown to bind Cdc13 by two-hybrid analysis (Grandin et al., 1997), a finding recently confirmed by large-scale two-hybrid screening (Uetz et al., 2000), we wanted to know whether Cdc13 function was necessary for the association between Stn1 and Ten1 observed above. To this end, the association between Stn1 and Ten1 was assessed in ts cdc13-1 mutant cells after shifting the culture to the restrictive temperature of 37°C for 4 h prior to preparation of cell extracts. Importantly, under such conditions, Stn1–myc could still bind Ten1–GFP and vice versa (Figure 3C).
Ten1 associates with Cdc13 by two-hybrid analysis
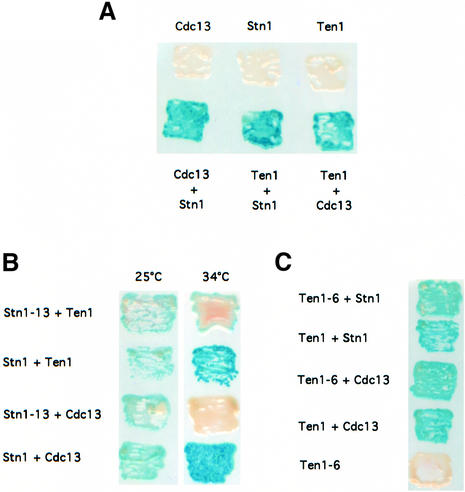
To know whether Ten1 could associate physically with Cdc13, we expressed simultaneously TEN1 and CDC13 from plasmids used to detect protein–protein interactions in a yeast two-hybrid system (Fields and Song, 1989). Y190 S.cerevisiae strains simultaneously expressing TEN1 and CDC13 or TEN1 and STN1 were found to be positive in the X-gal assay, yielding an intense blue colour (Figure 4A). These interactions were confirmed by quantitative assays performed in liquid cultures, using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate to measure β-galactosidase activity (Table II). These data demonstrate that Ten1 interacts with both Stn1 and Cdc13 by two-hybrid analysis, thus supporting the existence within the cell of a putative complex made of Cdc13, Stn1 and Ten1.
Fig. 4. Two-hybrid interactions between Ten1, Stn1 and Cdc13. (A) Ten1 associates with Cdc13, as well as with Stn1, in a yeast two-hybrid system. Strains simultaneously expressing pAS2-TEN1 and pACT2-STN1 (bottom row, Ten1 + Stn1) or pAS2-TEN1 and pACT2-CDC13 (bottom row, Ten1 + Cdc13) were positive for β-galactosidase activity, measured as described in Materials and methods, like cells co-expressing pAS2-CDC13 and pACT2-STN1 (bottom row, Cdc13 + Stn1) used here as a positive control. On the other hand, cells simultaneously expressing pAS2-TEN1 and pACT2 alone (top row, Ten1), pAS2 alone and pACT2-CDC13 (top row, Cdc13) or pAS2 alone and pACT2-STN1 (top row, Stn1) scored negative in the X-gal assay. Patches of cells replica-plated on nitrocellulose membrane were incubated for 2 h at 30°C and then photographed. For each one of the six pAS2/pACT2 pairs shown here, three different strains were monitored (four transformants for each). (B) The Stn1-13 mutant protein fails to interact with either wild-type Ten1 or wild-type Cdc13 at restrictive temperature. The stn1-13 allele was expressed from pACT2 together with pAS2-TEN1 or pAS2-CDC13, as explained above. Both pairs scored negative for β-galactosidase activity (white patches) at 34°C, but not at 25°C (permissive temperature), while the corresponding controls expressing wild-type STN1 were positive at both temperatures (blue patches). (C) The Ten1-6 mutant protein still interacts with both wild-type Stn1 and wild-type Cdc13. The ten1-6 allele was expressed from pAS2 together with pACT2-STN1 or pACT2-CDC13, as explained in (A). Both pairs scored positive in the X-gal assay (blue patches) at 30°C (ten1-6 is not a ts allele), similarly to the corresponding controls expressing wild-type TEN1.
