Abstract
RNAs in Physarum mitochondria contain extra nucleotides that are not encoded by the mitochondrial genome, at least in the traditional sense. While it is known that insertion of non-encoded nucleotides is linked to RNA synthesis, the exact nature of this relationship remains unclear. Here we demonstrate that the efficiency of editing is sensitive not only to the concentration of the nucleotide that is inserted, but also to the concentration of the nucleotide templated just downstream of an editing site. These data strongly support a co-transcriptional mechanism of Physarum RNA editing in which non-encoded nucleotides are added to the 3′ end of nascent RNAs. These results also suggest that transcription elongation and nucleotide insertion are competing processes and that recognition of editing sites most likely involves transient pausing by the Physarum mitochondrial RNA polymerase. In addition, the pattern of nucleotide concentration effects, the context of editing sites and the accuracy of the mitochondrial RNA polymerase argue that the mechanism of Physarum editing is distinct from that of other co-transcriptional editing systems.
Keywords: mitochondrial RNA polymerase/pausing/stuttering/transcription elongation
Introduction
RNA editing entails the revision of genomic information at the RNA level. This can occur via two general mechanisms: the insertion/deletion of nucleotides or the alteration of one base to another (reviewed in Cattaneo, 1991; Bass, 1993). Editing is widespread, occurring in a variety of single cell eukaryotes, viruses, Drosophila, plants, snails, worms and mammals, and examples of nuclear, cytoplasmic, mitochondrial and plastid editing have been described (reviewed in Smith et al., 1997). Perhaps not surprisingly, there are almost as many different editing mechanisms as there are systems that are known to edit their RNAs (Gott and Emeson, 2000).
One of the more complex patterns of RNA editing is found in the acellular slime mold Physarum polycephalum (reviewed in Gott, 2000). Nearly all mitochondrial RNAs in Physarum are subject to extensive editing, with highly specific nucleotide insertions occurring, on average, every 25 nucleotides (nt) in mRNAs and every 40 nt in rRNAs and tRNAs (Miller et al., 1993). Cytidine (C) insertion is by far the most frequent event (∼90%), although the addition of uridine (U), adenosine (A) and guanosine (G) residues and apparent C to U changes have also been described (Mahendran et al., 1991, 1994; Gott et al., 1993; Antes et al., 1998; Wang et al., 1999; Horton and Landweber, 2000). Insertional editing is predicted to be essential for survival, given that editing is required for the production of long open reading frames and generation of conserved features within the structural RNAs in these mitochondria (Gott, 2000). In support of this, >98% of insertion sites within steady-state RNAs are accurately and efficiently edited at every stage of the Physarum life cycle (Rundquist and Gott, 1995).
Mechanistic studies using isolated Physarum mitochondria have led to a number of insights into the means by which non-encoded nucleotides are added to Physarum RNAs. We have determined that nascent RNAs are the substrates for nucleotide insertion (Visomirski-Robic and Gott, 1995) and that editing occurs within the last 14 nt of nascent transcripts (Visomirski-Robic and Gott, 1997a). Although unedited RNA can be made if the concentration of the nucleotide to be inserted at a particular site is limited, once made, unedited RNA can not be chased into an edited species (Visomirski-Robic and Gott, 1997b). These findings argue that there is only a limited window of opportunity in which editing can occur, and that the editing process has a 5′→3′ polarity (Visomirski-Robic and Gott, 1997b).
Although our previous experiments led to the conclusion that extra nucleotides are added very close to the site of transcription, we were not able to determine whether nucleotides are inserted internally or at the 3′ end of the growing RNA chain. However, in studies in which unedited and partially edited RNAs were produced due to limiting nucleotide concentrations, sites that were edited to the greatest extent appeared to correspond to positions of polymerase stalling, suggesting a kinetic relationship between editing and transcription (Visomirski-Robic, 1997). Based on precedents in transcription elongation, attenuation and termination (Landick et al., 1996; von Hippel, 1998), slowing transcription just at the point at which editing occurs would be predicted to affect nucleotide insertion to a much greater extent than reducing elongation rates at other locations. We therefore reasoned that it might be possible to ascertain the point of nucleotide insertion by systematically altering nucleotide concentrations. If non-encoded residues are added to the 3′ end of nascent RNAs, reducing the concentration of the encoded nucleotide immediately 3′ of an editing site should enhance editing at that site, while if nucleotides are added internally, a different pattern involving nucleotides templated downstream might emerge. Thus, by examining nucleotide concentration effects, it should be possible to discriminate between internal insertion (via cleavage– ligation or transesterification pathways) and co-transcriptional models of Physarum editing.
Here we demonstrate that the concentration of the nucleotide encoded immediately 3′ of an editing site significantly affects editing efficiency at that site and that the pattern of nucleotide concentration effects is not consistent with internal insertion. These data indicate that editing and transcription are mechanistically linked and strongly support a co-transcriptional model for Physarum mitochondrial editing in which non-encoded nucleotides are added to the 3′ end of nascent RNA transcripts.
Results
Effect of CTP concentration on C insertion
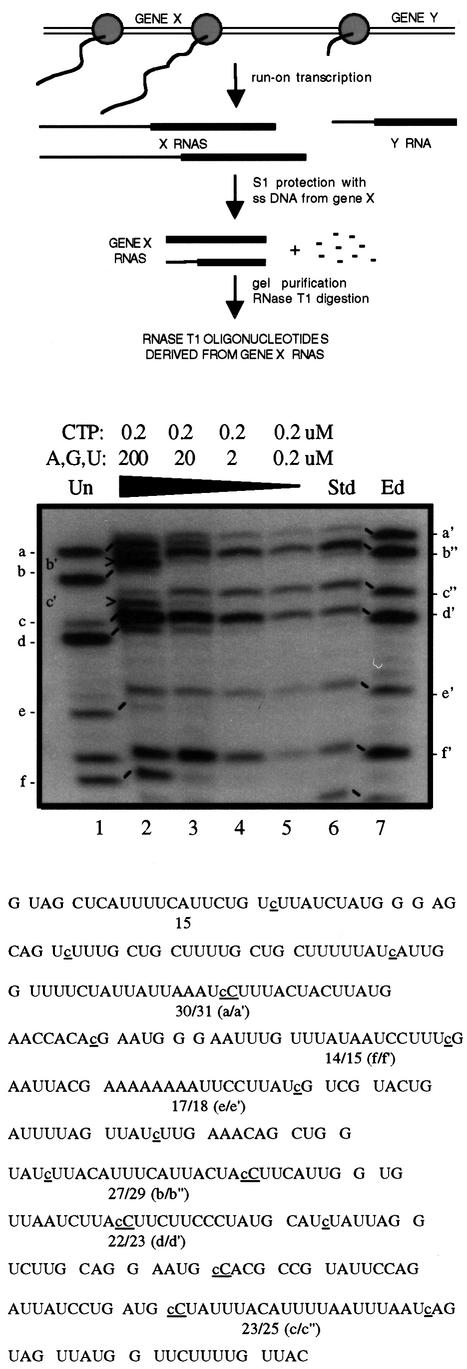
Since the only systems that currently support Physarum insertional editing in vitro are those derived from transcription complexes initiated on mitochondrial DNA in vivo (Visomirski-Robic and Gott, 1995; Cheng and Gott, 2000), we used an S1 protection protocol to assay for editing of nascent RNAs (Figure 1). In these experiments, run-on transcripts are 32P-labeled and the RNA of interest is isolated from the entire population of nascent mitochondrial RNAs via S1 nuclease protection. Hybrid-protected RNAs are then digested with RNase T1 and the resulting fragments are separated in either one or two dimensions to determine whether they contain added nucleotides. The identity and specificity of the added nucleotides are diagnosed by subsequent secondary analysis of the T1 fragments (Visomirski-Robic and Gott, 1997a). In the experiments shown here, we have examined the extent of editing at previously characterized C insertion sites within two genes, cytochrome c oxidase subunit I (coI; Gott et al., 1993) and α-atpase (Mahendran et al., 1991). Editing sites within the coI mRNA (Figure 1) are identified by letters to facilitate comparisons with previously published work (Visomirski-Robic and Gott, 1997b), while the insertion sites within the α-atpase mRNA (Figures 2 and 3; Table I; Supplementary data, available at The EMBO Journal Online) are numbered in a 5′ to 3′ direction, from editing site 1 (es1) to editing site 54 (es54).
