Abstract
A scan of the X chromosome of a European Drosophila melanogaster population revealed evidence for the recent action of positive directional selection at individual loci. In this study we analyze one such region that showed no polymorphism in the genome scan (located in cytological division 2C10–2E1). We detect a 60.5-kb stretch of DNA encompassing the genes ph-d, ph-p, CG3835, bcn92, Pgd, wapl, and Cyp4d1, which almost completely lacks variation in the European sample. Loci flanking this region show a skewed frequency spectrum at segregating sites, strong haplotype structure, and high levels of linkage disequilibrium. Neutrality tests reveal that these data are unlikely under both the neutral equilibrium model and the simple bottleneck scenarios. In contrast, newly developed maximum-likelihood ratio tests suggest that strong selection has acted recently on the region under investigation, causing a selective sweep. Evidence that this sweep may have originated in an ancestral population in Africa is presented.
ENVIRONMENTAL changes constitute significant challenges to both plant and animal life, since all life history aspects can potentially be affected. At the molecular level, mutations that confer adaptations increase in frequency, whereas those that have rather detrimental effects are removed from the population through purifying selection. In addition to selection, neutral processes such as random genetic drift, population substructure, or population size bottlenecks may also account for substantial changes in allele frequencies. However, these latter processes depend on special demographic conditions (e.g., small population size or restricted gene flow between populations) to produce similar effects as selection.
Previous studies provided convincing evidence for local adaptation of Drosophila melanogaster, which originated in sub-Saharan Africa and subsequently colonized many parts of the world (Lachaise et al. 1988). They suggested that numerous beneficial mutations were fixed during the habitat expansion after the last glaciation (∼10,000 years ago; David and Capy 1988) in the process of adapting to local environments (Glinka et al. 2003; Kauer et al. 2003).
At the DNA level, positive Darwinian selection is often associated with a phenomenon known as genetic hitchhiking (Maynard Smith and Haigh 1974): neutral variants in the proximity of a beneficial mutation rise in frequency as a consequence of selection. The result of this process is largely determined by the effects of recombination (rec) and the strength of selection (Kaplan et al. 1989). Thus, in regions of reduced crossing over (e.g., centromeric and telomeric regions of Drosophila), levels of heterozygosity are generally lower than in the middle of chromosome arms (Aguadé et al. 1989; Stephan and Langley 1989; Begun and Aquadro 1992). Similar patterns of reduced variation can also be caused by background selection (Charlesworth et al. 1993), where neutral variants are removed due to linkage to deleterious mutations that are selected against. However, the effects of background selection are primarily limited to regions of restricted recombination (Charlesworth et al. 1993).
In a recent study Glinka et al. (2003) investigated the evolutionary history of an African and a European D. melanogaster population on the basis of X chromosomal data. They identified several loci that are devoid of polymorphic sites within the European population, whereas levels of heterozygosity in the African population and divergence to its congener D. simulans appear to be relatively normal. They proposed that some of the loci with reduced levels of variation are not evolving neutrally; i.e., they are targets of natural selection. These loci in the European population served as a starting point for further investigation of the adaptation of D. melanogaster to temperate zones. One of these loci is a 348-bp fragment within the fifth intron of wings apart-like (wapl; denoted “fragment 10” in Glinka et al. 2003), a gene involved in heterochromatin organization and sister-chromatid adhesion (Verni et al. 2000); it is located at cytological position 2D5 on the X chromosome. In this article we analyze 12 additional fragments in the vicinity of wapl in a European and an African sample to examine whether the pattern of variation around this locus is consistent with the recent action of positive selection.
MATERIALS AND METHODS
Fly strains:
Intraspecific data were collected from 24 highly inbred D. melanogaster lines derived from two populations: 12 lines from a European population (Leiden, The Netherlands) and 12 lines from Africa (Lake Kariba, Zimbabwe). The European lines were kindly provided by A. J. Davis, and the African ones by C. F. Aquadro. For interspecific comparisons we used a single inbred D. simulans strain (Winters, CA; kindly provided by H. A. Orr).
Molecular methods:
We used the publicly available DNA sequence of the D. melanogaster genome (FlyBase Consortium 2003; http://www.flybase.org) for primer design. We amplified and sequenced 12 fragments of noncoding DNA from six intergenic regions and six introns around wapl. Genomic DNA was isolated from 15 females of each inbred line using the Puregene DNA isolation kit (Gentra Systems, Minneapolis). Standard PCR (25 μl) contained 1 μl template DNA, 2.5 μl of 10× buffer, 1 μl MgCl2 (2 mm), 0.25 μl of dNTPs (0.2 mm of each dNTP), 2 μl of each primer (10 μm), 16.12 μl distilled water, and 0.13 μl Taq polymerase (5 units/μl). PCR conditions were as follows: 4 min at 94°, 30 cycles of 30 sec at 94°, 30 sec at primer-specific temperatures, 30 sec at 72°, and a final extension step of 4 min at 72°. Afterward PCR fragments were scored on 1.5% agarose gels. Following purification of PCR products (using Exosap-It, USB, Cleveland), sequencing reactions were conducted for both strands with the DYEnamic ET terminator cycle sequencing kit (Amersham Biosciences, Buckinghamshire, UK). Sequences were run on a MegaBACE 1000 automated capillary sequencer and analyzed using Cimarron 3.12 base calling software (both from Amersham Biosciences). Finally, sequences were aligned, checked manually, and assembled into contigs with Seqman (DNAstar, Madison, WI). When D. simulans sequences could not be obtained, we used the publicly available DNA sequence of the D. simulans genome. In the case of a gap in the D. simulans sequence, we used the published D. yakuba sequence as the outgroup at the corresponding position (http://species.flybase.net/blast/).