Table II. Quantitation of the two-hybrid interactions between Ten1, Stn1 and Cdc13.
| Binding domain of Gal4 (pAS2) | Activation domain of Gal4 (pACT2) | Specific activity |
||
|---|---|---|---|---|
| 25°C | 30°C | 34°C | ||
| Gal4-Ten1 | Gal4-Stn1 | 70.6 ± 5.7 | ||
| Gal4-Ten1 | Gal4-Cdc13 | 130.9 ± 3.2 | ||
| Gal4-Cdc13 | Gal4-Stn1 | 91.7 ± 5.0 | ||
| Gal4 | Gal4-Stn1 | 0 | ||
| Gal4 | Gal4-Cdc13 | 0.1 ± 0.1 | ||
| Gal4-Ten1 | Gal4 | 0.3 ± 0.3 | ||
| Gal4-Ten1 | Gal4-Stn1-13 | 60.0 ± 2.7 | 15.5 ± 0.4 | 2.9 ± 0.3 |
| Gal4-Cdc13 | Gal4-Stn1-13 | 28.4 ± 4.0 | 3.2 | 1.0 ± 0.1 |
| Gal4-Ten1 | Gal4-Stn1 | 16.5 | 36.8 ± 6.7 | 31.6 |
| Gal4-Cdc13 | Gal4-Stn1 | 51.4 | 70.5 ± 10.5 | 52.2 |
| Gal4-Ten1-6 | Gal4-Stn1 | 132.8 ± 8.4 | ||
| Gal4-Ten1-6 | Gal4-Cdc13 | 133.7 ± 5.5 | ||
| Gal4-Ten1-6 | Gal4 | 1.2 ± 0.2 | ||
| Gal4-Ten1 | Gal4-Stn1 | 68.0 ± 16.0 | ||
| Gal4-Ten1 | Gal4-Cdc13 | 116.9 ± 8.0 | ||
The telomeric defect of stn1 mutants is associated with a defect in Ten1 binding
stn1-13 mutant cells exhibit a ts growth defect at 37°C and abnormally long telomeres at all temperatures between 25 and 37°C (Grandin et al., 1997). Two other ts stn1 mutants, stn1-101 and stn1-154, exhibited a tighter arrest at 37°C than stn1-13 (see Materials and methods), but shared with it the characteristic of possessing long telomeres even at permissive growth temperatures. All three Stn1 mutant proteins were found by co-immunoprecipitation (Figure 3B) and by two-hybrid analysis (Figure 4B; Table II) to be defective in their physical association with wild-type Ten1. Interestingly, we found that Stn1-13 was also defective in its association with wild-type Cdc13 in the two-hybrid system. In both cases, the defective interactions were found to be sensitive to temperature, as is the stn1-13 mutation (Figure 4B; Table II).
Increasing association between two telomeric proteins interacting directly but weakly within the cell by expression of a fusion (hybrid) protein has been proposed to mimic and enhance the function achieved through the interaction between these two proteins (Evans and Lundblad, 1999). A Ten1–Stn1 in-frame fusion protein, placed under the control of the TEN1 promoter in a single-copy plasmid, was fully functional because it rescued inviability of stn1Δ cells (Figure 3D) as well as that of ten1Δ cells (data not shown). Expression of a Ten1–Stn1-13 or a Ten1–Stn1-154 fusion protein totally prevented the abnormal telomere elongation conferred by the stn1-13 or stn1-154 mutations, respectively (Figure 3D). Importantly, both the Ten1–Stn1-13 and Ten1–Stn1-154 fusions also restored normal growth at restrictive temperatures (Figure 3D). In these experiments, the abundance of Ten1 was only twice that in wild-type cells, as the plasmid used for expression of the fusion protein was a single-copy plasmid, while the abundance of Stn1 was the same as in wild-type cells because the fusions were expressed in an stn1Δ background.
Altogether, these experiments suggest that a defect in the physical association between Stn1 and Ten1 might be responsible for the long telomere phenotype encountered in the stn1 mutant cells. The nature of the mutations present in the stn1 alleles documented here is shown in Table I.
ten1 mutants with deregulated telomere length
The isolation of the ten1-3, ten1-6 and ten1-13 mutant alleles, which displayed very elongated telomeres, confirmed that Ten1 is involved in telomere length maintenance (Figure 5A). Telomere lengthening in these ten1 mutants was progressive (Figure 5A). All three mutant cells exhibited no morphological or growth defects at temperatures between 25 and 37°C (data not shown). In all three ten1 mutants, telomere length deregulation resulted from a mutation in a single residue (Table I).