Fig. 1. Effects of NTP concentration on C insertion within the coI gene. Top: schematic representation of the editing assay. Middle: separation of RNase T1 fragments from hybrid-protected coI mRNAs synthesized under varying NTP concentrations in isolated mitochondria. Lanes 1 and 7, digestion patterns of unedited (lane 1) and edited (lane 7) S1-protected control transcripts. Lanes 2–6, RNAs synthesized in isolated mitochondria in the presence of the indicated nucleotides. Lane 2, 0.2 µM [α-32P]CTP and 200 µM ATP, GTP, UTP; lane 3, 0.2 µM [α-32P]CTP and 20 µM ATP, GTP, UTP; lane 4, 0.2 µM [α-32P]CTP and 2 µM ATP, GTP, UTP; lane 5, 0.2 µM [α-32P]CTP and 0.2 µM ATP, GTP, UTP; and lane 6, 0.2 µM [α-32P]GTP and 200 µM ATP, CTP, UTP. Each reaction was pulse-labeled for 3 min, then chased for 12 min with 200 µM NTPs. Note that due to differences in salt concentrations, the control digests migrate with a slightly increased mobility relative to the mitochondrial samples in this experiment. Bottom: sequence of the region of the coI mRNA analyzed, with individual RNase T1 fragments indicated. Inserted C residues are underlined, lower case letters. Oligonucleotides containing sites of nucleotide insertion, which differ in length in the two control RNAs, are labeled with lower case letters; each prime (′) designates an added nucleotide. Oligonucleotides b′ and c′, which are RNase T1 fragments containing one, rather than two, added nucleotides (Visomirski-Robic and Gott, 1997b), are indicated by arrowheads. Note that band f′ consists of a non-edited 15 nt oligonucleotide as well as the edited version of oligonucleotide f. Because RNA synthesis is extremely limited under these conditions and only RNAs that were extended to the end of the S1 probe were isolated for RNase T1 analysis, fragments near the 3′ end of the hybrid-protected RNA are more heavily labeled than the 5′ fragments, which are less likely to have been fully extended.
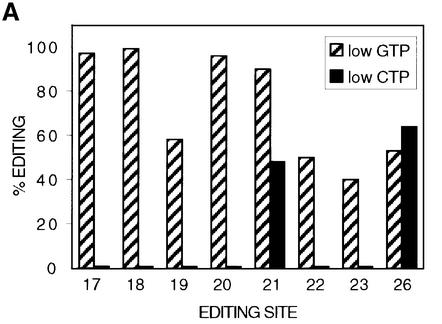
Fig. 2. C insertion editing in mtTEC is sensitive to the concentration of CTP. (A) Quantification of the extent of editing at C insertion sites within the α-atpase mRNA under low GTP (hatched bars) or low CTP (solid bars) conditions. RNAs synthesized by mtTEC in the presence of 20 µM [α-32P]GTP, 500 µM CTP, ATP and UTP or 20 µM [α-32P]CTP, 500 µM GTP, ATP and UTP were hybrid protected and digested with RNase T1 as described for Figure 1, and the resulting oligonucleotides separated in two dimensions. The RNA fingerprints from which the data are derived are presented as Supplementary data. The extent of editing at sites 24 and 25 is not included in the graph because the edited and unedited versions of the RNase T1 oligonucleotides containing these sites do not resolve well on RNA fingerprints. (B) Sequence of the region of the α-atpase mRNA analyzed, encompassing editing sites 17–26 of the α-atpase mRNA, with individual RNase T1 oligonucleotides indicated. Sites of C insertion (es17–es26) are underlined.
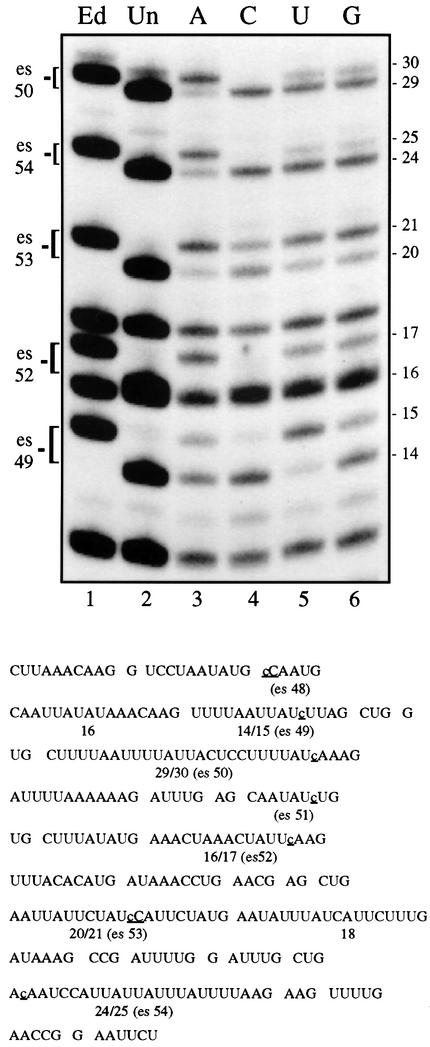
Fig. 3. The effect of nucleotide concentration on C insertion is context dependent. Lanes 1 and 2, digestion patterns of edited (lane 1) and unedited (lane 2) S1-protected control transcripts. Lanes 3–6, RNase T1 products from S1 nuclease-protected α-atpase transcripts synthesized by mtTEC under varying NTP concentrations. Lane 3, 20 µM ATP, 30 µM [α-32P]GTP, 500 µM CTP, UTP; lane 4, 20 µM CTP, 30 µM [α-32P]GTP, 500 µM ATP, UTP; lane 5, 20 µM UTP, 30 µM [α-32P]GTP, 500 µM CTP, ATP; lane 6, 30 µM [α-32P]GTP, 500 µM CTP, UTP, ATP. Editing sites (es #) within RNase T1 fragments resolved on this gel are indicated at the left. Bottom: sequence of the region analyzed, encompassing editing sites 48–54 of the α-atpase mRNA, with individual RNase T1 oligonucleotides indicated. Sites of C insertion are underlined.
