Abstract
In geographic areas where pied and collared flycatchers (Ficedula hypoleuca and F. albicollis) breed in sympatry, hybridization occurs, leading to gene flow (introgression) between the two recently diverged species. Notably, while such introgression is observable at autosomal loci it is apparently absent at the Z chromosome, suggesting an important role for genes on the Z chromosome in creating reproductive isolation during speciation. To further understand the role of Z-linked loci in the formation of new species, we studied genetic variation of the two species from regions where they live in allopatry. We analyzed patterns of polymorphism and divergence in introns from 9 Z-linked and 23 autosomal genes in pied and collared flycatcher males. Average variation on the Z chromosome is greatly reduced compared to neutral expectations based on autosomal diversity in both species. We also observe significant heterogeneity between patterns of polymorphism and divergence at Z-linked loci and a relative absence of polymorphisms that are shared by the two species on the Z chromosome compared to the autosomes. We suggest that these observations may indicate the action of recurrent selective sweeps on the Z chromosome during the evolution of the two species, which may be caused by sexual selection acting on Z-linked genes. Alternatively, reduced variation on the Z chromosome could result from substantially higher levels of introgression at autosomal than at Z-linked loci or from a complex demographic history, such as a population bottleneck.
AS incipient species diverge from each other under neutrality, a gradual loss of shared polymorphisms and accumulation of fixed differences are expected due to random genetic drift within each species (Wakeley and Hey 1997). Furthermore, levels of polymorphism and divergence across loci are expected to be correlated (Hudson et al. 1987). However, different parts of the genome may diverge at different rates. For example, if some loci are involved in adaptive evolution, or are linked to regions under selection, then these loci will show different patterns of variation from those that are evolving neutrally (Nachman 1997; Wang et al. 1997; Nachman et al. 1998; Fay and Wu 2000; Andolfatto 2001; Hey and Kliman 2002; Broughton and Harrison 2003; Schlötterer 2003). A useful approach to separate effects of selection from effects of demographic processes is to analyze patterns of polymorphism and divergence at several unlinked loci. Typically, selection will affect patterns of variation locally, near the loci under selection, whereas demographic processes affect the evolution of the entire genome.
Such multilocus DNA sequence data analysis is now a well-established tool to study selective forces acting in closely related species with a complex demographic history (Hey and Machado 2003). This approach has been effectively implemented to analyze regions associated with reproductive isolation in closely related Drosophila species (Begun 1996; Wang and Hey 1996; Kliman et al. 2000; Noor et al. 2001; Betancourt et al. 2002; Machado et al. 2002; Yi et al. 2003). These studies have demonstrated the existence of barriers to gene flow in regions of the Drosophila genome associated with regions important in speciation.
The pied flycatcher (Ficedula hypoleuca) and the collared flycatcher (F. albicollis) are two closely related, sexually dimorphic old-world flycatcher species. The role of selection in speciation has been investigated through analyzing interactions between the two species in sympatry. In regions where the pied (F. hypoleuca) and collared (F. albicollis) flycatcher have overlapping breeding areas (sympatric areas), a moderate level of gene flow has been observed between the two species at autosomal loci, whereas introgression of the Z chromosome is apparently absent (Saetre et al. 2003). A likely explanation for this pattern is that the Z chromosome is enriched with genes involved in species-specific adaptations. Accordingly, individuals with Z-linked genes introgressed from the other species would be removed by selection. In addition to the large role of the Z chromosome in reproductive isolation, it is also likely that Z-linked genes have diverged faster than the autosomes in these species because of sexual selection on Z-linked genes (Saetre et al. 2003). In the sympatric areas in Central Europe and on two islands in the Baltic Sea, there is evidence for a sexually selected character displacement on male plumage characteristics that increases assortative mating (Saetre et al. 1997). These male plumage characters are shown to be affected by Z-linked genes (Saetre et al. 2003) and have also been shown to be targets of sexual selection in allopatric populations (see, e.g., Saetre et al. 1997 and references therein). After two species have begun to diverge, traits involved in mate selection can contribute to reproductive isolation. It therefore seems likely that the emergence of reproductive isolation between the two flycatcher species has been accelerated by sexual selection.
To investigate this hypothesis we studied the evolutionary divergence of Z-linked and autosomal loci in pied and collared flycatchers that have been living in allopatry. We used pied flycatchers from Spain and collared flycatchers from Italy, which according to mtDNA divergence and a standard clock separated ∼2 million years ago (Saetre et al. 2001). These populations are geographically isolated from each other and are from the areas suggested to have housed the original populations isolated during the Pleistocene glaciations from where the pied and collared flycatchers recolonized northern Europe after the last glaciation period ∼10,000 years ago (Saetre et al. 2001). The birds from these allopatric populations are morphologically distinct from their conspecific sympatric populations, both in song and in male plumage characteristics (Saetre et al. 2003; Haavie et al. 2004). The geographical isolation of the two species means that these traits, which are involved in prezygotic isolation between the two species in sympatry, are likely to have evolved independently in the two populations, and that the patterns of genetic variation in each species have not been confounded by recent introgression. Relative rates of divergence at mtDNA and autosomal loci between these and other flycatcher species are in agreement with the geographic isolation hypothesis (Saetre et al. 2001, 2003). Current introgression is apparently restricted to narrow zones of secondary contact (steep clines) in central and eastern Europe and in the Baltic Isles (Saetre et al. 2003).
Here we present analyses of polymorphism and divergence from multiple introns from both Z-linked and autosomal loci in the pied and collared genomes. In total 20 introns located within 9 Z-linked genes (∼10 kb in total) and 25 introns from 23 autosomal genes (∼11 kb in total) in pied (n = 9) and collared (n = 9) flycatcher males were sequenced. In addition, one male red-breasted flycatcher (F. parva) is included as an outgroup species (see Saetre et al. 2001 for a justification). The evolutionary divergence of Z-linked genes is likely to have had significant effects on the development of barriers to gene flow between the pied and collared flycatchers (Saetre et al. 2003) and the results may shed light on the selective forces that result in the development of reproductive isolation.
MATERIALS AND METHODS
Samples:
Male collared flycatchers (n = 9) and pied flycatchers (n = 9) were trapped at breeding grounds in Abruzzo National Park, Italy and near Madrid, Spain, respectively. Additionally, one male red-breasted flycatcher caught in Northern Moravia, Czech Republic, is included in the analysis and referred to as the outgroup species.
DNA preparation:
Twenty-five microliters of blood from each male were collected by brachial vein puncture and suspended in 1 ml Queen's lysis buffer (Seutin et al. 1991). DNA was extracted from the blood samples by overnight incubation of 400 μl blood/buffer mix at 50° with 500 μl of 0.1 m NaCl, 10 mm Tris-Cl (pH 8.0), 5% SDS, and 50 μl of proteinase K solution (10 mg/ml). Each sample was extracted with two rounds of phenol/chloroform treatment before DNA was recovered by ethanol precipitation, dried, and redissolved in TE buffer (10 mm Tris, 1 mm EDTA, pH 7.5).
Introns from Z- and A-linked genes:
On the basis of chicken (Gallus gallus) sequences in GenBank, primers were designed as described in Primmer et al. (2002), to amplify introns from different genes widespread on the chicken Z chromosome and from autosomal genes. A large number of primer pairs were tested but many were later discarded due to amplification failures or because insertion-deletion polymorphisms (INDELs) or repetitive sequence made sequence alignment impossible. Finally, a total of 20 introns from 9 Z-linked genes and 25 introns from 23 autosomal genes that could be reliably aligned in both directions (see below) were chosen for further analysis. Even though there is extensive conservation of synteny between chicken and mammalian genomes for both chicken macrochromosomes and microchromosomes (Burt et al. 1999; Groenen et al. 2000) and between chicken and emu (Shetty et al. 1999), the intrachromosomal structure on the Z chromosome does not seem to be stable (Nanda and Schmid 2002). Hence, the exact intrachromosomal positions of the flycatcher genes are not necessarily the same as those in the model organism (supplemental Tables S1 and S2 at http://www.genetics.org/supplemental/). However, the chromosomal location of each gene (autosomal or Z-linked) in the flycatchers was confirmed by typing female F1-hybrids, where fixed differences between species would be identifiable from heterozygous sites at autosomal loci but not at Z-linked loci. As some of the Z-linked genes have a W-linked gametologue we have used only males (the homogametic sex; ZZ) in the comparative sequence analyses. Details of all loci, primers, and PCR conditions are available as supplemental information (Z-linked, S1; and autosomal, S2 at http://www.genetics.org/supplemental/).
PCR:
The introns were amplified in all 19 individuals in 10-μl reactions containing 2.5 mm MgCl2, 0.2 mm of each dNTP, 0.32 μm of each primer, 1 μg of bovine serum albumin (BSA), 0.3 units of HotStar DNA polymerase (QIAGEN, Valencia, CA), 1× PCR buffer (QIAGEN), and 20 ng DNA. On a PTC 225 (MJ Research, Watertown, MA) 35–40 cycles of amplification with 94° for 30 sec, 52–67° for 30 sec, and 72° for 1 min were preceded by 15 min predenaturation at 95° and followed by a prolonged 10-min extension step at 72°.