Fig. 5. Telomerase-dependent telomere elongation associated with ten1 mutations is not due to a defect in Stn1 binding. (A) All three ten1 mutant strains selected for telomere length deregulation (ten1::KAN YCp-ten1-3, lanes 2–5; ten1::KAN YCp-ten1-6, lanes 6–9; ten1::KAN YCp-ten1-13, lanes 10–13) exhibited dramatic telomere elongation compared with wild-type cells (wt, lane 1). All strains were grown at 30°C for various numbers of generations (one passage or restreak is typically performed after ∼25 generations). (B) Disruption of the RNA subunit of telomerase, TLC1, in ten1-6 and ten1-13 resulted in total suppression of telomere elongation (left panel, compare lanes 2 and 3 with lanes 5 and 6, respectively). Lanes 1 (wt cells) and 4 (tlc1::TRP1 cells) served as controls. The ten1 tlc1Δ mutant cells shown in the left panel are at a pre-survival stage, as attested by the presence of non-Y′ bands (indicated by arrowheads near lane 1 of the left panel), contrary to those shown in the middle panel, which have already entered senescence (same strains as in the left panel), attested by the disappearance of non-Y′ bands. Accelerated senescence in the ten1-6 tlc1::TRP1 and ten1-13 tlc1::TRP1 double mutants, compared with tlc1::TRP1 singles, is illustrated in the right panel. (C) The Ten1-3–HAHis and Ten1-13–HAHis mutant proteins still interact physically with wild-type Stn1. Protein extracts were prepared from cells of the indicated genotype and IP–western blots performed as described in the legend to Figure 3. (D) Expression of Stn1–Ten1-3 or Stn1–Ten1-6 fusion protein (under the control of STN1 natural promoter) does not abolish the telomere elongation phenotype conferred by the ten1 mutation. ten1Δ YEp195-TEN1 was transformed with either one of the plasmids indicated. The plasmid containing wild-type TEN1 was then shuffled out on 5-FOA medium. The resulting strains were then grown for ∼60 generations at 30°C and the length of their telomeres measured. Expression of either ten1-3 or ten1-6 alone produced dramatic telomere elongation (lanes 2 and 4, respectively) compared with wild-type cells (wt, lane 1). Expression of the Stn1–Ten1-3 (lane 3) or Stn1–Ten1-6 (lane 5) fusion had no effect on ten1-3- or ten1-6-induced telomere elongation.
We next investigated whether deregulation of telomere length in these mutants was dependent or not on telomerase. Indeed, telomere length can be regulated not only by telomerase, but also by telomerase-independent mechanisms that rely on Rad52-dependent homologous recombination (Lundblad and Blackburn, 1993; Teng and Zakian, 1999). To this end, a tlc1Δ ten1Δ YEp195-TEN1 strain was constructed by crossing, and transformed with a YCp111 (LEU2) plasmid harbouring either ten1-3, ten1-6 or ten1-13. Transformants were further grown on 5-FOA medium to shuffle out the plasmid containing wild-type TEN1. We could not recover any 5-FOA-resistant tlc1Δ ten1Δ YCp111-ten1-3 colony. In addition, the tlc1Δ ten1Δ YCp111-ten1-6 or ten1-13 that were recovered grew poorly. This indicated a severe synthetic defect between TEN1 and TLC1 and, possibly, an accelerated senescence (Figure 5B, right panel). Indeed, the disappearance of non-Y′ telomeres associated with extreme telomere shortening in these strains strongly suggested that survivors had been recovered (Figure 5B, middle panel, lanes 5 and 6). Since these survivors could not be used to interpret the nature of the telomere length regulation mechanisms taking place in the ten1 tlc1Δ double mutants, we set out to obtain cells at a pre-survival stage. To achieve this, a TEN1+/ten1Δ TLC1+/tlc1Δ YEp195-TEN1 zygote was transformed with YCp111-ten1-6 (or -13) and the resulting transformants further grown on 5-FOA medium to shuffle out the plasmid containing wild-type TEN1. Following sporulation, the desired ten1 tlc1Δ spores were selected and further grown for telomere length measurement in parallel with the ten1 TLC1+ and tlc1Δ sister cells. Under such conditions, we could obtain double mutants that, according to the criteria described above, had not yet undergone senescence. Telomere elongation no longer took place in these ten1 tlc1 double mutants (Figure 5B, left panel, lanes 2 and 3), thus indicating that it is dependent on telomerase.