Table I. Efficiency of editing at sites of C insertion within the α-atpase mRNA at various nucleotide concentrations.
| Edit site | 20 µM [NTP] | Extent edited | 3′ flanking nucleotides |
|---|---|---|---|
| 17 | ( G) | +++ | cGUUGAUUCUAUGUUA |
| (A ) | + | ||
| (C ) | - | ||
| 18 | ( G) | +++ | cGGACGUGGUCAAAGG |
| (A ) | ++ | ||
| (C ) | - | ||
| 19 | (A ) | +++ | cAAACAGGUAAAACUA |
| ( G) | ++ | ||
| (C ) | - | ||
| 20 | ( G) | +++ | cGAUACUAUUCUUAAU |
| (A ) | ++ | ||
| (C ) | - | ||
| 22 | ( G) | ++ | cUAUUGUGUGUAUGUU |
| (A ) | + | ||
| (C ) | - | ||
| 23 | ( G) | ++ | cUUGAAUAUUCAAACU |
| (A ) | ++ | ||
| (C ) | - | ||
| 33 | (U+G) | +++ | cUUUUAUGCUCAUUCA |
| ( G) | ++ | ||
| 35 | (AUG)a | +++ | cACUAAUUUCGGUGGA |
| (AUG) | +++ | ||
| (A+G) | +++ | ||
| (A ) | +++ | ||
| (A ) | +++ | ||
| (U ) | ++ | ||
| 40 | (U+G) | +++ | cUAUUCUUUAAAGGUA |
| (U ) | +++ | ||
| (AUG) | +++ | ||
| (A ) | ++ | ||
| (A+G) | ++ | ||
| ( G) | ++ | ||
| ( G) | ++ | ||
| (A ) | + | ||
| 49 | (U ) | +++ | cUUAGCUGGUGCUUUU |
| (U+G)b | +++ | ||
| (A ) | ++ | ||
| (A+G)b | + | ||
| ( G)b | + | ||
| (C+G)b | - | ||
| (C ) | - | ||
| 50 | (A+G)b | +++ | cAAAGAUUUUAAAAAA |
| (A ) | +++ | ||
| ( G)b | + | ||
| (U+G)b | + | ||
| (U ) | + | ||
| (C+G)b | - | ||
| (C ) | - | ||
| 52 | (A ) | +++ | cAAGUUUACACAUGAU |
| (A+G)b | +++ | ||
| ( G) | ++ | ||
| (U+G)b | ++ | ||
| (U ) | ++ | ||
| (C+G)b | - | ||
| (C ) | - | ||
| 54 | (A+G)b | +++ | cAAUCCAUUAUUAUUU |
| (A ) | +++ | ||
| (U ) | + | ||
| (U+G)b | + | ||
| ( G)b | + | ||
| (C+G)b | - | ||
| (C ) | - |
aATP, UTP and GTP at 10 µM.
bGTP at 30 µM.
Key: –, <10% editing; +, 10–35% editing; ++, 35–65% editing; +++, >65% editing.
Under our standard labeling conditions, nascent RNAs synthesized in isolated mitochondria are fully edited (Figure 1, lane 6; Visomirski-Robic and Gott, 1995). However, we have shown previously that insertion of non-encoded C residues is significantly reduced at low CTP concentrations (Visomirski-Robic and Gott, 1997b). This effect is illustrated in lane 2 of Figure 1, where labeled RNAs were synthesized using 0.2 µM CTP and relatively high levels (200 µM) of the other three nucleotides, and then protected using antisense ssDNA corresponding to a 346 nt region of the coI mRNA. Under these conditions, the efficiency of C insertional editing is reduced relative to our standard conditions (see, for example, bands a/a′, c/c′′ and f/f′ in Figure 1, lanes 2 versus 6). The extent of editing also varies between sites, as shown by the presence of partially edited versions of bands b and c, which each contain two C insertion sites. Since editing and transcription are linked in Physarum mitochondria (Visomirski-Robic and Gott, 1997b), one possible explanation for the different extents of editing is that the transcription/editing machinery has a longer dwell time at sites that are more highly edited. This model predicts that a slower elongation rate should lead to an increase in the efficiency of C insertion under conditions where CTP is limiting.
Editing can be ‘rescued’ by reducing the concentration of transcription substrates
To determine whether reducing the rate of transcription would enhance editing under low CTP conditions, the concentrations of the other three nucleotides were reduced while keeping that of CTP at 0.2 µM (Figure 1, lanes 3–5). Significantly, as the concentration of ATP, UTP and GTP in the reaction is lowered in the presence of 0.2 µM CTP, the extent of editing increases at each site, with full editing observed when the concentration of each of the four nucleotides in the reaction is 0.2 µM (Figure 1, lane 5) (compare with our standard editing conditions in lane 6). A similar increase in editing efficiency was also observed when the concentration of the other three nucleotides was lowered in the presence of either 2 or 20 µM CTP (data not shown). Thus, the low efficiency of C insertion under low CTP conditions is not simply due to use of a concentration of CTP that is significantly below the Km for the editing enzyme, but instead depends on the concentration of each of the nucleotides in the reaction.
The results in Figure 1 indicated that editing efficiency can be affected by changing parameters of the transcription reaction, and suggested that experiments in which the concentrations of individual nucleotides were altered might yield information regarding the precise point at which non-encoded nucleotides are inserted within the nascent transcript. However, a drawback to using isolated mitochondria for these experiments is that even after preincubation to deplete nucleotide pools, mitochondria still contain significant levels of endogenous nucleotides, particularly ATP and GTP (L.M.Visomirski-Robic, unpublished data). We therefore decided to pursue these observations in a recently developed in vitro system that allows the precise manipulation of nucleotide concentrations (Cheng and Gott, 2000). These preparations of partially purified mitochondrial transcription elongation complexes (mtTECs) yield run-on transcripts derived from the endogenous mitochondrial DNA and retain editing activity, but contain significantly reduced nucleotide pools.
C insertion in mtTEC preparations is sensitive to the concentration of CTP
Before using isolated transcription complexes to investigate the effects of altering the concentration of individual nucleotides, we first examined whether C insertion by mtTECs responds to alterations in CTP concentration in a manner similar to that of isolated mitochondria. To test this, the extent of editing of hybrid-protected transcripts generated under a variety of conditions was analyzed. The data for two experiments using different CTP concentrations (20 and 500 µM) are summarized in Figure 2. In these experiments, editing was examined in a region of the highly expressed α-atpase gene that includes 10 C insertion sites (editing sites 17–26). Because these editing sites fall within RNase T1 fragments of similar size, oligonucleotides derived from these hybrid-protected transcripts were separated in two dimensions and compared with RNase T1 fingerprints of unedited and edited control transcripts (see Supplementary data). The efficiency of nucleotide insertion at eight of these sites was then determined by quantification of phosphoimager scans of individual RNA fingerprints (Figure 2).
Although RNAs synthesized by mtTECs are not completely edited (Cheng and Gott, 2000), most sites in this region of the α-atpase gene are edited to a considerable extent under conditions where the concentrations of CTP, ATP and UTP are high (500 µM) and GTP is low (20 µM) (Figure 2, hatched bars). Sites 17, 18, 20 and 21 are efficiently edited and sites 19, 22, 23 and 26 are edited to ∼40–55% under these conditions. Consistent with the data shown here, substantial editing is also observed in other regions of the α-atpase mRNA under a variety of conditions when CTP is present at high concentrations (Cheng and Gott, 2000 and see below).
To determine whether editing in mtTECs was affected by lowering CTP concentrations, a parallel experiment was carried out using low levels of CTP (20 µM) and high levels (500 µM) of the other three nucleotides. As expected, a different pattern of editing was seen under these conditions. Most C insertion sites in this (Figure 2, solid bars) and other (see below) regions of the α-atpase mRNA are not edited when CTP concentrations are low. Indeed, of the sites shown in Figure 2, only sites 21 and 26 are edited to an appreciable extent (50–65%) at 20 µM CTP. Thus, editing at C insertion sites is also sensitive to the concentration of CTP in these mtTEC preparations.
