Abstract
The precise position of the replication origin (OR) of mtDNA was determined for insect species belonging to four different orders (four species of Drosophila, Bombyx mori, Triborium castaneum, and Locusta migratoria, which belong to Diptera, Lepidoptera, Coleoptera, and Orthoptera, respectively). Since the free 5′ ends of the DNA strands of mtDNA are interpreted as the OR, their positions were mapped at 1-nucleotide resolution within the A + T-rich region by using the ligation-mediated PCR method. In all species examined, the free 5′ ends were found within a very narrow range of several nucleotides in the A + T-rich region. For four species of Drosophila, B. mori, and T. castaneum, which belong to holometabolous insects, although the OR's were located at different positions, they were located immediately downstream of a series of thymine nucleotides, the so-called T-stretch. These results strongly indicate that the T-stretch is involved in the recognition of the OR of mtDNA at least among holometabolous insects. For L. migratoria (hemimetabolous insect), on the other hand, none of the long stretches of T's was found in the upstream portion of the OR, suggesting that the regulatory sequences involved in the replication initiation process have changed through insect evolution.
METAZOAN mitochondrial DNA (mtDNA) is a closed-circular, double-stranded molecule, ranging in size from 15 to 20 kb (Wolstenholme 1992). It contains a distinct replication origin (OR) on each of the DNA strands. The position of the OR and mode of replication have been examined in detail in mammalian mtDNA. The round of replication begins from the OR of the H-strand and DNA synthesis proceeds unidirectionally. When the synthesis of the H-strand reaches two-thirds of the genome, the synthesis of the L-strand is initiated from the OR of the L-strand located two-thirds of the genomic distance away from the OR of the H-strand. Synthesis of the L-strand also proceeds unidirectionally (Shadel and Clayton 1997; Taanman 1999).
Initiation of mtDNA replication is controlled by the interaction between nuclear-encoded proteins and regulatory sequences existing on the mtDNA (Clayton 1982; Shadel and Clayton 1997; Taanman 1999). Several regulatory sequences have been identified in the noncoding region of the vertebrate mtDNA. These are located around the OR and are widely conserved among vertebrate species. Immediately upstream of the OR of the H-strand, conserved sequence blocks (CSBs) are present and are suggested to be responsible for the initiation of the H-strand replication (Tapper and Clayton 1981; Chang and Clayton 1985; Chang et al. 1985; Cairns and Bogenhagen 1986; King and Low 1987; Kang et al. 1997). CSBs are thought to be involved in generating the 3′ ends of the RNA primers, which are required for the DNA synthesis of the H-strand (Shadel and Clayton 1997; Taanman 1999). Around the OR of the L-strand, the sequence that could form a stem-loop configuration is conserved among vertebrate species and is also suggested to be required for the initiation of replication (Martens and Clayton 1979; Tapper and Clayton 1981; Hixson et al. 1986). In vitro replication studies have suggested that this structure serves as the recognition structure for mtDNA primase, which provides a short RNA primer for L-strand synthesis, and DNA synthesis is initiated near the base of the stem-loop structure utilizing the 3′ ends of the RNA primer (Hixson et al. 1986).
In invertebrate species, Drosophila, a member of the protostome species, is the only organism in which the position of the OR and mode of replication have been examined. In several Drosophila species, replicative intermediates have been observed by electron microscopy (Goddard and Wolstenholme 1978, 1980). In Drosophila mtDNA, the leading strand and lagging strand are termed minor and major coding strands according to the relative numbers of the gene encoded on the respective DNA strand (Garesse 1988). The OR for the minor coding strand (ON) is located somewhere in the middle portion of the large noncoding region called the “A + T-rich region” due to its extremely high adenine and thymine contents. However, observation using electron microscopy did not allow determination of the precise location of the ON. Synthesis of the minor coding strand proceeds unidirectionally, and the major coding strand synthesis begins after 97% of the minor coding strand synthesis is completed (Goddard and Wolstenholme 1978, 1980). The position of the OR for the major coding strand (OJ) is unknown.
Since the A + T-rich region contains the ON, this region is thought to be involved in controlling of mtDNA replication in Drosophila (Wolstenholme 1992). However, the regulatory sequences involved in replication initiation have not been identified; thus, the role of the control region in the replication initiation process of invertebrate mtDNA is poorly understood. Comparative sequence analyses of the A + T-rich region have been carried out, and the conserved sequences among Drosophila species were also found (Clary and Wolstenholme 1987; Monforte et al. 1993; Lewis et al. 1994; Inohira et al. 1997; Brehm et al. 2001; Tsujino et al. 2002). These conserved sequences are proposed to play a role in regulation of mtDNA replication. However, they differ from conserved regulatory sequences reported in the vertebrate mtDNA. These observations indicate that regulatory sequences of mtDNA are different in invertebrate and vertebrate species and therefore suggest that the regulatory systems have changed through the evolution of animals.
Regulatory sequences are expected to exist near the OR as in the case of vertebrate mtDNA (Martens and Clayton 1979; Tapper and Clayton 1981; Walberg and Clayton 1981). Thus, the OR mapping on the nucleotide level is a necessary step for identifying potential regulatory sequences in the replication initiation process. In this study, we first attempted to determine the position of the OR for four Drosophila species (Drosophila yakuba, D. obscura, D. albomicans, and D. virilis). Since replication of Drosophila mtDNA proceeds unidirectionally from the OR (Goddard and Wolstenholme 1978, 1980), nascent DNA strands possess the free 5′ ends at the OR. Therefore we determined the position of the free 5′ ends of the DNA strand at 1-nucleotide resolution within the A + T-rich region for both strands, using the ligation-mediated PCR (LMPCR) method (Mueller et al. 1992; Kang et al. 1997). Then, the nucleotide sequences around the OR were compared among the four Drosophila species to identify potential regulatory sequences that may be involved in the replication initiation process. The relationships between the OR and conserved structure previously reported in the A + T-rich region of mtDNA in Drosophila are also discussed (Clary and Wolstenholme 1987; Monforte et al. 1993; Lewis et al. 1994; Inohira et al. 1997; Brehm et al. 2001; Tsujino et al. 2002). In addition, we also examined the position of the OR for three other insect species belonging to different orders: silkworm, Bombyx mori (Lepidoptera); red flour beetle, Triborium castaneum (Coleoptera); and migratory locust, Locusta migratoria (Orthoptera). Including Drosophila, which belongs to Diptera, the flanking nucleotide sequences of the OR were compared among species belonging to the four different orders that are phylogenetically distantly related to one another. Our data provide new insights into the evolutionary changes of the regulatory sequences involved in replication of insect mtDNA.