Mutant proteins of Ten1 conferring deregulated telomere length still bind Stn1 and Cdc13
None of the three ten1 mutations documented above affected the binding with Stn1 (Figures 3B, upper panel, lanes 5 and 6, for Ten1-6–HAHis and 5C for the Ten1-3–HAHis and Ten1-13–HAHis proteins). It should be noted that Ten1-6–HAHis no longer associated with Stn1-13–myc (Figure 3B, upper panel, lanes 7 and 8), which was logical since Stn1-13–myc did not bind wild-type Ten1–HAHis (Figure 3B, upper panel, lanes 3 and 4). This provided a control for these experiments. Physical association between the Ten1-6 mutant protein and wild-type Stn1 was confirmed by two-hybrid analysis (Figure 4C; Table II). Interestingly, the Ten1-3 (not shown) and Ten1-6 mutant proteins also interacted with wild-type Cdc13 by two-hybrid analysis (Figure 4C; Table II).
Importantly, wild-type Ten1–HAHis, as well as the Ten1-3–HAHis and Ten1-6–HAHis mutant proteins, still bound wild-type Stn1–myc when CDC13 was simultaneously overexpressed within the cell (data not shown). This suggested that overproduction of Cdc13 did not affect the stability of the Stn1–Ten1 complex and that Cdc13 could not displace Stn1 from Ten1 by direct titration.
To confirm that telomere length deregulation in ten1-3 and ten1-6 mutant cells was not the consequence of a defect of binding between Stn1 and Ten1, we used the fusion protein approach described above. We observed that ten1Δ cells expressing YEp-STN1-ten1-3 or YEp-STN1-ten1-6 fusion constructs exhibited telomeres elongated to the same extent as those in the ten1 mutants alone (Figure 5D). The functionality of these fusions was attested by the fact that they rescued inviability of both ten1Δ cells (Figure 5D) and stn1Δ cells (not shown). Therefore, these experiments confirm the view that a defect of binding between the Ten1-3 or Ten1-6 mutant proteins and wild-type Stn1 is not the cause of ten1-associated telomere elongation.
Discussion
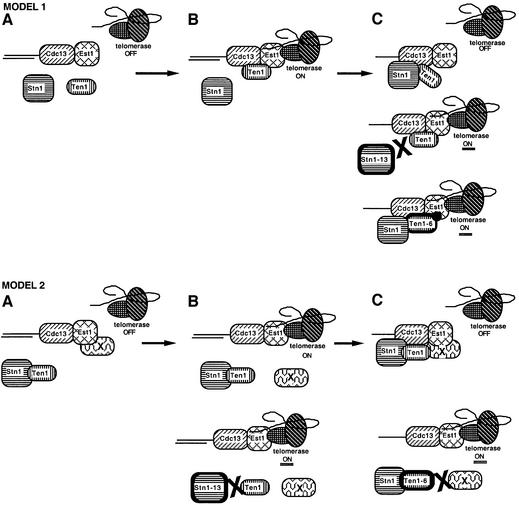
The results presented here provide genetic and biochemical evidence that Ten1, a novel essential protein, interacts with the telomere maintenance machinery in S.cerevisiae. We find that Ten1 is involved both in telomere length regulation, as attested by the long telomere phenotype conferred by some ten1 mutant alleles, and in telomere end protection, suggested by the presence of lethal, Rad9-recognized, telomeric DNA damage in other ten1 mutants. Interestingly, both Stn1 and Cdc13, also previously implicated in both telomere length regulation and telomere end protection (Garvik et al., 1995; Grandin et al., 1997), were found here to associate physically with Ten1. We therefore propose the existence of a putative Cdc13–Stn1–Ten1 complex functioning both in recruiting telomerase at the telomere ends and in the physical protection of telomere ends. A molecular working model for the Cdc13–Stn1–Ten1 complex taking into account the features of the various ten1 and stn1 alleles documented here is presented in Figure 6.