A curious feature of the data in Figure 2 is the bimodal nature of CTP effects at individual editing sites. Both of the sites that are efficiently edited at low CTP concentrations [sites 21 (UUAUAcCAAUG) and 26 (CAAcCG)] are sites where a non-encoded C is added next to an encoded C residue, whereas none of the other editing sites analyzed in Figure 2 is found in this context. Of course, in these experiments we can not distinguish which is the inserted C and which is the templated C. However, coupled with the finding that a reduction in the concentration of transcription substrates enhances editing (Figure 1), these observations suggest that flanking nucleotides can influence the Physarum editing reaction.
The effect of NTP concentration on editing efficiency is context dependent
To test directly for effects of neighboring residues on C insertion, we have reduced the concentration of individual nucleotides and examined the extent of editing in a variety of contexts. In the experiment shown in Figure 3, four separate reactions were carried out using mtTECs, each labeled with 30 µM [α-32P]GTP in the presence of a different nucleotide present at low concentration, but equivalent results were also obtained at higher GTP concentrations (Table I). Here a different 287 nt region of the α-atpase mRNA was hybrid protected and subjected to RNase T1 analysis, then run on a denaturing gel to facilitate the side-by-side comparison of the efficiency of editing at five different C insertion sites (editing sites 49, 50, 52, 53 and 54).
Strikingly, no two conditions yield the same pattern of editing. Similarly to what was seen in the previous experiment (Figure 2), when the concentration of CTP is held low (Figure 3, lane 4), most sites are completely unedited, except for an intermediate level (∼40%) of editing observed at site 53. In contrast, under conditions where the concentration of CTP is high (lanes 3, 5 and 6), all C insertion sites are at least partially edited, as has been observed previously (Cheng and Gott, 2000 and Figure 2). However, the efficiency of editing varies considerably between sites, depending on the identity of the nucleotide present at low concentration (Table I). When the concentration of ATP is low (Figure 3, lane 3), sites 50, 54 and 52 are edited with high efficiency (70–90%), while extensive editing at site 49 is only seen under low UTP conditions (lane 5). Thus, while each editing site can be almost fully edited under defined reaction conditions, it is clear that optimal editing conditions vary from site to site.
If the data in Figure 3 are examined relative to the context of each of these editing sites, an intriguing pattern emerges. Consistent with our previous results, the only site in this region that is edited to a significant extent at low CTP concentrations (site 53) is the one that involves the insertion of a C next to an encoded C residue. Remarkably, the sites that are highly edited only under low ATP conditions (sites 50, 54 and 52) are all followed by encoded A residues. Similarly, site 49, which is followed by two U residues, is edited to the greatest extent when UTP is low. Finally, although none of the insertion sites in the region examined in Figure 3 has Gs immediately 3′ of the editing site, three of the C insertion sites examined in Figure 2 (editing sites 17, 18 and 20) precede a G residue. These sites are the most extensively edited sites in this region when GTP concentrations are limiting (Figure 2). Therefore, there is a strong inverse correlation between the extent of editing at a given site and the concentration of the nucleotide that is templated immediately 3′ of that site.
Using a combination of gel analyses and fingerprinting of RNAs synthesized by mtTECs, we have examined nucleotide concentration effects at 21 of the 54 C insertion sites within the α-atpase gene, including 13 unambiguous sites of C insertion (Table I). Under most conditions, each site is edited with a low to moderate efficiency (generally 10–50%). However, in every case where the concentration of the next templated nucleotide has been reduced, we see a substantial increase in the efficiency of editing at that site (to 70–95%). Reactions containing low concentrations of CTP present a special case, since CTP is needed for both editing and transcription, but the overall pattern is maintained. In these instances, low CTP concentrations result in essentially no editing for C insertion sites that are flanked by residues other than C (Table I), but 40–60% editing for sites that are adjacent to an encoded C residue (Figures 2 and 3 and data not shown). Thus, at sites where the templated nucleotide is also a C, editing efficiencies at low CTP concentrations approach those observed at high CTP concentrations in other contexts, but are relatively insensitive to the concentration of the other nucleotides. Interestingly, although we can not distinguish between the added and the encoded C residues in our current assays, these results suggest that in most cases the added C is likely to be the first, rather than the second C residue present in the RNA.
Comparable neighboring nucleotide effects have also been observed in isolated mitochondria when the concentration of individual nucleotides is varied in the context of low CTP, although in these experiments we could not definitively rule out the involvement of nucleotides encoded farther downstream (Visomirski-Robic, 1997). Because we have demonstrated previously that nucleotide insertion occurs within 14 nt of the 3′ end of nascent RNAs (Visomirski-Robic and Gott, 1997a), effects of pausing would be expected to occur somewhere within this window. However, no discernible pattern of nucleotide concentration effects other than the 3′ flanking nucleotide is apparent upon alignment of the regions downstream of C insertion sites (see Table I). For example, although the effects of ATP concentration on nucleotide insertion at site 54 could, in theory, be due to the presence of encoded As at downstream position 1, 2, 6, 9 or 12, the data for editing site 35 indicate that reducing the concentration of nucleotides encoded within a window of 6–14 nt away is not necessary for efficient editing. Similarly, windows between 2 and 6 nt downstream of editing sites are contradicted by data from multiple sites. Finally, the effects of limiting GTP concentration on editing at site 20 argue against pausing within the entire window between 2 and 19 nt downstream (Figure 2A; Table I), since there are no encoded G residues in this 3′ region. We therefore conclude that the effects observed are due to the identity of the nucleotide immediately 3′ of editing sites.
Physarum mitochondrial RNA polymerase is not prone to ‘stuttering’
The data presented above are most consistent with a co-transcriptional model of nucleotide insertion in Physarum mitochondria (see Discussion). The other known instance of co-transcriptional editing, insertion of G residues within paramyxoviral RNAs, is a result of ‘polymerase stuttering’ at a single AnGn run during RNA synthesis (Vidal et al., 1990). The viral polymerase also adds poly(A) tails to mRNAs by stuttering on a run of Us in the template (Jacques and Kolakofsky, 1991). A similar pseudo-templated transcription mechanism is likely to be responsible for the insertion of A residues at a single site within the Ebola virus glycoprotein (GP) RNA (Volchkov et al., 1995; Sanchez et al., 1996). Remarkably, the bacteriophage T7 RNA polymerase also adds extra nucleotides at this ‘slippery’ site (Volchkov et al., 1995). Based on the context of Physarum editing sites, the majority of which are not found within homopolymer tracts, it did not seem likely that a similar mechanism is used for the insertion of non-encoded nucleotides in Physarum mitochondria. However, because many other polymerases tend to slip and this tendency is often biologically relevant (Jacques and Kolakofsky, 1991; Uptain et al., 1997), it was of interest to determine whether the Physarum mitochondrial RNA polymerase (mtRNAP) is prone to such transcriptional errors.
