Abstract
Two malic enzyme alleles, Men113A and Men113G, occur at approximately equal frequency in North American populations of Drosophila melanogaster, while only Men113A occurs in African populations. We investigated the population genetics, biochemical characteristics, and selective potential of these alleles. Comparable levels of nucleotide polymorphism in both alleles suggest that the Men113G allele is not recently derived, but we find no evidence in the DNA sequence data for selection maintaining the polymorphism. Interestingly, the alleles differ in both Vmax and Km for the substrate malate. Triglyceride concentration and isocitrate dehydrogenase (IDH) and glucose-6-phosphate dehydrogenase (G6PD) activities are negatively correlated with the in vivo activities of the Men alleles. We examined the causality of the observed correlations using P-element excision-derived knockout alleles of the Men gene and found significant changes in the maximum activities of both IDH and G6PD, but not in triglyceride concentration, suggesting compensatory interactions between MEN, IDH, and G6PD. Additionally, we found significantly higher than expected levels of MEN activity in knockout heterozygotes, which we attribute to transvection effects. The distinct differences in biochemistry and physiology between the naturally occurring alleles and between the engineered alleles suggest the potential for selection on the Men locus.
SIGNIFICANT progress has been made in characterizing population variability at the level of DNA sequences, but the central challenge of elucidating rules that connect genotype and phenotype remains. In many cases, genotype may give rise to phenotype through interactions among multiple genes and metabolite intermediates. Presumably, these interactions are at least partly the product of natural selection. The challenge, however, is not to simply infer the presence of selection, but to elucidate its action and distribution across such multiple gene networks. In this study we begin an analysis of the variation and functional interactions among three oxidative enzymes known to be the primary source of the reduced form of the metabolic cofactor NADP.
A single amino acid polymorphism was identified in cytosolic malic enzyme (Men) as part of a study of geographic variation of single-nucleotide polymorphisms associated with the genes of central metabolic enzymes in Drosophila melanogaster (Sezgin et al. 2004). The polymorphism is an alanine-to-glycine substitution at amino acid 113 and the two alleles defined by the amino acid substitution occur in approximately equal frequency in all 10 North American populations examined. Allele frequency clines were detected at a number of other metabolic genes, but these Men alleles and associated haplotypes showed no clinal changes with latitude. While the presence of clinal change in allele frequency can be interpreted as evidence for selection on a locus, the lack of such a pattern does not indicate that a polymorphism is necessarily neutral. Here we examine other aspects of the population genetics, and the biochemistry, of the two malic enzyme alleles for evidence of selection or the potential for selection between the two alleles. In the second part of this article, we compare these findings with those from synthetic Men alleles produced in the laboratory by P-element excision, again with the goal of determining the potential for selection between alleles of the Men gene.
Malic enzyme oxidizes malate to pyruvate with the concurrent reduction of NADP to NADPH, a major reductant in lipid synthesis (Wise and Ball 1964). It has been estimated that MEN produces ∼30% of the available NADPH in larval D. melanogaster, with the remainder coming from cytosolic isocitrate dehydrogenase (IDH) activity (∼20%) and the oxidative enzymes of the pentose shunt (glucose-6-phosphate dehydrogenase, G6PD, and 6-phosphogluconate, 6PGD, combined ∼40%) (Geer et al. 1979a,b). Induction studies have shown significant interactions between these loci, presumably to maintain a constant NADPH/NADP ratio and supply of reduced cofactor for lipogenesis (Geer et al. 1976, 1978, 1981; Wilton et al. 1982). This explanation is, however, likely an oversimplification. Apparently compensatory changes in enzyme activity have been documented between the oxidative pentose shunt enzymes and MEN; extreme reduction in MEN activity is associated with an increase in G6PD and 6PGD activity (Wilton et al. 1982; Geer and Laurie-Ahlberg 1984). However, no such compensation has been found between IDH and MEN; reduction in MEN is associated with reduction, not increase, in IDH activity (Wilton et al. 1982; Geer and Laurie-Ahlberg 1984). Similarly, an increase in dietary carbohydrate increases G6PD, 6PGD, and MEN activity, but decreases IDH activity (Geer et al. 1978; Geer and Laurie-Ahlberg 1984). Finally, reduction in IDH activity was associated with an increase in G6PD and 6PGD, but a decrease in MEN activity (Bentley et al. 1983).
This project first examines biochemical characteristics of the Men alleles to determine the possible phenotypic effects of the observed naturally occurring polymorphism. In vitro biochemical characteristics do not, however, necessarily correspond to in vivo differences in physiology and fitness (Cavener and Clegg 1981; Dykhuizen and Dean 1990). Furthermore, large differences in enzyme activity unrelated to the observed polymorphism (i.e., chromosome effects) may mask or inflate true allelic differences (Laurie-Ahlberg et al. 1981, 1982). Studying polymorphisms in different genetic backgrounds will to some extent randomize chromosome differences between alleles, but this may not suffice to characterize allelic differences if they are small. Thus, to complement our examination of the naturally occurring alleles, we examined an independent set of laboratory-derived alleles in which phenotypic variation was exclusively attributable to changes in enzyme concentration and for which background genetic variation was minimized.
MATERIALS AND METHODS
Flies: basic lines:
Isothird chromosome lines were a subsample of a set of nonlethal, third chromosomes extracted from isofemale lines established in 1997 (see Duvernell and Eanes 2000; Verrelli and Eanes 2001). Inbred lines used as common genetic backgrounds were either obtained from the Bloomington Drosophila Stock Center at Indiana University (6326, line no. 6326) or developed in the Eanes lab by 20 generations of full-sib mating (VT83i, HFL8i). Inbred lines developed in the Eanes lab were cytologically and enzymatically screened for genetic homozygosity. Line 6326 is isogenic for the second and third chromosomes (Hoskins et al. 2001). A line containing a deficiency that covers the entire malic enzyme gene, Df(3R)kar3l, was obtained from the Bloomington Drosophila Stock Center at Indiana University (line no. 6160). Samples of flies from east Africa, Z(H) and Z(S), were kindly donated by Chung-I Wu (University of Chicago). Thirteen D. simulans lines, full-sib inbred for 14 generations (see Duvernell and Eanes 2000), were also included in our sequence analysis. All flies were maintained on standard cornmeal media. All assays for enzyme activity, protein, and triglyceride content were conducted on male flies aged 5 days after emergence at 25°.
Naturally occurring Men alleles for activity, protein, and triglyceride analysis:
Genetic background effects were minimized by replacing the X and second chromosomes of Men113A and Men113G (hereafter simply MenA and MenG) third chromosome lines with those from inbred genetic backgrounds. Four lines of each genotype were placed in the VT83i background and one of each genotype was placed in the 6326 background.
P-element excision series for Men:
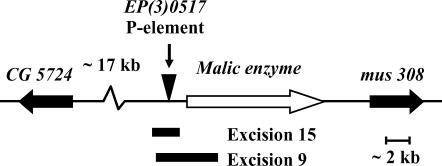
P-element-mediated deletion (Tsubota and Schedl 1986; Salz et al. 1987) was used to generate activity variant alleles for the Men gene. The EP(3)0517 element (Rorth 1996) is inserted 473 bases 5′ to the Men start codon (Figure 1). Crosses were carried out to create dysgenic males, using the EP(3)0517 element line and the Bloomington Drosophila Stock no. 2030 as a source of transposase. Recovered excision lines were screened for MEN activity. Four chromosome lines, two with wild-type levels of MEN, designated MenEx3+ and MenEx5+, and two with no apparent MEN activity (knockout lines), designated MenEx9− and MenEx15−, were placed in a common genetic background (HFL8i). The excision site for each of these four lines was amplified and then sequenced using oligonucleotide primers flanking the initial insertion site.
Figure 1.
The genomic region surrounding the Men gene including the insertion site of the EP(3)0517 P element and the pair of P-element excision-mediated deletions used in this study. The Men gene is shown by the open arrow. The two flanking genes are shown by the solid arrows. The regions covered by the deletions are indicated by the solid bars. Excision 15 removed 2418 bases, including 160 bp of the 5′ end of the cDNA UTR. Excision 9 removed 4080 bp, including 1925 bp of the cDNA, which covers the entire first exon.