Fig. 6. Speculative models for the functions of the Cdc13–Stn1–Ten1 complex at telomere ends. Two models are proposed to account for the data described here, on the basis of previously published data (see text). The principal actors in both models are represented in (A). Stn1 and Ten1, which physically associate together as well as with Cdc13, a single-stranded telomeric DNA binding protein, act as inhibitors of Cdc13-mediated telomerase recruitment (telomerase contains the catalytic subunit Est2, and TLC1, the RNA template; Est3, also shown here, is a regulator of telomerase). Est1, another regulator of telomerase, which also binds single-stranded telomeric DNA, associates physically with TLC1 and Cdc13. In model 1, Ten1 binding to Cdc13–Est1 might represent the signal allowing interactions between Cdc13–Est1 and telomerase (telomerase ON, B). Binding of Stn1 to Cdc13 and Ten1 might then result in termination of telomere elongation (telomerase OFF, top panel in C). Ten1 could possibly be required to recruit Stn1 in this latter step. Subsequently, Stn1 and Ten1 release from the Cdc13–Stn1–Ten1 complex followed by Ten1 rebinding, or, alternatively, release of Stn1 alone from this complex, might represent the signal allowing interactions between Cdc13, Est1 and telomerase for a new round of elongation of telomeric DNA (not shown). Abolition of the physical interaction between Stn1-13 and both Cdc13 and Ten1 is proposed to explain telomere lengthening in stn1-13 mutant cells (middle panel in C), while the Ten1-6 mutant protein, which is still capable of binding Cdc13 and Stn1, might confer a situation favourable to an increased association between itself and Cdc13–Est1 (bottom panel in C). In model 2, Stn1 and Ten1 are always in complex together and Ten1 is viewed basically as a cargo to convey Stn1 to Cdc13 or, alternatively, as a regulator of Stn1 function; protein X is an as yet unidentified telomeric protein that associates with Est1 (A). Release of protein X from Est1 allows interactions between Cdc13–Est1 and telomerase (top panel in B). Binding of Stn1–Ten1 to Cdc13 and of protein X to both Est1 and Ten1 might terminate the reaction of telomerase recruitment (telomerase OFF, top panel in C). Subsequently, Stn1 and Ten1 release from the Cdc13–Stn1–Ten1 complex accompanied by release of protein X from Est1 and Ten1 might represent the signal allowing interactions between Cdc13, Est1 and telomerase for a new round of telomeric DNA elongation (not shown). Failure of the Stn1-13 mutant protein to bind Ten1 would prevent termination of telomerase recruitment (telomerase ON, bottom panel in B), while a defect in the interaction between the Ten1-6 mutant protein and protein X would prevent re-association with Cdc13–Est1 and inhibition of the interaction between Cdc13–Est1 and telomerase (telomerase ON, bottom panel in C).
Ten1 has a function in telomere length regulation in association with Stn1 and Cdc13
Three sets of data demonstrate that Ten1 functions in regulating telomere length. First, Ten1 was found by co-immunoprecipitation to bind Stn1 in vivo when both proteins were overproduced (Figure 3). Ten1 could also associate with Cdc13 (and Stn1) in a two-hybrid interaction (Figure 4; Table II). Secondly, the ten1-3, ten1-6 and ten1-13 mutations were found to confer abnormal telomere lengthening in a telomerase-dependent manner (Figure 5). Thirdly, TEN1 was isolated as a partial suppressor of ts stn1 mutations when expressed at a low level. In addition, Ten1 and Stn1 could cooperate in partially substituting for Cdc13 essential function (Figure 1).
We note that overexpression of TEN1 alone could not rescue the cdc13-1 defect at all, while overexpression of STN1 could, albeit weakly (Figure 1B), unless under the control of the strong GAL1 promoter (Grandin et al., 1997). The nature of the defect in the Cdc13-1 mutant protein, possibly involving a defect in physically interacting with Stn1 rather than with Ten1, might be at the origin of this difference. Alternatively, particularities of the structure of the putative Cdc13–Stn1–Ten1 complex, unknown for the moment, might result in a more efficient recruitment of Ten1 following STN1 overexpression than the converse. We also note that overexpression of CDC13 or inactivation of Cdc13 did not affect the binding between Stn1 and Ten1, which indicates that Cdc13 is unlikely to mediate the association between Stn1 and Ten1.
An important observation was that the Stn1-13 mutant protein was defective in physically associating with both Ten1 and Cdc13 (Figures 3B and 4B; Table II). Two other Stn1 mutant proteins studied here, Stn1-101 and Stn1-154, displayed similar telomere length deregulation to Stn1-13 and also the same defect in Ten1 binding. Most importantly, expression of a Ten1–Stn1-13 or a Ten1–Stn1-154 fusion protein totally abolished the stn1-induced telomere length defect (Figure 3D). Such hybrid proteins are thought to mimic natural interactions between two proteins that are either in close proximity or even bound together, as was found to be the case for Cdc13 and Est1 (Evans and Lundblad, 1999; Qi and Zakian, 2000). Therefore, it is very probable that the function of Stn1 directly involves physical association with Ten1 (Figure 6, model 2), although it is equally possible that Ten1 possesses an additional function in promoting telomerase recruitment independently of Stn1 (Figure 6, model 1). Stn1 possibly functions as a negative regulator of Cdc13 (Grandin et al., 1997), alone or in association with Ten1, while Ten1 might function, alternately, both in promoting telomerase recruitment via interactions with Cdc13 and Est1, and in inhibiting telomerase functioning via recruitment of Stn1 to Cdc13 (Figure 6).