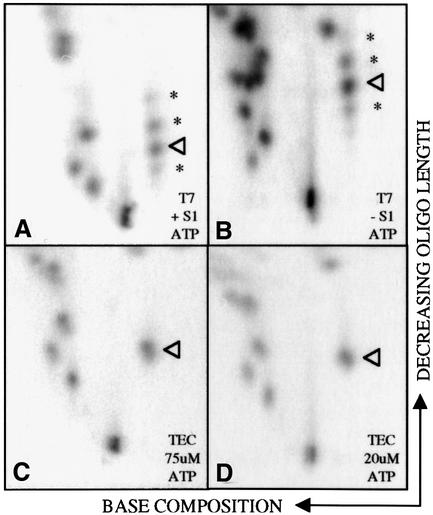
In the course of our analyses of RNA fingerprints used to generate the graphs in Figure 2, we found that when control transcripts were synthesized by T7 RNA polymerase under conditions of low ATP concentrations, a number of unexpected spots were present (* in Figure 4A). Based on their mobility, these extra RNase T1 fragments appeared to have a base composition similar to that of the oligonucleotide UCAAAAAAAAAG (arrowhead in Figure 4A), but were of different lengths (both shorter and longer). We hypothesized that these oligonucleotides were likely to result from slipping by T7 RNA polymerase during synthesis of the run of nine As at low ATP concentrations, since these spots were not observed in control transcripts synthesized in the presence of low concentrations of UTP, CTP or GTP (see Supplementary data). Secondary analyses of these extra RNase T1 fragments isolated from RNA fingerprints (data not shown) or a denaturing gel (see Supplementary data) are consistent with the insertion of extra or fewer A residues at this site. To ascertain whether our hybrid protection protocol would introduce bias, we also fingerprinted a control T7 transcript that had not been subjected to S1 digestion (Figure 4B). As anticipated based on the sequence of the control transcript, there are a number of additional spots present in this fingerprint relative to that in Figure 4A. Importantly, however, comparison of the S1-protected (Figure 4A) and undigested (Figure 4B) T7 transcripts showed that the distribution of ‘unexpected’ oligonucleotides (denoted by *) was not changed by S1 digestion.
Fig. 4. The Physarum mtRNAP does not stutter at a homopolymer tract. RNA fingerprints from an unedited T7 control transcript synthesized in the presence of 75 µM [α-32P]ATP, 500 µM CTP, GTP and UTP after (A) and before (B) S1 nuclease protection. The fingerprinted region is the same as that shown in the Supplementary data for Figure 2, but because all of the spots of interest fall within the lower third of the fingerprint, only this region is shown here to allow for enlargement. Mobility in the first dimension is determined largely by base composition, whereas migration in the second dimension is influenced primarily by size. Extra spots are indicated by asterisks. (C and D) Fingerprints of RNAs synthesized by mtTEC in the presence of 75 µM [α-32P]ATP, 500 µM CTP, GTP and UTP (C) or 20 µM [α-32P]ATP, 500 µM CTP, GTP and UTP (D). The sequence of the fingerprinted region is given in the bottom panel of Figure 2. The oligonucleotide indicated by the arrowhead has the sequence UCAAAAAAAAAG. Secondary analyses of the extra oligonucleotides are presented as Supplementary data.
Interestingly, despite the high sequence similarity between the T7 and Physarum mtRNAPs (A.Rhee, unpublished data), these additional RNase T1 oligonucleotides were not observed in RNAs synthesized by the Physarum polymerase at the same (Figure 4C) or even lower (Figure 4D) concentrations of ATP. Thus, although low overall nucleotide concentrations enhance nucleotide insertion at editing sites, reducing the rate of transcription does not lead to the incorporation of extra nucleotides at non-editing locations, even within homopolymer tracts, in Physarum mitochondrial transcripts.
Discussion
We have previously demonstrated that editing occurs within 14 nt of the 3′ end of nascent RNA and that once unedited RNA is synthesized, it is not chased into edited RNA (Visomirski-Robic and Gott, 1997a,b). These data indicated that there is a limited window in which editing can occur and that editing is coupled to transcription. Consistent with these results, when crude preparations of mtTECs are isolated via gel filtration, editing activity is found in the high molecular weight fractions (>20 × 106 kDa) that contain the transcription activity (Cheng and Gott, 2000). Here we further define the relationship between transcription and editing by demonstrating that the concentration of transcription substrates has a dramatic effect on the insertion of non-encoded nucleotides by the Physarum editing machinery. Specifically, we find that under transcription conditions that result in the production of partially edited and unedited RNAs, editing efficiency can be increased at each insertion site simply by decreasing the concentration of the next templated nucleotide.
The sensitivity of Physarum editing to the concentration of the residue encoded just downstream of insertion sites (Figures 2 and 3; Table I) is reminiscent of nucleotide concentration effects observed in studies of transcriptional pausing, attenuation, arrest and termination. For example, pausing by T7 RNA polymerase is enhanced by decreasing the concentration of the next required nucleotide, although, interestingly, limitation for a particular nucleotide does not induce pausing to the same degree at every occurrence of that residue (Levin and Chamberlin, 1987). Similarly, the pause half-life at the attenuation sites within the pyrBI (Turnbough et al., 1983) and trp (Landick and Yanofsky, 1984) operons increases when the concentration of the next nucleotide to be added is reduced, but is relatively insensitive to levels of the other three nucleotides. Experiments using mutant templates have demonstrated that pausing at the trp attenuator depends on the availability of the nucleotide that is immediately downstream of the pause site (Fisher et al., 1985). Transcription arrest by RNA polymerase II (pol II) within the adenovirus major late transcription unit also depends on limiting the concentration of a single nucleotide (Wiest and Hawley, 1990), and this nucleotide dependence changes with the identity of the nucleotide just downstream of the arrest site (Wiest et al., 1992). Finally, the concentration of the next nucleotide to be incorporated is an important determinant of termination efficiency for pol II (Kerppola and Kane, 1990) and Escherichia coli RNA polymerase at intrinsic terminators (McDowell et al., 1994; Wilson and von Hippel, 1994) and at the λ tR′ terminator (Rees et al., 1997). In each case, the effects of nucleotide concentration on termination efficiency have been attributed to a kinetic competition between elongation and transcript release (see also von Hippel and Yager, 1991). Based on these precedents, the effects we observe are most likely the result of a direct competition between transcription elongation and insertional editing, leading to the conclusion that non-encoded nucleotides are added to the growing 3′ end of Physarum mitochondrial RNAs.
While relative nucleotide concentrations also affect editing efficiency in other co-transcriptional editing systems, our data indicate that co-transcriptional nucleotide insertion in Physarum occurs through a distinct mechanism. Unlike what is seen in Physarum mitochondria, the frequency of G insertions into paramyxovirus RNA is increased at low GTP concentrations (Vidal et al., 1990). The differences between the two systems can readily be explained by the context of their respective insertion sites. In paramyxoviruses, editing is ‘pseudo-templated’, i.e. part of the viral template is read more than once (Hausmann et al., 1999). Because the same nucleotide is the substrate for both editing and transcription, low GTP concentrations are thought to enhance the chance of polymerase slippage, increasing the frequency of G insertions at the editing site (Vidal et al., 1990). However, while such a mechanism could account for the effect of low CTP concentrations at C insertion sites next to an encoded C in Physarum mitochondria, the pattern of C insertion at the majority of Physarum editing events, where neither of the flanking nucleotides is a C residue, does not resemble that observed in viral systems.
Our finding that non-encoded nucleotides are added to the 3′ end of nascent RNA has several additional implications for the mechanism of editing in Physarum. First, there must be a pause in elongation at editing sites to allow non-templated nucleotide addition. Secondly, the 3′ end of the RNA must be positioned to allow access to the editing activity. And thirdly, transcriptional elongation must resume from a nucleotide that is not paired (at least in a conventional manner) to the template. Each of these issues, which have obvious ramifications for transcription in general, is discussed in more detail below.
