Abstract
Mark-recapture experiments showed that D. melanogaster has high dispersal capabilities. Consistent with a highly migratory species, only very low levels of differentiation were described for D. melanogaster populations from the same continent. We reinvestigated the population structure in D. melanogaster using 49 polymorphic markers in 23 natural populations. While European and American D. melanogaster populations showed very low differentiation, Asian D. melanogaster populations were highly structured. Despite the high differentiation of Asian flies, we confirm that all non-African populations are derived from a single colonization event. We propose that the availability of D. melanogaster populations with high and low population structure provides a novel tool for the identification of ecologically important adaptations by hitchhiking mapping.
HITCHHIKING mapping is a population genetics approach for the identification of genomic regions carrying a favorable mutation (Schlötterer 2003). In the wake of recent genome projects, hitchhiking mapping provides a hitherto unmatched opportunity for the identification and characterization of ecologically relevant alleles (Schlötterer 2002; Feder and Mitchell-Olds 2003; Luikart et al. 2003).
Apart from some studies in humans and maize, Drosophila has been the primary target of systematic screens for the identification of recently selected alleles (Harr et al. 2002; Glinka et al. 2003; Kauer et al. 2003b; Orengo and Aguade 2004; Schöfl and Schlötterer 2004). Drosophila melanogaster is particularly well suited for the identification of ecologically relevant alleles. Like humans, D. melanogaster originated in sub-Saharan Africa and colonized the rest of the world only recently (David and Capy 1988). This habitat expansion presumably required numerous adaptations, which have left their traces in the genome (Harr et al. 2002).
Despite the overwhelming evidence for a large number of adaptive mutations, the identification of the genomic region affected by a selective sweep is significantly complicated by demographic history. Non-sub-Saharan African D. melanogaster populations are most likely derived from a single founder event (Baudry et al. 2004), which was probably associated with a significant reduction in population size. Genetic drift during the early phase of the colonization could have led to a genetic signature in the genome that is almost indistinguishable from a selective sweep (Barton 1998). Thus, one of the major challenges for the identification of genomic regions carrying a beneficial mutation is the distinction between demography and selection (Nielsen 2001; Wall et al. 2002; Depaulis et al. 2003; Jensen et al. 2005).
Recent theoretical work has suggested that the genomic signature of a selective sweep strongly depends on the population structure (Santiago and Caballero 2005). In the case of low population differentiation, all populations show the same signature of a selective sweep: a valley of reduced variation and a surplus of rare, derived alleles (i.e., negative Tajima's D; Tajima 1989). For highly differentiated populations, however, only the population in which the beneficial mutation arose shows this characteristic pattern. The other populations, which acquire the beneficial mutation by migration, show a characteristically different pattern: only a very narrow genomic region around the target of selection has low variation and a negative Tajima's D. With an increasing distance from the target of selection, Tajima's D increases above neutral expectations (Santiago and Caballero 2005). Hence, the analysis of multiple differentiated populations will provide a highly characteristic pattern of a selective sweep, with a very narrow genomic region carrying the beneficial mutation.
Until the discovery of the high differentiation between sub-Saharan African and cosmopolitan D. melanogaster populations (Begun and Aquadro 1993), D. melanogaster was often considered a completely panmictic population. Even when a large number of highly informative markers were used, the differentiation among non-African D. melanogaster populations was found to be extremely low with FST values in the range of 0.004–0.152 (Caracristi and Schlötterer 2003).
In this report we analyzed a large collection of Asian D. melanogaster populations. We find that Asian D. melanogaster are derived from the same colonization event as other non-African populations, but we detected extremely high levels of differentiation (up to 0.3) between them. This hitherto unappreciated high population structure in Asian D. melanogaster populations opens completely new possibilities for studying adaptation in natural D. melanogaster populations.
MATERIALS AND METHODS
Table 1 provides details about the populations used. Analyses were based on offspring from wild-caught females and on collections of isofemale lines that had been in culture for a number of generations (see Table 1). To account for inbreeding in the isofemale lines, we estimated gene diversity by using a resampling method that randomly discards one allele per locus (Dieringer and Schlötterer 2003). For FST and Bayesian analysis of population structure (BAPS) (Corander et al. 2004) analyses, we used the nondiscarded data set, as neither FST estimates nor the BAPS analysis is affected by the inbreeding in isofemale lines (J. Corander, personal communication).