TEN1 is an essential gene whose failure activates the Rad9 DNA damage checkpoint
Like cdc13-1 and stn1-13 mutants (Garvik et al., 1995; Grandin et al., 1997), ten1-16 and ten1-31 mutants activated the Rad9-dependent DNA damage checkpoint (Figure 2B). Moreover, ten1-31 mutant cells, like cdc13-1 mutant cells, accumulated single-stranded DNA in telomeric regions of chromosomes (Figure 2C). This characteristic of these ten1 mutants, shared with cdc13-1 and stn1-13 mutants (Garvik et al., 1995; Grandin et al., 1997), was expected, given the physical and genetic interactions between Ten1, Stn1 and Cdc13 uncovered here. It is probable that this abnormally high level of single-stranded DNA at telomeres represents the damage recognized by the Rad9 checkpoint, as suggested previously (Garvik et al., 1995). The involvement of proteins capable of recognizing single-stranded DNA, such as RPA, in sensing the telomeric DNA damage experienced by these ten1, stn1 and cdc13 mutants, not addressed in the present study or in any previous one, should be the focus of future experiments. Also unexplored at the moment is the possibility that the ten1 mutants described here might exhibit genetic interactions with mutations in DNA replication proteins. Indeed, it is now becoming evident that conventional replication and telomere maintenance are intimately linked (Diede and Gottschling, 1999). Since Cdc13 has been shown recently to interact physically with DNA polymerase α (Qi and Zakian, 2000), it is possible that Ten1, alone or in complex with Stn1, cooperates with Cdc13 in these mechanisms.
We note that the ten1 and tlc1Δ mutations were very synthetic lethal (Figure 5B), as were the stn1-13 and tlc1Δ mutations at 34 or 37°C (N.Grandin and M.Charbonneau, unpublished results). This led to an accelerated senescence and appearance of survivors, as demonstrated by the disappearance of the non-Y′ telomeres (Figure 5B). These results suggest that telomerase and the Cdc13–Stn1–Ten1 complex play two distinct, but perhaps complementary, roles in telomere protection. It is tempting to speculate that Ten1 and Stn1 function together with Cdc13 in a putative ‘cap’ protein complex that might serve at all times to protect telomeres against inappropriate homologous recombination, for instance. In this hypothesis, the putative Cdc13–Stn1–Ten1 complex might represent the functional equivalent of the Oxytricha single-stranded telomere DNA binding proteins (Froelich-Ammon et al., 1998; Horvath et al., 1998). It is not yet known whether Stn1 and Ten1 interrupt their binding with Cdc13 during the telomere replication process, an Est1-mediated reaction (Evans and Lundblad, 1999; Qi and Zakian, 2000). The telomeric phenotypes of stn1 and ten1 mutants support the view that both Stn1 and Ten1 might negatively regulate telomerase recruitment by Cdc13 (Grandin et al., 1997; this study), a model that suggests temporary interruption of the association between Stn1/Ten1 and Cdc13 (Figure 6). However, since neither Stn1 nor Ten1 appears to be able to bind telomeric DNA in vivo or in vitro (our unpublished data), it is not yet known how Stn1 and Ten1 can assume their function of protection during telomerase recruitment. Perhaps the putative regulation of Cdc13 by Stn1–Ten1 does not necessitate dissociation of the ternary complex. Alternatively, telomere ends might not need physical protection by the Cdc13–Stn1–Ten1 complex during telomere replication.
Materials and methods
Plasmids, strains, screening and mutagenesis
General plasmids and media used in this study were as described previously (Grandin et al., 1997). DNA manipulations were performed according to standard procedures (Ausubel et al., 1998). A diploid strain heterozygous for YLR010c [genotype FY; accession No. 10692D; strain name FWEF004(HE)], in which one copy of YLR010c had been disrupted by the kanMX4 marker gene between nucleotides 49 and 435, was purchased from Euroscarf (Frankfurt, Germany). TEN1 was isolated as a low copy suppressor of the growth defect of stn1-13 cells at 37°C (Grandin et al., 1997) using a genomic YCp50 library (Rose and Broach, 1991). Details on TEN1 isolation, as well as the procedures for construction and selection of ten1 and stn1 alleles, can be found in the Supplementary data (available at The EMBO Journal Online).