Data analysis:
Most statistical analyses were performed using DnaSP 4.0 (Rozas et al. 2003). We estimated nucleotide diversity using π (Tajima 1983) and θ (Watterson 1975). Expected numbers of segregating sites were calculated by performing coalescent simulations. Furthermore, we determined the number of haplotypes (h), haplotype diversity (Hd; Nei 1987), and divergence (K) between D. melanogaster and D. simulans. Linkage disequilibrium (LD) was determined per fragment in terms of ZnS (Kelly 1997), which is the average of r2 (Hill and Robertson 1968) over all pairwise comparisons. To test the neutral equilibrium model, we used Tajima's D (Tajima 1989), Fay and Wu's H (Fay and Wu 2000), and the multi-locus-HKA statistic (Hudson et al. 1987). The latter was calculated using the program HKA, kindly provided by J. Hey. Significance of the test statistics was assessed by comparing the observed values to those obtained from 10,000 neutral coalescent simulations. Simulated data were generated using the observed θ-values.
Estimation of the parameters of a selective sweep model:
We computed the likelihood of a selective sweep model vs. the neutral model for our polymorphism data using a recently developed composite likelihood ratio (CLR) test (Kim and Stephan 2002). Briefly, in this test the maximum likelihood of observing a given number of derived variants at a polymorphic site under the selective sweep model (L1 in Kim and Stephan 2002) is compared to that expected under the standard neutral model (L0). L1 and L0 are based on the frequency spectrum and the spatial distribution of polymorphic sites where the derived variants occur with given frequencies in a population sample. The resulting likelihood ratio was compared to the cumulative frequency distribution of likelihood ratios obtained from 10,000 simulations of neutral data sets. Significance was determined at the 5% level (one-tailed test). Since levels of heterozygosity were greatly reduced over a considerable stretch in the European sample (see results), we used a modified version of test A of Kim and Stephan (2002), where neutral data sets were generated conditioned on the observed number of segregating sites. Results were evaluated by a recently proposed goodness-of-fit test (GOF; Jensen et al. 2005), where GOF values obtained from polymorphism data were compared to those estimated from 1000 data sets simulated under a selection scenario.
In addition, we applied the test of Kim and Nielsen (2004) to compute the likelihood of a selective sweep model and estimate the strength of selection. In contrast to Kim and Stephan (2002), this test takes LD into account. The strength of directional selection required to cause the reductions in nucleotide diversity observed in our data was estimated as α =1.5Nes, where Ne is the effective population size and s is the selection coefficient. For both tests, we estimated the local population recombination rate (R) as 2Neρ, where the recombination rate (per site per generation) is ρ = 0.48 × 10−8 (following Comeron et al. 1999, using the computer program “Recomb-rate,” kindly provided by J. M. Comeron). We assumed Ne = 0.3 × 106 and θ = 0.0044 for our European sample and Ne = 106 and θ = 0.0127 for the African sample (Glinka et al. 2003).
Position of selected site:
We estimated the approximate position of the putative selected site using both the composite likelihood ratio approach by Kim and Stephan (2002) and the test by Kim and Nielsen (2004). Input files were prepared with parameter settings (Ne, R, θ, and α) as mentioned above. The current frequency of the beneficial allele was set to 1 and, given the observed pattern of variation (see results), a very recent fixation of the beneficial allele was assumed (τ = 0). Two-locus sampling probability tables under the selective sweep model and the neutral model were kindly provided by Y. Kim (personal communication) and R. Hudson (http://home.uchicago.edu/∼rhudson1/), respectively.
Demographic modeling of the European population:
Since demographic processes, such as a population bottleneck and subsequent expansion, can leave a signature in the genome that resembles that of selection, we tested the likelihood of such a scenario, given our data. We used a coalescent-based method (Ramos-Onsins et al. 2004) that simplifies the bottleneck model to three parameters: θ (population mutation rate), Tb (time of occurrence of the bottleneck), and Sb (strength of the bottleneck; Galtier et al. 2000). The likelihood that the 60.5-kb reduction in heterozygosity was caused by a bottleneck was estimated by comparison to 100,000 genealogies (500,000 for method II; see Table 3) simulated with θ = 0.0066 (the average level of heterozygosity estimated from fragments 4–9 in the African sample) and various combinations of Tb and Sb (both measured in units of 3Ne generations), chosen across a range of bottleneck times reported by Ometto et al. (2005). The probability of observing a 60.5-kb region of reduced diversity in the European sample was estimated using only the fraction of genealogies for which either exactly or at most 43 segregating sites were observed in the entire region and for which the wapl fragment was invariant. The probability of our data under the bottleneck situation was then estimated as the proportion of simulations that yielded at most one segregating site in the fragments located in the monomorphic region (i.e., loci 4–9). The simulations were done both with and without recombination between adjacent fragments (with ρ = 0.48 × 10−8 rec/bp/generation).
TABLE 3.