TABLE 1.
Fly stocks used
| Abbreviation of population/location | Location | Country | Date of collection | Provided/ collected by: | No. of lines | F1/isofemale |
|---|---|---|---|---|---|---|
| Europe | ||||||
| Fia | Harjavalta | Finland | August 1996 | 19 | Isofemale | |
| Kata | Katowice | Poland | October 2000 | J. Gorczyca | 30 | F1 |
| WRa | Weil am Rhein | Germany | September 2000 | B. Harr | 30 | F1 |
| Texa | Texel | Netherlands | 1997 | D. Slezak | 30 | F1 |
| Kbha | Copenhagen | Denmark | 1998 | V. Loeschcke | 30 | F1 |
| Kr | Crete | Greece | August 1998 | A. Hans | 9 | F1 |
| Naa | Naples | Italy | 2000 | C. Schlötterer | 32 | F1 |
| Africa | ||||||
| Kea | Kenya | K. Yoon | 24 | Isofemale | ||
| ZiHa | Harare | Zimbabwe | C.-I Wu, C. Aquadro | 15 | Isofemale | |
| America | ||||||
| Pea | Penn State, PA | United States | 1998 | M. Dermitzakis | 30 | F1 |
| SCa | West End, NC | United States | September 2000 | G. Gibson | 30 | Isofemale |
| GRa | Groth Winery, Napa Valley, CA | United States | October 1996 | J. F. McDonald | 30 | Isofemale |
| NJa | Rockaway, NJ | United States | Summer 1999 | E. Weiss | 31 | F1 |
| Bea | La Milpa | Belize | Summer 1999 | E. Weiss | 19 | F1 |
| Asia | ||||||
| KL | Kuala Lumpur | Malyasia | October 2002 | A. Das | 20 | Isofemale |
| KK | Kota Kinabalu | Malaysian Borneo | October 2002 | A. Das | 20 | Isofemale |
| MAN | Manila | Philippines | October 2002 | A. Das | 14 | Isofemale |
| CEB | Cebu | Philippines | October 2002 | A. Das | 20 | Isofemale |
| CM | Chiang Mai | Thailand | October 2002 | A. Das | 25 | Isofemale |
| BKK | Bangkok | Thailand | October 2002 | A. Das | 20 | Isofemale |
| H.Chu | Hsin Chu | Taiwan | July 2002 | S.-C. Tsaur | 9 | Isofemale |
| Ci | Jiamusi | Heilongiiang, China | August 2002 | E. Li | 30 | F1 |
| Mys | Mysore | India | 2003 | H. A. Ranganath | 9 | Isofemale |
Data for 48 loci were taken from Caracristi and Schlötterer (2003).
Genomic DNA was isolated for each line using a single female fly by the high salt extraction method (Miller et al. 1988). We typed 49 microsatellite loci, 48 of which were already described in Caracristi and Schlötterer (2003). One additional locus located on chromosome 2L (forward primer 5′-GCCCTCTGCGATGTGCGTTG-3′ and reverse primer 5′-TTCGCTTCACAGCCTTACTC-3′) was also included. Ten-microliter PCR reactions were carried out with 50 ng genomic DNA, 32P-labeled forward primer, 1.5 mm MgCl2, 200 μm dNTPs, 1 μm of each primer, and 0.5 units Taq polymerase. A typical cycling profile consisted of 30 cycles with 50 sec at 94°, 50 sec at 50°–57° (depending on the primer pair), and 50 sec at 72°. All PCR reactions were run with an initial denaturing step of 3 min at 94° and a final extension of 45 min at 72° for quantitative terminal transferase activity of the Taq polymerase. PCR products were separated on 7% denaturing polyacrylamide gels (32% formamide, 5.6 m urea) and visualized by autoradiography. PCR products were sized by loading a “slippage ladder” next to the amplified microsatellites (Schlötterer and Zangerl 1999). While we used most genotypes of the African, European, and American populations described in Caracristi and Schlötterer (2003), some loci were regenotyped for some populations.