Sequencing:
PCR products were purified with ExoSap-IT (United States Biochemical, Cleveland) and cycle sequenced with either BigDye terminator chemistry or the DYEnamic ET Terminator cycle sequencing kit (Amersham Biosciences, Arlington Heights, IL), depending on whether they were analyzed on an ABI 377 automated sequencer (Perkin-Elmer, Norwalk, CT) or on a MegaBACE 1000 (Amersham Biosciences), respectively. The sequences were aligned and edited in the programs AutoAssembler 2.1 (Applied Biosystems) and Sequence Navigator 1.0 (Applied Biosystems) or with Sequencher 4.1 (Gene Codes, Ann Arbor, MI) and modified manually. Each base in the study was called, using at least single-fold coverage sequencing reads for each strand. The only exceptions were for a few short regions where sequence repeats or INDELs made the reads in one direction of poor quality, and bases were called using information primarily from multiple sequences of one strand.
Data analysis:
Diploid sequences from each individual in all three species at each locus were first separated into two “pseudo-haplotypes” on the basis of the ambiguity codes produced by Sequencher using a Perl script. This was done by randomly assigning the two alleles at each polymorphic site to one of two sequences to produce two haplotypes of unknown phase. The sequences derived from all three species at each locus were then aligned using ClustalW (Thompson et al. 1994) and improved by manual adjustment. A number of additional Perl programs were used during the analysis to extract polymorphism information from the sequences and to format them for use in data analysis programs.
Most polymorphism and divergence analyses, including tests of neutrality based on the allele frequency spectrum, were performed using the DnaSP program (Rozas and Rozas 1999). The H-test (Fay and Wu 2000) was performed using the htest program (http://crimp.lbl.gov/htest.html). Haplotype frequency estimation was performed using the expectation-maximization (EM) algorithm implemented in Arlequin (Schneider et al. 2000). DnaSP was used to calculate RM, the minimum number of recombination events in each sample. The program MEGA (Kumar et al. 2001) was used to construct neighbor-joining trees using inferred haplotypes. Multilocus Hudson-Kreitman-Aguadé (HKA) tests (Hudson et al. 1987) were performed using the HKA program (http://lifesci.rutgers.edu/∼heylab/). Fitting of the isolation model of speciation to the data was performed using the method described in Wakeley and Hey (1997) and Wang et al. (1997), using the program WH (http://lifesci.rutgers.edu/∼heylab/).
RESULTS
The data set:
Details of all loci in the study are shown in Table 1. These include 20 loci from 9 Z-linked genes (9657 bp in total) and 25 loci from 23 autosomal genes (11,539 bp in total). The majority of sequence is noncoding although some exon sequence is included in the analysis. Two collared flycatcher-exclusive nonsynonymous polymorphisms were found: one in the Z-linked locus GHR-exon 9 and one in the autosomal locus MPP-4 (see supplemental information supplemental Table S3 and supplemental Table S4 at http://www.genetics.org/supplemental/ for more details).
TABLE 1.
Sample sequences
| Cytological position
|
Length
|
|||
|---|---|---|---|---|
| Locusa | Total | Exon | Intron | |
| ALDOB-4 | Z | 228 | 38 | 190 |
| ALDOB-6 | Z | 516 | 40 | 476 |
| ALDOB-7 | Z | 153 | 15 | 138 |
| BRM-12 | Z | 1604 | 29 | 1575 |
| BRM-15 | Z | 368 | 78 | 290 |
| CHDZ-15 | Z | 501 | 37 | 464 |
| CHDZ-18 | Z | 308 | 69 | 239 |
| CHDZ-20 | Z | 941 | 28 | 913 |
| CHDZ-24 | Z | 460 | 76 | 384 |
| GHR-5.1 | Z | 246 | 0 | 246 |
| GHR-5.2 | Z | 507 | 0 | 507 |
| GHR-exon 9 | Z | 102 | 102 | 0 |
| PKCI-2 | Z | 448 | 39 | 409 |
| PRLR-UN | Z | 335 | 168 | 167 |
| PTCH-6 | Z | 579 | 42 | 537 |
| SPINZ-2 | Z | 991 | 96 | 895 |
| VLDLR-7 | Z | 575 | 123 | 452 |
| VLDLR-8 | Z | 191 | 114 | 77 |
| VLDLR-9 | Z | 409 | 33 | 376 |
| VLDLR-12 | Z | 196 | 72 | 124 |
| ACLY-16 | A | 443 | 3 | 440 |
| ACTB-2 | A | 544 | 50 | 494 |
| ACTB-3 | A | 379 | 44 | 335 |
| ALAS1-8 | A | 403 | 0 | 403 |
| CEPU-1 | A | 582 | 0 | 582 |
| CHC-5 | A | 355 | 0 | 355 |
| CKB-2 | A | 465 | 0 | 465 |
| ENO1-8 | A | 177 | 49 | 128 |
| FAS-Y | A | 901 | 0 | 901 |
| FN1-31 | A | 443 | 0 | 443 |
| FTH-C | A | 520 | 0 | 520 |
| GAPD-3 | A | 210 | 77 | 133 |
| GAPD-11 | A | 147 | 0 | 147 |
| GH1-2 | A | 611 | 0 | 611 |
| LAMA-2 | A | 224 | 124 | 100 |
| LHCGR-7 | A | 634 | 0 | 634 |
| MPP-4 | A | 246 | 61 | 185 |
| ODC1-9 | A | 454 | 115 | 339 |
| RHO-1 | A | 404 | 0 | 404 |
| RPL30-3 | A | 1065 | 0 | 1065 |
| RPL37A-3 | A | 610 | 3 | 607 |
| RPL5-4 | A | 556 | 4 | 552 |
| RPL7A-4 | A | 414 | 83 | 331 |
| TGFB2-5 | A | 402 | 0 | 402 |
| TM-5 | A | 376 | 0 | 376 |
The locus identifiers consist of the gene abbreviation followed by the specific intron/exon identified in the chicken gene (see supplemental Tables A1 and A2 at http://www.genetics.org/supplemental/).
Description of polymorphism and divergence:
Summaries of polymorphism on Z-linked and autosomal loci are shown in Figure 1 and detailed in Tables 2 and 3, respectively (loci from the same gene are combined). A number of estimates of the population mutation rate, θ, are presented (θ = 4Neμ for autosomal loci and 3Neμ for Z-linked loci, where Ne is the effective population size and μ is the neutral mutation rate). These are based on the number of segregating sites, average number of pairwise differences, and allele frequency distribution weighted by derived alleles (Watterson 1975; Nei 1987; Fay and Wu 2000). Levels of variation in the pied flycatcher are slightly lower than those in the collared flycatcher at both Z-linked and autosomal loci (the variation in the pied is 93% of collared flycatcher levels at Z-linked loci and 76% of collared flycatcher levels at autosomal loci, on the basis of θW calculated from silent sites: 0.0015 and 0.0014 on the Z chromosome and 0.0040 and 0.0030 on autosomes in the collared flycatcher and the pied flycatcher, respectively). As divergence from the outgroup is about the same in both species, indicating that mutation rate is unlikely to vary between lineages, the lower variation probably reflects a lower effective population size in the pied flycatcher.
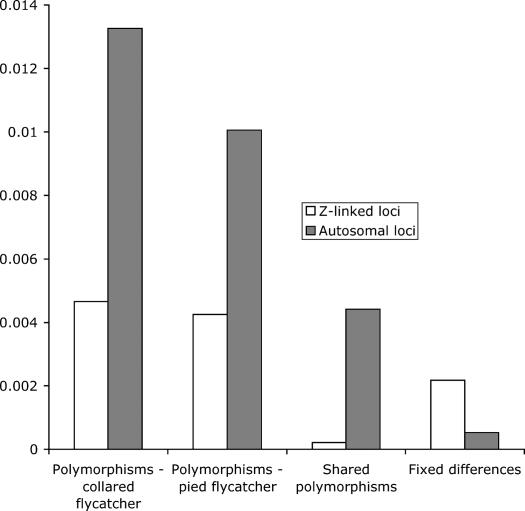
Figure 1.
Descriptive statistics of polymorphism and divergence in collared and pied flycatchers at 9 Z-linked loci (9657 bp) and 23 autosomal loci (11539 bp). For both classes of loci the observed number of segregating sites divided by the total length of loci is displayed. The number of polymorphisms segregating in each species is shown alongside the number of shared polymorphisms and the number of fixed differences between species (per base pair).
TABLE 2.