MATERIALS AND METHODS
Insect species:
D. yakuba and D. obscura belong to the subgenus Sophophora and are members of the D. melanogaster and the D. obscura species groups, respectively. D. albomicans and D. virilis belong to the subgenus Drosophila and are members of the D. immigrans and the D. virilis species groups, respectively. The isofemale line of D. yakuba was derived from Nairobi (Kenya), D. obscura was from Tuebingen (Germany), and D. albomicans was from San-hu-tang (Taiwan). The strain of D. virilis is an inbred strain. Silkworm B. mori, Red flour beetle T. castaneum, and migratory locust L. migratoria belong to Lepidoptera, Coleoptera, and Orthoptera, respectively. The B. mori strain is kinshu × showa, which was purchased from Ueda Sanshu. The T. castaneum strain has been maintained in a laboratory for >25 years. L. migratoria was collected from Tokyo and Tateyama, Japan.
Isolation of mtDNA:
Mitochondrial DNA was extracted according to the method described by Tamura and Aotsuka (1988), omitting the alkaline lysis procedure. In the case of Drosophila, live adult flies (1.0 g) were homogenized in 40 ml of a chilled buffer containing 0.25 m sucrose, 10 mm EDTA, and 30 mm Tris-HCl (pH 7.5). The homogenate was centrifuged at 1000 × g for 5 min at 4°, and the supernatant was retained. The centrifugation process was repeated three to four times. The resulting supernatant was centrifuged at 12,000 × g for 10 min at 4°, and the pellet was resuspended in 4 ml of a buffer containing 0.15 m NaCl, 10 mm EDTA, and 10 mm Tris-HCl (pH 8.0). Then, 100 μl of 20% SDS was added to the mixture and incubated at room temperature for 30 min. DNA was extracted twice with phenol and once with phenol/chloroform and then precipitated with ethanol. The pellet was dried and dissolved in 30 μl of TE buffer containing 10 mm Tris-HCl (pH 8.0) and 0.1 mm EDTA. Then, DNA was treated with RNase A (20 μg/ml) at 37° for 30 min. In the case of T. castaneum, live adults were used, and for B. mori and L. migratoria, eggs from one female were used for mtDNA extraction.
Ligation-mediated PCR:
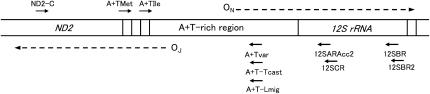
LMPCR was performed according to previously described methods (Mueller et al. 1992; Kang et al. 1997). The primers used in this study are shown in Figure 1. A unidirectional linker was prepared by hybridizing M13F20 (5′-GTTGTAAAACGACGGCCAGT-3′) and M13FC (5′-ACTGGCCG-3′). Primer extension was performed in 30 μl of reaction mixture containing 10 mm Tris-HCl (pH 8.5), 50 mm KCl, 2 mm MgCl2, 0.001% gelatin, 200 μm of each dNTP, 0.3 pmol of primer 1, mtDNA (∼15–80 ng), and 1.25 units of Taq DNA polymerase. The reaction conditions were 2 min at 95°, 30 min at 54°–64°, and 10 min at 72°. In the case of Drosophila species, the ND2-C primer (5′-GCTCTTAGTATTCATCCTAAATG-3′) was used as primer 1 for the major coding strand. The annealing temperature for primer extension was 54°. For the minor coding strand, the 12SBR primer (5′-AGCGACGGGCGATGTGTACA-5′) was used as primer 1 and the annealing temperature was 64°. In the case of B. mori, T. castaneum, and L. migratoria, the 12SBR2 primer (5′-GAAAGCGACGGGCAATATGT-3′) was used as primer 1 for the minor coding strand. The annealing temperature for primer extension was 60°. After the primer extension reaction, 0.00625 units of KOD polymerase (TOYOBO) was added to the mixture and incubated for 10 min at 68° to obtain the primer extension products with blunt ends. To ligate the unidirectional linker to the blunt end of the primer extension product, 43 μl of ligation mix was added to the mixture, which was then incubated at 16° for >12 hr. The ligation mix contained 52 mm Tris-HCl (pH 7.5), 14 mm MgCl2, 35 mm dithiothreitol (DTT), 19 μg/ml bovine serum albumin (BSA), 100 pmol of unidirectional linker, and 350 units of T4 DNA ligase. After the ligation reaction, DNA was purified according to the method of Boom et al. (1990). DNA was eluted in 20 μl of TE buffer.
Figure 1.
The locations of primers on the minor coding strand (top strand) and the major coding strand (bottom strand) of insect mtDNA around the OR. Arrows indicate primers with their names. The direction of replication from the OR of the minor (ON) and major (OJ) coding strands is indicated by arrows with dashed lines. ND2 is the gene of NADH dehydrogenase subunit 2.
To amplify the DNA, PCR was carried out in 30 μl of 10 mm Tris-HCl (pH 8.5), 50 mm KCl, 2 mm MgCl2, 0.001% gelatin, 200 μm of each dNTP, 5 pmol of the M13F20 primer, 5 pmol of primer 2, 0.5 μl of the purified primer extension product, and 1.88 units of Taq DNA polymerase. The reaction was carried out for 30 sec at 95°, for 1 min at 55°–65°, and for 1–2 min at 72° for 35 cycles, followed by an extension for 7 min at 72°. In the case of Drosophila species, the A + TMet primer (5′-GGTATGAACCCAGTAGTAGCTTA-3′) was used as primer 2 for the major coding strand. The annealing temperature was 55°. Using the PCR product obtained via the A + TMet primer as a template, the second PCR was further carried out with 5-carboxyfluorescein-labeled A + TIle primer (5′-GCATGATTTACCCTATCAAG-3′) as primer 3 to label the PCR products. The annealing temperature was 55°. For the minor coding strand, the 12SARAcc2 primer (5′-CCGCGAYTGCTGGCACCAAT-3′) for Drosophila, 12SBR for B. mori, and 12SCR (5′-ATAACCGCRACTGCTGGCAC-3′) for T. castaneum and L. migratoria were used as primer 2. The annealing temperatures were 64° for Drosophila, 60° for B. mori and T. castaneum, and 65° for L. migratoria. Using the PCR product obtained via primer 2, the second PCR was further carried out using the following 5-carboxyfluorescein-labeled primers as primer 3 to label the PCR products: The A + Tvar-yak primer (5′-CTAAATCTGATAACTTATTCCC-3′) was used for D. yakuba, the A + Tvar-obs primer (5′-ATAAATTTTATAGAACATTAACTT-3′) for D. obscura, the A + Tvar-alb primer (5′-AATTATTGTTCCCCTTATAGAC-3′) for D. albomicans, the A + Tvar-vir primer (5′-AAATTAATACCAATAACCCTA-3′) for D. virilis, the 12SCR primer for B. mori, the A + T-Tcast primer (5′-GCTCAGAAAATAATTTCTCTGC-3′) for T. castaneum, and the A + T-Lmig primer (5′-AGAATAATACTGCCCTCAGC-3′) for L. migratoria. The annealing temperatures were 50° for D. obscura and D. virilis, 55° for D. yakuba and D. albomicans, 60° for B. mori, and 58° for T. castaneum and L. migratoria. After the labeling reaction, 0.00625 unit of KOD polymerase was added to the PCR reaction mixture and incubated for 10 min at 68° to obtain the PCR products with blunt ends.
