Abstract
Through simultaneous interactions with Hsp70 and Hsp90 via separate tetratricopeptide repeat (TPR) domains, the cochaperone protein Hop/Sti1 has been proposed to play a critical role in the transfer of client proteins from Hsp70 to Hsp90. However, no prior mutational analysis demonstrating a critical in vivo role for the TPR domains of Sti1 has been reported. We used site-directed mutagenesis of the TPR domains combined with a genetic screen to isolate mutations that disrupt Sti1 function. A single amino acid alteration in TPR2A disrupted Hsp90 interaction in vivo but did not significantly affect function. However, deletion of a conserved residue in TPR2A or mutations in the carboxy-terminal DP2 domain completely disrupted Sti1 function. Surprisingly, mutations in TPR1, previously shown to interact with Hsp70, were not sufficient to disrupt in vivo functions unless combined with mutations in TPR2B, suggesting that TPR1 and TPR2B have redundant or overlapping in vivo functions. We further examined the genetic and physical interaction of Sti1 with a mutant form of Hsp90, providing insight into the importance of the TPR2A domain of Sti1 in regulating Hsp90 function.
THE essential molecular chaperone Hsp90 accounts for 1–2% of all cytosolic proteins and is critical for the activity of diverse cellular proteins that are involved in a variety of cellular processes, including development, cell cycle, and steroid hormone signaling (Pratt and Toft 2003). Because Hsp90 interacts with a number of oncogenic signaling proteins, including Akt, Raf-1, Bcr-Abl, mutant p53, and HER-2/Erb2, it is a promising anticancer target and the Hsp90 inhibitor 17-AAG is currently in clinical trials (Workman 2003).
The interaction of Hsp90 with client proteins is dependent on a number of cochaperone proteins (Pratt and Toft 2003; Prodromou and Pearl 2003). Studies of the assembly of steroid receptors with Hsp90 and cochaperones have identified five proteins that are required for efficient maturation of Hsp90 client proteins in a purified system: Hsp70, Hsp40/Ydj1, Hsp90, Hop/Sti1, and p23/Sba1. These proteins cooperate in an ordered pathway that involves sequential ATP-dependent interactions of the client protein with the chaperones Hsp70 and Hsp90. The cochaperone proteins Hsp40/Ydj1, Hop/Sti1, and p23/Sba1 regulate the function of Hsp70 and/or Hsp90. On the basis of the results of a number of in vitro studies, the current model of Hsp90 interaction with client proteins purports that Hsp40/Ydj1 and Hsp70 interact with the client first, followed by Hop/Sti1 binding. Because Hop/Sti1 can simultaneously interact with both Hsp70 and Hsp90 through separate tetratricopeptide repeat (TPR) domains, Hop/Sti1 is proposed to have a critical function in the transfer of client proteins from Hsp70 to Hsp90. Hop/Sti1 release allows ATP binding and this is accompanied by Hsp90-client interaction and dimerization of the amino-terminal ATPase domain of Hsp90, which is stabilized by the cochaperone p23.
The main structural features of Hop/Sti1 (Hop in mammalian cells, Sti1 in Saccharomyces cerevisiae) are its three TPR domains (Odunuga et al. 2004; Smith 2004). TPR domains are composed of 34 amino acid helix-turn-helix motifs. Many diverse proteins contain TPR domains and the specificity of the domains is determined by side chain residues that project into a central binding groove. Separate TPR domains of Hop/Sti1 specifically interact with Hsp70 and Hsp90, which share a conserved carboxy-terminal EEVD sequence that binds the central groove of the TPR domain. The amino-terminal TPR1 domain bound the carboxy-terminal 10-kDa fragment of Hsp70, while the TPR2A domain interacted with the carboxy-terminal 12-kDa fragment of Hsp90 (Demand et al. 1998; Young et al. 1998). TPR1 of Hop cocrystallized with an ocatapeptide containing terminal EEVD residues of Hsp70 and TPR2A of Hop cocrystallized with a peptapeptide containing the terminal EEVD residues of Hsp90 (Scheufler et al. 2000). In each case, basic amino acid residues within the TPR domain formed salt bridges with the acidic EEVD residues of the Hsp70- or Hsp90-based peptides, forming what was called a carboxylate clamp. The structure or binding partner of TPR2B is unknown. Hop/Sti1 also contains regions that contain asparatate-proline (DP) residue repeats: DP1 after TPR1 and DP2 after TPR2B (Chen and Smith 1998; Carrigan et al. 2004).