FACS analysis and tubulin staining
DNA content was analysed by flow cytometry, as described previously (Hutter and Eipel, 1979). Briefly, cells were fixed overnight, at 4°C, in 70% ethanol, treated with RNase (1 mg/ml) and pepsin (5 mg/ml), stained with propidium iodide (50 µg/ml) and analysed in a Becton Dickinson FACScan analyser. For immunostaining of tubulin of mitotic spindles, rat anti-tubulin (clone YOL1/34, Sera-Lab) was used. Nuclei were simultaneously stained with 4′,6-diamidino-2-phenylindole (DAPI).
Two-hybrid experiments
The pACT2 and pAS2 vectors and the Y190 strain used in the two-hybrid system of Fields and Song (1989) have been described in Durfee et al. (1993). Y190 cells harbouring the pAS2 and pACT2 constructs were streaked on selective minimal medium. Patches of cells were then replica-plated from the culture plates on to nitrocellulose membrane and assayed for β-galactosidase activity using 5-bromo-4-chloro-3-indolyl-β-d- galactopyranoside (X-Gal; Sigma). The β-galactosidase activity of strains expressing the different constructs was quantitated in liquid cultures using the ONPG (Boehringer) protocol, as described in Ausubel et al. (1998).
Measurement of telomere length
Genomic DNA was prepared, digested with XhoI and separated by electrophoresis in a 0.9% agarose gel in TBE, as described in Grandin et al. (1997). Southern membranes were hybridized to a 270 bp TG1–3 32P-labelled probe representing S.cerevisiae telomeric sequences or with a 2.5 kb Y′ 32P-labelled probe representing subtelomeric sequences from chromosome V-R (Garvik et al., 1995). Under such conditions, telomere tracts of wild-type cells appear as a broad band of ∼1.1–1.2 kb, which represents the average length of most chromosomes, those containing Y′ subtelomeric regions (Louis and Borts, 1995). From non-Y′ chromosomes, XhoI cutting typically generates the fragments indicated by arrowheads near lane 1 of the left panel of Figure 5B, best revealed using the TG1–3 probe (Figure 5B) but also visible using the Y′ probe (in all other figures) due to the presence of a few TG1–3 sequences in the Y′ probe. In senescing cells, the disappearance of the non-Y′ fragments attests to the fact that survivors have arisen by homologous recombination. Consequently, the presence of these fragments in senescing cells was used as a marker of the state before the acquisition of survivors (Figure 5B). Results were analysed using a Storm PhosphorImager (Molecular Dynamics).
Detection of single-stranded DNA
To detect single-stranded TG1–3 DNA, genomic DNA was prepared and digested with XhoI, as described in Grandin et al. (1997), run in a 0.7% agarose gel and subjected to non-denaturing Southern hybridization, using either a TG1–3 or a Y′ 32P-labelled probe, as described in Wellinger et al. (1993). Alternatively, single-stranded DNA was detected using the in-gel hybridization method described by Dionne and Wellinger (1996).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Leland Hartwell, Dan Gottschling, Stephen Elledge and Victoria Lundblad for the gifts of strains and plasmids. We thank Suzy Markossian and Armelle Roisin for operating the semi-automated DNA sequencer. This work was supported by grants from the Association pour la Recherche contre le Cancer, the Centre National de la Recherche Scientifique, programme Génome and the Comités Départementaux de l’Ardèche, la Loire et la Haute-Savoie de la Ligue Nationale contre le Cancer.
References
- Ausubel F.A., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1998) Current Protocols in Molecular Biology. John Wiley and Sons, New York, NY.