Determination of the positions of the free 5′ ends:
The positions of the free 5′ ends in all seven insect species were determined by the following method. The amplified PCR products obtained with the labeled primer were electrophoresed in a 5% acrylamide/6 m urea gel by using an ABI prism 377 sequencer (Perkin-Elmer, Norwalk, CT). A sequence ladder obtained with the same primer was applied in parallel with the PCR products to determine the positions of the free 5′ ends. The sequence reaction was performed with the DYEnamic Direct cycle sequence kit (Amersham, Arlington Heights, IL), using the DNA fragment containing the A + T-rich region as a template. The minor coding strand in D. obscura, D. albomicans, D. virilis, B. mori, T. castaneum, and L. migratoria and the major coding strand in D. albomicans were also determined by the following method. The amplified PCR product was cloned into the pUC118 or pUC119 plasmid vector and sequenced (Sambrook and Russell 2001).
Nucleotide sequence of the A + T-rich region:
For sequencing of the entire A + T-rich region of D. albomicans, mtDNA was digested with XbaI, and the DNA fragment containing the A + T-rich region was cloned into the plasmid vector pUC and sequenced (Sambrook and Russell 2001). For D. obscura, the entire A + T-rich region was amplified by PCR with primers 5′-CTGCATGATTTACCCTAT-3′ and 5′-GGAATTCTAAGCCAAAATAAAACTT-3′ and cloned into the pUC vector and sequenced. For L. migratoria, the DNA fragment containing the entire A + T-rich region and part of the 12S rRNA gene was amplified with the 12SBR and 5′-CAAGATAACCCTTTAATCAGGCA-3′ primers, and the nucleotide sequence of this DNA fragment was determined by direct sequencing with the 12SBR and (5′-TAGGGTATCTAATCCAGT-3′) primers. Sequence reactions were performed by using the ABI PRISM BigDye Terminator cycle sequencing kit (Perkin-Elmer) according to the supplier's instructions. Then nucleotide sequences were determined on an ABI PRISM 377 DNA sequencer (Perkin-Elmer). The nucleotide sequences determined in this study were deposited with the EMBL/GenBank Data Libraries under accession nos. AB198740, AB198741, and AB212084.
Nucleotide sequences of the A + T-rich region were obtained from GenBank for the following species: D. yakuba (NC001322), D. virilis (X05914), D. subobscura subL (AJ132900), D. subobscura subR (AJ132899), D. madeirensis (AJ132902), D. guanche (AJ132901), D. teissieri (X54011), D. melanogaster (U37541), D. simulans (AB003095, AB003096), D. mauritiana (AB003097, AB003098), B. mori (AB070264), and T. castaneum (AJ312413).
RESULTS
Positions of the free 5′ ends of the minor and major coding strands in insect mtDNA:
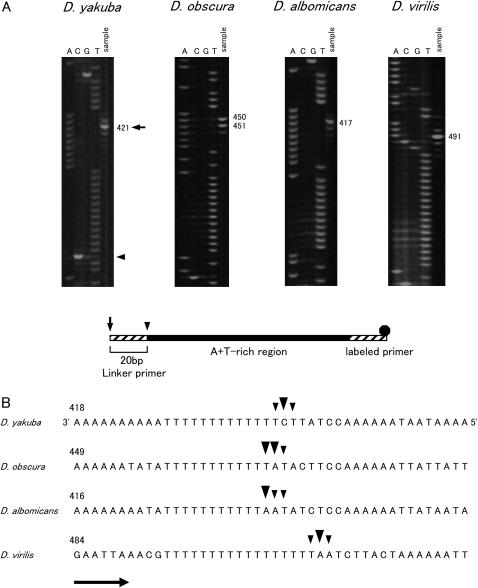
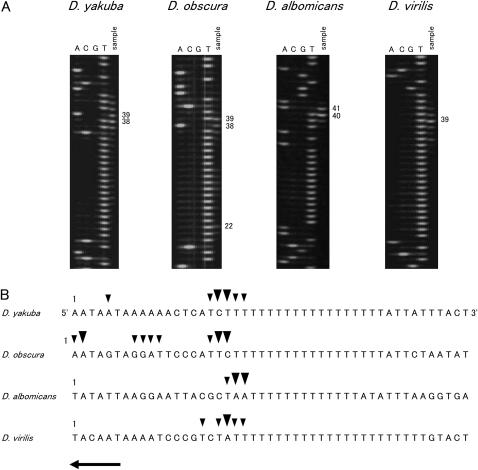
The positions of the free 5′ ends of the minor coding strand were determined in D. yakuba, D. obscura, D. albomicans, and D. virilis using LMPCR. Figure 1 shows the direction of the replication and the location of the primers used for LMPCR. Primer extension was carried out with the 12SBR primer and mtDNA as a template. Then, the DNA was amplified by PCR with the 12SARAcc2 primer. In each of the PCRs, the M13F20 primer was used as a linker primer. Only the nascent DNA strands initiated from the ON in the A + T-rich region and passing beyond the 3′ portion of the 12S rRNA gene were amplified by this primer set. DNA fragments of ∼850, 900, 660, and 730 bp were amplified for D. yakuba, D. obscura, D. albomicans, and D. virilis, respectively. The results indicate that the free 5′ ends were located in the middle portion of the A + T-rich region in all four species. Since the 12SARAcc2 primer was relatively far from the free 5′ ends (Figure 1), internal primers were used to determine the positions of the free 5′ ends more accurately. Thus, a second PCR was carried out with the labeled A + Tvar primers. The resultant PCR product was resolved by a denaturing gel with a sequence ladder that was applied in parallel to determine the positions of the free 5′ ends. Figure 2A shows the electrophoretogram and the schematic structure of the LMPCR product. The positions of the free 5′ ends (indicated by an arrowhead) were 20 bp downstream of the sites where signals were observed (indicated by an arrow) because linker primers were attached to the 5′ ends of the LMPCR products. The free 5′ ends are indicated above the nucleotide sequence in Figure 2B. The free 5′ ends were designated as minor or major ends according to the relative intensity of the bands. The free 5′ ends were mapped at three sites located adjacently for all four species examined. The major free 5′ ends were located 441 bp from the tRNAIle gene for D. yakuba, 470 and 471 bp for D. obscura, 437 bp for D. albomicans, and 511 bp for D. virilis in the middle portion of the A + T-rich region. The free 5′ ends were located at similar positions relative to the tRNAIle gene for D. yakuba and D. albomicans; however, for D. obscura and D. virilis they were located ∼30 and 70 bp downstream of the former two species, respectively.
Figure 2.