Recent evidence indicates that the interaction of Sti1/Hop with Hsp70 and Hsp90 is more complex than the respective TPR-EEVD interactions. Sti1 regulates the ATPase activity of both Hsp90 and Hsp70 (Prodromou et al. 1999; Richter et al. 2003; Wegele et al. 2003), but the domains responsible for these functions are unknown. The Hop–Hsp70 interaction is not strictly dependent upon the presence of the terminal EEVD residues since Hsp70 mutants lacking the carboxy-terminal 34 amino acids were still able to co-immunoprecipitate with Hop (Carrigan et al. 2004). In addition, mutation of residues outside of TPR1 affected the in vitro interaction of Hop with Hsp70, indicating that multiple domains of Hop are required for Hsp70 binding (Chen and Smith 1998; Odunuga et al. 2003; Carrigan et al. 2004). The Hop–Hsp90 interaction was specifically disrupted by mutations in basic residues in TPR2A (Carrigan et al. 2004), and deletion of the terminal MEEVD residues of yeast Hsp90 (Hsp82-1-704) disrupted Sti1–Hsp90 interaction (Abbas-Terki et al. 2001). However, the in vivo importance of this interaction is unclear since Hsp82-1-704 was able to support near-wild-type levels of growth when present as the only Hsp90 in the cell (Louvion et al. 1996).
Despite interest in determining how Sti1 regulates Hsp70 and Hsp90 function, no prior in vivo mutational analysis targeting specific domains of Sti1 has been completed, in part because disruption of sti1 in yeast causes only minor growth defects (Nicolet and Craig 1989). In contrast to a proposed critical role in transfer of client proteins from Hsp70 to Hsp90, deletion of STI1 does not have dramatic effects on glucocorticoid receptor (GR) complexes isolated out of yeast (Chang et al. 1997). However, consistent with an in vivo role in maturation of Hsp90 client proteins, deletion of STI1 affects the activity of heterologous Hsp90 substrates such as v-src and the GR (Chang et al. 1997), as well as the native yeast transcription factor HSF (Liu et al. 1999) and protein kinase Stell (Lee et al. 2004). Moreover, deletion of STI1 in combination with deletion in genes encoding Hsp90 or Hsp90 cochaperones causes a lethal phenotype or enhanced temperature-sensitive growth defects (Chang et al. 1997; Liu et al. 1999; Abbas-Terki et al. 2002). We recently found that the combination of deletion of STI1 along with a deletion of YDJ1, which encodes an Hsp40 required for Hsp90 client protein activity, is lethal (Flom et al. 2005). This synthetic lethality provided us with a powerful tool to study the in vivo importance of the various domains of Sti1 and also to select mutations that disrupt Sti1 function.
In this article we examine the effect of STI1 mutation on in vivo functions. We used the YDJ1–STI1 synthetic lethality to determine the effects of site-directed mutations of the TPR domains and also conducted a genetic screen to isolate mutations in STI1 that failed to support viability of a sti1ydj1 strain. We describe the effect of mutation of TPR1, TPR2A, and TPR2B, alone and in combination, on growth, assays of Hsp90 client protein activity, and Hsp90 interaction. We also describe the effect of mutation or deletion of the DP2 region of Sti1. Our results indicate that TPR2A and the DP2 region have a critical role in Sti1 function and suggest that TPR1 and TPR2B have overlapping or redundant functions. These results raise interesting questions about the structure of Hop/Sti1 and how Hop/Sti1 physically interacts with Hsp70.