Sequence analysis:
The primary sequence of the Men gene was initially recovered, prior to the whole-genome sequence (Adams et al. 2000), using the enriched cDNA method described by Verrelli and Eanes (2000). Oligonucleotide primers were designed from the cDNA sequence to amplify the complete coding region of the Men gene from genomic DNA. Amplifications were conducted under the following conditions: 10 ng of genomic DNA was amplified in 10 mm Tris (pH 8.3), 50 mm KCL, 0.01% gelatin, 1.5 mm MgCl2, 2 units of platinum Taq polymerase (Life Technologies, Rockville, MD), and 100 nm of each primer. The amplification products were purified (Prep-A-gene DNA purification kit; Bio-Rad, Hercules, CA) and double-stranded templates (∼500 ng per reaction) were sequenced manually (Sequenase, v.2.0; United States Biochemical, Cleveland), using primers spaced at ∼300-bp intervals. The primary sequence was determined for both strands and all ambiguous sites were verified on both strands. The excision sites of the alleles created through P-element excision were sequenced by the DBS sequencing facility, University of California (Davis, CA), using Big Dye Terminator version 3.1 (ABI) and then sequenced on a 3730 DNA Analyzer (ABI). Base calls from the chromatograms were checked, and the 2-kb fragments assembled, using Sequencher (Gene Codes, Ann Arbor, MI). All sequences have been deposited in GenBank (accession nos. DQ148977–DQ149022).
DnaSP 4.0 (Rozas et al. 2003) was also used to calculate θ and π, to test for differences in θ using a coalescence simulation, to carry out the Tajima (1989) and Fu and Li (1993) tests, and to generate standardized estimates of linkage disequilibrium (R2).
Fly homogenization:
Flies were homogenized in grinding buffer (100 mm Tris-HCl, 0.15 mm NADP, pH 7.4) at a “concentration” of one fly per 200 μl of buffer and spun at 13,000 rpm for 5 min at 4° to pellet all solids. In general, all assays were conducted using samples of five flies (five flies in 1 ml grinding buffer). In a few cases, insufficient flies were available and fewer flies were assayed.
Soluble protein content:
Soluble protein was measured using a commercially available kit (Bio-Rad kit no. 500-0006) following the manufacturer's protocol. The assays contained either 25 μl homogenate and 1 ml reagent (Beckman spectrophotometer) or 10 μl homogenate and 100 μl reagent (Molecular Devices spectrophotometer) and were incubated at room temperature for 10 min. Reactions were measured at OD595 and total soluble protein concentrations (micrograms per milliliter) were determined by comparison with bovine serum albumen standards. Each sample was assayed twice for protein concentration and the mean was used in analysis. Enzyme activities and triglyceride concentrations (below) were standardized by soluble protein content to account for differences in mass or body size between individual flies and for possible differences in the degree of homogenization between samples.
Soluble triglyceride content:
Soluble triglyceride was measured using a commercially available kit (Infinity triglyceride assay; Thermo Electron, Arlington, TX; catalog no. 2780), following the manufacturer's protocol. The assays contained either 25 μl homogenate and 1 ml reagent (for samples assayed on the Beckman spectrophotometer) or 10 μl homogenate and 100 μl reagent (the Molecular Devices spectrophotometer) and were incubated at 37° for 10 min. Reactions were measured at OD500 and total soluble triglyceride concentrations (micrograms per milliliter) were determined by comparison with a commercially available standard (Sigma-Aldrich, St Louis; catalog no. T2522). Each sample was assayed twice and the mean was used in analysis. Results are reported as micrograms triglyceride per microgram soluble protein.
Enzyme activity measurements:
Enzyme activity assays were carried out using either a Beckman DU 640 or a Molecular Designs SpectraMax 384 Plus 96-well plate spectrophotometer. Assays on the Beckman spectrophotometer used 25 μl of fly homogenate and 400 μl assay buffer. The initial reaction rate was calculated by sampling absorbance (OD340) every 12 sec over 3 min. Assays on the Molecular Designs spectrophotometer used 10 μl of fly extract and 100 μl of assay buffer and absorbance was measured every 9 sec over 3 min. All activity assays were conducted at 25°. In all experiments, samples were assayed twice and the average was used in analysis. The estimates of thermal stabilities, substrate Km, and cofactor Km and the initial examination of the MEN deletion series activities were determined using the Beckman spectrophotometer. Enzyme activity is expressed as micromoles NADP+ reduced per minute per microgram soluble protein times 10,000.
The assay buffers for the three enzymes assayed in this study were as follows: MEN, 100 mm Tris-HCl, 0.34 mm NADP, 50 mm MnCl2, 50 mm malate, pH 7.4; G6PD, 100 mm Tris-HCl, 0.32 mm NADP, 3.5 mm d-glucose-6-phosphate, pH 7.4; and IDH, 100 mm Tris-HCl, 0.10 mm NADP, 0.84 mm MgSO4, 1.37 mm dl-isocitrate, pH 8.6. Initial values for appropriate pH, substrate, and cofactor concentrations for the reactions were taken from the literature and modified to give maximum enzyme activity.
Thermal stability:
Allele-specific enzyme thermostabilities were estimated by following MEN activity decline over time at 50° (Verrelli and Eanes 2001). Two replicate copies from three isochromosomal lines of each haplotype were assayed for MEN activity on a Beckman spectrophotometer. A single aliquot of fly homogenate was immediately placed on ice for use as a reference. Ten aliquots were placed in a 50° heat block, and at 1-min intervals a single aliquot was removed and placed on ice. All aliquots were subsequently kept on ice until their maximum activities were measured. The activity of each sample was compared to that of the reference sample to determine the proportion of activity remaining at each time point. The decline in enzyme activity with time was treated as a first-order exponential decay process and denaturing constants (Kd) were determined using the relationship 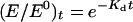 , where (E/E0)t is the proportion of initial enzyme activity remaining at time t and Kd is the denaturation rate (Hall 1985). The slope of the line from the linear regression of ln(E/E0)t on time is an estimate of Kd. A mean Kd was calculated for each line from the two replicates and a mean Kd for each allele was calculated from the three lines.
, where (E/E0)t is the proportion of initial enzyme activity remaining at time t and Kd is the denaturation rate (Hall 1985). The slope of the line from the linear regression of ln(E/E0)t on time is an estimate of Kd. A mean Kd was calculated for each line from the two replicates and a mean Kd for each allele was calculated from the three lines.
Estimation of Michaelis-Menton constants (Km):
Allele-specific Michaelis-Menton constants for both malate and NADP were determined using the two-substrate concentration method (Duggleby 1979; Cornish-Bowden 1995). Reaction rates were determined in parallel for four isochromosomal lines of each allele using the Beckman spectrophotometer. Individual cuvette use was randomized with respect to line. Initial estimates of the apparent Km values for both NADP and malate were obtained at 25° using nine substrate concentrations from 0.5 Km to 20 Km (initial guesses were from Geer et al. 1980). Fits of initial rate vs. concentration did not appear to deviate from Michaelis-Menton kinetics over this range of substrate concentrations. From these initial estimates, the Km of each line was determined from 12 replicate initial velocity estimates made at each of two substrate concentrations approximating 0.5 Km and Vmax conditions (Duggleby 1979; Cornish-Bowden 1995). Homogenate concentrations were adjusted so that all lines showed effectively identical Vmax values during Km estimation. Km estimation under this design is robust if preliminary estimates of Km are close to the true values (Cornish-Bowden 1995). Km values, and their associated standard errors, were estimated from initial rate data by nonlinear least squares using the computer program Leonora (Cornish-Bowden 1995). For estimation of the Km of malate, cuvette homogenate concentrations were held such that <5% of the substrate was consumed during a 3-min reaction. For estimation of the Km of NADP, the low concentration of NADP required for estimation of the initial rate at ∼0.5 Km precluded this control, and instead homogenate concentration was held such that a plot of absorbance against time was linear for the duration of a 4-min assay.
Data analysis:
In all cases, crosses were replicated in multiple vials (two vials in examination of the wild Men alleles, four vials in the later experiments using the excision Men alleles) and each vial was sampled twice. Initial ANOVAs were conducted to determine the presence or absence of a genotype effect and Tukey's honestly significant difference (HSD) multiple-comparison tests were conducted to determine whether or not individual crosses could be grouped within genotype classes. Grouped data were then analyzed using ANOVA. Planned pairwise comparisons were conducted using the method of Sokal and Rolf (1994, p. 230), which calculates a sum-of-squares for the difference between two means and the group mean (from the ANOVA) and tests this value over the within-group mean square from the ANOVA.