Results of bottleneck simulations
| Tba | Sbb | Method Ic | Method IId | Method IIIe |
|---|---|---|---|---|
| 0.0100 | 0.340 | 0.180 | <0.001 | 0.090 |
| 0.400 | 0.210 | 0.005 | 0.140 | |
| 0.0122f | 0.371f | 0.150 | <0.001 | 0.082 |
| 0.0125f | 0.350f | 0.130 | <0.001 | 0.073 |
| 0.0128f | 0.345f | 0.120 | <0.001 | 0.069 |
| 0.0200 | 0.340 | 0.050 | <0.001 | 0.030 |
| 0.400 | 0.060 | <0.001 | 0.031 | |
| 0.0264f | 0.380f | 0.028 | <0.001 | 0.013 |
| 0.0267f | 0.400f | 0.028 | <0.001 | 0.014 |
| 0.0300 | 0.340 | 0.016 | <0.001 | 0.006 |
| 0.400 | 0.018 | <0.001 | 0.008 | |
| 0.0500 | 0.340 | 0.002 | <0.001 | 0.0007 |
| 0.400 | 0.003 | <0.001 | 0.0012 |
Age of the bottleneck, measured in 3Ne generations.
Strength of the bottleneck.
Probability of observing at most one segregating site in loci 4–9 under the assumption of no recombination between fragments, conditioned on the observation of 43 segregating sites in the entire region and zero polymorphisms in wapl.
Probability of observing at most one segregating site in loci 4–9 under the assumption of intergenic recombination (with ρ = 0.48 × 10−8 rec/bp/generation), conditioned on the observation of 43 segregating sites in the entire region and zero polymorphisms in wapl.
Probability of observing at most one segregating site in loci 4–9 under the assumption of intergenic recombination, conditioned on the observation of at most 43 segregating sites in the entire region and zero polymorphisms in wapl.
RESULTS
Nucleotide variation around the wapl fragment:
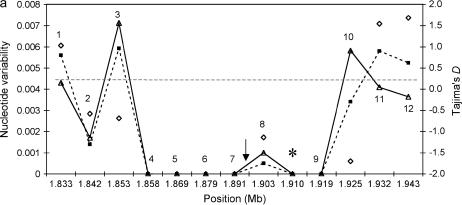
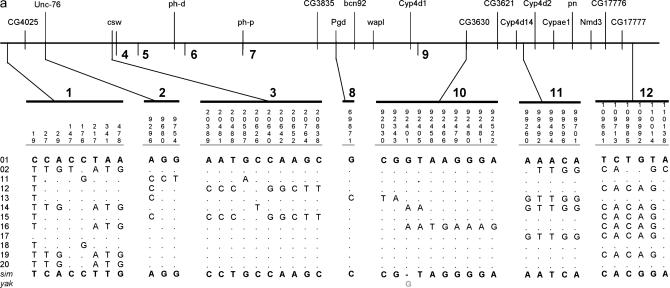
To determine the extent of the low-variability region around the wapl fragment in the European D. melanogaster sample (Glinka et al. 2003), we sequenced 12 new fragments of noncoding DNA (six introns and six intergenic regions with an average length of 492 bp and an average distance between fragments of 9 kb) around the wapl locus in the European lines. The entire region encompasses a total of 110 kb (Figure 1a). Of the 12 fragments, only 7 were polymorphic (Table 1). With the exception of one singleton in fragment 8, no intraspecific variation could be detected in a region comprising 60.5 kb. The pattern of polymorphism is illustrated in Figure 2a. The remaining fragments showed an average heterozygosity level (θ) of 0.004, consistent with a mean θ-value of 0.0044 for the European population (Glinka et al. 2003).
Figure 1.
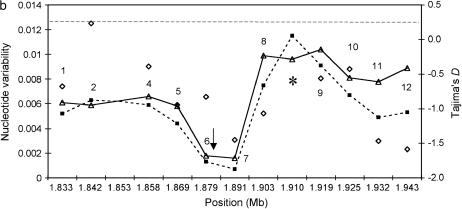
Intraspecific variation around the wapl fragment. (a) The European sample. (b) The African sample. Solid lines and triangles correspond to θ, dashed lines and squares indicate π. Diamonds indicate Tajima's D, and the shaded dashed line represents chromosome-wide average heterozygosity as reported by Glinka et al. (2003). Arrows indicate estimated positions of the target of selection following tests of Kim and Stephan (2002) and Kim and Nielsen (2004); Asterisk indicates the position of the wapl fragment. Absolute genomic positions of fragments are given in megabases, according to release 3 of the annotated D. melanogaster genome. For fragment 3 of the African lines, sequences could not be obtained.
TABLE 1.
Polymorphism of the European sample and divergence
| Fragment | L | K | Sobs | θobs | θexp | Sexp | h | Hd | ZnS |
|---|---|---|---|---|---|---|---|---|---|
| 1ir | 598 | 0.13 | 8 | 0.0043 | 0.0011–0.0099 | 2–18 | 6 | 0.879 | 0.452 |
| 2in | 590 | 0.05 | 3 | 0.0017 | 0.0005–0.0101 | 1–18 | 3 | 0.530 | NA |
| 3in | 465 | 0.04 | 10 | 0.0071 | 0.0007–0.0107 | 1–15 | 4 | 0.531 | 1.0* |
| 4in | 600 | 0.06 | 0 | 0.0000 | 0.0011–0.0099** | 2–18 | 1 | 0.000 | NA |
| 5ir | 465 | 0.02 | 0 | 0.0000 | 0.0007–0.0107* | 1–15 | 1 | 0.000 | NA |
| 6ir | 287 | 0.05 | 0 | 0.0000 | 0.0000–0.0115* | 0–10 | 1 | 0.000 | NA |
| 7in | 422 | 0.03 | 0 | 0.0000 | 0.0008–0.0102* | 1–13 | 1 | 0.000 | NA |
| 8in | 319 | 0.04 | 1 | 0.0010 | 0.0000–0.0114 | 0–11 | 2 | 0.167 | NA |
| 9ir | 460 | 0.06 | 0 | 0.0000 | 0.0007–0.0101* | 1–14 | 1 | 0.000 | NA |
| 10in | 569 | 0.05 | 10 | 0.0058 | 0.0006–0.0099 | 1–17 | 4 | 0.455 | 1.0* |
| 11ir | 405 | 0.09 | 5 | 0.0041 | 0.0008–0.0106 | 1–13 | 3 | 0.530 | 0.867 |
| 12ir | 546 | 0.06 | 6 | 0.0036 | 0.0006–0.0103 | 1–17 | 3 | 0.621 | 0.829* |
L, number of sites studied; K, divergence between D. melanogaster and D. simulans; Sobs, observed number of segregating sites; θobs, observed heterozygosity; θexp, expected heterozygosity (95% confidence intervals); h, number of haplotypes; Hd, haplotype diversity; ZnS, linkage disequilibrium; ir, intergenic region; in, intron; and NA, not available. The expected number of segregating sites (Sexp) is calculated according to Tajima (1983) for n = 12 lines. Fragments significantly devoid of segregating sites are in italics. *, significant at the 0.05 level; **, significant at the 0.01 level.