Polymorphism summaries of Z-linked genes
| Gene | Speciesa | Lb | Kc | Sd | Sexpe | sf | πg (SD) | θWh (SD) | θHi | DTj | DFLk | FFLl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALDOB | IC | 897 | 18 | 1 | 11.4 | 0 | 0.0002 (0.0001) | 0.0003 (0.0003) | 0.0019* | −0.53 | 0.64 | 0.40 |
| SP | 897 | 16 | 4 | 7.5 | 1 | 0.0011 (0.0003) | 0.0013 (0.0008) | 0.0011 | −0.42 | 0.16 | 0.00 | |
| BRM | IC | 1971 | 53 | 11 | 33.6 | 6 | 0.0013 (0.0002) | 0.0016 (0.0007) | 0.0017 | −0.81 | −0.89 | −1.04 |
| SP | 1971 | 51 | 13 | 24.0 | 0 | 0.0027 (0.0002) | 0.0019 (0.0008) | 0.0021 | 1.56 | 1.65* | 1.96* | |
| CHDZ | IC | 2210 | 43 | 1 | 27.3 | 1 | 0.0001 (0.0000) | 0.0001 (0.0001) | 0.0000 | −1.16 | −1.56 | −1.69 |
| SP | 2210 | 45 | 7 | 21.2 | 0 | 0.0011 (0.0001) | 0.0009 (0.0005) | 0.0005 | 0.57 | 1.39 | 1.37 | |
| GHR | IC | 855 | 12 | 4 | 7.6 | 1 | 0.0017 (0.0002) | 0.0014 (0.0008) | 0.0010 | 0.84 | 0.16 | 0.41 |
| SP | 855 | 13 | 0 | 6.1 | 0 | 0 (0) | 0 (0) | 0 | — | — | — | |
| PKCI | IC | 448 | 3 | 3 | 1.9 | 2 | 0.0013 (0.0013) | 0.0020 (0.0013) | 0.0003 | −0.90 | −1.33 | −1.43 |
| SP | 448 | 3 | 1 | 1.4 | 0 | 0.0010 (0.0002) | 0.0007 (0.0007) | 0.0025 | 0.87 | 0.64 | 0.81 | |
| PRLR | IC | 335 | 0 | 0 | 0 | 0 | 0 (0) | 0 (0) | 0.0002 | — | — | — |
| SP | 335 | 2 | 0 | 0.9 | 0 | 0 (0) | 0 (0) | 0 | — | — | — | |
| PTCH | IC | 579 | 12 | 0 | 7.6 | 0 | 0 (0) | 0 (0) | 0 | — | — | — |
| SP | 579 | 12 | 3 | 5.6 | 0 | 0.0019 (0.0003) | 0.0015 (0.0010) | 0.0046* | 0.68 | 1.03 | 1.10 | |
| SPINZ | IC | 991 | 20 | 4 | 12.7 | 1 | 0.0012 (0.0002) | 0.0012 (0.0007) | 0.0007 | 0.14 | 0.16 | 0.18 |
| SP | 991 | 20 | 10 | 9.4 | 0 | 0.0044 (0.0005) | 0.0029 (0.0012) | 0.0043 | 1.75 | 1.54* | 1.93* | |
| VLDLR | IC | 1369 | 9 | 21 | 5.7 | 6 | 0.0050 (0.0003) | 0.0045 (0.0018) | 0.0043 | 0.48 | 0.33 | 0.46 |
| SP | 1371 | 10 | 3 | 4.7 | 0 | 0.0005 (0.0002) | 0.0006 (0.0004) | 0.0001 | −0.78 | 1.03 | 0.63 | |
| Total | IC | 9655 | 170 | 45 | 107.8 | 17 | 0.0015 (0.0003) | 0.0014 (0.0009) | 0.0013 | −0.04 | −0.30 | −0.26 |
| SP | 9657 | 172 | 41 | 80.9 | 1 | 0.0010 (0.0002) | 0.0009 (0.0006) | 0.0015 | 1.12 | 1.83** | 1.96* |
**P < 0.02, *P < 0.05.
IC, Italian collared flycatcher; SP, Spanish pied flycatcher.
Number of sites surveyed (same in both species).
Divergence with outgroup.
Number of segregating sites.
The expected number of segregating sites based on Ne estimated from autosomal loci.
Number of singleton sites.
Average pairwise sequence difference per nucleotide (Nei 1987).
Expected heterozygosity per nucleotide (Watterson 1975).
TABLE 3.
Polymorphism summaries of autosomal genes
| Gene | Speciesa | Lb | Kc | Sd | Sexpe | sf | πg (SD) | θWh (SD) | θHi | DTj | DFLk | FFLl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACLY | IC | 417 | 7 | 1 | 2.5 | 1 | 0.0003 (0.0002) | 0.0007 (0.0007) | 0.0000 | −1.16 | −1.56 | −1.69 |
| SP | 417 | 6 | 2 | 1.9 | 2 | 0.0005 (0.0003) | 0.0014 (0.0010) | 0.0000 | −1.51 | −2.13 | −2.29 | |
| ACTB | IC | 923 | 13 | 3 | 4.6 | 1 | 0.0010 (0.0002) | 0.0009 (0.0006) | 0 | −0.09 | −0.15 | −0.16 |
| SP | 923 | 12 | 8 | 3.8 | 2 | 0.0020 (0.0003) | 0.0025 (0.0009) | 0 | −0.67 | 0.20 | −0.07 | |
| ALAS1 | IC | 403 | 15 | 15 | 5.3 | 6 | 0.0104 (0.0009) | 0.0108 (0.0046) | 0.0150 | −0.14 | −0.64 | −0.58 |
| SP | 403 | 17 | 7 | 5.4 | 2 | 0.0045 (0.0006) | 0.0051 (0.0025) | 0.0025 | −0.39 | 0.02 | −0.11 | |
| CEPU | IC | 582 | 5 | 8 | 1.8 | 4 | 0.0022 (0.0004) | 0.0040 (0.0019) | 0.0002 | −1.56 | −0.42 | −0.90 |
| SP | 582 | 5 | 7 | 1.6 | 2 | 0.0035 (0.0005) | 0.0035 (0.0017) | 0.0022 | 0.01 | 0.71 | 0.60 | |
| CHC | IC | 355 | 4 | 7 | 1.4 | 5 | 0.0036 (0.0010) | 0.0057 (0.0028) | 0.0073 | −1.25 | 0.02 | −0.41 |
| SP | 355 | 5 | 6 | 1.6 | 3 | 0.0038 (0.0007) | 0.0049 (0.0025) | 0.0164* | −0.76 | 0.57 | 0.22 | |
| CKB | IC | 465 | 5 | 7 | 1.8 | 2 | 0.0039 (0.0007) | 0.0044 (0.0022) | 0.0049 | −0.35 | 0.71 | 0.48 |
| SP | 465 | 5 | 4 | 1.6 | 1 | 0.0036 (0.0003) | 0.0025 (0.0015) | 0.0030 | 1.35 | 1.15 | 1.42 | |
| ENO1 | IC | 177 | 2 | 1 | 0.7 | 0 | 0.0027 (0.0005) | 0.0016 (0.0016) | 0.0053 | 1.17 | 0.64 | 0.89 |
| SP | 177 | 2 | 1 | 0.6 | 1 | 0.0006 (0.0005) | 0.0016 (0.0016) | 0.0000 | −1.16 | −1.56 | −1.69 | |
| FAS | IC | 901 | 17 | 2 | 6.0 | 0 | 0.0006 (0.0002) | 0.0007 (0.0005) | 0.0001 | −0.33 | 0.87 | 0.65 |
| SP | 900 | 16 | 0 | 5.1 | 0 | 0 (0) | 0 (0) | 0 | 0 | 0 | 0 | |
| FN1 | IC | 443 | 5 | 8 | 1.8 | 2 | 0.0054 (0.0006) | 0.0053 (0.0025) | 0.0021 | 0.08 | 0.20 | 0.20 |
| SP | 443 | 6 | 5 | 1.9 | 2 | 0.0030 (0.0007) | 0.0033 (0.0018) | 0.0013 | −0.29 | 0.39 | 0.23 | |
| FTH | IC | 520 | 7 | 8 | 2.5 | 3 | 0.0044 (0.0008) | 0.0045 (0.0021) | 0.0033 | −0.08 | −0.42 | −0.38 |
| SP | 520 | 8 | 4 | 2.5 | 3 | 0.0016 (0.0004) | 0.0022 (0.0013) | 0.0018 | −0.93 | −0.83 | −1.01 | |
| GAPD | IC | 357 | 0 | 6 | 0 | 1 | 0.0054 (0.0009) | 0.0049 (0.0020) | 0 | 0.31 | 0.57 | 0.58 |
| SP | 357 | 1 | 2 | 0.3 | 2 | 0.0006 (0.0004) | 0.0016 (0.0012) | 0 | −1.51 | −0.63 | −1.00 | |
| GH1 | IC | 611 | 21 | 9 | 7.4 | 5 | 0.0027 (0.0006) | 0.0043 (0.0020) | 0.0004 | −1.33 | 0.93 | 0.31 |
| SP | 611 | 20 | 9 | 6.4 | 1 | 0.0045 (0.0010) | 0.0043 (0.0020) | 0.0050 | 0.15 | 0.93 | 0.83 | |
| LAMA | IC | 224 | 7 | 0 | 2.5 | 0 | 0 (0) | 0 (0) | 0 | 0 | 0 | 0 |
| SP | 224 | 6 | 1 | 1.9 | 1 | 0.0005 (0.0004) | 0.0013 (0.0013) | 0.0084* | −1.16 | 0.64 | 0.21 | |
| LHCGR | IC | 634 | 8 | 6 | 2.8 | 3 | 0.0024 (0.0004) | 0.0028 (0.0014) | 0.0008 | −0.42 | −0.19 | −0.31 |
| SP | 634 | 8 | 7 | 2.5 | 3 | 0.0024 (0.0004) | 0.0032 (0.0016) | 0.0006 | −0.87 | −0.66 | −0.86 | |
| MPP | IC | 246 | 1 | 4 | 0.4 | 1 | 0.0055 (0.0010) | 0.0047 (0.0028) | 0.0070 | 0.47 | 0.16 | 0.29 |
| SP | 246 | 1 | 2 | 0.3 | 0 | 0.0034 (0.0005) | 0.0024 (0.0018) | 0.0081 | 1.02 | 0.87 | 1.06 | |
| ODC1 | IC | 454 | 12 | 10 | 4.2 | 1 | 0.0074 (0.0006) | 0.0064 (0.0029) | 0.0060 | 0.58 | 1.54* | 1.50 |
| SP | 454 | 13 | 6 | 4.1 | 1 | 0.0051 (0.0005) | 0.0038 (0.0020) | 0.0045 | 1.04 | 0.57 | 0.83 | |
| RHO | IC | 404 | 7 | 7 | 2.5 | 1 | 0.0058 (0.0007) | 0.0050 (0.0025) | 0.0053 | 0.51 | 0.71 | 0.77 |
| SP | 404 | 7 | 7 | 2.2 | 1 | 0.0062 (0.0007) | 0.0050 (0.0025) | 0.0042 | 0.81 | 0.71 | 0.88 | |
| RPL30 | IC | 1065 | 17 | 21 | 6.0 | 4 | 0.0054 (0.0007) | 0.0057 (0.0023) | 0.0083 | −0.23 | 0.92 | 0.68 |
| SP | 1065 | 18 | 13 | 5.7 | 7 | 0.0026 (0.0007) | 0.0036 (0.0015) | 0.0090* | −0.98 | −0.53 | −0.81 | |
| RPL37a | IC | 610 | 12 | 11 | 4.2 | 3 | 0.0057 (0.0006) | 0.0052 (0.0023) | 0.0086 | 0.30 | 0.59 | 0.61 |
| SP | 610 | 13 | 9 | 4.1 | 3 | 0.0042 (0.0006) | 0.0043 (0.0020) | 0.0101* | −0.07 | 0.35 | 0.27 | |