RESULTS
Naturally occurring Men alleles:
Sequence variation:
We amplified and sequenced all 1737 bases of the Men coding region from 33 D. melanogaster isochromosomal and 13 D. simulans inbred lines. Polymorphic sites are shown in Figure 2. We observed 16 polymorphic sites in D. melanogaster, only one of which was a nonsynonymous change, the previously reported (Sezgin et al. 2004) G/C polymorphism at position 338 that results in the alanine/glycine amino acid polymorphism (Figure 2, boldface type). The sequence and three-dimensional (3D) structure of the pigeon cytosolic malic enzyme and the human mitochondrial malic enzyme are known (Yang et al. 2000, 2002; Yang and Tong 2000). Alignment of the D. melanogaster sequence with the human and pigeon sequences and comparison with the known pigeon (cytosolic MEN) structure indicates that the D. melanogaster polymorphic site is in a α-helical segment of the molecule, in the region of the active site. The D. melanogaster MEN sequence is 55% identical to the pigeon sequence. Given that the human and pigeon MEN sequences are 59% identical, and all major structural features are conserved between them (Yang et al. 2002), we expect that the pigeon MEN structure is a reasonable predictor of the structure of the D. melanogaster molecule.
Figure 2.
Nucleotide polymorphism in the amino acid coding region of the Men gene of (A) Drosophila melanogaster and (B) D. simulans. The variable site (338) responsible for the amino acid polymorphism in D. melanogaster is in boldface type. The first 11 sequences have the MenG (glycine-coding) allele, while the remaining sequences all have the MenC (alanine) allele. D. simulans sequences are indicated by a final “s.”
In our sample, the Men gene shows normal levels of synonymous polymorphism for D. melanogaster and D. simulans genes (Table 1) (Moriyama and Powell 1996). African samples showed somewhat higher levels of polymorphism than North American samples (Table 1) although the difference was not significant at the P < 0.05 level using a coalescence simulation (Rozas et al. 2003). African lines carried the MenA allele, as did all the D. simulans lines, suggesting that MenA is the ancestral allele. Approximately half, 13 of 24, of the North American D. melanogaster lines sequenced were MenA, reflecting the approximately equal frequencies reported for these alleles across North American populations sampled by Sezgin et al. (2004). There are no fixed synonymous differences between the MenA and MenG alleles, although three positions, 1323, 1428, and 1548, are fixed in almost all MenA lines; only a single MenG line, DPF 90.2, shares the “MenA” sequence (Figure 2). The linkage disequilibrium between the amino acid-altering polymorphism and these three sites is statistically significant at the P < 0.05 level by a Bonferroni corrected chi-square test. The level of polymorphism within the MenG alleles is not significantly different from that within the North American D. melanogaster MenA alleles (Table 1). All of the Tajima (1989) and Fu and Li (1993) tests conducted on the D. melanogaster (combined or exclusively on the North American sequences) or D. simulans sequences were not statistically significant.
TABLE 1.
Levels of silent polymorphism in the Men gene across sampled populations of Drosophila melanogaster and D. simulans
| n | No. sites | S | π | θ | |
|---|---|---|---|---|---|
| D. melanogaster | 33 | 433.81 | 16 | 0.0082 | 0.0091 |
| African | 9 | 433.76 | 10 | 0.0097 | 0.0085 |
| North American | 24 | 433.83 | 10 | 0.0073 | 0.0062 |
| MenG | 11 | 433.83 | 7 | 0.0055 | 0.0055 |
| MenA | 13 | 433.83 | 4 | 0.0020 | 0.0030 |
| D. simulans | 13 | 433.23 | 17 | 0.0130 | 0.0127 |
MEN thermal stability and Michaelis-Menton constants (Km):
Table 2 summarizes thermal stability and Km measures for the two alleles. Thermal stability values from three MenG and three MenA isochromosomal lines did not differ between the two alleles or between lines of either allele (F1,4 = 2.90, P < 0.16, F4,6 = 0.93, P < 0.50, respectively). Similarly, the Km for the NADP cofactor from four isochromosomal lines of each allele did not differ between alleles or lines (F1,6 = 0.01, P < 0.92, F4,6 = 0.87, P < 0.53). The Km for malate from four isochromosomal lines of each allele did, however, differ between alleles and lines (F1,6 = 8.00, P < 0.03, F6,8 = 77.6, P < 0.0001).
TABLE 2.
Men genotype values (±SE) for thermal stability and Km for both cofactor and substrate
| Thermostabilitya | Km NADPb | Km malateb | |
|---|---|---|---|
| MenG | 0.13 ± 0.005 | 0.003 ± 0.0000 | 0.12 ± 0.018* |
| MenA | 0.11 ± 0.011 | 0.003 ± 0.0002 | 0.07 ± 0.005* |
P < 0.05.
Absolute value of Kd.
In micromoles per liter.
MEN Vmax:
We assumed that the allele-specific activity under saturating substrate levels is a relative Vmax estimate. Our interests were to determine whether the two Men allelic proteins differed in enzymatic activity under saturating substrate conditions (Vmax) and whether the Vmax observed in the MenA/G heterozygous genotype equaled the average of the mean activity of the MenA and MenG homozygous genotypes (this is our expectation if any difference in activity between the two alleles is simply additive, but see Kacser and Porteous 1987 for a cautionary note about this simplification). The four VT83i background-replaced MenA and MenG lines were crossed to the single 6326 background-replaced MenA and MenG lines to produce sets of MenA and MenG homozygous and heterozygous flies (e.g., w; VT83i/6326; MenA1/MenA5 or w; VT83i /6326; MenA1/MenG5). This cross scheme produces MenA and MenG homozygotes in four different third chromosome combinations and heterozygotes in eight, all in a pseudo-outcrossed second chromosome background (VT83i/6326), effectively randomizing any background effects on Vmax. This scheme eliminates “line” effects (individual chromosomes are never homozygous) and replaces them with possible “cross” effects (the combination of any pair of third chromosomes).
The ANOVA of the Vmax estimates with four genotypes (G/G, A/A, G/A, and A/G) indicated a significant genotype effect (F3,31 = 36.93, P < 0.001). A Tukey's HSD test found no significant Vmax difference between the reciprocal heterozygotes and these were combined for further analysis. The subsequent two-way ANOVA of polymorphism and cross effects on the combined data again indicated a significant amino acid polymorphism effect (F2,32 = 10.32, P < 0.002). There were no significant differences between crosses within a genotype (F13,32 = 1.40, P < 0.26), but there were differences between vials within a cross (F16,32 = 5.37, P < 0.001). The Vmax estimates were 0.38 ± 0.01 (mean ± SE across the four crosses) for MenG flies, 0.26 ± 0.01 for MenA flies, and 0.33 ± .01 for the heterozygotes (combined, eight crosses). We found a significant difference in Vmax between the MenA and MenG activities (F1,32 = 116.21, P < 0.001), but no significant difference between the heterozygote Vmax and the mean of the homozygote Vmax-values (F1,32 = 2.25, P > 0.10). The lack of significant difference between the heterozygotes and the average of the homozygotes suggests that the Vmax of the two alleles is additive in the heterozygous genotypes.
Intergenic and physiological effects:
Substrate concentrations in vivo are generally believed to be low, on the order of the Km (Fersht 1977; Hochachka and Somero 2004), and under such conditions Vmax, the reaction rate at saturating substrate concentration, does not reflect the true reaction rate. Instead, the ratio Vmax/Km provides a more accurate indicator of the relative in vivo reaction velocities of alleles differing in both Vmax and Km (Hall and Koehn 1983; Watt 1985; Watt and Dean 2000). Given our estimates of Vmax and malate Km, Vmax/Km = 3.26 ± 0.158 (mean ± SE across the four crosses using the mean Km-value calculated above) for MenG flies, 3.87 ± 0.186 for MenA flies, and 3.59 ± 0.165 for the (combined) heterozygotes. The two Men alleles differ in Vmax/Km by ∼19% (t14 = −2.50, P < 0.013). Note that because of the inverse relationship in Vmax and Km observed between the two alleles, the genotype with the higher Vmax, MenG, has the lower predicted in vivo activity.