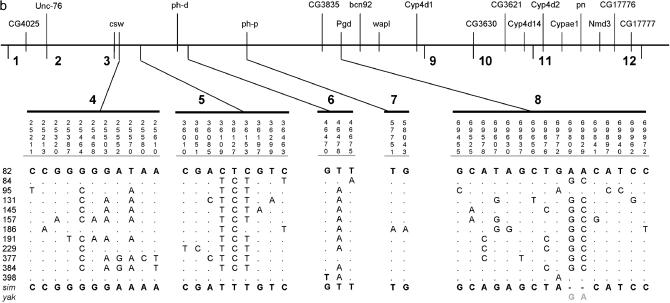
Figure 2.
Polymorphism data for the 2C10–2E1 region. (a) The European sample. (b) The African sample. D. melanogaster lines 01 (European sample) and 82 (African sample) are taken as references. For the African sample (b), we present polymorphism data only for fragments 4–8 due to space limitations. sim, D. simulans; yak, D. yakuba; (−) no sequence data available.
Furthermore, we analyzed the corresponding nucleotide variation in the African sample. Estimated levels of heterozygosity for 11 fragments are listed in Table 2. The pattern of polymorphism for loci 4–8 is illustrated in Figure 2b. For fragment 3 we were not able to obtain sequences. With the exception of two low-variation fragments (i.e., loci 6 and 7), the pattern of nucleotide diversity is consistently higher for the African lines (than for the European ones) with an average θ of 0.0065 for fragments 4–9 (Figure 1b). However, this value is ∼50% lower than the average level of heterozygosity reported for the entire X chromosome in Africa (θ = 0.0127; Glinka et al. 2003).
TABLE 2.
Polymorphism of the African sample
| Fragment | L | n | Sobs | θobs | h | Hd | ZnS |
|---|---|---|---|---|---|---|---|
| 1ir | 557 | 11 | 10 | 0.0061 | 8 | 0.927 | 0.791* |
| 2in | 562 | 12 | 10 | 0.0059 | 11** | 0.985** | 0.103 |
| 3in | NA | NA | NA | NA | NA | NA | NA |
| 4in | 551 | 12 | 11 | 0.0066 | 9* | 0.939 | 0.288 |
| 5ir | 518 | 12 | 9 | 0.0058 | 8 | 0.894 | 0.253 |
| 6ir | 563 | 12 | 3 | 0.0018** | 4 | 0.561 | NA |
| 7in | 515 | 12 | 2 | 0.0013** | 2 | 0.167 | NA |
| 8in | 534 | 12 | 16 | 0.0099 | 10* | 0.955 | 0.224 |
| wapl | 346 | 12 | 10 | 0.0096 | 6 | 0.879 | 0.239 |
| 9ir | 493 | 11 | 15 | 0.0104 | 8 | 0.927 | 0.259 |
| 10in | 462 | 11 | 10 | 0.0074 | 8* | 0.945 | 0.195 |
| 11ir | 500 | 11 | 11 | 0.0075 | 8 | 0.945 | 0.127 |
| 12ir | 475 | 12 | 12 | 0.0084 | 4 | 0.636 | 0.534 |
n, number of lines analyzed. For other abbreviations, see Table 1 legend.
Standard neutrality tests for the European sample:
Tajima's D (Tajima 1989) was highly negative for two distal loci (2 and 3), one proximal fragment (10) directly flanking the invariant region, and the locus showing a singleton (8; Figure 1a). For fragment 10, the observed value is significantly lower than the neutral expectation (P = 0.038). Similar to Tajima's D statistic, Fay and Wu's H (Fay and Wu 2000) was negative for three polymorphic fragments (H = −0.67, −1.67, and −0.36 for fragments 3, 8, and 10, respectively), indicating an excess of high-frequency-derived variants. However, these values were not significant. In contrast, we observed positive values of Tajima's D for three fragments located more distal to the invariant region (i.e., loci 1, 11, and 12). For locus 12, this value is significant (P < 0.05).