In order for a non-encoded nucleotide to be added at the 3′ end of the growing RNA chain, the progress of the Physarum mtRNAP along the DNA template must be temporarily halted. Pausing by the polymerase could be mediated through interactions with the template, nascent RNA and/or trans-acting factors (see Artsimovitch and Landick, 2000; Marr and Roberts, 2000). Based on work on both bacterial (Kassavetis and Chamberlin, 1981) and eukaryotic RNA polymerases (Kadesch and Chamberlin, 1982), a reduction in the concentration of available nucleotides would be expected to reduce the rate of RNA synthesis, potentially increasing the efficiency of pause site recognition at many sites along the template. Importantly, however, the effects of lowered nucleotide concentrations on insertional editing that we observe are not due simply to an overall reduction in the rate of RNA synthesis, since editing efficiency is clearly context dependent and differs even between adjacent sites. In addition, unlike T7 RNA polymerase, neither the Physarum mtRNAP nor the editing activity adds extra nucleotides at a long A run at low ATP concentrations (Figure 4A versus C and D), conditions that should increase the dwell time within this region. During the course of our work, we have characterized a large number of nascent transcripts synthesized under a variety of substrate concentrations in both isolated Physarum mitochondria (Visomirski-Robic and Gott, 1995, 1997a,b; Figure 1 of this work) and by transcription elongation complexes formed in vivo (Cheng and Gott, 2000; Figures 2–4; Table I). Our data indicate that the Physarum transcription and editing machineries are not error prone, even under conditions of severely limiting nucleotide concentrations. These results indicate that simply increasing the dwell time of the polymerase is not sufficient to cause nucleotide insertion and suggest that there are specific signals at or around editing sites. The nature of these signals has not yet been determined.
Co-transcriptional addition of non-encoded nucleotides also requires the specific positioning of the 3′ end of nascent RNAs in the editing active site. Although the enzymic activity responsible for the insertion of extra nucleotides into Physarum mitochondrial RNAs has not been identified, the three most likely possibilities are that non-templated nucleotide addition occurs in the context of (i) a slightly altered polymerization site of the mtRNAP, (ii) a second ‘insertion’ site within the mtRNAP or (iii) the active site of an editing enzyme associated with transcription complexes. Both DNA polymerases and aminoacyl tRNA synthetases have two distinct active sites, one that carries out the forward reaction, with the other required for proofreading or ‘editing’ (Silvian et al., 1999). During removal of misincorporated nucleotides by DNA polymerases, the nascent DNA strand is unwound by a few base-pairs to enable the polymerase to position the 3′ end in the editing site (Cowart et al., 1989). Rather than removing an incorrect nucleotide, however, the active site of the Physarum polymerase/editing enzyme would have to add an extra nucleotide in a highly specific manner. In this regard, editing could be viewed as a form of defined nucleotide ‘misincorporation’ (Thomas et al., 1998; von Hippel, 1998). The third possibility, addition of non-encoded nucleotides by a separate editing enzyme, would likely require backtracking of the mtRNAP in the vicinity of editing sites to allow access to the 3′ end of the nascent RNA. Lowered nucleotide concentrations would be expected to slow polymerase elongation (Kassavetis and Chamberlin, 1981), potentially enhancing backtracking (Komissarova and Kashlev, 1997) and affecting partitioning of the polymerase between different conformational states (Erie et al., 1993; Yin et al., 1999). These changes in the Physarum mitochondrial transcription complex would be predicted to allow the 3′ end of the nascent RNA to enter the active site of the editing enzyme and afford the editing activity a greater opportunity to add non-encoded nucleotides at those sites.
Once the non-encoded nucleotide is added to the growing RNA chain, this nucleotide must be extended in a template-directed manner, potentially requiring an altered conformation of the mtRNAP to align the unpaired nucleotide at the 3′ end of the transcript with the next (templated) NTP substrate. This scenario is in contrast to paramyxoviral editing, where the viral polymerase extends the RNA chain from an inserted nucleotide that is complementary to the template (Hausmann et al., 1999). Since we find that the Physarum mtRNAP is not prone to slippage, this suggests that the RNA–DNA hybrid may actually be stabilized in some way in Physarum mitochondrial transcription complexes. One model is that either the polymerase itself or a specific editing/elongation factor stabilizes the unpaired nucleotide in an everted conformation that would allow extension of the 3′ end of the RNA in a template-directed fashion. A second possibility is that the 3′ end of the RNA is subject to ‘scrunching’ at insertional editing sites, similar to what is proposed to occur with the 3′ end of tRNAs associated with CCA-adding enzyme (Shi et al., 1998) or the DNA within the active site pocket of T7 RNA polymerase (Cheetham and Steitz, 1999).
In addition to the activity that inserts the non-encoded nucleotides, additional trans-acting factors may be required to allow recognition of pause sites, specification of the added nucleotide and/or resumption of elongation after non-templated nucleotide addition. Many proteins are known to influence transcription elongation, some of which exert their influence via the RNA polymerase. Such factors include the prokaryotic elongation factors GreA (Borukhov et al., 1992) and GreB (Borukhov et al., 1993), eukaryotic elongation factors such as TFIIS (Reines, 1992), TFIIF (Flores et al., 1989), elongin (Bradsher et al., 1993) and ELL (Shilatifard et al., 1996), and proteins involved in termination, antitermination and the recognition of specific pause sites (Barik et al., 1987; Liu et al., 1996; Burns et al., 1998; Yarnell and Roberts, 1999; Bae et al., 2000; Pasman and von Hippel, 2000). If there are separate editing/elongation factors in Physarum, they are likely to be tightly associated with the mitochondrial transcription machinery, since preliminary experiments have demonstrated that editing activity is not removed by rinsing mtTECs with 0.2% Sarkosyl, and the two activities remain associated upon further fractionation on glycerol gradients (Y.-W.Cheng, unpublished data).
Clearly, factors other than nucleotide concentration play a role in editing in vivo. Editing activity associated with mtTECs is more sensitive to nucleotide concentrations than are reactions in isolated mitochondria, which synthesize fully edited RNAs under most experimental conditions (Visomirski-Robic and Gott, 1995). This difference may be due to the absence of nucleotide pools in mtTEC preparations (Cheng and Gott, 2000), which could affect both elongation rates and pausing efficiencies. However, because qualitatively similar flanking nucleotide effects are also observed in isolated mitochondria (Visomirski-Robic, 1997), we feel that conclusions based on findings in mtTECs accurately reflect the editing process in Physarum mitochondria. In support of this, the incomplete editing observed in mtTECs is not due to the lack of site-specific factors, because under appropriate conditions every site can be extensively edited (see Figures 2 and 3; Table I). Instead, we think it likely that, by manipulating nucleotide concentrations in these experiments, we have mimicked the effects of other events, such as pausing by the Physarum mtRNAP, which actually influence editing efficiency in vivo.
Materials and methods
All experimental procedures were carried out as described for mitochondria by Visomirski-Robic and Gott (1995) and for mtTEC by Cheng and Gott (2000).