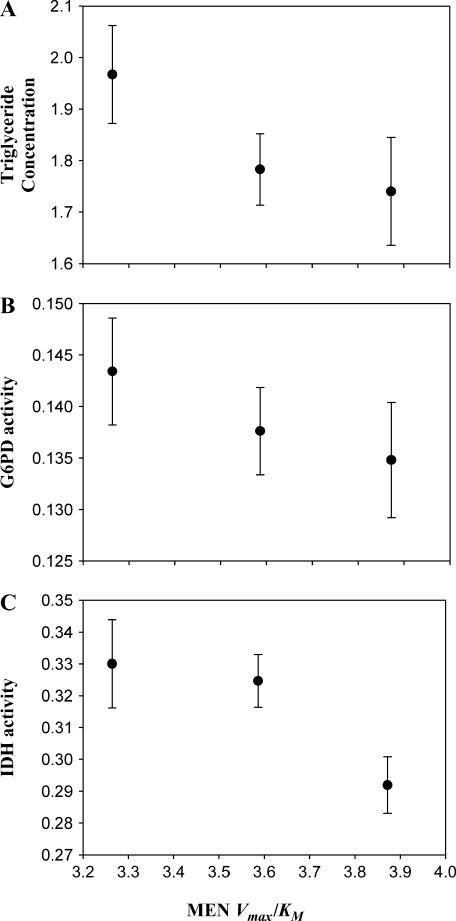
We examined whether the predicted genotypic difference in MEN activity was associated with detectable changes in adult physiology. As an assessment, we measured total triglycerides, a major lipid component highly correlated with total lipid content (Clark and Keith 1988). We also examined the induction by MEN activity of two other NADPH-generating enzymes, cytosolic IDH and G6PD. Figure 3 shows plots of triglyceride concentration and IDH and G6PD activities against the predicted in vivo activities (Vmax/Km) for each Men genotype. Using the same pairwise comparison of means that we used for the Vmax data, we found that the triglyceride concentration and IDH and G6PD activity all differ significantly between the MenG and MenA homozygotes (F1,32 = 7.19, P < 0.025; F1,32 = 13.86, P < 0.025; and F1,32 = 5.90, P < 0.05; respectively), whereas the MenA/G heterozygotes do not differ from the mean of the homozygotes in triglyceride concentration or IDH or G6PD activity (F1,32 = 1.28, P > 0.05; F1,32 = 3.27, P > 0.05; and F1,32 = 0.40, P > 0.05; respectively). Triglyceride concentration and IDH and G6PD activity all increase as MEN genotype activity decreases (Figure 3). The negative relationship between MEN, IDH, and G6PD might be expected as a physiological response to maintain a pool of NADPH at a constant concentration. The decrease in triglyceride concentration was unexpected.
Figure 3.
Relationship between triglyceride concentration (micrograms per microgram protein), IDH activity or G6PD activity (10,000 × μm NADPH/min/μg protein), and predicted in vivo MEN activity (Vmax/Km) of the Men genotypes. The genotypes are, left to right, MenG/G, MenG/A, and MenA/A. Error bars indicate one standard error.
P-element excision-derived Men alleles:
Sequence characterization:
Using the EP(3)0517 element, we recovered and characterized a chromosome set of 28 excision lines derived from P-element transposition 5′ to the Men gene. We identified two lines with essentially no MEN activity (knockout lines) and selected two lines with relatively normal levels of activity (full-activity lines). Approximately 1 kb of DNA, centered on the original P-element insertion site, was sequenced from each line. The sequences of the excision chromosomes with normal activity (MenEx3+ and MenEx5+) were identical to the standard D. melanogaster genome sequence (both have the MenG allele). The knockout lines (MenEx9− and MenEx15−) possessed deletions covering portions of the Men gene (Figure 1). MenEx15− had a 2418-bp deletion, roughly centered on the P-element insertion site, removing 160 bp of the 5′-UTR of the Men gene. MenEx9− had a 4080-bp deletion that included the entire first exon (44 aa) of the Men gene. Annotation is based on the D. melanogaster genome sequence (Adams et al. 2000) (http://www.fruitfly.org). Both deletion lines were homozygous viable, but clearly possessed poor viability and low fertility. An initial screening of MEN activity of these lines found that MenEx9− and MenEx15− had ≪10% the activity of either MenEx3+ or MenEx5+. Closer examination of the activity of these two alleles (below) indicated that they show <2% normal MEN activity, indicating that they are, as expected from the sequence data, clean knockout alleles.
MEN Vmax:
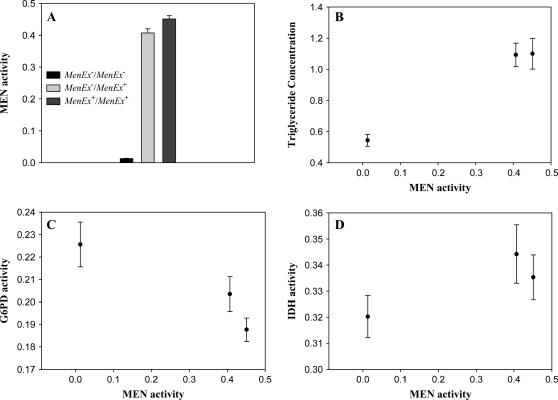
The four excision lines (in a common VT83i genetic background) were intercrossed to create flies with varying levels of MEN activity. Our intention was to use these crosses to create individuals with 100% (MenEx5+/MenEx5+, MenEx5+/MenEx3+, MenEx3+/MenEx3+), 50% (MenEx5+/MenEx15−, MenEx5+/MenEx9−, MenEx3+/MenEx15−, MenEx3+/MenEx9−), and 0% (MenEx15−/MenEx15−, MenEx15−/MenEx9−, MenEx9−/MenEx9−) relative MEN activities. We detected a significant genotype effect in MEN activity among the 10 groups (F9,39 = 219.13, P < 0.0001). When the crosses were grouped in the three activity classes (0, 50, and 100%), we again detected a significant difference in MEN activity (F2,39 = 975.50, P < 0.0001, Figure 4A). The Men+/Men+ flies had average MEN activities of 0.45 ± 0.01 (mean ± SE across all crosses); the Men−/Men+ flies, of 0.41 ± 0.01; and the Men−/Men− flies, of 0.01± 0.0001. The “0” and 100% groups differed significantly in activity (F1,32 = 3282.48, P < 0.001) with the Men−/Men− flies showing <2% the activity of the normal-activity flies. The 50% group, however, showed ∼90% the activity of the 100% group, significantly higher activity than the average of the 0 and 100% groups (F1,32 = 861.10, P < 0.001). This higher than expected amount of MEN activity is presumably from an upregulation of the functional copy of the gene because the Men−/Men− flies show essentially no activity. The upregulation could be the result of intrinsic autoregulation of MEN activity per se or of interallelic transvection, an interchromosomal interaction between a chromosome with a nonfunctional gene and the paired chromosome with an intact, functional, copy of the gene (Henikoff and Comai 1998; Wu and Morris 1999).
Figure 4.
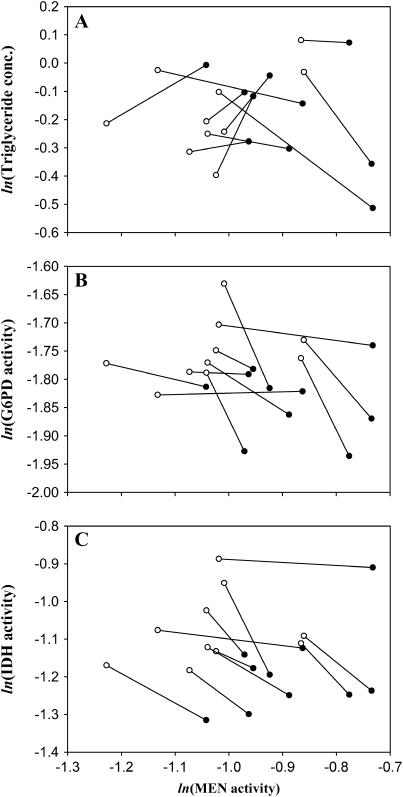
(A) MEN Vmax from crosses of the EP(3)0517 excision series. Crosses were expected to generate three different activity classes: “0%,” “50%,” and “100%.” The 0% and 100% groups are significantly different. The 50% flies show significantly greater 50% activity, but are significantly lower in activity than the 100% flies. Error bars indicate one standard error. (B–D) Graphs of triglyceride concentration (micrograms per microgram protein), G6PD activity, and IDH activity, respectively, against Men activity (10,000 × μm NADPH/min/μg protein).