Measures of genetic variation, such as heterozygosity and number of alleles, were calculated using the MS-Analyzer (MSA) software, version 4.0 (Dieringer and Schlötterer 2003). When more than a single population was typed for one continent, estimates of variability were calculated for each population separately and subsequently averaged. This treatment was chosen to avoid the Wahlund effect (Wahlund 1928). The proportion of shared alleles was calculated by the MSA software (Dieringer and Schlötterer 2003). The obtained distance matrix was converted into a dendrogram using the FITCH program, which is part of the PHYLIP package (Felsenstein 1991), and graphically displayed with TREEVIEW (Page 1996). The statistical significance of the nodes of the dendrogram were evaluated by bootstrapping loci (Efron and Gong 1983). To estimate population differentiation, pairwise Θ-values were determined as an unbiased estimate of FST (Weir and Cockerham 1984) using the MSA software. The significance of pairwise FST values was tested by permuting genotypes among populations (10,000 times), as this method does not rely on Hardy–Weinberg assumptions (Goudet et al. 1996). To account for multiple testing, we used the Bonferroni method (Sokal and Rohlf 1995). Bayesian clustering and admixture analysis was performed with the BAPS3.1 software (Corander et al. 2004) using the default settings.
RESULTS AND DISCUSSION
We genotyped 49 microsatellites, 26 autosomal and 23 X-linked, in 10 populations and supplemented already published data of 13 populations (Caracristi and Schlötterer 2003) for one additional locus. Consistent with the sub-Saharan origin of D. melanogaster, the populations from Zimbabwe and Kenya harbored the most variation (H = 0.77). Among the non-African populations analyzed, American D. melanogaster were most variable (H = 0.58), followed by European (H = 0.53) and Asian (H = 0.48) populations. Notably, even the difference between Asian and European populations was statistically significant (P = 0.043, Wilcoxon sign ranks test).
Using a Bayesian method (Corander et al. 2004), we identified 11 genetically distinct groups. Four of the identified clusters consisted of more than one population. The first cluster contained both African populations. The second cluster contained all populations from America, including the population from Belize, which was previously shown to harbor a relatively high proportion of African alleles (Caracristi and Schlötterer 2003). The third cluster combined all European populations. Finally, the populations from continental China and Taiwan were also clustered. All remaining Asian populations formed separate genetic entities. The conclusion that Asian populations are highly differentiated is further corroborated by a pairwise FST analysis (supplemental Table 1 at http://www.genetics.org/supplemental/). The population from Chiang Mai (Thailand) was the most differentiated one with an average pairwise FST value of 0.26. Interestingly, the two populations from mainland China and Taiwan were the Asian populations with the least differentiation from the European/American populations. With an average of 0.059, the differentiation between Taiwan and America was particularly low.
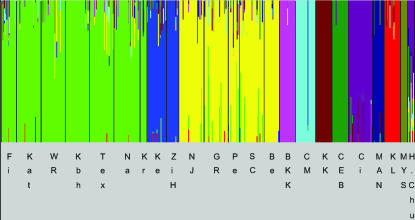
As this low level of differentiation between the Taiwanese population and America may be indicative of recent gene flow, we performed a Bayesian admixture analysis. Figure 1 depicts the 11 genetically distinct groups, with one color specific for each group. A thin vertical line consisting of only one color represents individuals without admixture. In admixed individuals, the line is partitioned into colored segments that represent the individual's estimated membership fraction in the corresponding clusters. The different colors occurring in one line (individual) represent the admixture proportions. While in most populations, the inferred admixture is low, the population from Taiwan has a significant proportion of alleles with coancestry in American populations. Apart from the Taiwanese population, Asian D. melanogaster populations showed the lowest degree of admixture (Figure 1).
Figure 1.
Estimated population structure. Genetically distinct groups are indicated by distinct colors. (Bottom) The populations constituting a group. Each individual is represented by a thin line, which is partitioned into colored segments that represent the individual's estimated membership fraction in the corresponding clusters. See Table 1 for abbreviations of populations. Europe: Fi, Kat, WR, Kbh, Tex, Na, Kr; Africa: Ke, ZiH; America: NJ, GR, Pe, Sc, Be; Asia: BKK, CM, KK, CEB, Ci, MAN, KL, MYS, H.Chu.