Mapping of the free 5′ ends for determining the position of the ON of Drosophila mtDNA. (A) The nascent DNA strands of the minor coding strand were amplified with 5-carboxyfluorescein-labeled primer, and amplified product was resolved in a 5% acrylamide/6 m urea gel (sample lane). Sequence ladders were applied in parallel (lanes A, C, G, and T). The nucleotide positions with major signals are indicated on the right. The schematic structure of LMPCR product is shown below. Primers, the portion of the A + T-rich region, and the fluorescent dye are shown as a hatched box, solid box, and solid octagon, respectively. Arrows and arrowheads indicate the 5′ ends of the PCR products and free 5′ ends of the nascent DNA strands. (B) Location of the free 5′ ends marking the ON. The site of the nucleotide with the free 5′ end is corrected by 20 bases for the length of the linker from the site where the signal was observed in A. Large arrowheads indicate the sites where major signals were observed and small arrowheads show the sites where minor signals were observed. Arrows indicate the direction of replication. The sequences of the template strands are in the middle portion of the A + T-rich region. The sequences shown for D. yakuba and D. virilis are from Clary and Wolstenholme (1985, 1987), respectively. The numbering above the sequence begins with the nucleotide that is next to the tRNAIle gene and proceeds through the A + T-rich region. Nucleotide position 1 for the sequence of D. yakuba corresponds to position 16019 of the sequence for the same species according to Clary and Wolstenholme (1985). The actual length of the T-stretch in the major coding strand for the strains in this study is 18 bp for D. virilis.
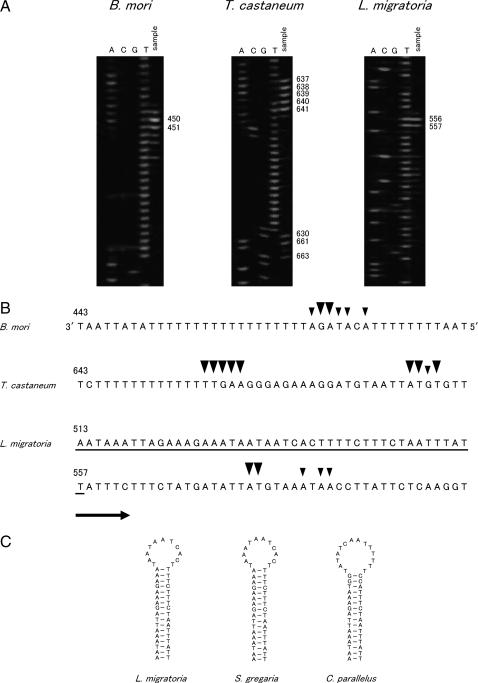
We further determined the positions of the free 5′ ends of the minor coding strand for three insect species belonging to different orders. Primer extension was carried out with the 12SBR2 primer (Figure 1). Then, DNA was amplified with the 12SBR primer for B. mori and with the 12SCR primer for T. castaneum and L. migratoria. DNA fragments of ∼760, 800, and 860 bp were obtained for B. mori, T. castaneum, and L. migratoria, respectively. To determine the positions of the free 5′ ends, sequential PCR was repeated using these PCR products as a template with the labeled 12SCR, A + T-Tcast, and A + T-Lmig primers for B. mori, T. castaneum, and L. migratoria, respectively. Figure 3 shows the electrophoretogram (Figure 3A) and the sites where the free 5′ ends were detected (Figure 3B). In each of the species, several signals were detected at adjacent sites. The major free 5′ ends were mapped at 470 and 471 bp from the tRNAMet gene for B. mori (in the case of B. mori, the A + T-rich region is flanked by the tRNAMet gene instead of by the tRNAIle gene); at 657–661, 680, 681, and 683 bp from the tRNAIle gene for T. castaneum; and at 576 and 577 bp from the tRNAIle gene for L. migratoria within the A + T-rich region. The positions of the free 5′ ends in the A + T-rich region are also indicated in Figure 4. The free 5′ ends were located near the 12S rRNA gene for B. mori and at the middle portion of the A + T-rich region for four species of Drosophila, T. castaneum, and L. migratoria. Therefore, the results of this study revealed that the ON is located within the A + T-rich region for insect species belonging to four different orders, Diptera, Lepidoptera, Coleoptera, and Orthoptera.
Figure 3.
Mapping of the free 5′ ends for determining the position of the ON of insect mtDNA. (A) The nascent DNA strands of the minor coding strand were amplified, and LMPCR product was resolved in a 5% acrylamide/6 m urea gel (sample lane) with the sequence ladders, which were applied in parallel (Lanes A, C, G, and T). (B) Location of the free 5′ ends marking the ON. The sequences of the template strands are the portion that is next to the 12S rRNA gene for B. mori and the middle portion for T. castaneum and L. migratoria in the A + T-rich region. The sequences shown for B. mori and T. castaneum are from Yukuhiro et al. (2002) and Friedrich and Muqim (2003), respectively. See Figure 2 for an explanation of symbols. (C) Possible stem-loop structures formed immediately upstream of the ON of L. migratoria mtDNA. The nucleotide sequence of L. migratoria, which potentially forms the stem-loop structure, is underlined in B. The possible stem-loop structures of S. gregaria and C. parallelus are from Zhang et al. (1995).
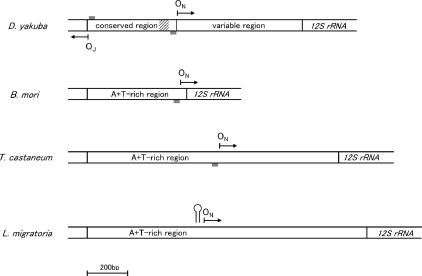
Figure 4.
The positions of the OR of mtDNA for insect species belonging to four different orders. The directions of replication from the ON and the OJ are indicated by arrows. Shaded boxes denote the T-stretch. A hatched box indicates the 50-bp portion highly conserved among Drosophila species in the A + T-rich region. The possible stem-loop structure is indicated immediately upstream of the ON of L. migratoria.
The positions of the free 5′ ends of the major coding strand in insect mtDNA have not previously been mapped. The positions of the free 5′ ends were determined in D. yakuba, D. obscura, D. albomicans, and D. virilis. Under the assumption that replication is initiated within the A + T-rich region (Figure 1), we carried out primer extension with the ND2-C primer. The DNA was then amplified with the A + TMet primer. Using this PCR product as a template, sequential PCR was repeated with labeled A + TIle primer. Figure 5 shows the electrophoretogram (A) and the sites where the free 5′ ends were detected (B). In each of the species, several signals were detected at adjacent sites. The major free 5′ ends were located 18 and 19 bp from the tRNAIle gene for D. yakuba; 2, 18, and 19 bp for D. obscura; 20 and 21 bp for D. albomicans; and 19 bp for D. virilis. The free 5′ ends were near the tRNAIle gene (Figure 4) and were located at similar positions in all four species.
Figure 5.
Mapping of the free 5′ ends for determining the position of the OJ of Drosophila mtDNA. (A) The nascent DNA strands of the major coding strand were amplified, and LMPCR product was resolved in a 5% acrylamide/6 m urea gel (sample lane) with the sequence ladders, which were applied in parallel (Lanes A, C, G, and T). (B) Location of the free 5′ ends marking the OJ. The sequences of the template strands are the portion that is next to the tRNAIle gene. The actual lengths of the T-stretch in the minor coding strands for the strains in this study are 17 and 24 nucleotides for D. yakuba and D. virilis, respectively. See Figure 2 for explanation of symbols.