Intergenic and physiological effects:
Figure 4, B–D, shows triglyceride concentration, and IDH and G6PD activities, plotted against the estimated MEN Vmax of the excision genotypes. Recall that the third chromosomes of all genotypes in this experiment are identical, derived from the single P-element-containing parent chromosome, EP(3)0517, and that the remainder of the genome has been replaced with a common background (HFL8i). No differences in Km between the genotypes are expected, and Vmax should accurately reflect the in vivo activities. Both triglyceride and G6PD significantly differed across the three MEN-activity groups (F2,39 = 18.45, P < 0.0001 and F2,39 = 4.79, P < 0.02, respectively). The triglyceride levels of the knockout and “100%” flies were significantly different, while there was no significant difference between levels in the two high-activity groups (Tukey's HSD test, P < 0.05, Figure 4B). The homozygous Men knockouts had approximately half the triglyceride levels of the 50 and 100% MEN-activity groups. The steep decline in triglyceride concentration in parallel with decline in MEN activity is consistent with the expected central role of MEN (along with IDH and G6PD) in producing reduced NADP for fatty acid synthesis. Similarly, a Tukey's HSD test indicates that G6PD levels are significantly higher in the Men knockouts than in the two high-activity classes (P < 0.05). IDH activity did not vary significantly with MEN activity (F2,39 = 1.26, P < 0.31), although there was an apparent trend toward reduction in IDH activity in the MEN knockouts, consistent with earlier reports in the literature (Wilton et al. 1982; Geer and Laurie-Ahlberg 1984).
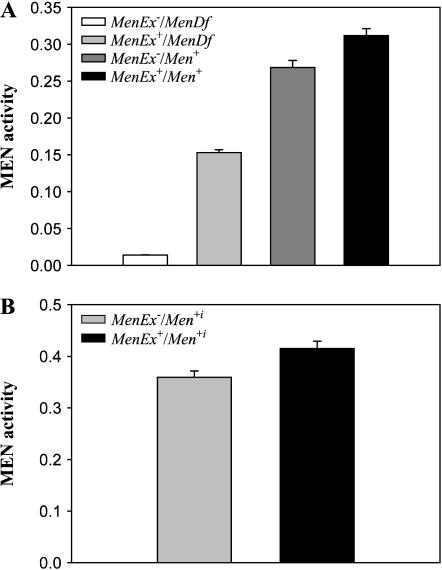
To examine whether the much higher than expected levels of MEN activity in the knockout/full-activity heterozygotes were the result of transvection or upward autoregulation the Men gene, we crossed the four Men excision lines to a third chromosome line containing a large-scale deficiency [Df(3R)kar3l] that covers the Men gene (Voelker et al. 1981) and a second line with a wild-type, MenG, copy of the Men gene (CT68). These crosses yielded four groups of genotypes: normal activity (expected 100% relative activity, MenEx+/Men+: MenEx3+/CT68 and MenEx5+/CT68), deletion heterozygote (expected 50% relative activity, MenEx−/Men+: MenEx9−/MenCT68, MenEx15/MenCT68), deficiency heterozygote [expected 50% relative activity, MenEx+/MenDf: MenEx3/Df(3R)kar3l, MenΔ5/Df(3R)kar3l], and knockout (expected 0% relative activity, MenEx−/MenDf: MenEx9−/Df(3R)kar3l, MenEx15−/Df(3R)kar3l).
Transvection appears to result from homology-dependent chromosome pairing (Henikoff and Comai 1998). It is generally thought that chromosomes with large deletions or rearrangements might not pair and therefore would not show transvection (Henikoff and Comai 1998). If the higher than expected levels of MEN activity observed in the knockout/full-activity heterozygotes were the result of transvection, then the excision-deletion group should show higher levels of MEN activity (elevated by transvection) than the deficiency group (incapable of transvection by the reasoning above). We detected a significant effect of genotype on MEN activity (F7,63 = 224.89, P < 0.0001). A Tukey's HSD pairwise comparison showed no difference within a genotypic group [i.e., MenEx3+/Df(3R)kar3l and MenEx5+/Df(3R)kar3l did not significantly differ in activity] and the data were therefore were combined by class. The combined data showed a significant genotype effect (F3,63 = 1090.46, P < 0.0001) and a Tukey's HSD pairwise comparison of MEN Vmax between the four classes found that knockout < deficiency heterozygote < deletion heterozygote < 100% (Figure 5A). The significantly higher levels of MEN activity in the deletion heterozygote group than in the deficiency heterozygote group indicate that having only a single functional copy of the Men gene does not necessarily result in upregulation of that locus. Additionally, pairwise comparisons indicated that the deletion heterozygote flies (MenEx−/MenEx+), but not the deficiency heterozygote flies [Df(3R)kar3l/Men+], had significantly greater than half the activity of the normal-activity flies (MenEx+/Men+; F1,40 = 248.00, P < 0.001). Together, these two results suggest that the increased activity in deletion flies is the result of transvection. This is not, however, a perfect test in that the comparison between the Df(3R)kar3l/Men+ group and the MenEx−/Men+ group is confounded by potential trans-acting differences between the Df(3R)kar3l chromosome and the CT68 (Men+) chromosome (i.e., line effects) in addition to any effects from the Men deficiency. However, comparison of the activity in flies that combine Df(3R)kar3l and other third chromosomes consistently found that the Df(3R)kar3l lines had half the activity of their counterparts (data not shown), supporting our conclusion that the higher levels of MEN activity in the MenEx−/Men+ flies are the result of transvection.
Figure 5.
(A) Mean MEN activity estimates from crosses of EP(3)0517 excision series with a line carrying a large chromosomal deficiency [Df(3R)kar3l] that covers the Men gene and a line carrying a wild-type copy of the gene. The four groups are MenEx−/MenDf, MenEx+/MenDf, MenEx−/Men+, and MenEx+/Men+, reading from left to right. (B) Mean MEN activity from crosses of MenEx− (left) and MenEx+ (right) to 10 different Men+ lines. Error bars indicate one standard error.
To test for possible differences in the level of transvection across different chromosomes, we crossed the MenEx9− knockout line and the MenEx3+ normal-activity line to 10 different isothird chromosome lines (Men+i, i = 1–10) that shared a common genetic background (X and second chromosomes from line 6326). All lines carried the MenG allele (the same as the normal-activity excision lines) to avoid differences in Km between alleles from possibly preventing us from accurately predicting the amounts of in vivo MEN activity. The MenEx−/Men+i and MenEx+/Men+i flies differed in activity (F9,175 = 23.86, P < 0.001). Across the 10 isothird lines, the activities of the MenEx−/Men+i genotypes averaged 87% (±0.02) of those of the MenEx+/Men+i genotypes, with a range from 75 to 93% (Figure 5B). Individual third chromosome lines varied in MEN activity (a significant line effect was detected, F9,175 = 4.67, P < 0.0001), but there was no significant line-by-excision interaction (F9,175 = 0.97, P < 0.47), indicating that the amount of transvection did not significantly differ across the different third chromosome lines. The significant line effect allows us to estimate the amount of variation in MEN activity contributed by the different third chromosomes. Across all 10 lines the mean MEN activity of the MenEx+/Men+i flies was 0.041, with a line-specific standard deviation of 0.00346 or 11% of the mean activity.
The MenEx−/Men+i and MenEx+/Men+i samples were assayed for triglyceride concentration and IDH and G6PD activity (Figure 6, A–C). The data were analyzed for Men genotype and line effects following the same methods as the wild Men allele data. We found no consistent variation in triglyceride concentration with changes in MEN activity (F1,175 = 0.004, P < 0.95). A significant line effect on triglyceride concentration was observed (F1,175 = 2.13, P < 0.04), but there was no significant interaction between line and excision chromosome (F9,175 = 1.67, P < 0.11). For G6PD and IDH, we found highly significant increases in activity as MEN activity decreased across this small range (F1,175 = 8.41, P < 0.005 and F1,175 = 11.42, P < 0.001, respectively), similar to the results from the comparison between MenA and MenG alleles and the initial characterization of the Men excision set. The results also show a significant difference in IDH, but not in G6PD, activity among third chromosome lines (F9,175 = 5.86, P < 0.0001 and F9,175 = 1.39, P < 0.21, respectively). There were no significant line-by-excision chromosome interactions for IDH or G6PD activity (F9,175 = 0.64, P < 0.76 and F9,175 = 0.42, P < 0.92, respectively).