In vitro transcription of control RNAs
Control RNAs were produced using the Ambion Maxiscript in vitro transcription kit and linearized templates using 75 µM limiting nucleotide and 500 µM other nucleotides unless otherwise noted. Control RNAs were derived from the following regions: Figure 1, nt 85–1778 (edited, E), nt 82–1716 (unedited, U) of the coI gene; Figures 2 and 4, nt 6–785 (E), nt 6–759 (U) of the α-atpase gene; Figure 3, nt 877–1631 (E), nt 848–1577 (U) of the α-atpase gene.
mtTEC transcription and RNA isolation
Transcription reactions using mtTEC preparations were carried out in 20 mM Tris–HCl pH 8.0, 10 mM MgCl2, 100 µg/ml bovine serum albumin (BSA), 2 mM dithiothreitol (DTT), 500 µM unlabeled nucleotides using 0.2–0.3 mg/ml mtTECs for 40 min at 30°C; then chased for 10 min with 500 µM limiting nucleotide. The concentration of labeled nucleotide varied between experiments. See figure legends for details.
S1 nuclease digestion
S1 nuclease digestions were performed at 26°C for 1.5 h using S1 mapping buffer (0.75 M NaCl, 0.05 M sodium acetate pH 4.5, 4.5 mM ZnSO4) containing 200–300 U of S1 nuclease (Roche). ssDNAs were derived from the following regions: Figure 1, nt 443–906 of coI; Figures 2 and 4, nt 507–785 (E), nt 491–759 (U) of α-atpase; Figure 3, nt 1345–1631 (E), nt 1298–1577 (U) of α-atpase.
RNase T1 digestion
Gel-purified S1-protected RNAs were resuspended in 5 µl of dH2O, heated to 95°C for 2 min, then put on ice. After the addition of 1 µl of 1 mg/ml tRNA (Sigma) to each sample, RNAs were digested for 45 min at 37°C with 1 µl of 100 U/µl RNase T1 (Roche). RNase T1 fragments were separated in either one dimension [on 20% polyacrylamide, 7 M urea, 50 mM Tris, 50 mM boric acid, 1 mM EDTA pH 8.3 (TBE) gels] or in two dimensions (RNA fingerprints) as described by Visomirski-Robic and Gott (1995, 1997a). The extent of editing at each site was based on analyses of two to seven reactions. Because a few oligonucleotides that contain sites of editing do not completely resolve from other fragments (see es52 in Figure 3), the extent of editing at individual sites was assessed by a combination of methods, including quantification of phosphoimager scans, autoradiographs and secondary analyses of fingerprint spots and bands eluted from denaturing gels. The data in Table I are therefore presented in qualitative, rather than quantitative, terms. However, as can be seen in Figure 3 and the Supplementary data, the differences between levels of editing designated: –, <10%; +, 10–35%; ++, 35–65% and +++, >65% are readily apparent and highly reproducible.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Adam Majewski for providing some of the data presented in Table I and David Setzer, Donal Luse, Timothy Nilsen, Mark Caprara and the members of our laboratory for discussions, advice and critical reading of the manuscript. We also thank the reviewers of this manuscript for their insightful suggestions. This work was supported by grants to J.M.G. from NSF (MCB-9630672) and NIH (GM54663).
References
- Antes T., Costandy,H., Mahendran,R., Spottswood,M. and Miller,D. (1998) Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol. Cell. Biol., 18, 7521–7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I. and Landick,R. (2000) Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl Acad. Sci. USA, 97, 7090–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W., Xia,B., Inouye,M. and Severinov,K. (2000) Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl Acad. Sci. USA, 97, 7784–7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S., Ghosh,B., Whalen,W., Lazinski,D. and Das,A. (1987) An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell, 50, 885–899. [DOI] [PubMed] [Google Scholar]
- Bass B.L. (1993) RNA editing: new uses for old players in the RNA world. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 383–418.
- Borukhov S., Polyakov,A., Nikiforov,V. and Goldfarb,A. (1992) GreA protein: A transcription elongation factor from Escherichia coli. Proc. Natl Acad. Sci. USA, 89, 8899–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S., Sagitov,V. and Goldfarb,A. (1993) Transcript cleavage factors from E. coli. Cell, 72, 459–466. [DOI] [PubMed] [Google Scholar]
- Bradsher J.N., Tan,S., McLaury,H.-J., Conaway,J.W. and Conaway,R.C. (1993) RNA polymerase II transcription factor SIII. II. Functional properties and role in RNA chain elongation. J. Biol. Chem., 268, 25594–25603. [PubMed] [Google Scholar]
- Burns C.M., Richardson,L.V. and Richardson,J.P. (1998) Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J. Mol. Biol., 278, 307–316. [DOI] [PubMed] [Google Scholar]
- Cattaneo R. (1991) Different types of messenger RNA editing. Annu. Rev. Genet., 25, 71–88. [DOI] [PubMed] [Google Scholar]
- Cheetham G.M.T. and Steitz,T.A. (1999) Structure of a transcribing T7 RNA polymerase initiation complex. Science, 286, 2305–2309. [DOI] [PubMed] [Google Scholar]
- Cheng Y.-W. and Gott,J.M. (2000) Transcription and RNA editing in a soluble in vitro system from Physarum mitochondria. Nucleic Acids Res., 28, 3695–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart M., Gibson,K.J., Allen,D.J. and Benkovic,S.J. (1989) DNA substrate structural requirements for the exonuclease and polymerase activities of procaryotic and phage DNA polymerases. Biochemistry, 28, 1975–1983. [DOI] [PubMed] [Google Scholar]
- Erie D.A., Hajiseyedjavadi,O., Young,M.C. and von Hippel,P.H. (1993) Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science, 262, 867–873. [DOI] [PubMed] [Google Scholar]
- Fisher R.F., Das,A., Kolter,R., Winkler,M.E. and Yanofsky,C. (1985) Analysis of the requirements for transcription pausing in the tryptophan operon. J. Mol. Biol., 182, 397–409. [DOI] [PubMed] [Google Scholar]
- Flores O., Maldonado,E. and Reinberg,D. (1989) Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J. Biol. Chem., 264, 8913–8921. [PubMed] [Google Scholar]
- Gott J.M. (2000) RNA editing in Physarum polycephalum. In Bass,B. (ed.), RNA Editing: Frontiers in Molecular Biology. Oxford University Press, Oxford, UK, pp. 20–37.
- Gott J.M. and Emeson,R.B. (2000) Functions and mechanisms of RNA editing. Annu. Rev. Genet., 34, 499–531. [DOI] [PubMed] [Google Scholar]
- Gott J.M., Visomirski,L.M. and Hunter,J.L. (1993) Substitutional and insertional RNA editing of the cytochrome c oxidase subunit 1 mRNA of Physarum polycephalum. J. Biol. Chem., 268, 25483–25486. [PubMed] [Google Scholar]
- Hausmann S., Garcin,D., Delenda,C. and Kolakofsky,D. (1999) The versatility of paramyxovirus RNA polymerase stuttering. J. Virol., 73, 5568–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton T.L. and Landweber,L.F. (2000) Evolution of four types of RNA editing in myxomycetes. RNA, 6, 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques J.-P. and Kolakofsky,D. (1991) Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev., 5, 707–713. [DOI] [PubMed] [Google Scholar]
- Kadesch T.R. and Chamberlin,M.J. (1982) Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J. Biol. Chem., 257, 5286–5295. [PubMed] [Google Scholar]
- Kassavetis G.A. and Chamberlin,M.J. (1981) Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J. Biol. Chem., 256, 2777–2786. [PubMed] [Google Scholar]
- Kerppola T.K. and Kane,C.M. (1990) Analysis of the signals for transcription termination by purified RNA polymerase II. Biochemistry, 29, 269–278. [DOI] [PubMed] [Google Scholar]
- Komissarova N. and Kashlev,M. (1997) Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl Acad. Sci. USA, 94, 1755–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R. and Yanofsky,C. (1984) Stability of an RNA secondary structure affects in vitro transcription pausing in the trp operon leader region. J. Biol. Chem., 259, 11550–11555. [PubMed] [Google Scholar]
- Landick R., Turnbough,C.L. and Yanofsky,C. (1996) Transcription attenuation. In Neidhart,F. (ed.), Escherichia coli and Salmonella. ASM Press, Washington, DC, pp. 1263–1286.