MATERIALS AND METHODS
Strains, media and growth assays:
Standard yeast genetic methods were employed (Sherman et al. 1986; Gietz et al. 1995). Yeast cells were grown in either yeast extract–peptone–dextrose (YPD: 1% Bacto yeast extract, 2% peptone, and 2% dextrose) or defined synthetic complete media supplemented with 2% dextrose. Growth was examined by colony streaking or spotting 10-fold serial dilutions of yeast cultures on appropriate media, followed by incubation for 2 days at the indicated temperature. Radicicol (RD) was obtained from Sigma, dissolved in DMSO (10 mg/ml) and added to YPD plates after autoclaving. 5-FOA was obtained from Toronto Research Chemicals.
All S. cerevisiae strains are isogenic to W303. Strain JJ160 (ydj1∷HIS3/pRS316–YDJ1) has been described (Johnson and Craig 2000). A sti1∷MET2 disruption construct was created by replacing the STI1 sequence encoding amino acids 22–175 with a 2.2-kb fragment containing the MET2 gene. STI1 was disrupted in strain PJ53 (Johnson and Craig 2000) and sporulated to create strain JJ623 (MATa sti1∷MET2). A sti1∷MET2 ydj1∷HIS3/pRS316–YDJ1 strain (JJ609) was constructed by mating and sporulating confirmed deletion strains. We also constructed strain JJ816 (MATa hsc82∷LEU2 hsp82∷LEU2/YeP24–HSP82) and strain JJ832 (MATa hsc82∷LEU2 hsp82∷LEU2 sti1∷MET2/YeP24–HSP82).
Plasmids:
A 2.1-kb MunI–EcoRI fragment of Yep24–STI1 (Nicolet and Craig 1989) containing the complete STI1 gene was subcloned into the low-copy LEU2 plasmid pRS315 (Sikorski and Hieter 1989). Wild type (WT) and mutant STI1 were also cloned into the multi-copy URA3 plasmid pRS426 and the LYS2 plasmid pRS317. After transformation of indicated plasmids into strain JJ609, colonies were grown in media containing 5-FOA to counterselect for pRS316–YDJ1. Plasmids expressing WT HSP82 or hsp82–G313S under the GPD promoter were a gift of Susan Lindquist.
Plasmid mutagenesis:
A series of point mutants in STI1 was generated using site-directed mutagenesis (QuickChange kit, Stratagene, La Jolla, CA) or other PCR-based methods. Sequences of mutagenic primers are available on request. All mutant DNA sequences were verified by automated sequencing. pRS315–ΔSS was generated by cutting pRS315 with SmaI–Eco1CRI followed by religation of the blunt ends. An NcoI site was introduced at the initiation codon of Sti1 prior to amplification under mutagenic conditions and the 2.1-kb MunI–EcoRI fragment was cloned into pRS315–ΔSS. An STI1 mutant library was constructed by error-prone PCR using Taq polymerase and cloned into pRS315–ΔSS STI1–NcoI+. After mutagenesis, a DNA fragment corresponding to amino acids 25–585 (full length Sti1 is 589 amino acids) was cloned into pRS315–ΔSS STI1–NcoI+ lacking this region. Plasmid library DNA was transformed into strain JJ609 (sti1ydj1/pRS316–YDJ1) and colonies that failed to grow in the presence of 5-FOA were analyzed further. Approximately 1600 colonies were screened for growth on 5-FOA. Plasmid DNA was rescued from ∼100 of these strains and retransformed into the parent strain to check the phenotype. The sequences of mutant sti1 constructs are available upon request.