The positions of the free 5′ ends were also examined by sequencing the LMPCR products for the minor coding strand in D. obscura, D. albomicans, D. virilis, B. mori, T. castaneum, and L. migratoria and for the major coding strand in D. albomicans. We cloned and sequenced the DNA fragments obtained by LMPCR and confirmed that the sites where the free 5′ ends were mapped corresponded to those detected using the labeled primer (data not shown).
Conservation of the T-stretch immediately upstream of the free 5′ ends of insect mtDNA:
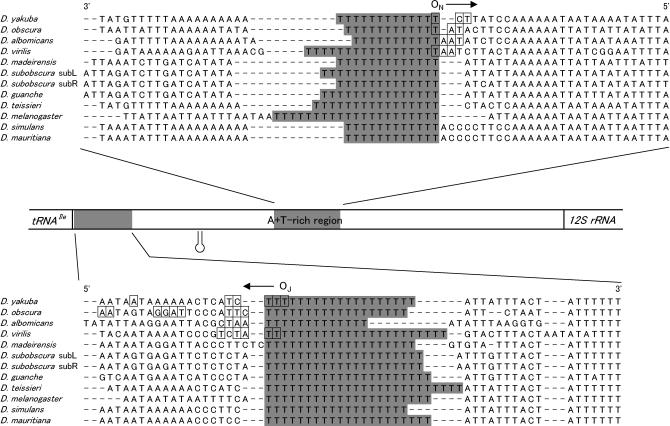
A consecutive group of thymine nucleotides, the so called “T-stretch,” is located immediately upstream of the sites where the free 5′ ends were found on the template strand in four Drosophila species examined (Figures 2B and 5B). The T-stretch is known to be conserved near the tRNAIle gene in the minor coding strand and at the center in the major coding strand of the A + T-rich region in all Drosophila species so far examined, although the position of the T-stretch in the major coding strand is not always confined to a narrow area (Clary and Wolstenholme 1985, 1987; Monnerot et al. 1990; Monforte et al. 1993; Lewis et al. 1994; Inohira et al. 1997; Brehm et al. 2001). Figure 6 shows the nucleotide sequences around the T-stretch for several Drosophila species that have been sequenced thus far. The nucleotide sequences are aligned with the T-stretches, and the nucleotides with boxes are the sites where the free 5′ ends were mapped for the four species in this study. The long T-stretch was also found immediately upstream of the ON for B. mori (18 bp in size) and T. castaneum (14 bp); however, none of the long stretches of T's was found in the upstream portion of the ON for L. migratoria (Figure 3B). The flanking nucleotide sequences of the ON were compared for four species of Drosophila, B. mori, T. castaneum, and L. migratoria; however, the portion excluding the T-stretch was highly variable among species belonging to different orders so that the nucleotide sequences were not reliably aligned. Therefore, none of the portions that are conserved among species belonging to different orders was found except for the T-stretch.
Figure 6.
The nucleotide sequences around the T-stretches and the positions of the OR of Drosophila mtDNA. The nucleotide sequences for several Drosophila species that were determined thus far were aligned with the T-stretches (shaded areas) on the major coding (top) and the minor coding (bottom) strands, respectively. The sites where the free 5′ ends were mapped for four Drosophila species in this study are indicated by boxes. The direction of replication is indicated by an arrow. The stem-loop structure previously proposed is also shown (Clary and Wolstenholme 1987; Monforte et al. 1993).
The length of the T-stretch varied among and within the species compared. The shortest T-stretch was 11 nucleotides on the major coding strand of D. albomicans and the longest was 25 nucleotides on the minor coding strand of D. teisseri (Figure 6). In D. subobscura, the length of the T-stretch on the major coding strand differed even within identical species, i.e., 15 nucleotides in strain subL and 13 nucleotides in strain subR.
DISCUSSION
Replication origin of insect mtDNA:
In this study, we reported for the first time the precise positions of the OR's of mtDNA by mapping the free 5′ ends at 1-nucleotide resolution for seven insect species that are phylogenetically distantly related (four species of Drosophila, B. mori, T. castaneum, and L. migratoria, which belong to Diptera, Lepidoptera, Coleoptera, and Orthoptera, respectively). In all species examined, the free 5′ ends of the minor coding strands were found in a very narrow range of several nucleotides in the A + T-rich region, located at the middle portion for four Drosophila species, T. castaneum, and L. migratoria and near the 12S rRNA gene for B. mori (Figures 2B, 3B, and 4). D. yakuba and D. virilis were used in a previous mapping study using electron microscopy, and the free 5′ ends were found in the middle of the A + T-rich region (Goddard and Wolstenholme 1980). Therefore, the results of the present study are consistent with the previous results.
The position of the OJ of Drosophila mtDNA has been unknown. However, we assumed that the OJ is also located within the A + T-rich region for the following reason. According to the strand-asynchronous, asymmetric model of vertebrate mtDNA, the replication of the L-strand is initiated when the synthesis of the H-strand passes beyond the L-strand origin, and the template strand for the L-strand replication becomes single stranded (Clayton 1982; Shadel and Clayton 1997; Taanman 1999). In Drosophila mtDNA, the replication of the major coding strand initiates after 97% of the minor coding synthesis is completed (Goddard and Wolstenholme 1978, 1980). If the replication mode is similar between both Drosophila and vertebrate mtDNAs, the OJ must be located 97% of the genomic distance away from the ON, that is, within the A + T-rich region. As expected, in this study, the free 5′ ends were found in the A + T-rich region near the tRNAIle gene (Figures 4 and 6), exactly 97% of the genomic distance away from the ON.
In animal mtDNA, a strand-asynchronous, asymmetric model of replication has been widely accepted (Clayton 1982; Shadel and Clayton 1997; Taanman 1999). However, recently in human and mouse mtDNA, a symmetrical model of replication has been proposed (Holt et al. 2000; Bowmaker et al. 2003). A symmetrical model differs greatly from a strand-asynchronous, asymmetric model in respect to the position of the OR and the direction of replication. In a symmetrical model of replication, the OR is located outside of the large noncoding region, the so-called D-loop region, and replication proceeds bidirectionally from the OR. Therefore, in mammalian mtDNA, controversy exists regarding the mode of replication. On the other hand, in the case of Drosophila, only a strand-asynchronous, asymmetric mode has been reported to date (Goddard and Wolstenholme 1978, 1980). The OR mapping in the present study also supported the finding that Drosophila mtDNA replicates with a strand-asynchronous, asymmetric mode.