For each fragment located in the monomorphic region, we estimated the probability of observing a locus of length L devoid of polymorphisms, given an expected heterozygosity of 0.0044 by comparison with values obtained from 10,000 coalescent simulations of the standard neutral model under the conservative assumption of zero recombination (see Hudson 1990). As shown in Table 1, all fragments under consideration represent a reduced number of segregating sites and, with the exception of fragment 8 (containing the singleton), this reduction is significant compared to the neutral expectation. In the center of the analyzed region (i.e., fragments 4–9) that encompasses 2553 nucleotides where a valley of reduced variation has been observed, we detected only one segregating site. The probability of this result, under the conservative assumption of no recombination, is significantly low (P < 0.00001) and incompatible with the standard neutral model. The possibility of selective constraints or a low regional mutation rate being the cause of the observed reduction in variation can be excluded, since levels of divergence between D. melanogaster and its sister species D. simulans observed in the 110-kb region investigated are on average normal (Table 1). Indeed, a multi-locus version of the HKA test (Hudson et al. 1987) revealed a significant departure from neutrality (χ2 = 30.15, P = 0.0015). This result still holds when the fragment with the largest contribution to the HKA statistic (i.e., locus 3) was removed from analysis. Only when, in addition, the next largest contribution (fragment 10) was removed, this result was no longer significant (P = 0.319).
The observed number of haplotypes for the fragments surrounding the invariant region varies from three to six per fragment, with haplotype diversity increasing with distance from the invariant region (Figure 3 and Table 1).
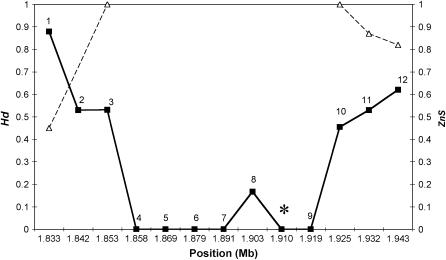
Figure 3.
Haplotype diversity and linkage disequilibrium in the European sample. Solid lines and squares denote Hd. Dashed lines and triangles indicate levels of linkage disequilibrium (ZnS). ZnS was estimated using parsimony informative sites only. Therefore, no value for fragment 2 was obtained. Asterisk indicates the position of the wapl fragment.
The observed values, however, did not depart significantly from neutral expectations. Yet, two fragments directly flanking the monomorphic region (i.e., loci 3 and 10) showed a considerable reduction in haplotype diversity (P = 0.09 and 0.05, respectively).
In addition, we detected significant LD (P < 0.05) within fragments 3, 10, and 12 (Table 1). For fragment 11, the observed value was marginally significant (P = 0.05). As expected, LD decays in both directions with distance from the valley of reduced variation. We did not detect LD among adjacent fragments. This may be due to recombination. For instance, between fragments 11 and 12, which are separated by ∼10 kb, at least one recombination event can be inferred applying the four-gamete rule (Hudson and Kaplan 1985).
Standard neutrality tests for the African sample:
Tajima's D (Tajima 1989) estimated from fragments 1 to 12 showed a general trend toward negative values, with the exception of fragment 2 (Figure 1b). However, only the D value estimated from locus 12 was significantly different from neutral expectations (P = 0.05). For the same fragment, Fay and Wu's H (Fay and Wu 2000) was significantly negative as well (H = −9.0, P = 0.03), indicating an excess of high-frequency-derived variants. H was also negative for fragments 6, 8, and 11 (H = −0.52, −0.18, and −0.56, respectively). However, these values are not significantly different from zero.
As for the European population, we estimated whether the observed number of polymorphic sites in regions 4–9 (Sobs= 56; see Table 2) is significantly different from expectations under the standard neutral model. Given an expected heterozygosity of 0.0127 (Glinka et al. 2003), 41–251 segregating sites would be expected. The observation of 56 SNPs is therefore not significantly different from neutral expectations (P = 0.07). However, it should be noted that the θ estimates for loci 6 and 7 are significantly low (Table 2). The multi-locus HKA test (Hudson et al. 1987) did not reveal any significant departure from neutrality for our fragments in the African sample (χ2 = 9.90, P = 0.45).
The observed number of haplotypes varies from 2 to 11 with generally high haplotype diversities (Table 2). We ran coalescent simulations using the estimated recombination rate (ρ = 0.48 × 10−8 rec/bp/generation) to infer significance. This is conservative, since recombination tends to increase both statistics. Of 11 loci analyzed, 4 show significantly increased numbers of haplotypes. This pattern is in accordance with previous studies reporting a chromosome-wide trend toward elevated numbers of haplotypes and high haplotype diversities in African D. melanogaster (Glinka et al. 2003; Ometto et al. 2005). With the exception of fragment 1, no significant linkage disequilibrium was detected in the region analyzed (Table 2).
Can the severe reduction in variation observed in the European sample be explained by a population size bottleneck?
To account for the possibility that a population size bottleneck has caused the reduction in heterozygosity observed in our European sample, we applied the approach of Ramos-Onsins et al. (2004), modified by Ometto et al. (2005). That is, on the basis of the parameter estimates of Ometto et al. (2005; see materials and methods), we simulated a drop in effective population size at various times in the past. For bottleneck times assayed across a range of times, i.e., Tb = 0.01, 0.02, 0.03, or 0.05, we simulated genealogies with two different strengths of the bottleneck, as suggested by Ometto et al. (2005). Under the assumption of no recombination between loci, the probability of our data being explained by a simple bottleneck scenario is low (Table 3). However, only P-values for reasonably old bottlenecks (Tb ≥ 0.02, i.e., >6000 years ago, assuming 10 generations/year) are significant. Note that according to Ometto et al. (2005) the X chromosomal nucleotide diversity of the European sample is best described by a combination of Tb = 0.0267 and Sb = 0.400, i.e., a bottleneck that has occurred ∼8000 years ago. If some recombination (ρ = 0.48 × 10−8 rec/bp/generation) was allowed between fragments, the probability of our data is significantly low for all simulated scenarios. When the condition of observing exactly 43 segregating sites was relaxed, our data still remained significant for the older bottleneck scenarios but were only marginally significant for more recent bottlenecks (Tb = 0.01–0.0128, i.e., between 3000 and 3840 years ago).