GR activity:
The rat GR was expressed from a multicopy plasmid under control of the constitutive GPD promoter. A plasmid expressing the GR and the corresponding GRE–lacZ reporter plasmid pUCΔ55-26X were generous gifts from Didier Picard, University of Geneva. β-Galactosidase assays were performed as described (Johnson and Craig 2000). Briefly, cultures were grown at 25° in selective media to midlog phase (OD600 = 0.4–0.8). To activate the GR, deoxycorticosterone (DOC) was added to the culture at the indicated final concentration (0–10 μM DOC) for a 1-hr incubation. β-Galactosidase units were calculated as 103 × OD420 divided by the OD600 × elapsed time (in minutes). Each experiment contained triplicate samples and the activity of each mutant was assayed at least two independent times.
v-src expression:
sti1 disruption strain JJ623 expressing various sti1 mutants was transformed with a multicopy plasmid expressing GAL1-v-src (pBv-src) or the control plasmid (pB656) (a gift from Frank Boschelli) (Dey et al. 1996). Yeast cultures were grown overnight at 30° in raffinose-uracil drop-out media until mid-log phase and 20% galactose was added to a final concentration of 2%. After 6 hr, cultures were harvested for growth assays or preparation of cell lysates for immunoblot analysis using an antibody that recognizes phosphotyrosine residues (Upstate Biologicals).
SDS–PAGE and immunoblot analysis:
Western blot analysis of Sti1 proteins was conducted using a monoclonal antibody raised against the carboxy-terminus of Sti1 (Chang et al. 1997) (a generous gift of David Toft) or a polyclonal antiserum against Sti1 raised against the KLH-conjugated peptide NH2-DEAESNYKKALELDASNKC-C00H (amino acids 91–108 of Sti1). The antibodies against Tim44 and Ssa1/2 were generous gifts of Elizabeth Craig. To prepare yeast cell lysates, 0.5 OD600 units of cells were resuspended in cold phosphate-buffered saline containing 1 mm phenylmethylsulfonylfluoride, and disrupted with glass beads in the presence of SDS and Triton X-100. Samples were resuspended in SDS-sample buffer, analyzed using a 10% SDS-acrylamide gel, transferred to nitrocellulose, and probed with antibodies against Sti1, Tim44 or phosphotyrosine residues. Chemiluminescence immunoblots were performed according to the manufacturer's suggestions (Pierce Chemical, Rockford, IL).
Sti1 immunoprecipitations:
Strain JJ623 expressing WT or mutant Sti1 was grown overnight at 30° in selective media to an OD600 of 1.2–1.7. Cells were harvested, washed once with water, and resuspended in 20 mm Tris, pH 7.5, 100 mm KCl, 5 mm MgCl2, 10 mm β-mercaptoethanol plus complete mini protease inhibitor mixture (Roche Applied Science) (Carrigan et al. 2005). Cells were disrupted with 8 × 30-sec pulses in the presence of glass beads. Cell lysate was cleared by centrifugation. Mouse monoclonal antibody specific for Sti1 (5 μl) was preadsorbed to Protein-A Sepharose and incubated with 600 μl of cell lysate for 2 hr at 4°. Resin-bound complexes were washed four times with 20 mm Tris–HCl pH 7.5, 50 mm KCl, 0.1% Tween-20, resuspended in 2× SDS–PAGE sample buffer, separated by gel electrophoresis, transferred to nitrocellulose, and immunoblotted with rabbit polyclonal antibodies specific for Sti1, Hsc82/Hsp82, and Ssa1/2. Additional immunoprecipitations were performed in strain JJ816 or strain JJ832 expressing plasmid borne WT Hsp82, Hsp82–G313S, and/or WT or mutant Sti1.