- Levin J.R. and Chamberlin,M.J. (1987) Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J. Mol. Biol., 196, 61–84. [DOI] [PubMed] [Google Scholar]
- Liu K.B., Zhang,Y.Y., Severinov,K., Das,A. and Hanna,M.M. (1996) Role of Escherichia coli RNA polymerase α subunit in modulation of pausing, termination and anti-termination by the transcription elongation factor NusA. EMBO J., 15, 150–161. [PMC free article] [PubMed] [Google Scholar]
- Mahendran R., Spottswood,M.R. and Miller,D.L. (1991) RNA editing by cytidine insertion in mitochondria of Physarum polycephalum. Nature, 349, 434–438. [DOI] [PubMed] [Google Scholar]
- Mahendran R., Spottswood,M.S., Ghate,A., Ling,M.-L., Jeng,K. and Miller,D.L. (1994) Editing of the mitochondrial small subunit rRNA in Physarum polycephalum. EMBO J., 13, 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr M.T. and Roberts,J.W. (2000) Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell, 6, 1275–1285. [DOI] [PubMed] [Google Scholar]
- McDowell J.C., Roberts,J.W., Jin,D.J. and Gross,C. (1994) Determin ation of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science, 266, 822–825. [DOI] [PubMed] [Google Scholar]
- Miller D., Mahendran,R., Spottswood,M., Costandy,H., Wang,S., Ling,M.-L. and Yang,N. (1993) Insertional editing in mitochondria of Physarum. Semin. Cell Biol., 4, 261–266. [DOI] [PubMed] [Google Scholar]
- Pasman Z. and von Hippel,P.H. (2000) Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry, 39, 5573–5585. [DOI] [PubMed] [Google Scholar]
- Rees W.A., Weitzel,S.E., Das,A. and von Hippel,P.H. (1997) Regulation of the elongation-termination decision at intrinsic terminators by antitermination protein N of phage lambda. J. Mol. Biol., 273, 797–813. [DOI] [PubMed] [Google Scholar]
- Reines D. (1992) Elongation factor-dependent transcript shortening by template-engaged RNA polymerase II. J. Biol. Chem., 267, 3795–3800. [PMC free article] [PubMed] [Google Scholar]
- Rundquist B.A. and Gott,J.M. (1995) RNA editing of the coI mRNA throughout the life cycle of Physarum polycephalum. Mol. Gen. Genet., 247, 306–311. [DOI] [PubMed] [Google Scholar]
- Sanchez A., Trappier,S.G., Mahy,B.W.J., Peters,C.J. and Nichol,S.T. (1996) The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl Acad. Sci. USA, 93, 3602–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P.-Y., Maizels,N. and Weiner,A.M. (1998) CCA addition by tRNA nucleotidyltransferase: polymerization without translocation? EMBO J., 17, 3197–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A., Lane,W.S., Jackson,K.W., Conaway,R.C. and Conaway,J.W. (1996) An RNA polymerase II elongation factor encoded by the human ELL gene. Science, 271, 1873–1876. [DOI] [PubMed] [Google Scholar]
- Silvian L.F., Wang,J. and Steitz,T.A. (1999) Insights into editing from an Ile-tRNA synthetase structure with tRNA-Ile and mupirocin. Science, 285, 1074–1077. [PubMed] [Google Scholar]
- Smith H.C., Gott,J.M. and Hanson,M.R. (1997) A guide to RNA editing. RNA, 3, 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- Thomas M.J., Platas,A.A., and Hawley,D.K. (1998) Transcriptional fidelity and proofreading by RNA polymerase II. Cell, 93, 627–637. [DOI] [PubMed] [Google Scholar]
- Turnbough C.L., Hicks,K.L. and Donahue,J.P. (1983) Attenuation control of pyrBI operon expression in Escherichia coli K-12. Proc. Natl Acad. Sci. USA, 80, 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain S.M., Kane,C.M. and Chamberlin,M.J. (1997) Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem., 66, 117–172. [DOI] [PubMed] [Google Scholar]
- Vidal S., Curran,J. and Kolakosky,D. (1990) A stuttering model for paramyxovirus P mRNA editing. EMBO J., 9, 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic L.M. (1997) Examination of insertional editing in Physarum polycephalum in an isolated mitochondrial system: a potential coupling of editing and transcription. PhD thesis, Case Western Reserve University.
- Visomirski-Robic L.M. and Gott,J.M. (1995) Accurate and efficient insertional RNA editing in isolated Physarum mitochondria. RNA, 1, 681–691. [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic L.M. and Gott,J.M. (1997a) Insertional editing of nascent mitochondrial RNAs in Physarum. Proc. Natl Acad. Sci. USA, 94, 4324–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visomirski-Robic L.M. and Gott,J.M. (1997b) Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis. RNA, 3, 821–837. [PMC free article] [PubMed] [Google Scholar]
- Volchkov V.E., Becker,S., Volchkova,V.A., Ternovoj,V.A., Kotov,A.N., Netesov,S.V. and Klenk,H.D. (1995) GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology, 214, 421–430. [DOI] [PubMed] [Google Scholar]
- von Hippel P.H. (1998) An integrated model of the transcription complex in elongation, termination, and editing. Science, 281, 660–665. [DOI] [PubMed] [Google Scholar]
- von Hippel P.H. and Yager,T.D. (1991) Transcript elongation and termination are competitive processes. Proc. Natl Acad. Sci. USA, 88, 2307–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.S., Mahendran,R. and Miller,D. (1999) Editing of cytochrome b mRNA in Physarum mitochondria. J. Biol. Chem., 274, 2725–2731. [DOI] [PubMed] [Google Scholar]
- Wiest D.K. and Hawley,D.K. (1990) In vitro analysis of a transcription termination site for RNA polymerase II. Mol. Cell. Biol., 10, 5782–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest D.K., Wang,D. and Hawley,D.K. (1992) Mechanistic studies of transcription arrest at the adenovirus major late attenuation site. J. Biol. Chem., 267, 7733–7744. [PubMed] [Google Scholar]
- Wilson K.S. and von Hippel,P.H. (1994) Stability of Escherichia coli transcription complexes near an intrinsic terminator. J. Mol. Biol., 244, 36–51. [DOI] [PubMed] [Google Scholar]
- Yarnell W.S. and Roberts,J.W. (1999) Mechanism of intrinsic transcription termination and antitermination. Science, 284, 611–615. [DOI] [PubMed] [Google Scholar]
- Yin H., Artsimovitch,I., Landick,R. and Gelles,J. (1999) Nonequilibrium mechanism of transcription termination from observations of single RNA polymerase molecules. Proc. Natl Acad. Sci. USA, 96, 13124–13129. [DOI] [PMC free article] [PubMed] [Google Scholar]