Estimation of selection parameters:
We applied the maximum-likelihood ratio tests of Kim and Stephan (2002) and Kim and Nielsen (2004) to estimate the significance of the reduction in variation observed in our data under both the standard neutral model and a selection model. Furthermore, we estimated the strength of selection. Since a large fraction of the genomic region under analysis shows highly reduced levels of heterozygosity in the European sample (i.e., the putative sweep region), we specified θ = 0.0044 for this analysis as reported by Glinka et al. (2003). Using the Kim and Stephan (2002) method we compared the likelihood ratio (LRKS = L1/L0) to those obtained from 10,000 neutral coalescent simulations. The probability of finding the likelihood ratio obtained from our data (LRKS = 16.30) under a neutral scenario is low (P = 0.037).
Since polymorphism patterns produced by a selective sweep can be confounded by those resulting from demographic events, e.g., population structure or a recent bottleneck, we applied the GOF test proposed by Jensen et al. (2005). We obtained ΛGOF = 467 with a Monte Carlo P-value estimate of 0.81. Therefore, the significant LRKS value is unlikely to be a false positive, i.e., the result of demographic forces alone. This result is supported by the Kim and Nielsen (2004) test, which also yielded a significantly large likelihood ratio (LRKN) of the selective sweep vs. the neutral model (LRKN = 17.09, P = 0.05 in comparison to 10,000 simulated neutral data sets).
Estimates of the strength of selection (α = 1.5Nes) are 661 and 552 for the Kim and Stephan (2002) and the Kim and Nielsen (2004) method, respectively. Assuming that the effective population size (Ne) of the European D. melanogaster population is approximately one-third that of the African Ne (Glinka et al. 2003), s is estimated to be 1.3 × 10−3 and 1.1 × 10−3 using the Kim and Stephan (2002) and the Kim and Nielsen (2004) test, respectively.
Next we consider the African sample. We applied only the Kim and Stephan test (2002) to the polymorphism data obtained from the African sample, since estimated levels of LD were rather low (see Table 2). We obtained a LRKS of 21.54, which was significant in comparison to 10,000 neutral data sets (P = 0.02). ΛGOF estimated by the GOF test (Jensen et al. 2005) was 912 (P = 0.87). As for the European sample, the polymorphism pattern observed in the African sample therefore cannot be explained by simple demographic events alone. The estimate of the strength of selection (α) produced by the test is 2076, which yields s = 1.4 × 10−3, assuming an effective population size of 106.
Position of selected site:
Applying the CLR test (Kim and Stephan 2002), we estimated the approximate position of the selected site. For the European sample, the likelihood ratio is maximized at position 57.7 kb, indicating a target of selection approximately in the middle of the analyzed region. In addition, we used the approach by Kim and Nielsen (2004). The results obtained by this method indicate that the target of selection is located at 57.9 kb, i.e., also within the fourth intron of the gene ph-p. Thus, the inclusion of LD results in an estimated position of the target of selection that is shifted slightly downstream, presumably due to the somewhat stronger LD found in the downstream flanking region (compared to the other flanking region). For the African sample, we obtained a somewhat different estimate for the target of selection. The Kim and Stephan (2002) likelihood ratio is maximized at position 49.8 kb, therefore pointing toward a target of selection closer to fragment 6 (see Figure 2), which is located between ph-d and ph-p.
DISCUSSION
Previous population genetic studies revealed a general trend toward lower nucleotide diversity in non-African D. melanogaster populations (Begun and Aquadro 1993, 1995; Schlötterer et al. 1997; Langley et al. 2000; Andolfatto 2001; Kauer et al. 2002; Glinka et al. 2003; Baudry et al. 2004). Current research has attempted to reveal the mechanisms that have led to this geographical pattern of genetic variation. In addition to the effect of demographic events, such as population bottlenecks, positive directional selection has been hypothesized to substantially contribute to reductions in heterozygosity at individual loci as opposed to the genome-wide effects of demography (e.g., Begun and Aquadro 1992; Hudson et al. 1994; Harr et al. 2002; Quesada et al. 2003; Schlenke and Begun 2004).
Evidence for a selective sweep in the wapl region:
In this article we describe several lines of evidence suggesting that positive selection has shaped the genetic variation in the wapl region of D. melanogaster. First, we analyzed a 110-kb region by sequencing 12 fragments of noncoding DNA in a European D. melanogaster sample and detected a stretch of ∼60 kb that is nearly devoid of nucleotide diversity. That is, all seven loci that were analyzed within that 60-kb region are monomorphic, with the exception of one fragment containing a singleton. In contrast, levels of heterozygosity in the African sample appear to be relatively normal, but lower than the chromosome-wide average reported by earlier studies (Glinka et al. 2003; Ometto et al. 2005). A total of 66 segregating sites was observed in the 60-kb region of the African sample (including the wapl fragment of Glinka et al. 2003). A similarly large invariant region (100 kb), as detected in our European sample, has thus far been found only in D. simulans, possibly caused by the fixation of a positively selected allele of a cytochrome P450 gene (Schlenke and Begun 2004).
The pattern of variation that we observed in our European data is unlikely under both the neutral equilibrium model and a simple bottleneck scenario in which a single population size reduction occurred ∼8000 years ago (or earlier). A more recent bottleneck (∼3000–4000 years ago) could be sufficient to explain the observed reduction in heterozygosity. However, such a recent bottleneck is unlikely for European Drosophila (Lachaise et al. 1988; Haddrill et al. 2005; Ometto et al. 2005).