RESULTS
Site-directed mutagenesis of the TPR domains of Sti1:
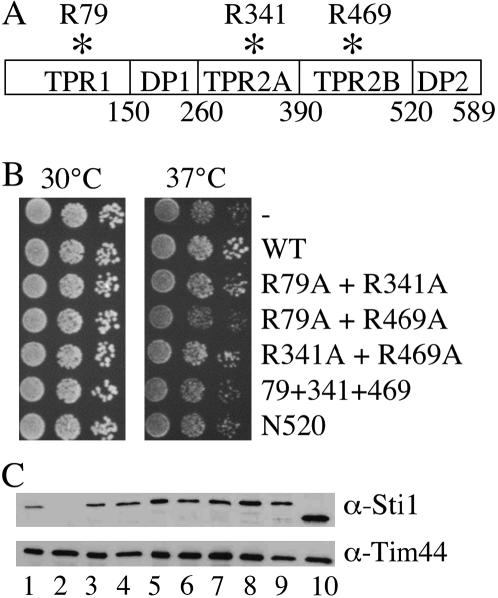
To determine the in vivo importance of the individual TPR domains of Sti1, we generated point mutations in Sti1 that mutate basic residues in the central binding groove of each TPR domain, singly or in combination. Similar site-directed mutagenesis of basic residues involved in TPR–EEVD interactions disrupted binding of Hsp90-cochaperones to Hsp70 and/or Hsp90 (Carrigan et al. 2004; Smith 2004). We changed a homologous conserved arginine residue to alanine in each of the three TPR domains (R79A in TPR1, R341A in TPR2A, and R469A in TPR2B) (Figure 1A). The homologous residues of mammalian Hop, R77 in TPR1, and R305 in TPR2A make contact with the carboxy-terminal EEVD residues of Hsp70 and Hsp90, respectively (Scheufler et al. 2000). We also constructed Sti1–N520, a truncation mutant that lacks the carboxy-terminal DP2 domain (residues 521–589).
Figure 1.
Targeted Sti1 mutants. (A) Schematic of the domains of Sti1. The position of targeted conserved basic residues in TPR1 (R79), TPR2A (R341), and TPR2B (R469) are indicated. (B) Growth of sti1 mutants in an sti1 strain. Plasmids expressing WT or mutant STI1 or vector as control (−) were transformed into strain JJ623 (sti1). Transformants were grown overnight and 10-fold serial dilutions were plated on selective media and incubated for 2 days at the indicated temperature. (C) Cell lysates from sti1 disruption strain expressing indicated mutants were analyzed by SDS–PAGE (10% acrylamide) and immunoblotted with a polyclonal antiserum specific for Sti1 or an antibody against the mitochondrial Tim44 protein as a loading control. Lane 1, Wt Sti1; lane 2, vector control; lane 3, R79A; lane 4, R341A; lane 5, R469A; lane 6, R79A + R341A; lane 7, R79A + R469A; lane 8, R341A + R469A; lane 9, R79A + R341A + R469A; lane 10, N520.
As an initial test to determine the in vivo effects of these mutations, we transformed the mutants into a strain containing a chromosomal deletion of STI1 (JJ623). We assayed the growth defect of the resultant strains by plating 10-fold serial dilutions at 30° and 37° (Figure 1B). The growth of the single TPR point mutants was indistinguishable from that of cells expressing wild type STI1 (not shown). However, the triple TPR mutation (R79A + R341A + R469A), R79A + R469A, and the N520 mutation conferred the slight growth defect of a sti1 null strain (Nicolet and Craig 1989), suggesting that these mutations disrupt the in vivo function of Sti1. To determine that each of these constructs make stable protein, we examined the expression of the Sti1 mutants using a polyclonal antibody against Sti1. All of the mutant Sti1 proteins accumulated to at least the wild-type level in vivo, except Sti1–N520, which appeared to accumulate at a slightly higher lever than wild-type Sti1 (Figure 1C).