| RPL5 | IC | 556 | 5 | 4 | 1.8 | 2 | 0.0016 (0.0003) | 0.0021 (0.0012) | 0.0003 | −0.72 | −0.83 | −0.94 |
| SP | 556 | 6 | 5 | 1.9 | 3 | 0.0020 (0.0005) | 0.0026 (0.0014) | 0.0018 | −0.77 | −0.47 | −0.66 | |
| RPL7a | IC | 414 | 4 | 4 | 1.4 | 0 | 0.0025 (0.0005) | 0.0028 (0.0016) | 0.0006 | −0.29 | 1.15 | 0.89 |
| SP | 414 | 4 | 2 | 1.3 | 1 | 0.0015 (0.0003) | 0.0014 (0.0011) | 0.0010 | 0.22 | −0.63 | −0.47 | |
| TGFB2 | IC | 402 | 5 | 6 | 1.8 | 4 | 0.0028 (0.0008) | 0.0043 (0.0022) | 0.0005 | −1.21 | −0.95 | −1.22 |
| SP | 402 | 5 | 3 | 1.6 | 1 | 0.0023 (0.0004) | 0.0022 (0.0014) | 0.0021 | 0.12 | −0.15 | −0.09 | |
| TM | IC | 376 | 2 | 5 | 0.7 | 1 | 0.0040 (0.0008) | 0.0039 (0.0021) | 0.0084 | 0.19 | 0.39 | 0.39 |
| SP | 376 | 1 | 6 | 0.3 | 1 | 0.0048 (0.0006) | 0.0046 (0.0024) | 0.0064 | 0.14 | 0.57 | 0.53 | |
| Total | IC | 11539 | 181 | 153 | 63.9 | 52 | 0.0036 (0.0001) | 0.0039 (0.0014) | 0 | −0.32 | −0.11 | −0.20 |
| SP | 11538 | 185 | 116 | 58.8 | 43 | 0.0027 (0.0001) | 0.0029 (0.0010) | 0 | −0.31 | −0.36 | −0.40 |
P < 0.05.
IC, Italian collared flycatcher; SP, Spanish pied flycatcher.
Number of sites surveyed (same in both species).
Divergence with outgroup.
Number of segregating sites.
The expected number of segregating sites based on Ne estimated from Z-linked loci.
Number of singleton sites.
Average pairwise sequence difference per nucleotide (Nei 1987).
Expected heterozygosity per nucleotide (Watterson 1975).
Genetic divergence from the outgroup on the Z chromosome is slightly higher than that on the autosomes (0.0176 and 0.0178 on the Z chromosome and 0.0157 and 0.0160 on the autosomes for the collared and pied flycatchers, respectively). A likely explanation for this is a higher mutation rate in males, because the male germline goes through many more rounds of cell divisions per generation than does the female germline (Haldane 1935). This is supported by a number of studies of evolutionary rates on the avian Z chromosome indicating a significant male bias in mutation rate (Ellegren and Fridolfsson 1997; Kahn and Quinn 1999; Carmichael et al. 2000) although the likely effect of different levels of ancestral polymorphism at Z-linked and autosomal loci makes an accurate estimate of the male bias difficult (Makova and Li 2002).
In contrast to patterns of divergence, levels of polymorphism on the Z chromosome are markedly lower than those on autosomes (Tables 2 and 3). When only silent sites are considered, θW is 0.0015 and 0.0014 on the Z chromosome and 0.0040 and 0.0030 on autosomes in the collared flycatcher and the pied flycatcher, respectively. Hence, levels of Z-linked variation are 38% of autosomal levels in the collared and 47% of those in the pied flycatcher. Under a standard neutral model assuming constant population size, random mating, and no migration (Watterson 1975) the higher mutation rate on Z would be expected to increase variation relative to the autosomes whereas the mode of inheritance of Z chromosomes would be expected to reduce it by a factor of 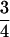 . We calculated the expected variation at each Z-linked locus on the basis of the effective population size estimated from average levels of polymorphism and divergence from the outgroup at autosomal loci and the mutation rate estimated from levels of divergence from the outgroup at each individual Z-linked locus, using the formula
. We calculated the expected variation at each Z-linked locus on the basis of the effective population size estimated from average levels of polymorphism and divergence from the outgroup at autosomal loci and the mutation rate estimated from levels of divergence from the outgroup at each individual Z-linked locus, using the formula
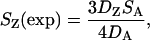 |
(1) |
where SZ(exp) is the expected number of segregating sites at a specific Z-linked locus, DZ is the divergence between the species (collared or pied flycatcher) and the outgroup (red-breasted flycatcher) at this locus, SA is the total number of segregating sites at autosomal loci, and DA is the total divergence between the species and the outgroup at autosomal loci. The corresponding formula for estimating the expected number of segregating sites at an autosomal locus from average polymorphism and divergence on Z is
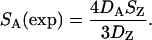 |
(2) |
We used these equations to calculate the expected number of segregating sites at each locus in both species in both Z-linked and autosomal loci. As shown in Table 2, levels of polymorphism on the Z chromosome in both flycatcher species are about half of those predicted from levels of polymorphism and divergence on autosomal loci. In 14 of 18 possible cases, variation on Z is lower than expected on the basis of autosomal data (signs test, P = 0.0154). Accordingly, variation at autosomal loci (Table 3) is higher than expected on the basis of Z-linked data in 40 out of 46 comparisons (P < 10−6). Hence, variation at Z-linked loci is significantly reduced compared to that at the autosomes under a standard neutral model (see also Figure 1).
Allele frequency spectra:
We performed four tests of neutrality based on allele frequency distributions: Tajima's (1989) D-, Fu and Li's (1993) D- and F-, and Fay and Wu's (2000) H-tests (Tables 2 and 3). For Fu and Li's statistics and Fay and Wu's test, the outgroup was used to determine the ancestral state of diallelic polymorphisms. In the collared flycatcher, both Z-linked and autosomal loci exhibit slightly negative values of Tajima's D and Fu and Li's statistics, indicating that the allele frequency spectrum closely matches the neutral expectations with a slight skew toward rare alleles. In the pied flycatcher, autosomal loci exhibit the same pattern. However, at Z-linked loci, these statistics are all positive and Fu and Li's D and F for all loci combined both display significant deviations from the neutral model, indicating a deficit of rare derived alleles. Fu and Li's statistics are strongly dependent on the number of singleton polymorphisms at a locus and this significant result reflects the lack of singletons at Z-linked loci in the pied flycatcher.
Significant H-test results were obtained from a small number of Z-linked and autosomal loci, indicating an excess of high-frequency derived alleles. However, it is unclear whether these results have any biological relevance, considering the large number of tests performed, and they are likely to represent random evolutionary variance under neutrality. The same is likely to be true for individual loci with significant Tajima's and Fu and Li's tests.