It should be noted that in our bottleneck simulations we did not account for the observation that the African population has been undergoing a size expansion (Glinka et al. 2003). To estimate θ from the observed number of segregating sites (see materials and methods), we assumed a constant population size. Under an expansion model, a higher θ-value would be estimated, given the observed number of segregating sites. Therefore, the θ-value used in the bottleneck simulations of the European population is probably too low. In other words, our method is conservative.
Using the methods of Kim and Stephan (2002) and Kim and Nielsen (2004), we showed that a selective sweep model fits the data significantly better than the neutral equilibrium model. Additional predictions of the selective sweep model were also verified in the data. First, we found a skew in the frequency spectrum of polymorphisms toward rare variants, as indicated by negative values of Tajima's D (Aguadé et al. 1989; Hudson 1990; Braverman et al. 1995; Fay and Wu 2000; Payseur and Nachman 2002). This test statistic is notably sensitive to the influx of new mutations that have occurred after a hitchhiking event (Fay and Wu 2000). We detected such an excess of low-frequency mutations in some of our fragments, consistent with previous studies (e.g., Langley et al. 2000). Second, we observed an excess of high-frequency-derived variants (Fay and Wu 2000; Kim and Stephan 2000). Visual inspection of our data revealed a high frequency of derived variants at three polymorphic loci (3, 8, and 10; Figure 2a). Negative values of Fay and Wu's H statistic confirm this observation and are in accordance with the selective sweep hypothesis. Third, under the hitchhiking model, strong transient LD is expected between neutral segregating sites located in the vicinity (on one side) of the target of selection (Thomson 1977; Kim and Stephan 2002; Kim and Nielsen 2004). Consistent with these predictions, we detected strong haplotype structure and high levels of LD in our data. In addition, haplotype diversity increased and LD decayed with distance from the monomorphic region (Figure 3).
Where did the selective sweep detected in the European population originate? Our analysis of the African sample may suggest that the sweep arose in an ancestral African population before the colonization of Europe. A similar transpopulation sweep (between Africa and Europe) has been detected by Li and Stephan (2005) in a different data set. The hypothesis of a transpopulation sweep in the wapl region needs to be verified by additional sequencing to establish the complete haplotypes in the region under consideration. An alternative hypothesis is that the sweeps in Africa and Europe are independent, as the estimated positions of the target sites of selection that differ by ∼8 kb may seem to indicate (see results). However, this may simply be a consequence of the fact that the target of selection is difficult to localize precisely in the European sample due to a lack of variation (see below).
Although most loci in the wapl region of the African sample do not show a severe reduction in nucleotide diversity, the π- and θ-values for two fragments located in the center of the region are significantly reduced. This reduction in variation and an associated skew in the frequency spectrum resulted in a significant Kim and Stephan test, which is unlikely to be the sole product of simple demographic forces (Jensen et al. 2005). The observed lack of further statistical evidence for selection may be attributed to the relatively old age of the hitchhiking event. Signatures of directional selection are difficult to identify with our methods if they are much older than ∼0.1Ne generations (Kim and Stephan 2000, 2002).
Estimating the strength and target site of selection:
Using the methods of Kim and Stephan (2002) and Kim and Nielsen (2004), we estimated the strength of selection and the approximate target site of selection. For the European sample, both methods produced selection coefficients in the order of 10−3, and the target of selection was located ∼2500 bp downstream from the center of the monomorphic region (see Figure 1) within the fourth intron of the gene ph-p. The method of Kim and Nielsen (2004) suggested that the beneficial mutation occurred an additional 200 bp downstream from the Kim and Stephan (2002) estimate. This result may be explained by the incorporation of LD into the test statistic and the fact that LD appears to be slightly stronger in the fragments located farther downstream (i.e., fragments 10–12) than in those on the other side of the valley of reduced polymorphism. However, it should be noted that it is difficult to precisely localize the putative target of selection using composite likelihood ratio tests for technical reasons and also, in this case, due to a strong reduction of variation over a large region. For the African sample, the Kim and Stephan (2002) test indicates that the position of the target of selection is 8 kb upstream from the estimate based on the European data. Since the valley of reduced variation in the African population is much narrower (see Figure 1), this should facilitate pinpointing the target site of selection.
Genes near the target site of selection:
The genomic region of reduced variation (in the European sample) from ph-d to Cyp4d1 harbors a relatively high density of genes coding for products with metabolic functions: CG3835, putatively involved in carbohydrate metabolism; Pgd, involved in the pentose-phosphate-shunt; bcn92, with putative oxidoreductase activity; and Cyp4d1, a cytochrome P450 gene putatively involved in steroid metabolism. Genes coding for metabolic enzymes have frequently been suggested as targets of positive selection (e.g., Begun and Aquadro 1994; Hudson et al. 1994; Mutero et al. 1994; Eanes 1999; Schlenke and Begun 2004). We are currently analyzing the genes whose products show enzymatic activity using both population genetic and functional methods.
Acknowledgments
We thank A. Wilken for excellent technical assistance, Y. Kim for helpful advice on his tests, L. Ometto for allowing us to use his bottleneck program, and D. Begun, H. Li, J. Parsch, and two reviewers for helpful comments on this manuscript. We are particularly grateful to a reviewer and D. Begun for pointing out the possible African origin of the sweep. This work was funded by the Deutsche Forschungsgemeinschaft (STE 325/6) and the Volkswagenstiftung (I/78815).