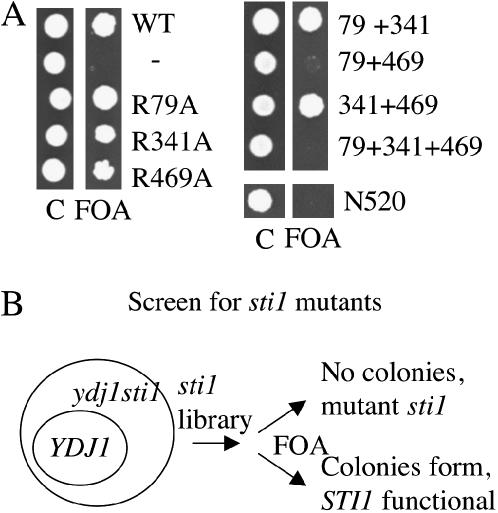
Next we took advantage of the synthetic lethality of YDJ1 and STI1 to analyze the effect of mutation on Sti1 function. Ydj1 is an Hsp40 molecular chaperone that has previously been shown to be required for the function of Hsp90 client proteins (Kimura et al. 1995; Dey et al. 1996; Johnson and Craig 2000). Deletion of YDJ1 causes slow, temperature-sensitive growth (Caplan and Douglas 1991). A strain containing chromosomal deletions of STI1 and YDJ1 is inviable in the absence of a plasmid expressing WT YDJ1 or STI1 (Flom et al. 2005). We tested the ability of available sti1 mutants to support viability of a sti1ydj1/pRS316–YDJ1 strain grown in the presence of 5-FOA, which counterselects for the URA3-based pRS316–YDJ1 plasmid. Each of the single point mutants, and the combination mutants R79A + R341A and R341A + R469A, was able to support viability in the presence of 5-FOA, indicating that these mutations do not disrupt the functions of Sti1 that are essential in the absence of YDJ1. However, the R79A + R469A mutant, the triple mutant (R79A + R341A + R469A), and N520 were unable to support viability of the sti1ydj1 strain (Figure 2A). These results indicate that the carboxy-terminal DP2 region is essential for Sti1 function in the absence of YDJ1 and also suggest that TPR1 and TPR2B may have overlapping or redundant in vivo functions, since a phenotype was observed upon combination of these mutations, but not in the presence of the single mutations.
Figure 2.
Viablity of sti1 mutants in an sti1ydj1 strain. (A) Strain JJ609 (sti1ydj1/pRS316–YDJ1) was transformed with plasmids containing WT STI1 or sti1 mutants. After overnight incubation at 30°, equal amounts of cells were plated onto selective media (C) to maintain WT YDJ1 or media containing 5-FOA to counterselect for YDJ1 (FOA). Plates were incubated at 25° for 5 days. (B) Genetic screen to isolate sti1 mutants that fail to support viability of a sti1ydj1 strain. A plasmid DNA library containing random mutations in STI1 was transformed into strain JJ609. Transformants were patched onto media to maintain YDJ1 or onto media containing 5-FOA to counterselect for YDJ1. Colonies that failed to grow in the presence of 5-FOA were selected for further study.
Screen for STI1 mutants defective in vivo:
Next we conducted a genetic screen to identify additional mutations that disrupt in vivo functions of Sti1. We used error-prone PCR to generate random mutations in STI1 and transformed this mutant STI1 plasmid library into the sti1ydj1/pRS316–YDJ1 strain (Figure 2B). We selected mutants that failed to grow in the presence of 5-FOA for further analysis. Plasmids expressing mutant STI1 were rescued out of yeast and retransformed into the parent strain to confirm the phenotype. Resultant mutant sti1 plasmids were sequenced fully and the mutants obtained in this screen are described in Figure 3 and Table 1.
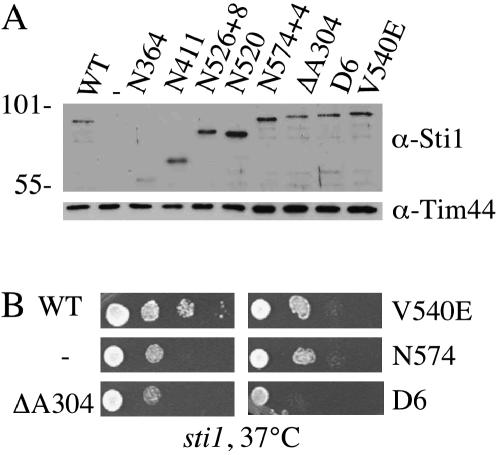
Figure 3.
Sti1 mutants obtained in genetic screen. (A) Mutant sti1 plasmids obtained in the genetic screen were transformed into strain JJ623 (sti1). Equal amounts of lysates from sti1 cells expressing WT STI1 or indicated sti1 mutants were separated by SDS–PAGE (10% acrylamide) and immunoblotted with Sti1-specific antibodies. (B) Growth of serial dilutions of the same strains after a 2-day incubation at 37° on selective medium.