Figure 6.
Comparison in response to changes in MEN activity across 10 different different MenG third chromosome lines. MenEx+ and MenEx− flies were crossed to 10 different MenG (Men+i) lines to produce MenEx−/Men+i and MenEx+/Men+i heterozygotes. Each pair of points shows the change in (A) triglyceride concentration (micrograms per microgram protein), (B) G6PD activity, or (C) IDH activity with change in MEN activity (10,000 × μm NADPH/min/μg protein) in a different third chromosome background (Men+i). Values from MenEx−/Men+i flies are shown in open circles, and those from MenEx+/Men+i flies are in closed circles. Pairs of values for each Men+i are connected by a line. Log values are plotted so that the slope of each line is an estimate of the control (A) or elasticity (B and C) coefficient in each line. Note the large difference in MEN values (x-axis) reflecting line-specific differences in MEN activity. Note also the relative constancy in the sign of slope of the lines (direction of interaction) in B and C, but not in A.
DISCUSSION
This study examines the potential of an amino acid polymorphism in D. melanogaster malic enzyme to be under selection, examining the population genetics and biochemistry of the two alleles as well as the interaction between the two alleles and IDH and G6PD activity and triglyceride concentration. To complement our description of the two natural alleles, we used P-element excision-derived synthetic alleles to examine the effects of differences in MEN activity on IDH and G6PD activity and on triglyceride concentration. The strength of this approach is our ability to contrast the results from our examination of naturally occurring alleles with those from these laboratory-derived alleles.
The presence of both MenA and MenG alleles across all North American populations (Sezgin et al. 2004) suggests the possibility that the polymorphism may be under selection. There is, however, no DNA sequence-based evidence for selection maintaining this polymorphism; all Tajima's and Fu and Li's tests were not significant and Sezgin et al. (2004) found no evidence for geographic clinal variation in the frequency of the two alleles (which could have indicated selection between them). Tajima's and Fu and Li's tests are, however, relatively weak tests for selection and not all polymorphisms under selection will necessarily show clinal patterns of allele frequency. Further, similar amounts of synonymous polymorphism within samples of the two alleles (Table 1) suggest that the MenG allele is not a recent mutation. There is also apparent partitioning of silent polymorphisms between the two allele classes (Figure 2). This partitioning and the apparent age and widespread distribution of the amino acid polymorphism are intriguing, but do not implicate any one factor in the maintenance, or possible selective significance, of the amino acid polymorphism.
Under standard in vitro characterizations, the Men glycine/alanine amino acid polymorphism appears to result in differences in both Vmax and Km for malate. Comparison with the structure of the cytosolic malic enzyme from pigeon (from Yang et al. 2002) suggests that the amino acid polymorphism is structurally close to the active site of the molecule, particularly close to a tyrosine residue that is thought to interact with the substrate in the active site and is conserved across MEN proteins (Yang and Tong 2000). While assigning functional consequences to amino acid substitutions in a 3D structure of any enzyme is complex, we propose that the close proximity of the amino acid polymorphism to the enzyme active site may be responsible for the observed difference in both Vmax and Km for malate. Also, the polymorphism is in a helical region of the protein and glycine/alanine substitutions are known to affect helical stability (Ganter and Plückthun 1990; Chakrabartty et al. 1991), suggesting that the differences between the two alleles could be a product of differing protein stability. We found no evidence of any difference in thermal tolerance between the two alleles, however, suggesting that stability, at least as reflected in thermal stability, is not the mechanism conferring the observed differences.
Vmax is a function of enzyme concentration and the catalytic rate constant, kcat. In addition to possible modification of kcat by the amino acid substitution, the observed difference in Vmax could also result from, or be modified by, a linked regulatory polymorphism resulting in a greater amount of MENG (i.e., upregulation of transcription of the MenG allele). The structure of the regulatory region of the Men gene is not known but the linkage disequilibrium associated with the amino acid polymorphism and sites 1.2 kb away suggests that association between that site and a regulatory polymorphism is conceivable. Further, the amino acid change itself could result in differences in protein turnover (half-life) altering protein concentration and resulting in differences in Vmax.
The differences in Vmax and malate Km predict a 19% difference in in vivo activity (Vmax/Km) between alleles. This difference in Vmax/Km is correlated with significant differences in triglyceride concentration and IDH and G6PD activity; all three increase with decreasing MEN activity across this range (Figure 3). Additionally, in all three measures from the MenG/A heterozygotes are intermediate to those from the homozygous flies, suggesting that overdominance for activity between the two alleles does not occur. The IDH and G6PD results are consistent with a model of compensation to maintain a constant pool of reduced cofactor, NADPH, for lipogenesis (Geer et al. 1976). The observed increase in triglyceride concentration with decrease in MEN activity was unexpected given the role of MEN in lipogenesis. It is possible that this increase results from the observed increase in G6PD and IDH activities in effect overcompensating, leading to a significant increase in available NADPH and the standing concentration of triglyceride.
To complement our analysis of the naturally occurring Men alleles, we used P-element-mediated excision to create a set of Men alleles in isogenic backgrounds. This allele set included two lines containing deletion knockout Men alleles and two lines containing full-, or normal-, activity Men alleles. Because the alleles are derived from a single parent chromosome and result from small deletions spanning the promoter and/or beginning of the coding region they must differ only in the amount of transcript produced (not Km) and therefore Vmax accurately reflects the amount of in vivo activity of each allele. Additionally, because the lines are entirely isogenic they are free of differences in trans-acting effects and any observed differences in triglyceride concentration or IDH or G6PD activity can be directly attributed to the allele-specific differences in MEN activity.
Interactions between MEN, triglyceride, IDH, and G6PD are apparent in comparison of the excision alleles. Triglyceride concentration is halved with the complete loss of MEN activity, consistent with the proposal that MEN is a major source of NADPH for lipogenesis (Wise and Ball 1964; Geer et al. 1979a). There is no apparent difference in triglyceride between the two high-MEN-activity groups, in contrast to the negative correlation between MEN and triglyceride seen between the naturally occurring alleles. G6PD activity increases as MEN activity decreases, which is consistent with activating the pentose shunt to supply reduced NADPH for lipogenesis and also consistent with our results from the Men polymorphism. There is an apparent trend toward an initial increase in IDH with a small decrease in MEN followed by a larger decrease in IDH activity with decreasing MEN activity, although this was not statistically significant (Figure 4D). The initial increase in IDH activity is consistent with the results from the wild Men alleles. The final decrease in IDH activity with the Men complete knockout does not fit a model of activity compensation, but is consistent with some earlier observations of the interaction across large-scale differences in activity between these loci in larvae (Wilton et al. 1982; Bentley et al. 1983; Geer and Laurie-Ahlberg 1984). Gromnicki and Bentley (1991), however, found compensatory changes in activity between these enzymes. The interaction between IDH and MEN is apparently complex, at least over large-scale changes in activity.
It appears that we have identified a clear case of transvection at an enzyme locus; the MenEx−/Men+ heterozygotes had significantly higher than expected MEN activity irrespective of whether the Men deletions are placed in trans with the precisely excised allele (Figures 4 and 5A) or with the 10 wild chromosome alleles (Figure 5B). Our finding that Df(3R)kar3l/Men+ (large deficiency) heterozygotes have only half the activity of wild-type flies and significantly less than levels of activity in the MenEx−/Men+ (small deletion) heterozygotes (Figure 5A) suggests that upregulation of the functional gene in response to some physiological cue is not the source of the observed higher than expected activity. The consistency in levels of upregulation in crosses to different third chromosomes (Figure 5B) indicates that the apparent transvection effect is robust to genetic background. The 5′ upstream region of Men has no recognizable coding regions for >17 kb, yet possesses several small highly conserved noncoding regions (in comparison with other Drosophila species, data not shown) that could be enhancer sites. Both P-element excision-derived deletions remove >2 kb upstream of the Men transcription start site and can be assumed to have removed the Men promoter, likely a requisite for transvection (Morris et al. 1999, 2004).