Relationship between the OR and conserved sequences of the A + T-rich region of Drosophila mtDNA:
Sequence analyses of the A + T-rich region of Drosophila mtDNA thus far showed that the A + T-rich region is divided into two parts according to the extent of sequence conservation (Clary and Wolstenholme 1987; Monforte et al. 1993; Lewis et al. 1994). Half of the A + T-rich region (∼430 bp) flanked by the tRNAIle gene is conserved among species and termed the conserved region, while the remaining part flanked by the 12S rRNA gene is highly variable and termed the variable region (Monforte et al. 1993). The OR mapping in the present study showed that the OR's are located near both ends of the conserved region of the A + T-rich region (Figure 4). With respect to the ON, an ∼50-bp portion (corresponding to position 357–411 in the D. yakuba sequence) located upstream of the T-stretch in the major coding strand (which serves as the template strand for the replication of the minor coding strand) is the most conservative portion of the A + T-rich region (Figure 4; see also Clary and Wolstenholme 1987; Monforte et al. 1993; Inohira et al. 1997; Brehm et al. 2001). The degree of conservation of this 50-bp portion is approximately the same as that of the 12S rRNA gene of Drosophila mtDNA (Monforte et al. 1993), suggesting that this portion is subject to a strong functional constraint. The round of replication of Drosophila mtDNA begins from the ON (Goddard and Wolstenholme 1978, 1980); therefore, the total replication rate of mtDNA is determined by synthesis of the minor coding strand. Taking this fact into account, an ∼50-bp conserved portion may play some role in the regulation of replication initiation.
Within the conserved region of the A + T-rich region of Drosophila, two conserved structures were proposed to be involved in the replication initiation process. A possible stem-loop structure that is located in the A + T-rich region has been conserved among several Drosophila species and has been speculated to act as the OJ (Clary and Wolstenholme 1987; Monforte et al. 1993; Lewis et al. 1994; Inohira et al. 1997; Brehm et al. 2001; Tsujino et al. 2002). The precise mapping of the OJ of Drosophila mtDNA in the present study revealed that the OJ was located near the tRNAIle gene and was ∼230–240 bp away from the previously proposed stem-loop structures (Figure 6). Therefore, a direct involvement of this stem-loop structure in replication initiation of Drosophila mtDNA is not supported by the present results.
Another conserved structure proposed to serve as the OR was the T-stretch (Lewis et al. 1994). Lewis et al. analyzed the nucleotide sequence of the A + T-rich region of D. melanogaster mtDNA. The A + T-rich region of this species is ∼4.6 kb long and is organized into two large arrays of tandemly repeated sequences, type I (corresponding to the variable region) and type II (corresponding to the conserved region) repeats. Type I repeats are tandemly repeated five times and located in half of the A + T-rich region adjacent to the 12S rRNA gene, and type II repeats are tandemly repeated four times and once partially and located adjacent to the tRNAIle gene. If the recognition sequence for replication initiation is included in the repeated sequence, the OR's must also be situated tandemly in D. melanogaster mtDNA. However, the ON has been found only at the central portion of the A + T-rich region by electron microscopy (Goddard and Wolstenholme 1978). The prominent T-stretch in the major coding strand is located at the central intervening portion between the type I and II repeats and is excluded from the repeated sequences. The T-stretch in the minor coding strand is located near the tRNAIle gene and is also excluded from the repeated sequence. Therefore, the T-stretch has been proposed to be involved in the replication initiation process (Lewis et al. 1994). The precise mapping of the OR of Drosophila mtDNA in the present study revealed that the T-stretches are located immediately upstream of the OR of both strands in all four species (Figures 2B and 5B). Furthermore, the T-stretches of several other Drosophila species that are not used in the present mapping study are also located at the center in the major coding strand and near the tRNAIle gene in the minor coding strand (Figure 6; see also Clary and Wolstenholme 1985, 1987; Monnerot et al. 1990; Monforte et al. 1993; Lewis et al. 1994; Inohira et al. 1997; Brehm et al. 2001; Tsujino et al. 2002). Therefore, the T-stretches are conserved at the positions where the OR's are located for four Drosophila species that are used in the present mapping study. This strongly suggests that the T-stretch is involved in the recognition of the OR of Drosophila mtDNA.
Possible function of the T-stretch of Drosophila mtDNA:
Thymine nucleotide stretches are also found at the OR's of other systems. They are located immediately upstream of the L-strand origin of the mammalian mtDNA (Clayton 1982), upstream of the OR of colicin E1 (Tomizawa et al. 1977), and in the core origin of simian virus 40 (Deb et al. 1986). In the case of simian virus 40, mutational analysis showed that the consecutive thymine nucleotides play an essential role in replication initiation (Deb et al. 1986). It has been shown in in vitro experiments that the consecutive thymine nucleotides induce a bending of the DNA (Kornberg and Baker 1992; Sinden 1994). In simian virus 40, DNA bending of the T-stretch was shown to be required for binding of a protein that is essential for the initiation of replication (Sinden 1994). The T-stretch is thought to serve as a structural signal for the recognition of proteins involved in replication initiation. In Drosophila, the T-stretches most likely also play an important role as a recognition sequence in replication initiation of mtDNA.
In the case of mammalian mtDNA, the T-stretch is located in the loop portion of the possible stem-loop structure in human, bovine, and mouse mtDNA and varies in size from 6 to 11 bp (Anderson et al. 1982). In human mtDNA, initiation of DNA synthesis requires a DNA primase, responsible for generating short RNA molecules with 5′ ends that map to the T-stretch. DNA synthesis initiates from the sites near the base of the stem of the secondary structure, utilizing the free 3′ ends of primer RNA (Hixson et al. 1986). In the case of Drosophila, a stem-loop structure conserved among Drosophila species was not predicted by a computer program such as MFOLD version 3.1 (Zuker et al. 1999). Therefore, the existence of the stem-loop structure around the OR remains ambiguous in Drosophila mtDNA. However, the fact that mammals and Drosophila share the T-stretch immediately upstream of the OR suggests that the T-stretch of Drosophila mtDNA may be involved in primer RNA synthesis for replication initiation. Precise mapping of both the 5′ and 3′ ends of RNA may provide important clues for understanding the function of the T-stretch and the replication initiation process. Although studies examining the protein-DNA interaction in the A + T-rich region are essential for understanding the replication initiation process of Drosophila mtDNA, only a few studies have been performed (Potter et al. 1980; Pardue et al. 1984). The identification of potentially functional sequences, such as the T-stretch, may provide useful information for analyzing the DNA-protein interaction of the Drosophila A + T-rich region in future studies.