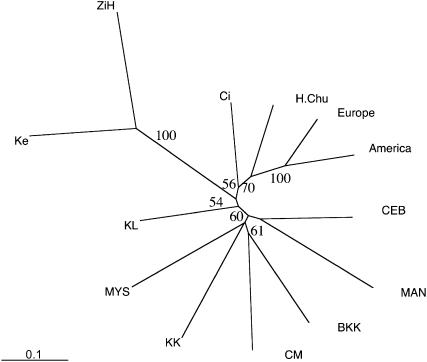
In the light of previous reports about a highly diverged Far Eastern race (David and Capy 1988), it may be possible that the high divergence between Asian and European/American populations reflects multiple, independent colonization events. We used genealogical inference based on the proportion of shared alleles to understand the colonization history of non-African flies. Multiple colonization events would result in a star-like topology with a very short branch between the African and the non-African node. In the tree shown in Figure 2, however, this branch is the longest internal branch of the tree, suggesting that most divergence occurred between these two nodes. As this long branch is shared among all non-African populations in our data set, we could exclude that multiple, independent colonization events led to the high differentiation among Asian and between Asian and European/American populations. Rather, all non-African populations probably originated from a common population that was quite diverged from contemporary sub-Saharan D. melanogaster populations. Given the demographic history of D. melanogaster, it is likely that high rates of genetic drift associated with the founder event (bottleneck) during the habitat expansion of sub-Saharan D. melanogaster caused the divergence between the African and non-African populations. The high proportion of alleles (58%) shared between Asian and European samples also supports a common ancestral population, which is diverged from African D. melanogaster. Similarly, a high proportion of nucleotide polymorphism (haplotypes) is shared between Asian and European populations (Baudry et al. 2004; V. Nolte and C. Schlötterer, unpublished results).
Figure 2.
Dendrogram based on the proportion of shared alleles. For more clarity, American and European populations were combined into a single operational taxonomic unit each. Bootstrap values indicate the statistical support for the corresponding node. Only bootstrap values >50 are shown. See Table 1 for abbreviations of populations.
We also performed bottleneck analyses by comparing observed gene diversity to the expected one on the basis of the observed number of alleles (Cornuet and Luikart 1996). However, no conclusive results were obtained. Some Asian populations showed significant evidence for population expansion, others provided significant support for population contraction, and no significant deviation from a constant size was also detected (data not shown).
In D. melanogaster and D. simulans, X-linked and autosomal loci show significant differences between African and non-African populations (Andolfatto 2001; Kauer et al. 2002; Kauer et al. 2003b; Schöfl and Schlötterer 2004) as well as among African populations (Kauer et al. 2003a). Hence, we also analyzed autosomal and X-linked loci separately. While we found an overall agreement between the two data sets (supplemental Table 2 and supplemental Figure 1 at http://www.genetics.org/supplemental/), we noted that the Chinese population grouped differently in both data sets. While for autosomal loci a grouping with the European and American populations was supported by a bootstrap support of 69%, for X-linked loci the Chinese population clustered with the Asian populations (supplemental Figure 1 at http://www.genetics.org/supplemental/). The reason for this discrepancy between X-linked and autosomal loci in the Chinese population is not clear and more data are required to distinguish between chance and directed (e.g., selection) effects.
Interestingly, previous studies using other genetic markers provided conflicting evidence for Asian flies. While a large survey of allozymes did not find higher differentiation among Asian populations (Singh and Rhomberg 1987), mtDNA data do indicate higher levels of differentiation among Asian populations (Hale and Singh 1991; Solignac 2004).
Well-designed experiments based on the recapture of D. melanogaster mutants released at an orchard in Maryland showed a large dispersal potential and released flies were able to move to potential breeding sites (Coyne and Milstead 1987). As the authors failed to find mutant flies in the subsequent year, they concluded that D. melanogaster does not overwinter and recolonizes the study site every year. Overall, these data strongly suggested that D. melanogaster is a highly migratory species. Consistent with these direct observations, indirect measurements also found low levels of population differentiation in American populations (Singh and Rhomberg 1987; Caracristi and Schlötterer 2003) (supplemental Table 1 at http://www.genetics.org/supplemental/). As European D. melanogaster also show low levels of differentiation, it could be assumed that they also have high dispersal capabilities. This picture contrasts with our observations in Asian D. melanogaster, where we observed high levels of differentiation, most likely caused by genetic drift. Thus, our data suggest that Asian D. melanogaster disperse to a much lower extent than European/American flies. Interestingly, the pronounced population structure in Asian D. melanogaster is paralleled by D. ananassae, which also shows strong population structure for populations collected in Southeast Asia (Das et al. 2004). Thus, the high population structure of Southeast Asian Drosophila may be a more general phenomenon.