TABLE 1.
Summary of growth phenotypes of sti1 mutants
| Mutant | Viability in sti1ydj1 strain | Growth in presence of 39 μM RD |
|---|---|---|
| WT (N589) | +++ | +++ |
| sti1 | − | − |
| R79A | +++ | +++ |
| K75E | +++ | +++ |
| ΔA48 | ++ | +++ |
| R341A | +++ | +++ |
| R341E | +++ | +++ |
| ΔA304 | − | − |
| R469A | +++ | +++ |
| R465E | +++ | +++ |
| ΔA438 | + | ++ |
| R79A + R341A | +++ | +++ |
| R79A + R469A | − | − |
| R341A + R469A | +++ | +++ |
| R79A + R341A + R469A | − | − |
| D6 | − | − |
| N364 | − | − |
| N411 | − | − |
| N520 | − | − |
| N526 + 8 | − | − |
| N574 + 4 | − | − |
| V540E | − | − |
Consistent with our results that indicate that the carboxy-terminal DP2 region of Sti1 is essential in the absence of YDJ1, the majority of mutants that we isolated were truncation mutants. Four isolated truncation mutants named by the last encoded amino acid are shown in Figure 3A. N364 is a truncation within TPR2A, N411 contains intact TPR1 and 2A, and N526, which is a frameshift mutation that contains the additional point mutant K168E, has all three TPR domains intact but lacks the DP2 region. N574 contains a frameshift mutation that deletes the last 15 amino acids of Sti1 and replaces them with four other amino acids. We also isolated three mutations containing amino acid alterations in Sti1. Sti1–ΔA304 contains a three-nucleotide deletion that results in the deletion of alanine residue 304 (ΔA304) in TPR2A. The corresponding conserved residue in human Hop, A267, is in helix 2A of TRP2A, and the nearby residue N264 contacts the backbone of the bound EEVD-containing peptide of Hsp90 (Scheufler et al. 2000). This mutant is predicted to alter alignment of the remainder of the TPR domain with the EEVD-containing peptide. The D6 mutant contains four amino acid alterations, two in TPR2A and two in TPR2B: F339S, A387V, E448V, and L509P. F339S in TRP2A is in the vicinity of the EEVD-binding cleft. The other residues are not conserved between Sti1 and human Hop and not located near residues that compose the carboxylate-clamp structure. The V540E mutation alters a conserved residue in the DP2 region that is part of the DPEV sequence targeted in the initial mutagenesis of this region of Hop (Chen and Smith 1998; Carrigan et al. 2004). The isolation of V540E and N574 provide independent confirmation of the critical importance of the DP2 region in the in vivo functions of Sti1 and further indicate that the last 15 amino acids of this domain are critical for Sti1 function.
To determine the expression level of these proteins, we transformed the mutants into a sti1 disruption strain and detected Sti1 in yeast cell lysates using a polyclonal antibody that recognizes sequences in TPR1 (Figure 3A). With the exception of N364, which accumulates at very low levels, the remaining mutants are present at levels similar to those of WT Sti1, indicating that the loss of viability in the sti1ydj1 strain is not due to altered protein expression levels. We also examined whether these mutants display the slight temperature-sensitive growth defect of a sti1 strain. As shown in Figure 3B, cells expressing Sti1–ΔA304, V540E, or N574 exhibit the slight 37° growth defect of a sti1 disruption strain, while the D6 mutant exhibits slightly worse growth than the sti1 strain. None of the sti1 mutants exhibited growth defects at 30° (not shown).