Evolution of the nucleotide sequence around the OR of insect mtDNA:
In this study, flanking nucleotide sequences of the ON were compared among species belonging to four different insect orders to understand the evolutionary changes of the regulatory sequences involved in the replication initiation process of insect mtDNA. From phylogenetic analysis using the morphological and molecular data, Orthoptera (hemimetabolous insect) is thought to have diverged first from the other three insect orders. Then, the Coleoptera diverged relatively early in holometabolous insect evolution, and Lepidoptera and Diptera diverged most recently (Carmean et al. 1992; Wheeler et al. 2001). For Drosophila species (Diptera), B. mori (Lepidoptera), and T. castaneum (Coleoptera), the long T-stretch was located immediately upstream of the OR, while for L. migratoria (Orthoptera), none of the long stretches of T's was found in the upstream portion of the OR (Figures 3 and 6). The former three species that belong to holometabolous insects shared the common feature immediately upstream of the OR; therefore, the long T-stretch seems to be conserved in holometabolous insects. To verify this idea, we searched for the T-stretch within the A + T-rich region of insect mtDNA for the species in which nucleotide sequences have been so far determined, since the long T-stretches would be found in the A + T-rich region if they are essential for mtDNA replication initiation of a wide variety of holometabolous insect species. Table 1 shows the longest T-stretch found on each of the DNA strands within the A + T-rich region. The length of the T-stretch ranged from 4 to 25 bp for the species examined. The minimum length of the T-stretch that is indispensable for the replication initiation of mtDNA is not known, while the lengths of the T-stretches found immediately upstream of the OR's were >10 bp (Table 1). Therefore, in this study, the T-stretch >10 bp was considered as participating in the replication initiation of insect mtDNA. In the major coding strand shown in Table 1, the T-stretch (11–25 bp) was found for all holometabolous and some species of hemimetabolous insects examined. This observation also suggests that the T-stretch is conserved immediately upstream of the ON at least among holometabolous insects. In the minor coding strand shown in Table 1, several species of holometabolous insects do not possess the long T-stretch, whereas all the species of the suborder Brachycera thus far examined possess the T-stretch, ranging in size from 13 to 23 bp. Therefore, immediately upstream of the OJ, the T-stretch may be conserved at least among the species of Brachycera.
TABLE 1.
Length of T-stretch in the A + T-rich region of insect mtDNA
| Length of T-stretch (bp)
|
|||
|---|---|---|---|
| Order | Species | Major coding | Minor coding |
| Holometabola | |||
| Diptera | Drosophila | 11–17a | 13–23a |
| Ceratitis capitata | 21 | 21 | |
| Chrysomya chloropyga | 23 | 19 | |
| Cochliomyia hominivorax | 25 | 23 | |
| Anopheles quadrimaculatus | 17 | 9 | |
| Anopheles gambiae | 17 | 6 | |
| Culex torrentium | 19 | 9 | |
| Lepidoptera | Bombyx mori | 18a | 11 |
| Cydia pomonella | 11 | 6 | |
| Pandemis heparana | 11 | 9 | |
| Hymenoptera | Apis mellifera | 15 | 7 |
| Coleoptera | Tribolium castaneum | 14a | 9 |
| Pyrocoelia rufa | 12 | 8 | |
| Crioceris duodecimpunctata | 17 | 12 | |
| Hemimetabola | |||
| Thysanoptera | Thrips imagines | 20 | 10 |
| Hemiptera | Rhopalosiphum padi | 11 | 6 |
| Triatoma dimidiate | 6 | 9 | |
| Phthiraptera | Heterodoxus macropus | 7 | 6 |
| Orthoptera | Locusta migratoria | 4 | 8 |
| Ametabola | |||
| Thysanura | Tricholepidion gertschi | 5 | 6 |
| Collembola | Gomphiocephalus hodgsoni | 9 | 5 |
| Tetrodontophora bielanensis | 4 | 6 | |
Ascertained the location of the T-stretch immediately upstream of the OR.
On the other hand, several hemimetabolous and ametabolous insects species did not possess the long T-stretch within the A + T-rich region on the major coding strand (Table 1). We ascertained that none of the long T-stretches was found in the upstream portion of the ON of L. migratoria mtDNA (Figure 3B). These results suggest that a sequence other than the T-stretch may be involved in the replication initiation process in the species that do not possess the long T-stretch in the A + T-rich region. It is worth noting that a potential stem-loop structure existed 20 bases upstream of the ON of L. migratoria, which was also located at a similar position on mtDNA of desert locust Schistocerca gregaria and the meadow grasshopper Chorthippus parallelus (Figures 3 and 4) (Zhang et al. 1995). This possible stem-loop structure is a favorable candidate that may be involved in replication initiation since its conformation is highly similar to that of the stem-loop structure located around the L-strand origin of vertebrate mtDNA. To examine whether or not a stem-loop structure is conserved immediately upstream of the OR, the determination of the precise position of the OR for additional species is necessary, especially for the species that do not possess the long T-stretch within the A + T-rich region.
In this study, we revealed that the nucleotide sequences upstream of the OR of mtDNA are conserved among insect species that are phylogenetically distantly related, while a difference in flanking nucleotide sequence of the OR of insect mtDNA was also found. Furthermore, no sequence with high similarity to those of conserved regulatory sequences among vertebrate species, such as CSBs, was found around the OR of insect mtDNA, although the T-stretches are located immediately upstream of the OR of insect mtDNA and of the L-strand origin of mammalian mtDNA. Therefore, the sequence properties of the flanking portion of the OR are different between insect and vertebrate mtDNA; namely, the replication system of mtDNA has evolved through the changes in the regulatory sequences.
Acknowledgments
We express our sincere appreciation to Etsuko T. Matsuura, Masaki Kuro-o, Claire T. Saito, Claus Schnarrenberger, G. Curt Fiedler, and the anonymous reviewers for their critical reading of the manuscript and helpful advice. We also thank Tetsuhiro Moriya for his contribution to the determination of the nucleotide sequences of the A + T-rich region, Susumu Izumi and Tsunaki Asano for providing silkworms, and Takahisa Miyatake for providing red flour beetles. This work was partly supported by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan (no. 06304002) to K.T. and from the Japan Society for promotion of Science (no. 15255006) to T.A.