- Bertuch A. and Lundblad,V. (1998) Telomeres and double-strand breaks: trying to make the ends meet. Trends Cell Biol., 8, 339–342. [DOI] [PubMed] [Google Scholar]
- Bourns B.D., Alexander,M.K., Smith,A.M. and Zakian,V.A. (1998) Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol., 18, 5600–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede S.J. and Gottschling,D.E. (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell, 99, 723–733. [DOI] [PubMed] [Google Scholar]
- Dionne I. and Wellinger,R.J. (1996) Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl Acad. Sci. USA, 93, 13902–13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T., Becherer,K., Chen,P.-L., Yeh,S.-H., Yang,Y., Kilburn,A.E., Lee,W.-H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type I catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Evans S.K. and Lundblad,V. (1999) Est1 and Cdc13 as comediators of telomerase access. Science, 286, 117–120. [DOI] [PubMed] [Google Scholar]
- Evans S.K. and Lundblad,V. (2000) Positive and negative regulation of telomerase access to the telomere. J. Cell Sci., 113, 3357–3364. [DOI] [PubMed] [Google Scholar]
- Fields S. and Song,O. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Froelich-Ammon S.J., Dickinson,B.A., Bevilacqua,J.M., Schultz,S.C. and Cech,T.R. (1998) Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev., 12, 1504–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson,M. and Hartwell,L. (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol., 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Reed,S.I. and Charbonneau,M. (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev., 11, 512–527. [DOI] [PubMed] [Google Scholar]
- Greider C.W. (1996) Telomere length regulation. Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (1999) Sir-Ku-itous routes to make ends meet. Cell, 97, 829–832. [DOI] [PubMed] [Google Scholar]
- Horvath M.P., Schweiker,V.L., Bevilacqua,J.M., Ruggles,J.A. and Schultz,S.C. (1998) Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Evans,S.K., Weilbaecher,R.G. and Lundblad,V. (2000a) The Est3 protein is a subunit of yeast telomerase. Curr. Biol., 10, 809–812. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Weilbaecher,R.G., Walterscheid,M. and Lundblad,V. (2000b) Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl Acad. Sci. USA, 97, 6457–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter K.J. and Eipel,H.E. (1979) DNA determination of yeast by flow cytometry. J. Gen. Microbiol., 113, 369–375. [DOI] [PubMed] [Google Scholar]
- Krauskopf A. and Blackburn,E.H. (1998) Rap1 protein regulates telomere turnover in yeast. Proc. Natl Acad. Sci. USA, 95, 12486–12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay T.S., Morris,D.K., Sah,J., Balasubramanian,B. and Lundblad,V. (1996) Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics, 144, 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.-J. and Zakian,V.A. (1996) The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J. and Cech,T.R. (1998) Telomerase and chromosome end maintenance. Curr. Opin. Genet. Dev., 8, 226–232. [DOI] [PubMed] [Google Scholar]
- Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- Louis E.J. and Borts,R.H. (1995) A complete set of marked telomeres in Saccharomyces cerevisiae for physical mapping and cloning. Genetics, 139, 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V. and Blackburn,E.H. (1993) An alternative pathway for yeast telomere maintenance rescues est1– senescence. Cell, 73, 347–360. [DOI] [PubMed] [Google Scholar]
- Lundblad V. and Szostak,J.W. (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell, 57, 633–643. [DOI] [PubMed] [Google Scholar]
- Lustig A.J. (1998) Mechanisms of silencing in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev., 8, 233–239. [DOI] [PubMed] [Google Scholar]
- Nugent C.I. and Lundblad,V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Hughes,T.R., Lue,N.F. and Lundblad,V. (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- Qi H. and Zakian,V.A. (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated Est1 protein. Genes Dev., 14, 1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Rose M.D. and Broach,J.R. (1991) Cloning genes by complementation in yeast. In Guthrie,C. and Fink,G.R. (eds), Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA, pp. 195–230. [DOI] [PubMed]
- Singer M.S. and Gottschling,D.E. (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science, 266, 404–409. [DOI] [PubMed] [Google Scholar]
- Stavenhagen J.B. and Zakian,V.A. (1998) Yeast telomeres exert a position effect on recombination between internal tracts of yeast telomeric DNA. Genes Dev., 12, 3044–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S.-C. and Zakian,V.A. (1999) Telomere–telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Virta-Pearlman V., Morris,D.K. and Lundblad,V. (1996) Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev., 10, 3094–3104. [DOI] [PubMed] [Google Scholar]
- Weinert T.A. and Hartwell,L.H. (1988) The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science, 241, 317–322. [DOI] [PubMed] [Google Scholar]
- Wellinger R.J., Wolf,A.J. and Zakian,V.A. (1993) Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell, 72, 51–60. [DOI] [PubMed] [Google Scholar]
- Wotton D. and Shore,D. (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev., 11, 748–760. [DOI] [PubMed] [Google Scholar]
- Zakian V.A. (1996) Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet., 30, 141–172. [DOI] [PubMed] [Google Scholar]
- Zhou J., Hidaka,K. and Futcher,B. (2000) The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol. Cell. Biol., 20, 1947–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]