HKA tests:
We used multilocus HKA tests to determine whether levels of polymorphism and divergence are correlated between loci and species, as predicted under neutrality. All tests used only the polymorphism and divergence data from the collared and the pied flycatcher, and introns from the same gene were combined. To determine significance, the test statistic was compared to a distribution generated from 10,000 coalescent simulations. When only Z-linked loci were compared, significant heterogeneity was observed between loci (χ2 = 24.56, P = 0.023). Significant deviations were not observed between autosomal loci (χ2 = 16.44, P = 0.975). When combined autosomal and combined Z-linked loci from the same species were used, the test statistic was also not significant (χ2 = 1.27, P = 0.441).
The expected values of number of segregating sites and divergence between the species for the Z-linked locus from the significant HKA test are shown in Table 4. The VLDLR locus shows the greatest deviation from the neutral model: in the collared flycatcher the observed number of segregating sites is twice that expected whereas in the pied flycatcher it is less than half. When this locus is removed, the test is no longer significant (χ2 = 16.23, P = 0.143). In addition, a number of loci show smaller deviations from neutrality with respect to the observed number of segregating sites. In the collared flycatcher, ALDOB and CHDZ have lower than expected values, whereas in the pied flycatcher GHR has a low value and the values for PTCH and SPINZ are higher than expected.
TABLE 4.
HKA results for Z-linked genes
| Segregating sites
|
||||||
|---|---|---|---|---|---|---|
| Collared flycatcher
|
Pied flycatcher
|
Divergence
|
||||
| Gene | Obs | Exp | Obs | Exp | Obs | Exp |
| ALDOB | 1 | 3.75 | 4 | 3.33 | 6 | 3.92 |
| BRM | 11 | 12.61 | 13 | 11.21 | 13 | 13.17 |
| CHDZ | 1 | 5.11 | 7 | 4.55 | 7 | 5.34 |
| GHR | 4 | 2.73 | 0 | 2.42 | 4 | 2.85 |
| PKCI | 3 | 1.70 | 1 | 1.52 | 1 | 1.78 |
| PRLR | 0 | 0.68 | 0 | 0.61 | 2 | 0.71 |
| PTCH | 0 | 1.70 | 3 | 1.52 | 2 | 1.78 |
| SPINZ | 4 | 6.14 | 9 | 5.45 | 5 | 6.41 |
| VLDLR | 21 | 10.57 | 3 | 9.39 | 7 | 11.04 |
Obs, observed; Exp, expected.
Shared and fixed polymorphisms:
Variable sites within the two flycatcher species can be divided into four categories, those that are polymorphic only in the collared flycatcher, those that are polymorphic only in the pied flycatcher, those that are polymorphic in both (shared polymorphisms), and fixed differences. The numbers of sites in these different classes are shown in Table 5. This reveals a striking pattern: Z-linked loci have a large number of fixed differences but very few shared polymorphisms (Figure 1, Table 5), whereas the reverse is true for autosomal loci. Some shared polymorphisms are expected because of mutations on the same site in both species. However, closely related species are expected to show higher levels of shared polymorphisms because they have persisted since the time of divergence. The proportion of shared polymorphisms generated by multiple parallel mutations is expected to be very low (Clark 1997).
TABLE 5.
Fixed and shared polymorphisms
| Sx-IC | Sx-SP | Sfixed | Sshared | Saverage | Length (bp) | |
|---|---|---|---|---|---|---|
| Z-linked genes | ||||||
| ALDOB | 1 | 4 | 4 | 0 | 5.83 | 897 |
| BRM | 10 | 12 | 6 | 1 | 13.14 | 1971 |
| CHDZ | 1 | 7 | 5 | 0 | 6.72 | 2210 |
| GHR | 4 | 0 | 3 | 0 | 4.11 | 855 |
| PKCI | 3 | 1 | 0 | 0 | 1.06 | 448 |
| PRLR | 0 | 0 | 2 | 0 | 2.00 | 579 |
| PTCH | 0 | 3 | 0 | 0 | 1.78 | 991 |
| SPINZ | 3 | 9 | 0 | 1 | 5.47 | 335 |
| VLDLR | 21 | 3 | 1 | 0 | 7.39 | 1371 |
| Autosomal genes | ||||||
| ACLY | 1 | 2 | 1 | 0 | 1.17 | 417 |
| ACTB | 3 | 8 | 0 | 0 | 1.67 | 923 |
| ALAS1 | 13 | 5 | 0 | 2 | 4.97 | 403 |
| CEPU | 5 | 4 | 0 | 3 | 2.01 | 582 |
| CHC | 3 | 2 | 0 | 4 | 4.25 | 355 |
| CKB | 4 | 1 | 0 | 3 | 2.42 | 465 |
| ENO1 | 1 | 1 | 0 | 0 | 0.72 | 177 |
| FAS | 2 | 0 | 3 | 0 | 3.28 | 901 |
| FN1 | 5 | 2 | 0 | 3 | 2.07 | 443 |
| FTH | 6 | 2 | 0 | 2 | 2.40 | 520 |
| GAPD | 5 | 1 | 0 | 1 | 1.31 | 357 |
| GH1 | 4 | 4 | 0 | 5 | 3.46 | 611 |
| LAMA | 0 | 1 | 0 | 0 | 0.94 | 224 |
| LHCGR | 2 | 3 | 0 | 4 | 1.54 | 634 |
| MPP | 2 | 0 | 0 | 2 | 1.26 | 246 |
| ODC1 | 6 | 2 | 0 | 4 | 3.70 | 454 |
| RHO | 6 | 6 | 1 | 1 | 4.72 | 404 |
| RPL30 | 15 | 7 | 0 | 6 | 7.07 | 1065 |
| RPL37a | 6 | 4 | 1 | 5 | 5.66 | 610 |
| RPL5 | 3 | 4 | 0 | 1 | 1.48 | 556 |
| RPL7a | 3 | 1 | 0 | 1 | 0.86 | 414 |
| TGFB2 | 5 | 2 | 0 | 1 | 1.38 | 402 |
| TM | 2 | 3 | 0 | 3 | 1.79 | 376 |
| Z-linked total | 43 | 39 | 21 | 2 | 47.49 | 9657 |
| A-linked total | 102 | 65 | 6 | 51 | 60.12 | 11539 |
S-values indicate numbers of variable sites, with subscripts indicating those exclusive (x) to the Italian collared flycatcher (Sx-IC) or Spanish pied flycatcher (Sx-SP), fixed differences between the species (Sfixed) or shared between the two species (Sshared), and the average pairwise differences (Saverage) between the two species.
Under the isolation model of speciation (Wakeley and Hey 1997), as two populations diverge, shared polymorphisms are gradually lost and become fixed differences due to random drift. The isolation model assumes that an ancestral population has split into two separate populations at a certain time in the past, which then evolve independently according to the assumptions of a standard neutral model. The model invokes three separate values of the population mutation parameter, θ: one for the ancestral population, θA, and one for each of the descendant populations, θ1 and θ2, and a time, T, since the speciation event, measured in units of 2N1 generations (Table 6)(Wakeley and Hey 1997; Wang et al. 1997).
TABLE 6.
Isolation model fitting
| Z and A (C.I.) | Z | A | |
|---|---|---|---|
| θ1a | 15.928 (0.003–549.548) | 13.67 | 13.68 |
| θ2 | 12.168 (0.003–211.823) | 12.24 | 8.94 |
| θA | 152.115 (7.46–321.333) | 39.95 | 78.90 |
| Tb | 0.361 (0.023–1.262) | 0.63 | 0.26 |
Estimates of θ for the collared flycatchers (1) and the pied flycatchers (2) and for their common ancestral population.
The estimate of time since the speciation event.
We performed isolation model fitting on the data, considering both all Z-linked loci and all autosomal loci as single loci. The loci were grouped in this way to test whether autosomal and Z-linked loci had significantly different patterns of polymorphism and divergence. Deviations from the isolation model were tested by comparing the true value of the WH statistic (Wakeley and Hey 1997) with values obtained from 10,000 coalescent simulations. This indicated that levels of divergence and polymorphism in the two species do not lead to a rejection of the isolation model of speciation (WH = 64.0, P = 0.43). Considering the collared flycatcher as species 1 and the pied flycatcher as species 2, the parameter estimates were θA = 152.1 (95% C.I.'s: 7.5–321.3), θ1 = 15.9 (0–549.5), θ2 = 12.2 (0–211.8), and T = 0.4 (0–1.3). Hence the data do not cause us to reject a model where θ can vary over time but that does not include selection or gene flow.
Haplotype diversity:
As sequencing was performed using a diploid template from male birds, the phase of heterozygous sites was unknown. We used the EM algorithm (Excoffier and Slatkin 1995) to estimate the frequencies of haplotypes found at each locus. Tables showing all polymorphic sites and estimated haplotypes at each locus are available as supplemental information (Z-linked, supplemental Table S3; and autosomal, supplemental Table S4; http://www.genetics.org/supplemental/). The majority of loci are shorter than 1 kb and do not contain enough segregating sites to reliably estimate the rate of recombination using the γ-statistic (Hey and Wakeley 1997). Using the four-gamete test (Hudson and Kaplan 1985), patterns of polymorphism at 5 Z-linked and 15 autosomal loci showed evidence for recombination.