This difference in dispersal of Asian and other non-African D. melanogaster has important implications for the design of experiments screening for genes involved in adaptation to non-African habitat. The population bottleneck associated with the out-of-Africa habitat expansion could generate a signature that closely resembles the signature of a selective sweep. The high similarity of American and European populations strongly limits the joint analysis of multiple populations as an effective means for distinguishing between a population bottleneck and a beneficial mutation that spread after the colonization event—the signature of the selective sweep has been found to be very similar among different populations (Harr et al. 2002).
Asian populations offer two conceptual advances for the identification of selective sweeps:
If a genomic region shows the signature of a selective sweep in multiple European and American populations, but not in multiple Asian populations, this could potentially indicate that this signature has not been caused by the bottleneck associated with the out-of-Africa founder event. Indeed, a sequence variation survey of six fragments with low variability in Europe found two of these fragments to be highly variable in Asia (V. Nolte and C. Schlötterer, unpublished results). Nevertheless, such differences between Asian and European flies are informative only in an analytical framework that considers the amount of genetic drift between Asian and European flies.
A recent theoretical study showed that the signature of a selective sweep differs for populations with high and low levels of genetic differentiation (Santiago and Caballero 2005). Thus, if the same beneficial mutation spreads through Asian and European/American populations, it would be possible to compare the signature of the sweep. On the basis of the difference in signature it could be possible to distinguish a sweep from a bottleneck effect.
Most important, due to the fact that in the continent to which the allele is exported by migration a smaller window of reduced variation is expected, a higher mapping precision should be achieved when hitchhiking mapping is applied. Nevertheless, as the theoretical study by Santiago and Caballero (2005) focused on extremely large FST values—higher than the ones observed for the Asian populations—more data are required to decide if the statistical power will be high enough to detect a selective sweep on the basis of the theoretical predictions of Santiago and Caballero (2005).
Acknowledgments
We are thankful to A. Schneider for help with genotyping. We are extremely grateful to all those who shared and collected flies. C. Vogl and B. Harr provided helpful comments and discussion on earlier versions of the manuscript. Comments from D. Rand and two anonymous reviewers helped to improve the manuscript. This work has been supported by Fonds zur Förderung der wissenschaftlichen Forschung funds to C.S.
References
- Andolfatto, P., 2001. Contrasting patterns of X–linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 18: 279–290. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Baudry, E., B. Viginier and M. Veuille, 2004. Non-African populations of Drosophila melanogaster have a unique origin. Mol. Biol. Evol. 21: 1482–1491. [DOI] [PubMed] [Google Scholar]
- Begun, D., and C. F. Aquadro, 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365: 548–550. [DOI] [PubMed] [Google Scholar]
- Caracristi, G., and C. Schlötterer, 2003. Genetic differentiation between American and European Drosophila melanogaster populations could be attributed to admixture of African alleles. Mol. Biol. Evol. 20: 792–799. [DOI] [PubMed] [Google Scholar]
- Corander, J., P. Waldmann, P. Marttinen and M. J. Sillanpaa, 2004. BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics 20: 2363–2369. [DOI] [PubMed] [Google Scholar]
- Cornuet, J. M., and G. Luikart, 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and B. Milstead, 1987. Long-distance migration of Drosophila dispersal of D. melanogaster alleles from a Maryland orchard. Am. Nat. 130: 70–82. [Google Scholar]
- Das, A., S. Mohanty and W. Stephan, 2004. Inferring the population structure and demography of Drosophila ananassae from multilocus data. Genetics 168: 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, J. R., and P. Capy, 1988. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4: 106–111. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., S. Mousset and M. Veuille, 2003. Power of neutrality tests to detect bottlenecks and hitchhiking. J. Mol. Evol. 57(Suppl. 1): S190–S200. [DOI] [PubMed] [Google Scholar]
- Dieringer, D., and C. Schlötterer, 2003. Microsatellite analyzer (MSA)—a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 3: 167–169. [Google Scholar]
- Efron, B., and G. Gong, 1983. A leisurely look at the bootstrap, the jackknife, and cross-validation. Am. Stat. 37: 36–48. [Google Scholar]
- Feder, M. E., and T. Mitchell-Olds, 2003. Evolutionary and ecological functional genomics. Nat. Rev. Genet. 4: 651–657. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1991. PHYLIP, Version 3.57c. University of Washington, Seattle.