The MenEx−/Men+i and MenEx+/Men+i flies were assayed for triglyceride concentration and IDH and G6PD activity to examine the consequences of the excision-associated difference in MEN activity in conjunction with a variety of background chromosomes (Figure 6, A–C). The difference in activity between the MenEx−/Men+i and MenEx+/Men+i flies was similar in magnitude to that between the naturally occurring MenA and MenG alleles, meaning that comparison of the synthetic allele genotypes is both biologically meaningful and directly comparable to that of the naturally occurring alleles. The results for G6PD and IDH activity are similar to those seen in our examination of the wild MenA and MenG alleles; both G6PD and IDH activity increase as MEN activity decreases (Figure 6, B and C), again suggesting compensation. In contrast, no consistent pattern of change in the average triglyceride concentration was detected across this upper range of MEN activity and in these 10 genetic backgrounds (Figure 6A). While the lack of consistent change in triglyceride concentration was unexpected, the concordance in results between the synthetic and natural Men allele experiments for IDH and G6PD activity supports our use of Vmax/Km, and not simply Vmax (which would have implied an opposite interaction), to estimate relative in vivo activity between the two naturally occurring Men alleles.
We can use this comparison of MenEx−/Men+i with MenEx+/Men+i flies to quantify the interactions between MEN activity and triglyceride concentration, IDH activity, and G6PD activity. A change in metabolite concentration in response to a change in enzyme activity can be quantified as a concentration control coefficient (Kacser and Burns 1981; Fell 1997), the ratio of change in concentration, S, to change in enzyme activity, E, scaled by S and E: 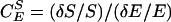 . Across small changes in enzyme,
. Across small changes in enzyme,  . Control coefficients can therefore be calculated as the slope of a plot of ln S vs. ln E across small changes in enzyme concentration (Kacser and Porteous 1987; Fell 1997). Control coefficients generally range from 0 to 1, but values >1, or <0, are possible in special circumstances. An enzyme with a coefficient near 1 has a large immediate influence over a given metabolite concentration. Figure 6A shows the changes in triglyceride concentration for each of the third chromosome crosses. The slope of the line connecting a pair of MEN values is an estimate the control of triglyceride concentration by malic enzyme,
. Control coefficients can therefore be calculated as the slope of a plot of ln S vs. ln E across small changes in enzyme concentration (Kacser and Porteous 1987; Fell 1997). Control coefficients generally range from 0 to 1, but values >1, or <0, are possible in special circumstances. An enzyme with a coefficient near 1 has a large immediate influence over a given metabolite concentration. Figure 6A shows the changes in triglyceride concentration for each of the third chromosome crosses. The slope of the line connecting a pair of MEN values is an estimate the control of triglyceride concentration by malic enzyme, 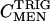 , in that comparison (Men+i background). The mean slope, and associated error, across all 10 lines gives an overall estimate of the control coefficient. The triglyceride results show no uniform pattern across lines; as MEN activity increased triglyceride concentration went up in 5 lines, but down in 5 lines (Figure 6A) and
, in that comparison (Men+i background). The mean slope, and associated error, across all 10 lines gives an overall estimate of the control coefficient. The triglyceride results show no uniform pattern across lines; as MEN activity increased triglyceride concentration went up in 5 lines, but down in 5 lines (Figure 6A) and 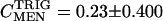 (mean ± SE). Overall, malic enzyme does not show significant control on triglyceride concentration across these 10 genetic backgrounds.
(mean ± SE). Overall, malic enzyme does not show significant control on triglyceride concentration across these 10 genetic backgrounds.
Elasticity coefficients (ɛ) quantify change in enzyme activity in response to change in some modifier (temperature, metabolite concentration, etc.) in a manner similar to that of a control coefficient. Strictly speaking, while the elasticity coefficient of an enzyme on itself, e.g., 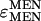 , is expected to be one (enzyme activity in exact proportion to enzyme concentration), the elasticity of one enzyme on another is expected to be zero; enzymes are not generally expected to directly interact (Westerhoff and van Dam 1987). The changes we observe in G6PD and IDH activity are most likely not direct, but mediated by the NADP/NADPH cofactor pool. Because our interest is in quantifying the possible responses between these loci, not necessarily the exact molecular mechanism, we can take the simplifying step of calculating the elasticity of IDH and G6PD with reference to MEN activity levels. We make this simplification with the understanding that we are quantifying the change in NADPH levels through changes in MEN activity and the associated responses of IDH and G6PD activity. In 9 of the 10 lines examined (Figure 6, B and C), G6PD activity increased as MEN activity decreased, with a mean elasticity coefficient (slope) of
, is expected to be one (enzyme activity in exact proportion to enzyme concentration), the elasticity of one enzyme on another is expected to be zero; enzymes are not generally expected to directly interact (Westerhoff and van Dam 1987). The changes we observe in G6PD and IDH activity are most likely not direct, but mediated by the NADP/NADPH cofactor pool. Because our interest is in quantifying the possible responses between these loci, not necessarily the exact molecular mechanism, we can take the simplifying step of calculating the elasticity of IDH and G6PD with reference to MEN activity levels. We make this simplification with the understanding that we are quantifying the change in NADPH levels through changes in MEN activity and the associated responses of IDH and G6PD activity. In 9 of the 10 lines examined (Figure 6, B and C), G6PD activity increased as MEN activity decreased, with a mean elasticity coefficient (slope) of 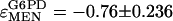 . Similarly, in all 10 lines examined IDH activity increased as MEN activity decreased, with a mean elasticity coefficient of
. Similarly, in all 10 lines examined IDH activity increased as MEN activity decreased, with a mean elasticity coefficient of 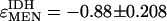 . The activities of MEN, IDH, and G6PD appear to be tightly interconnected, consistent with a need to maintain a stable pool of reduced NADPH. This interconnection is consistent in examination of both the naturally occurring and the excision series Men alleles. The relationship between MEN activity and triglyceride concentration is likely complicated by the interaction between MEN and these other sources of NADPH.
. The activities of MEN, IDH, and G6PD appear to be tightly interconnected, consistent with a need to maintain a stable pool of reduced NADPH. This interconnection is consistent in examination of both the naturally occurring and the excision series Men alleles. The relationship between MEN activity and triglyceride concentration is likely complicated by the interaction between MEN and these other sources of NADPH.
The glycine/alanine polymorphism in D. melanogaster malic enzyme is widespread in North America, but we find no population genetic evidence of selection maintaining its existence. However, to repeat a truism, an absence of evidence is not evidence of absence and sequence-based tests of selection are weak. In the absence of convincing sequence-based evidence, examination of the ability of genotypic differences to modify the physiology of the organism is a reasonable approach in examining selection potential. In sum, our results from the wild and engineered Men alleles indicate that MEN activity can affect the physiology of the fly and suggest that levels of MEN, IDH, and G6PD activity are coregulated to maintain the homeostasis of NADPH. Given the predicted difference in activity between the two naturally occurring Men alleles it seems unlikely that they are selectively neutral under all conditions. Further, the results presented here are from whole adult males fed ad lib on a complete diet and are likely minimum measurements of the effects of MEN activity. Future research, on both adult and larval flies, incorporating variation and stress in diet and environment, as well as direct modification of IDH and G6PD activities, will further clarify the interactions in this system and shed light on the extent of selection on this polymorphism.
Acknowledgments
Efe Sezgin, Andre Levy, Bengt Allen, Daniel Stoebel, and Daniel Dykhuizen all provided helpful discussion that shaped this work. This study was supported by U.S. Public Health Service grant GM-45247 to W.F.E. This is contribution no. 1142 from the Graduate Program in Ecology and Evolution, Stony Brook University, Stony Brook, New York.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Bentley, M. M., R. G. Meidinger and J. H. Williamson, 1983. Characterization of a low-activity allele of NADP+-dependent isocitrate dehydrogenase from Drosophila melanogaster. Biochem. Genet. 21: 725–733. [DOI] [PubMed] [Google Scholar]
- Cavener, D. R., and M. T. Clegg, 1981. Evidence for biochemical and physiological differences between enzyme genotypes in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 78: 4444–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabartty, A., J. A. Schellman and R. Baldwin, 1991. Large differences in helix propensities of alanine and glycine. Nature 351: 585–588. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., and L. E. Keith, 1988. Variation among extracted lines of Drosophila melanogaster in triacylglycerol and carbohydrate storage. Genetics 119: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden, A., 1995. Analysis of Biochemical Data. Oxford University Press, New York.