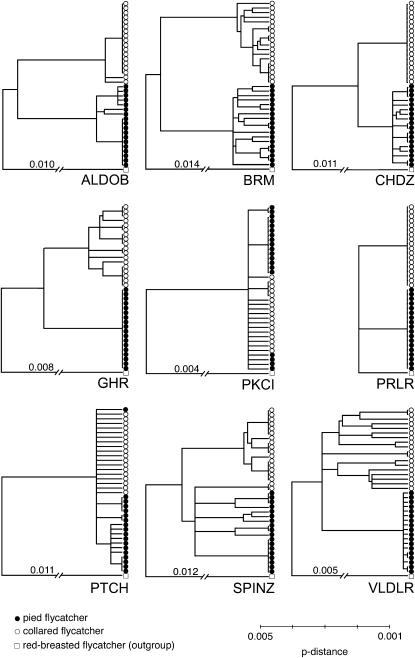
Haplotype estimation allowed us to construct neighbor-joining (NJ) trees for all loci. Note, however, that as there is evidence for recombination at many loci, such diagrams cannot be regarded as accurate genealogies and are meant to serve as an illustration of the variability in patterns of variation between loci. Figure 2 shows the trees for Z-linked loci, which exhibit significantly heterogeneous patterns of evolution as demonstrated by the HKA test. In particular, the VLDLR, GHR, and SPINZ loci stand out as having different genealogies in the two species. NJ trees for all autosomal loci are provided as supplemental information (S5 at http://www.genetics.org/supplemental/).
Figure 2.
Haplotype variation illustrated by neighbor-joining trees using p-distances of individual alleles at each of the nine Z-linked genes.
DISCUSSION
We analyzed patterns of evolution at Z-linked and autosomal loci in two flycatcher species. By using multiple unlinked loci from various genomic locations we attempt to distinguish between certain demographic factors that affect the evolution of the entire genome and other factors that may act on a subset of loci, such as gene flow (introgression) and natural selection. Our main findings are (a) significantly reduced levels of genetic variation at Z-linked loci and (b) significant heterogeneity in patterns of evolution between Z-linked loci compared to the expectations of a standard neutral model. Below we evaluate the factors that may have influenced patterns of variation in the two species
Reduced variation at Z-linked loci:
Random genetic drift is expected to reduce variation and fix shared polymorphisms between two diverging species faster in a small population than in a large population (Wakeley and Hey 1997). Hence, as Ne at Z-linked loci is three-fourths that of autosomal loci due to the mode of inheritance of the Z chromosome, both lower levels of variation and a faster rate of fixation of shared polymorphisms are expected to some extent. In this study we have shown that a significant number of Z-linked loci have lower levels of variation than predicted by this effect under the assumptions of a standard neutral model.
A smaller male population size will reduce the value of Ne at Z-linked loci even further. A female-biased operational sex ratio has been observed among breeding flycatchers (i.e., some males mate with more than one female). Some males are polygynous, having one primary and one secondary female in different territories, whereas others may not mate at all. In a study of the collared flycatchers ∼4% of the females were classified as secondary (Qvarnström et al. 2003), while in the pied flycatcher, on average 10–15% of the females have been classified as secondary (Lundberg and Alatalo 1992). Another way to study the operational sex ratio is by measuring extra pair paternity (EPP). The mean rate of EPP was found to be 14.5% in collared flycatchers and slightly less in pied flycatchers (Veen et al. 2001). Both polygyny and EPP have been suggested to represent a potentially important source of sexual selection on male secondary sexual characters in the collared flycatchers (Gustafsson et al. 1995; Sheldon and Ellegren 1999).
If we assume that there is no operational sex ratio bias and the mutation rate at Z-linked and autosomal loci is the same, then under a neutral model assuming random mating, constant population size, and no migration, the ratio of neutral variation on Z:A is predicted to be 0.75. In the most extreme scenario, where only one male fertilizes all the females in the population, this ratio approaches 0.5. Moreover, it is likely that the mutation rate on the Z chromosome is higher than that at autosomal loci due to a male-biased mutation rate, which would cause these proportions predicted under neutrality to be higher. In this study we find the Z:A to be 0.37 in the collared flycatcher and 0.46 in the pied flycatcher (θW based on silent sites only). This is lower than what even the most extreme female-biased operational sex ratio could produce (Z:A = 0.5). It is therefore likely that either selection or biased gene flow has contributed significantly to the difference in levels of variation between the Z chromosome and autosomes.
A study of levels of variation at Z-linked and autosomal loci in chicken (Sundström et al. 2004) found the ratio of neutral variation on Z:A to be 0.24 and concluded that this was strong evidence for selective sweeps operating on the Z chromosome. This ratio is lower than the values for both pied and collared flycatchers presented here. It is likely that a large part of this difference can be explained by the effects of poultry breeding, which entails both a smaller male effective population size and an artificial selection regime. Here we have demonstrated that levels of variation at Z-linked loci are also significantly reduced in populations of two wild bird species.
Isolation model fitting:
Under the isolation model of speciation, as two populations diverge, shared polymorphisms are gradually lost and become fixed differences due to random drift. This process occurs faster when effective population size is low. Hence, under this model, Z-linked loci are expected to exhibit fewer shared polymorphisms, as their effective population size is three-fourths of that of autosomal loci due to the mode of inheritance of the Z chromosome. However, a number of factors may cause populations to violate the assumptions of the isolation model. Such factors include unequal variance in mating success between males and females, gene flow between species, and selective sweeps.
The pied and collared flycatcher populations share polymorphisms at a number of loci analyzed in this study. However, shared polymorphisms are almost exclusively located in autosomal loci (51 shared polymorphisms compared with 6 fixed differences; Figure 1). In contrast, Z-linked loci have very few shared polymorphisms but a large number of fixed differences between species (2 shared polymorphisms compared with 21 fixed differences). Despite these differences, isolation model fitting indicates that these values are compatible with the isolation model of speciation. This suggests that patterns of polymorphism and divergence could result from a scenario without gene flow or selection but where historical changes in θ (possibly due to changes in Ne) generate a greater degree of stochastic variance in these patterns. This contrasts with our previous finding that under the stricter assumption of constant population size, variation at Z-linked loci is significantly reduced. We now consider the potential influence of demography and selection in generating the low levels of variation observed at Z-linked loci.
Population history:
It is believed that the pied and collared flycatchers and two other flycatchers in the Ficedula flycatcher species complex—the Atlas flycatcher (F. speculigera), located in North Africa, and the semicollared flycatcher (F. semitorquata), which breeds in the areas around the Black Sea—arose from a single ancestral population (Saetre et al. 2001). This ancestral population probably had a wide preglacial breeding range in the Old World, which became fragmented, followed by the expansion of the Ficedula species complex from glacial refugia. The results from the isolation model of speciation are in qualitative agreement with this population history scenario, suggesting that the two flycatcher species analyzed here are descendants of an ancestral, much larger population. However, as selection and gene flow may also have affected patterns of variation at a subset of loci, interpretation of ancestral population parameters is problematic.
Values of Tajima's D and Fu and Li's D and F in the collared flycatcher population are slightly negative, indicating an excess of rare variants, which could potentially reflect a bottlenecked expansion of this population from a preglacial ancestral population. However, in the pied flycatcher, whereas autosomal loci show slightly negative values of these statistics (in concordance with the collared sample), Z-linked loci generally have positive Tajima's D-values, indicating an excess of common variants. It is possible that a change in the population size affected patterns of variation at autosomal and Z-linked loci differently because the effective population size of the latter is smaller than that of autosomal loci. For example, a recent reduction in population size is expected to cause a larger initial increase in Tajima's D at loci with smaller Ne, and such effects may explain the differences in patterns of variation observed between human mtDNA and nuclear loci (Fay and Wu 1999).
Historical changes in population size violate the assumptions of the standard neutral model. Our comparison of levels of variation at Z-linked and autosomal loci assumes constant population size. Furthermore, certain demographic scenarios such as bottlenecks may increase the variance expected in patterns of polymorphism and divergence and thus reduce the power of the HKA test to reject neutrality (Hammer et al. 2004). Our findings of significant differences both between levels of variation at Z-linked and autosomal loci and in patterns of evolution between different Z-linked loci may therefore be caused at least in part by the demographic history of the two flycatcher species.
A further potential explanation for low levels of genetic variation at Z-linked loci is the presence of a greater degree of gene flow at autosomal loci compared with genes on the Z chromosome. Biased gene flow could occur between two incipient species in an ancestral population or could be ongoing. When polymorphisms are shared between two species because they have persisted in both populations since the time of divergence, we expect levels of linkage disequilibrium (LD) between them to have decayed, since they are relatively old. However, when shared polymorphisms arise by recent gene flow between populations, we expect entire haplotypes to be transferred and thus greater LD. As the regions involved in this study are generally too short to allow reliable comparisons of LD in shared and nonshared polymorphisms, it was not possible to perform the LD test of gene flow as described by Machado and Hey (2003). However, some insight can be gained by examination of the NJ trees (Figure 1 for Z-linked loci; autosomal loci are available as supplemental information, S5 at http://www.genetics.org/supplemental/) and of patterns of variation in the inferred haplotypes (haplotype tables are available as supplemental information, supplemental Table 3 and supplemental Table 4 at http://www.genetics.org/supplemental/). Z-linked loci have only two shared polymorphisms in total. It is thus highly unlikely that recent gene flow has occurred. The majority of polymorphisms at autosomal loci are shared between populations and at 17 of 25 loci there is evidence for haplotype sharing. In general, however, it is the ancestral haplotypes (or their close derivatives) that are shared, and there is no evidence for sharing of haplotypes that are more recently derived, by either mutation or recombination. Hence, we have no evidence to suggest that recent gene flow has contributed to patterns of extant variation.