- Glinka, S., L. Ometto, S. Mousset, W. Stephan and D. De Lorenzo, 2003. Demography and natural selection have shaped genetic variation in Drosophila melanogaster: a multi-locus approach. Genetics 165: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet, J., M. Raymond, T. de Meeüs and F. Rousset, 1996. Testing differentiation in diploid populations. Genetics 144: 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, L. R., and R. S. Singh, 1991. A comprehensive study of genic variation in natural populations of Drosophila melanogaster. IV. Mitochondrial DNA variation and the role of history vs. selection in the genetic structure of geographic populations. Genetics 129: 103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr, B., M. Kauer and C. Schlötterer, 2002. Hitchhiking mapping: a population based fine mapping strategy for adaptive mutations in D. melanogaster. Proc. Natl. Acad. Sci. USA 99: 12949–12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J. D., Y. Kim, V. B. Dumont, C. F. Aquadro and C. D. Bustamante, 2005. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170: 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M., B. Zangerl, D. Dieringer and C. Schlötterer, 2002. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics 160: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer, M., D. Dieringer and C. Schlötterer, 2003. a Nonneutral admixture of immigrant genotypes in African Drosophila melanogaster populations from Zimbabwe. Mol. Biol. Evol. 20: 1329–1337. [DOI] [PubMed] [Google Scholar]
- Kauer, M. O., D. Dieringer and C. Schlötterer, 2003. b A microsatellite variability screen for positive selection associated with the “out of Africa” habitat expansion of Drosophila melanogaster. Genetics 165: 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart, G., P. R. England, D. Tallmon, S. Jordan and P. Taberlet, 2003. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4: 981–994. [DOI] [PubMed] [Google Scholar]
- Miller, S. A., D. D. Dykes and H. F. Polesky, 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R., 2001. Statistical tests of selective neutrality in the age of genomics. Heredity 86: 641–647. [DOI] [PubMed] [Google Scholar]
- Orengo, D. J., and M. Aguade, 2004. Detecting the footprint of positive selection in a European population of Drosophila melanogaster: multilocus pattern of variation and distance to coding regions. Genetics 167: 1759–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R. D. M., 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Santiago, E., and A. Caballero, 2005. Variation after a selective sweep in a subdivided population. Genetics 169: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer, C., 2002. Towards a molecular characterization of adaptation in local populations. Curr. Opin. Genet. Dev. 12: 683–687. [DOI] [PubMed] [Google Scholar]
- Schlötterer, C., 2003. Hitchhiking mapping: functional genomics from the population genetics perspective. Trends Genet. 19: 32–38. [DOI] [PubMed] [Google Scholar]
- Schlötterer, C., and B. Zangerl, 1999. The use of imperfect microsatellites for DNA fingerprinting and population genetics, pp. 153–165 in DNA Profiling and DNA Fingerprinting, edited by J. T. Epplen and T. Lubjuhn. Birkhäuser, Basel, Switzerland.
- Schöfl, G., and C. Schlötterer, 2004. Patterns of microsatellite variability among X chromosomes and autosomes indicate a high frequency of beneficial mutations in non-African D. simulans. Mol. Biol. Evol. 21: 1384–1390. [DOI] [PubMed] [Google Scholar]
- Singh, R. S., and L. R. Rhomberg, 1987. A comprehensive study of genic variation in natural populations of Drosophila melanogaster. II. Estimates of heterozygosity and pattterns of geographic differentiation. Genetics 117: 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R. R., and F. J. Rohlf, 1995. Biometry. W. H. Freeman, New York.
- Solignac, M., 2004. Mitochondrial DNA in the Drosophila melanogaster complex. Genetica 120: 41–50. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlund, S., 1928. Zusammensetzung von Populationen und Korrelationserscheinungen vom Standpunkt der Vererbungslehre aus betrachtet. Hereditas 11: 65–105. [Google Scholar]
- Wall, J. D., P. Andolfatto and M. Przeworski, 2002. Testing models of selection and demography in Drosophila simulans. Genetics 162: 203–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, B. S., and C. C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]