- Duggleby, R. G., 1979. Experimental designs for estimating the kinetic parameters for enzyme-catalysed reactions. J. Theor. Biol. 81: 671–684. [DOI] [PubMed] [Google Scholar]
- Duvernell, D. D., and W. F. Eanes, 2000. Contrasting molecular population genetics of four hexokinases in Drosophila melanogaster, D. simulans and D. yakuba. Genetics 156: 1191–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen, D. E., and A. M. Dean, 1990. Enzyme activity and fitness: evolution in solution. Trends Ecol. Evol. 5: 257–262. [DOI] [PubMed] [Google Scholar]
- Fell, D., 1997. Understanding the Control of Metabolism (Frontiers in Metabolism). Ashgate Publishing, Burlington, VT.
- Fersht, A., 1977. Enzyme structure and mechanism. W. H. Freeman, San Francisco.
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganter, C., and A. Plückthun, 1990. Glycine to alanine substitutions in helices of glyceraldehydes-3-phosphate dehydrogenase: effects on stability. Biochemistry 29: 9395–9402. [DOI] [PubMed] [Google Scholar]
- Geer, B. W., S. N. Kamiak, K. R. Kidd, R. A. Nishimura and S. J. Yemm, 1976. Regulation of the oxidative NADP-enzyme tissue levels in D. melanogaster. I. Modulation by dietary carbohydrate and lipid. J. Exp. Zool. 195: 15–32. [DOI] [PubMed] [Google Scholar]
- Geer, B. W., C. G. Woodward and S. D. Marshall, 1978. Regulation of the oxidative NADP-enzyme tissue levels in Drosophila melanogaster. II. The biochemical basis of dietary carbohydrate and D-glycerate modulation. J. Exp. Zool. 203: 391–402. [DOI] [PubMed] [Google Scholar]
- Geer, B. W., D. Krochko and J. H. Williamson, 1979. a Ontogeny, cell distribution, and the physiological role of NADP-malic enzyme in Drosophila melanogaster. Biochem. Genet. 17: 867–879. [DOI] [PubMed] [Google Scholar]
- Geer, B. W., D. L. Lindel and D. M. Lindel, 1979. b Relationship of the oxidative pentose shunt pathway to lipid synthesis in Drosophila melanogaster. Biochem. Genet. 17: 881–895. [DOI] [PubMed] [Google Scholar]
- Geer, B. W., D. Krochko, M. J. Oliver, V. K. Walker and J. H. Williamson, 1980. A comparative study of the NADP-malic enzymes from Drosophila and chick liver. Comp. Biochem. Physiol. 65B: 25–34. [Google Scholar]
- Geer, B. W., J. H. Williamson, D. R. Cavener and B. J. Cochrane, 1981. Dietary modulation of glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in Drosophila, pp 253–281 in Current Topics in Insect Endocrinology and Nutrition, edited by G. Bhaskaran, S. Friedman and J. G. Rodriguez. Plenum, New York.
- Geer, B. W., and C. C. Laurie-Ahlberg, 1984. Genetic variation in the dietary sucrose modulation of enzyme activities in Drosophila melanogaster. Genet. Res. 43: 307–321. [DOI] [PubMed] [Google Scholar]
- Gromnicki, A. R., and M. M. Bentley, 1991. The isolation and characterization of a mutant allele at a new X-linked locus, mex, affecting NADP+-dependent enzymes in Drosophila melanogaster. Biochem. Genet. 29: 145–162. [PubMed] [Google Scholar]
- Hall, J. G., 1985. Temperature-related kinetic differentiation of glucosephosphate isomerase alleloenzymes isolated from the blue mussel, Mytilus edulis. Biochem. Genet. 23: 705–728. [PubMed] [Google Scholar]
- Hall, J. G., and R. K. Koehn, 1983. The evolution of enzyme catalytic efficiency and adaptive inference from steady-state kinetic data. Evol. Biol. 15: 53–96. [Google Scholar]
- Henikoff, S., and L. Comai, 1998. Trans-sensing effects: the ups and downs of being together. Cell 93: 329–332. [DOI] [PubMed] [Google Scholar]
- Hochachka, P. W., and G. N. Somero, 2004. Biochemical Adaptation. Oxford University Press, New York.
- Hoskins, R. A., A. C. Phan, M. Naeemuddin, F. A. Mapa, D. A. Ruddy et al., 2001. Single nucleotide polymorphism markers for genetic mapping in Drosophila melanogaster. Genome Res. 11: 1100–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser, H., and J. A. Burns, 1981. The molecular basis of dominance. Genetics 97: 639–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacser, H., and W. Porteous, 1987. Control of metabolism: What do we measure? Trends Biochem. Sci. 12: 5–14. [Google Scholar]
- Laurie-Ahlberg, C. C., J. H. Williamson, B. J. Cochrane, A. N. Wilton and F. I. Chasalow, 1981. Autosomal factors with correlated effects on the activities of the glucose 6-phosphate and 6-phosphogluconate dehydrogenases in Drosophila melanogaster. Genetics 99: 127–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Ahlberg, C. C., A. N. Wilton, J. W. Curtsinger and T. H. Emigh, 1982. Naturally occurring enzyme activity variation in Drosophila melanogaster. I. Sources of variation for 23 enzymes. Genetics 102: 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, E. N., and J. R. Powell, 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277. [DOI] [PubMed] [Google Scholar]
- Morris, J. R., P. K. Geyer and C.-T. Wu, 1999. Core promoter elements can regulate transcriptome on a separate chromosome in trans. Genes Dev. 13: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J. R., D. A. Petrov, A. M. Lee and C.-T. Wu, 2004. Enhancer choice in cis and in trans in Drosophila melanogaster: role of the promoter. Genetics 167: 1739–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth, J., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sánchez-DelBarrio, X. Messegyer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Salz, H. K., T. W. Cline and P. Schedl, 1987. Functional changes associated with structural alterations induced by mobilization of a P element inserted in the Sex-lethal gene of Drosophila melanogaster. Genetics 117: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin, E., D. D. Duvernell, L. M. Matzkin, Y. Duan, C. T. Zhu et al., 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168: 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rolf, 1994. Biometry. W. H. Freeman, New York.
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota, S., and P. Schedl, 1986. Hybrid dysgenesis-induced revertants of insertions at the 5′ end of the rudimentary gene in Drosophila melanogaster: transposon-induced control mutations. Genetics 114: 165–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli, B. C., and W. F. Eanes, 2000. Extensive amino acid polymorphism at the Pgm locus is consistent with adaptive protein evolution in Drosophila melanogaster. Genetics 156: 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrelli, B. C., and W. F. Eanes, 2001. Clinal variation for amino acid polymorphism at the Pgm locus in Drosophila melanogaster. Genetics 157: 1649–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker, R. A., S. Ohnishi, C. H. Langley, J. Gausz and H. Gyurkovics, 1981. Genetic and cytogenetic studies of malic enzyme in Drosophila melanogaster. Biochem. Genet. 19: 525–534. [DOI] [PubMed] [Google Scholar]
- Watt, W. A., 1985. Bioenergetics and evolutionary genetics: opportunities for new synthesis. Am. Nat 125: 118–143. [Google Scholar]
- Watt, W. A., and A. M. Dean, 2000. Molecular-functional studies of adaptive genetic variation in prokaryotes and eukarylotes. Annu. Rev. Genet. 34: 593–622. [DOI] [PubMed] [Google Scholar]
- Westerhoff, H. V., and K. van Dam, 1987. Thermodynamics and Control of Biological Free-Energy Transduction. Elsevier, New York.
- Wilton, A. N., C. C. Laurie-Ahlberg, T. H. Emigh and J. W. Curtsinger, 1982. Naturally occurring enzyme activity variation in Drosophila melanogaster. II Relationships among enzymes. Genetics 102: 207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, E. M., and E. G. Ball, 1964. Malic enzyme and lipogenesis. Proc. Natl. Acad. Sci. USA 52: 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. T., and J. R. Morris, 1999. Transvection and other homology effects. Curr. Opin. Genet. Dev. 9: 237–246. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and L. Tong, 2000. Structural studies of a human malic enzyme. Protein Pept. Lett. 7: 287–296. [Google Scholar]
- Yang, Z., D. Floyd, G. Loeber and L. Tong, 2000. Structure of the closed form of human malic enzyme and implications for catalytic mechanism. Nat. Struct. Biol. 7: 251–257. [DOI] [PubMed] [Google Scholar]
- Yang, Z. T., H. Zhang, H.-C. Hung, C.-C. Kuo, L.-C. Tsai et al., 2002. Structural studies of the pigeon cytosolic NADP+-dependent malic enzyme. Protein Sci. 11: 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]