References
- Aguadé, M., N. Miyashita and C. H. Langley, 1989. Reduced variation in the yellow- achaete-scute region in natural populations of Drosophila melanogaster. Genetics 122: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto, P., 2001. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Baudry, E., B. Viginier and M. Veuille, 2004. Non-African populations of Drosophila melanogaster have a unique origin. Mol. Biol. Evol. 21: 1482–1491. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365: 548–550. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1994. Evolutionary inferences from DNA variation at the 6-phosphogluconate dehydrogenase locus in natural populations of Drosophila: selection and geographic differentiation. Genetics 136: 155–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1995. Molecular variation at the vermilion locus in geographically diverse populations of Drosophila melanogaster and D. simulans. Genetics 140: 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman, J. M., R. R. Hudson, N. L. Kaplan, C. H. Langley and W. Stephan, 1995. The hitchhiking effect on the site frequency spectrum of DNA polymorphisms. Genetics 140: 783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., M. Kreitman and M. Aguadé, 1999. Natural selection on synonymous sites is correlated with gene length and recombination in Drosophila. Genetics 151: 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, J. R., and P. Capy, 1988. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4: 106–111. [DOI] [PubMed] [Google Scholar]
- Eanes, W. F., 1999. Analysis of selection on enzyme polymorphisms. Annu. Rev. Ecol. Syst. 30: 301–326. [Google Scholar]
- Fay, J. C., and C.-I Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase Consortium, 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31: 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier, N., F. Depaulis and N. H. Barton, 2000. Detecting bottlenecks and selective sweeps from DNA sequence polymorphism. Genetics 155: 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka, S., L. Ometto, S. Mousset, W. Stephan and D. De Lorenzo, 2003. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics 165: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill, P. R., K. R. Thornton, B. Charlesworth and P. Andolfatto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15: 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr, B., M. Kauer and C. Schlötterer, 2002. Hitchhiking mapping: a population-based fine-mapping strategy for adaptive mutations in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99: 12949–12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 1990. Gene genealogies and the coalescent process, pp. 1–44 in Oxford Surveys in Evolutionary Biology, edited by D. Futuyma and J. Antonovics. Oxford University Press, New York.
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., K. Bailey, D. Skarecky, J. Kwiatowski and F. J. Ayala, 1994. Evidence for positive selection in the superoxide dismutase (sod) region of Drosophila melanogaster. Genetics 136: 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J. D, Y. Kim, V. Bauer DuMont, C. F. Aquadro and C. D. Bustamante, 2005. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170: 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, N. L., R. R. Hudson and C. H. Langley, 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M., B. Zangerl, D. Dieringer and C. Schlötterer, 2002. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics 160: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M., D. Dieringer and C. Schlötterer, 2003. A microsatellite variability screen for positive selection associated with the “out of Africa” habitat expansion of Drosophila melanogaster. Genetics 165: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., and R. Nielsen, 2004. Linkage disequilibrium as a signature of selective sweeps. Genetics 167: 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., and W. Stephan, 2000. Joint effects of genetic hitchhiking and background selection on neutral variation. Genetics 155: 1415–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y., and W. Stephan, 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise, D., M. Cariou, J. R. David, F. Lemeunier, L. Tsacas et al., 1988. Historical biogeography of the Drosophila melanogaster species subgroup, pp. 159–225 in Evolutionary Biology, edited by M. K. Hecht, B. Wallace and G. T. Prance. Plenum, New York.
- Langley, C. H., B. P. Lazzaro, W. Phillips, E. Heikkinen and J. Braverman, 2000. Linkage disequilibrium and the site frequency spectra in the su(s) and su(wa) regions of the Drosophila melanogaster X chromosome. Genetics 156: 1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., and W. Stephan, 2005. Maximum-likelihood methods for detecting recent positive selection and localizing the selected site in the genome. Genetics 171: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Mutero, A., M. Pralavorio, J.-M. Bride and D. Fournier, 1994. Resistance- associated point mutations in insecticide-insensitive acetylcholinesterase. Proc. Natl. Acad. Sci. USA 91: 5922–5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Ometto, L., S. Glinka, D. De Lorenzo and W. Stephan, 2005. Inferring the impact of demography and selection on Drosophila melanogaster from a chromosome-wide DNA polymorphism study. Mol. Biol. Evol. 22: 2119–2130. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., and M. W. Nachman, 2002. Natural selection at linked sites in humans. Gene 300: 31–42. [DOI] [PubMed] [Google Scholar]
- Quesada, H., U. E. M. Ramirez, J. Rozas and M. Aguadé, 2003. Large-scale adaptive hitchhiking upon high recombination in Drosophila melanogaster. Genetics 165: 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins, S. E., B. E. Stranger, T. Mitchell-Olds and M. Aguadé, 2004. Multi-locus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics 166: 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sánchez-Del Barrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schlenke, T. A., and D. J. Begun, 2004. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc. Natl. Acad. Sci. USA 101: 1626–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer, C., C. Vogl and D. Tautz, 1997. Polymorphism and locus-specific effects on polymorphism at microsatellite loci in natural Drosophila melanogaster populations. Genetics 146: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W., and C. H. Langley, 1989. Molecular genetic variation in the centromeric region of the X chromosome in three Drosophila melanogaster populations. I. Contrasts between the vermilion and forked loci. Genetics 121: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, G., 1977. The effect of a selected locus on linked neutral loci. Genetics 85: 753–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verni, F., R. Gandhi, M. L. Goldberg and M. Gatti, 2000. Genetic and molecular analysis of wings apart-like (wapl), a gene controlling heterochromatin organization in Drosophila melanogaster. Genetics 154: 1693–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]