References
- Anderson, S., M. H. L. De Bruijn, A. R. Coulson, I. C. Eperon, F. Sanger et al., 1982. Complete sequence of bovine mitochondrial DNA: conserved feature of the mammalian mitochondrial genome. J. Mol. Biol. 156: 683–717. [DOI] [PubMed] [Google Scholar]
- Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-Van Dillen et al., 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowmaker, M., M. Y. Yang, T. Yasukawa, A. Reyes, H. T. Jacobs et al., 2003. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 278: 50961–50969. [DOI] [PubMed] [Google Scholar]
- Brehm, A., D. J. Harris, M. Hernández, V. M. Cabrera, J. M. Larruga et al., 2001. Structure and evolution of the mitochondrial DNA complete control region in the Drosophila subobscura subgroup. Insect Mol. Biol. 10: 573–578. [DOI] [PubMed] [Google Scholar]
- Cairns, S. S., and D. F. Bogenhagen, 1986. Mapping of the displacement loop within the nucleotide sequence of Xenopus laevis mitochondrial DNA. J. Biol. Chem. 261: 8481–8487. [PubMed] [Google Scholar]
- Carmean, D., L. S. Kimsey and M. L. Berbee, 1992. 18S rDNA sequences and the holometabolous insects. Mol. Phylogenet. Evol. 1: 270–278. [DOI] [PubMed] [Google Scholar]
- Chang, D. D., and D. A. Clayton, 1985. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl. Acad. Sci. USA 82: 351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D. D., W. W. Hauswirth and D. A. Clayton, 1985. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 4: 1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary, D. O., and D. R. Wolstenholme, 1985. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 22: 252–271. [DOI] [PubMed] [Google Scholar]
- Clary, D. O., and D. R. Wolstenholme, 1987. Drosophila mitochondrial DNA: conserved sequences in the A+T-rich region and supporting evidence for a secondary structure model of the small ribosomal RNA. J. Mol. Evol. 25: 116–125. [DOI] [PubMed] [Google Scholar]
- Clayton, D. A., 1982. Replication of animal mitochondrial DNA. Cell 28: 693–705. [DOI] [PubMed] [Google Scholar]
- Deb, S., A. L. Delucia, A. Koff, S. Tsui and P. Tegtmeyer, 1986. The adenine-thymine domain of the simian virus 40 core origin directs DNA bending and coordinately regulates DNA replication. Mol. Cell. Biol. 6: 4578–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich, M., and N. Muqim, 2003. Sequence and phylogenetic analysis of the complete mitochondrial genome of the flour beetle Tribolium castaneum. Mol. Phylogenet. Evol. 26: 502–512. [DOI] [PubMed] [Google Scholar]
- Garesse, R., 1988. Drosophila melanogaster mitochondrial DNA: gene organization and evolutionary considerations. Genetics 118: 649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, J. M., and D. R. Wolstenholme, 1978. Origin and direction of replication in mitochondrial DNA molecules from Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 75: 3886–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, J. M., and D. R. Wolstenholme, 1980. Origin and direction of replication in mitochondrial DNA molecules from genus Drosophila. Nucleic Acids Res. 8: 741–757. [PMC free article] [PubMed] [Google Scholar]
- Hixson, J. E., T. W. Wong and D. A. Clayton, 1986. Both the conserved stem-loop and divergent 5′-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J. Biol. Chem. 261: 2384–2390. [PubMed] [Google Scholar]
- Holt, I. J., H. E. Lorimer and H. T. Jacobs, 2000. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100: 515–524. [DOI] [PubMed] [Google Scholar]
- Inohira, K., T. Hara and E. T. Matsuura, 1997. Nucleotide sequence divergence in the A+T-rich region of mitochondrial DNA in Drosophila simulans and Drosophila mauritiana. Mol. Biol. Evol. 14: 814–822. [DOI] [PubMed] [Google Scholar]
- Kang, D., K. Miyako, Y. Kai, T. Irie and K. Takeshige, 1997. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 272: 15275–15279. [DOI] [PubMed] [Google Scholar]
- King, T. C., and R. L. Low, 1987. Mapping of control elements in the displacement loop region of bovine mitochondrial DNA. J. Biol. Chem. 262: 6204–6213. [PubMed] [Google Scholar]
- Kornberg, A., and T. A. Baker, 1992. DNA Replication, Ed. 2, pp. 1–52. W. H. Freeman, New York.
- Lewis, D. L., C. L. Farr, A. L. Farquhar and L. S. Kaguni, 1994. Sequence, organization, and evolution of the A+T-rich region of Drosophila melanogaster mitochondrial DNA. Mol. Biol. Evol. 11: 523–538. [DOI] [PubMed] [Google Scholar]
- Martens, P. A., and D. A. Clayton, 1979. Mechanism of mitochondrial DNA replication in mouse L-cells: localization and sequence of the light-strand origin of replication. J. Mol. Biol. 135: 327–351. [DOI] [PubMed] [Google Scholar]
- Monforte, A., E. Barrio and A. Latorre, 1993. Characterization of the length polymorphism in the A+T-rich region of the Drosophila obscura group species. J. Mol. Evol. 36: 214–223. [DOI] [PubMed] [Google Scholar]
- Monnerot, M., M. Solignac and D. R. Wolstenholme, 1990. Discrepancy in divergence of the mitochondrial and nuclear genomes of Drosophila teissieri and Drosophila yakuba. J. Mol. Evol. 30: 500–508. [DOI] [PubMed] [Google Scholar]
- Mueller, P. R., P. A. Garrity and B. Wold, 1992. Ligation-mediated PCR for genomic sequencing and footprinting, pp. 15.5.1–15.5.26 in Current Protocols in Molecular Biology, Vol. 2, edited by F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seideman et al. John Wiley & Sons, New York.
- Pardue, M. L., J. M. Fostel and T. R. Cech, 1984. DNA-protein interactions in the Drosophila virilis mitochondrial chromosome. Nucleic Acids Res. 12: 1991–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, D. A., J. M. Fostel, M. Berninger, M. L. Pardue and T. R. Cech, 1980. DNA-protein interactions in the Drosophila melanogaster mitochondrial genome as deduced from trimethylpsoralen crosslinking patterns. Proc. Natl. Acad. Sci. USA 77: 4118–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shadel, G. S., and D. A. Clayton, 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66: 409–435. [DOI] [PubMed] [Google Scholar]
- Sinden, R. R., 1994. DNA Structure and Function, pp. 58–94. Academic Press, San Diego.
- Taanman, J.-W., 1999. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410: 103–123. [DOI] [PubMed] [Google Scholar]
- Tamura, K., and T. Aotsuka, 1988. Rapid isolation method of animal mitochondrial DNA by the alkaline lysis procedure. Biochem. Genet. 26: 815–819. [DOI] [PubMed] [Google Scholar]
- Tapper, D. A., and D. A. Clayton, 1981. Mechanism of replication of human mitochondrial DNA: localization of the 5′ ends of nascent daughter strands. J. Biol. Chem. 256: 5109–5115. [PubMed] [Google Scholar]
- Tomizawa, J. I., H. Ohmori and R. E. Bird, 1977. Origin of replication of colicin E1 plasmid DNA. Proc. Natl. Acad. Sci. USA 74: 1865–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujino, F., A. Kosemura, K. Inohira, T. Hara, Y. F. Otsuka et al., 2002. Evolution of the A+T-rich region of mitochondrial DNA in the melanogaster species subgroup of Drosophila. J. Mol. Evol. 55: 573–583. [DOI] [PubMed] [Google Scholar]
- Walberg, M. W., and D. A. Clayton, 1981. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 9: 5411–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, W. C., M. Whiting, Q. D. Wheeler and J. M. Carpenter, 2001. The phylogeny of the extant hexapod orders. Cladistics 17: 113–169. [DOI] [PubMed] [Google Scholar]
- Wolstenholme, D. R., 1992. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 141: 173–216. [DOI] [PubMed] [Google Scholar]
- Yukuhiro, K., H. Sezutu, M. Itoh, K. Shimizu and Y. Banno, 2002. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol. Biol. Evol. 19: 1385–1389. [DOI] [PubMed] [Google Scholar]
- Zhang, D., J. M. Szymura and G. M. Hewitt, 1995. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 40: 382–391. [DOI] [PubMed] [Google Scholar]
- Zuker, M., D. H. Mathews and D. H. Tuner, 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, pp. 11–43 in RNA Biochemistry and Biotechnology, edited by J. Barciszewski and B. F. C. Clark. NATO ASI Series, Kluwer Academic Publishers, Dordrecht, The Netherlands/Norwell, MA.