Data from present-day hybrid zones in northern Europe show that the rate of introgression is higher on autosomal markers than on Z-linked ones (Saetre et al. 2003). Previous analyses of genotype data suggest that it is very unlikely that the Italian and Spanish populations investigated here are affected by introgression that currently occurs in flycatcher hybrid zones (Saetre et al. 2001, 2003). However, we cannot rule out the possibility that historic episodes of introgression (e.g., during warm interglacials) have contributed to elevating levels of polymorphism at autosomal loci relative to that at Z-linked loci.
The finding of slightly lower variation in the pied compared to the collared flycatcher could indicate a smaller effective population size in the Spanish pied flycatcher population. This has previously been suggested on the basis of a comparison of microsatellites and mtDNA in pied flycatcher populations in Europe (Haavie et al. 2000). The pied flycatcher has a patchy distribution on the Iberian Peninsula, living in isolated mountain forests separated by the Pyrenees from continental populations (Potti and Montalvo 1991). This might give a population structure that is vulnerable to factors such as environmental stochasticity, leading to a reduced genetic variation.
Natural selection and the evolutionary history of the Z chromosome:
The action of selective sweeps on the Z chromosome is supported by the HKA test, which indicates significant heterogeneity in patterns of evolution between different Z-linked loci, suggesting that selection has affected Z-linked loci to varying extents. Hence it is possible that natural selection has reduced levels of variation on the Z chromosome of both the pied and collared flycatcher compared to autosomal levels and that the signature of selection varies between loci and between species. It is, however, important to note the potential effect of demography before rejecting neutrality (see above).
Both selective sweeps and background selection affect larger genomic regions when the recombination rate is low (Maynard Smith and Haigh 1974; Charlesworth et al. 1993), and selection has been cited as a reason for the positive correlation observed between variation and recombination in various species (see Begun and Aquadro 1992; Stephan and Langley 1998; reviewed by Nachman 2001). Data from comparisons of physical and genetic maps in the chicken genome suggest that the average rate of recombination on the Z chromosome is ∼2.5 times lower than the genomic average (Smith and Burt 1998; Groenen et al. 2000; Schmid et al. 2000; Smith et al. 2000; Sundström et al. 2004). As flycatchers also possess a similar karyotype to the chicken, comprising a few macrochromosomes and many microchromosomes, it is likely that the recombination rate on Z chromosomes in flycatchers is also lower than the average autosomal rate. The effects of selection on linked variation could therefore potentially extend over larger genomic regions on the Z chromosome compared with autosomes.
As female birds only possess one Z chromosome, recessive deleterious mutations are exposed to natural selection and thus can be efficiently removed from a population. Hence, assuming that recessive deleterious mutations arise at similar rates at Z-linked and autosomal loci, the Z chromosome should exhibit lower average levels of deleterious polymorphism compared with autosomal loci. Neutral variants at Z-linked loci are therefore less likely to be linked to a deleterious mutant compared with neutral alleles on autosomes. We therefore predict that background selection should have a smaller impact on neutral variation on the Z chromosome than at autosomal loci (Charlesworth et al. 1993). It is therefore unlikely that the reduced levels of variation on the Z chromosome are due to the action of background selection; they are more consistent with the effects of recurrent selective sweeps. In contrast, positive selection is predicted to be more effective on Z-linked loci compared to autosomal loci because recessive positive mutations are not masked by dominance in hemizygous females (Charlesworth et al. 1987; Servedio and Saetre 2003). Thus, assuming that recessive positive mutations arise at a similar rate at Z-linked and autosomal loci, the Z chromosome is predicted to experience selective sweeps more often than autosomal chromosomes.
The sex chromosomes are known to possess a relative excess of genes involved in reproduction, sexual conflict, and male secondary sexual traits (Sperling 1994; Prowell 1998; Reinhold 1998; Ritchie and Phillips 1998; Civetta and Singh 1999; Wang et al. 2001; Gibson et al. 2002; Lercher et al. 2003; Saetre et al. 2003). Rice (1984) suggested that traits that are beneficial to one sex, but detrimental to the other are predicted to accumulate on the sex chromosomes. In addition, both theoretical predictions and experimental observations have shown that genes for such traits exhibit high rates of adaptive evolution (Civetta and Singh 1998; Begun and Whitley 2000; Singh and Kulathinal 2000; Wyckoff et al. 2000; Swanson and Vacquier 2002a,b; Torgerson et al. 2002; Meiklejohn et al. 2003; Torgerson and Singh 2003). In birds, Z-linked male sexual traits are transmitted directly from father to son, which may facilitate sexual selection on these traits (Reeve and Pfennig 2003). Furthermore, if a male sexual trait and the female preference for that trait become linked on the Z chromosome, then selection can act to rapidly fix the combination in a population (Servedio and Saetre 2003).
Summary:
Levels of silent variation at Z-linked loci in the pied and collared flycatcher, respectively, are 46 and 37% of levels observed at autosomal loci. This is lower than predicted under the most extreme female-biased operational sex ratio possible, where a relative level of 50% is expected. Furthermore, an HKA test indicates significant heterogeneity between patterns of polymorphism and divergence at Z-linked loci. These observations are not compatible with a standard neutral model. One potential explanation is the recurrent action of selective sweeps on the Z chromosome. However, it is also likely that the population sizes of these species have not remained constant, suggesting that a demographic scenario such as a population bottleneck could also play a major role in generating the observed patterns of variation.
Acknowledgments
The authors thank Centro Studi Ecologici Appenninici, J. Moreno, J. Haavie, and K. Räsänen for field assistance and Jody Hey and Göran Arnqvist for comments on the manuscript. Financial support was received from the Swedish Research Council, the Norwegian Research Council, and O. & L. Lamms Memorial Foundation.
References
- Andolfatto, P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., 1996. Population genetics of silent and replacement variation in Drosophila simulans and D. melanogaster: X/autosome differences? Mol. Biol. Evol. 13: 1405–1407. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and P. Whitley, 2000. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc. Natl. Acad. Sci. USA 97: 5960–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt, A. J., D. C. Presgraves and W. J. Swanson, 2002. A test for faster X evolution in Drosophila. Mol. Biol. Evol. 19: 1816–1819. [DOI] [PubMed] [Google Scholar]
- Broughton, R. E., and R. G. Harrison, 2003. Nuclear gene genealogies reveal historical, demographic and selective factors associated with speciation in field crickets. Genetics 163: 1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt, D. W., C. Bruley, I. C. Dunn, C. T. Jones, A. Ramage et al., 1999. The dynamics of chromosome evolution in birds and mammals. Nature 402: 411–413. [DOI] [PubMed] [Google Scholar]
- Carmichael, A. N., A. K. Fridolfsson, J. Halverson and H. Ellegren, 2000. Male-biased mutation rates revealed from Z and W chromosome-linked ATP synthase alpha-subunit (ATP5A1) sequences in birds. J. Mol. Evol. 50: 443–447. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., J. A. Coyne and N. H. Barton, 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130: 113–146. [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1998. Sex-related genes, directional sexual selection, and speciation. Mol. Biol. Evol. 15: 901–909. [DOI] [PubMed] [Google Scholar]
- Civetta, A., and R. S. Singh, 1999. Broad-sense sexual selection, sex gene pool evolution, and speciation. Genome 42: 1033–1041. [PubMed] [Google Scholar]
- Clark, A. G., 1997. Neutral behavior of shared polymorphism. Proc. Natl. Acad. Sci. USA 94: 7730–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren, H., and A. K. Fridolfsson, 1997. Male-driven evolution of DNA sequences in birds. Nat. Genet. 17: 182–184. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., and M. Slatkin, 1995. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 12: 921–927. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C.-I Wu, 1999. A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol. Biol. Evol. 16: 1003–1005. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C.-I Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, J. R., A. K. Chippindale and W. R. Rice, 2002. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc. R. Soc. Lond. Ser. B 269: 499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen, M. A. M., H. H. Cheng, N. Bumstead, B. F. Benkel, W. E. Briles et al., 2000. A consensus linkage map of the chicken genome. Genome Res. 10: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, L., A. Qvarnström and B. C. Sheldon, 1995. Trade-offs between life-history traits and a secondary sexual character in male collared flycatchers. Nature 375: 311–313. [Google Scholar]
- Haavie, J., G. P. Saetre and T. Moum, 2000. Discrepancies in population differentiation at microsatellites, mitochondrial DNA and plumage colour in the pied flycatcher—inferring evolutionary processes. Mol. Ecol. 9: 1137–1148. [DOI] [PubMed] [Google Scholar]
- Haavie, J., T. Borge, S. Bureš, L. Z. Garamszegi, H. M. Lampe et al., 2004. Flycatcher song in allopatry and sympatry—convergence, divergence and reinforcement. J. Evol. Biol. 17: 227–237. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1935. The rate of spontaneous mutation of a human gene. J. Genet. 31: 317–326. [DOI] [PubMed] [Google Scholar]
- Hammer, M. F., D. Garrigan, E. Wood, J. A. Wilder, Z. Mobasher et al., 2004. Heterogeneous patterns of variation among multiple human X-linked loci: the possible role of diversity-reducing selection in non-Africans. Genetics 167: 1841–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., and R. M. Kliman, 2002. Interactions between natural selection, recombination and gene density in the genes of Drosophila. Genetics 160: 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., and C. A. Machado, 2003. The study of structured populations—new hope for a difficult and divided science. Nat. Rev. Genet. 4: 535–543. [DOI] [PubMed] [Google Scholar]
- Hey, J., and J. Wakeley, 1997. A coalescent estimator of the population recombination rate. Genetics 145: 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, N. W., and T. W. Quinn, 1999. Male-driven evolution among Eoaves? A test of the replicative division hypothesis in a heterogametic female (ZW) system. J. Mol. Evol. 49: 750–759. [DOI] [PubMed] [Google Scholar]
- Kliman, R. M., P. Andolfatto, J. A. Coyne, F. Depaulis, M. Kreitman et al., 2000. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156: 1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., A. O. Urrutia and L. D. Hurst, 2003. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol. Biol. Evol. 20: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Lundberg, A., and R. V. Alatalo, 1992. The Pied Flycatcher. T. & A. D. Poyser, London.
- Machado, C. A., and J. Hey, 2003. The causes of phylogenetic conflict in a classic Drosophila species group. Proc. R. Soc. Lond. Ser. B 270: 1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, C. A., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19: 472–488. [DOI] [PubMed] [Google Scholar]
- Makova, K. D., and W.-H. Li, 2002. Strong male-driven evolution of DNA sequences in humans and apes. Nature 416: 624–626. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitchhiking effect of a favorable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Meiklejohn, C. D., J. Parsch, J. M. Ranz and D. L. Hartl, 2003. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 100: 9894–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M. W., 1997. Patterns of DNA variability at X-linked loci in Mus domesticus. Genetics 147: 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M. W., 2001. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17: 481–485. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W., V. L. Bauer, S. L. Crowell and C. F. Aquadro, 1998. DNA variability and recombination rates at X-linked loci in humans. Genetics 150: 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, I., and M. Schmid, 2002. Conservation of avian Z chromosomes as revealed by comparative mapping of the Z-linked aldolase B gene. Cytogenet. Genome Res. 96: 176–178. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Noor, M. A., K. L. Grams, L. A. Bertucci, Y. Almendarez, J. Reiland et al., 2001. The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution 55: 512–521. [DOI] [PubMed] [Google Scholar]
- Potti, J., and S. Montalvo, 1991. Male arrival and female mate choice in pied flycatchers (Ficedula hypoleuca) in central Spain. Ornis Scand. 22: 45–54. [Google Scholar]
- Primmer, C. R., T. Borge, J. Lindell and G. P. Sætre, 2002. Single-nucleotide polymorphism characterization in species with limited available sequence information: high nucleotide diversity revealed in the avian genome. Mol. Ecol. 11: 603–612. [DOI] [PubMed] [Google Scholar]
- Prowell, D. P., 1998. Sex linkage and speciation in Lepidoptera, pp. 309–319 in Endless Forms: Species and Speciation, edited by D. J. Howard and S. H. Berlocher. Oxford University Press, New York.
- Qvarnström, A., B. C. Sheldon, T. Pärt and L. Gustafsson, 2003. Male ornamentation, timing of breeding, and cost of polygyny in the collared flycatcher. Behav. Ecol. 14: 68–73. [Google Scholar]
- Reeve, H. K., and D. W. Pfennig, 2003. Genetic biases for showy males: Are some genetic systems especially conductive to sexual selection? Proc. Natl. Acad. Sci. USA 100: 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold, K., 1998. Sex linkage among genes controlling sexually selected traits. Behav. Ecol. Sociobiol. 44: 1–7. [Google Scholar]
- Rice, W. R., 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742. [DOI] [PubMed] [Google Scholar]
- Ritchie, M. G., and S. D. F. Phillips, 1998. The genetics of sexual isolation, pp. 291–308 in Endless Forms: Species and Speciation, edited by D. J. Howard and S. H. Berlocher. Oxford University Press, New York.
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an intergrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Saetre, G. P., T. Moum, S. Bureš, M. Král, M. Adamjan et al., 1997. A sexually selected character displacement in flycatchers reinforces premating isolation. Nature 387: 589–592. [Google Scholar]
- Saetre, G. P., T. Borge, J. Lindell, T. Moum, C. R. Primmer et al., 2001. Speciation, introgressive hybridization and nonlinear rate of molecular evolution in flycatchers. Mol. Ecol. 10: 737–749. [DOI] [PubMed] [Google Scholar]
- Saetre, G. P., T. Borge, K. Lindroos, J. Haavie, B. C. Sheldon et al., 2003. Sex chromosome evolution and speciation in Ficedula flycatchers. Proc. R. Soc. Lond. Ser. B 270: 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer, C., 2003. Hitchhiking mapping: functional genomics from the population genetics perspective. Trends Genet. 19: 32–38. [DOI] [PubMed] [Google Scholar]
- Schmid, M., I. Nandra, M. Guttenbach, C. Steinlein, H. Hoehn et al., 2000. First report on chicken genes and chromosomes 2000. Cytogenet. Cell Genet. 90: 169–218. [DOI] [PubMed] [Google Scholar]
- Schneider, S., D. Roessli and L. Excoffier, 2000. Arlequin Version 2.000: A Software for Population Genetic Data Analysis. University of Geneva, Geneva.
- Servedio, M. R., and G. P. Sætre, 2003. Speciation as a positive feedback loop between postzygotic and prezygotic barriers to gene flow. Proc. R. Soc. Lond. Ser. B 270: 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seutin, G., B. N. White and P. T. Boag, 1991. Preservation of avian blood and tissue samples for DNA analyses. Can. J. Zool. Rev. Can. Zool. 69: 82–90. [Google Scholar]
- Sheldon, B. C., and H. Ellegren, 1999. Sexual selection resulting from extrapair paternity in collared flycatchers. Anim. Behav. 57: 285–298. [DOI] [PubMed] [Google Scholar]
- Shetty, S., D. K. Griffin and J. A. M. Graves, 1999. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 7: 289–295. [DOI] [PubMed] [Google Scholar]
- Singh, R. S., and R. J. Kulathinal, 2000. Sex gene pool evolution and speciation: a new paradigm. Genes Genet. Syst. 75: 119–130. [DOI] [PubMed] [Google Scholar]
- Smith, J., and D. W. Burt, 1998. Parameters of the chicken genome (Gallus gallus). Anim. Genet. 29: 290–294. [DOI] [PubMed] [Google Scholar]
- Smith, J., C. K. Bruley, I. R. Paton, I. Dunn, C. T. Jones et al., 2000. Differences in gene density on chicken macrochromosomes and microchromosomes. Anim. Genet. 31: 96–103. [DOI] [PubMed] [Google Scholar]
- Sperling, F. A. H., 1994. Sex-linked genes and species differences in Lepidoptera. Can. Entomol. 126: 807–818. [Google Scholar]
- Stephan, W., and C. H. Langley, 1998. DNA polymorphism in Lycopersicon and crossing over per physical length. Genetics 150: 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström, H., M. T. Webster and H. Ellegren, 2004. Reduced variation on the chicken Z chromosome. Genetics 167: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. a The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. b Reproductive protein evolution. Annu. Rev. Ecol. Syst. 33: 161–179. [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position- specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson, D. G., and R. S. Singh, 2003. Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol. Biol. Evol. 20: 1705–1709. [DOI] [PubMed] [Google Scholar]
- Torgerson, D. G., R. J. Kulathinal and R. S. Singh, 2002. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol. Biol. Evol. 19: 1973–1980. [DOI] [PubMed] [Google Scholar]
- Veen, T., T. Borge, S. C. Griffith, G. P. Sætre, S. Bureš et al., 2001. Hybridization and adaptive mate choice in flycatchers. Nature 411: 45–50. [DOI] [PubMed] [Google Scholar]
- Wakeley, J., and J. Hey, 1997. Estimating ancestral population parameters. Genetics 145: 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. J., J. R. McCarrey, F. Yang and D. C. Page, 2001. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27: 422–426. [DOI] [PubMed] [Google Scholar]
- Wang, R. L., and J. Hey, 1996. The speciation history of Drosophila pseudoobscura and close relatives: inferences from DNA sequence variation at the period locus. Genetics 144: 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. L., J. Wakeley and J. Hey, 1997. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics 147: 1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Wyckoff, G. J., W. Wang and C.-I Wu, 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403: 304–309. [DOI] [PubMed] [Google Scholar]
- Yi, S., D. Bachtrog and B. Charlesworth, 2003. A survey of chromosomal and nucleotide sequence variation in Drosophila miranda. Genetics 164: 1369–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]