Additional mutagenesis of the TPR domains:
To further examine the in vivo importance of the EEVD-binding groove of the TPR domains we constructed the K75E mutation in TPR1, R341E in TRP2A, and R465E in TPR2B. These mutations reverse the charge on residues critical for EEVD-peptide interaction and thus may have a stronger effect on the TPR–EEVD interaction than changing the basic arginine residue to the uncharged alanine residue. Because we observed a dramatic disruption of activity upon deletion of A304 in TPR2A, we also constructed similar mutations in TPR1 (ΔA48) and TPR2B (ΔA438). We compared the effect of the different mutations in each TPR domain on the in vivo functions of Sti1. When expressed in a sti1 deletion strain, all mutants were expressed at similar levels (not shown). We examined the phenotype of these mutants in the sti1ydj1 strain (Table 1). Within each TPR domain the effect of changing a basic residue to an acidic residue had the same effect as the change to an alanine residue. Of all the TPR mutations, only ΔA304 exhibits the level of growth defects observed in the absence of STI1, although the ΔA438 mutant exhibited reduced growth in this assay.
RD sensitivity of sti1 mutant strains:
Yeast lacking STI1 are hypersensitive to Hsp90-inhibiting drugs such as geldanamycin, macbecin, or RD (Liu et al. 1999; Piper et al. 2003). As a further test to determine how sti1 mutation affects function, all mutants were transformed into the sti1 strain and transformants were streaked onto YPD plates containing 39 μM RD or DMS0 as a control. These results are summarized in Table 1. In the absence of STI1, very little growth was observed in the presence of RD, but transformation of a sti1 strain with wild-type STI1 restored growth. In summary, all mutations that are unable to support growth of the sti1ydj1 strain are also unable to support growth in the presence of RD. The close correlation between these phenotypes suggests that these assays measure related functions of Sti1. We also found that overexpression of sti1 mutants unable to support growth in the presence of RD does not alter the growth phenotype (not shown), suggesting that the mutants cause a significant loss of Sti1 activity rather than reduced activity.
sti1 mutants are defective in Hsp90 client protein activity:
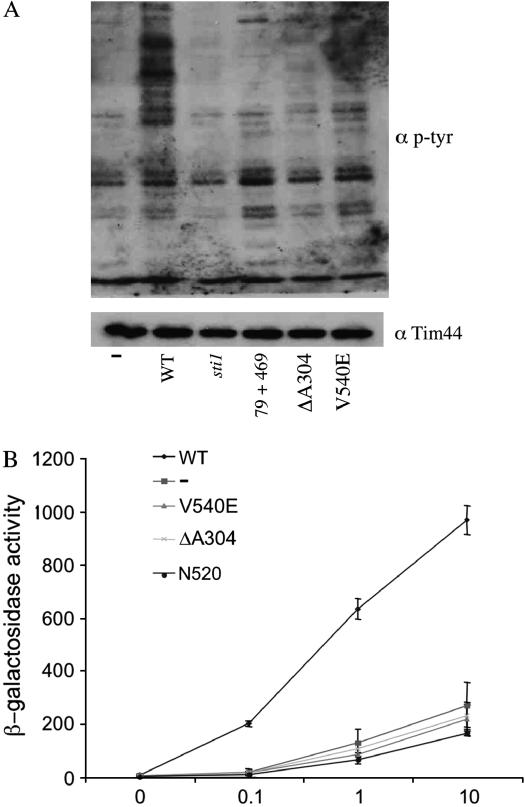
As an initial test to determine that sti1 mutants affect Hsp90 client protein activity, we assayed the function of the oncogenic tyrosine kinase v-src in yeast expressing WT or mutant STI1. Expression of v-src is toxic to WT yeast, but not to yeast containing mutations in Hsp90 or Hsp90 cochaperones (Nathan and Lindquist 1995; Dey et al. 1996). The activity of v-src expressed in yeast is measured with an antibody specific for phosphotryosine residues. Low levels of phosphotyrosine activity are observed in the absence of v-src, but a large increase is observed upon conditional expression of v-src. Previously it was shown that expression of v-src in sti1 cells resulted in very low levels of phosphotyrosine residues without affecting the accumulation of v-src protein (Chang et al. 1997). We assayed v-src activity in sti1 cells expressing WT Sti1, Sti1–R79A + R469A, Sti1–ΔA304, and Sti1–V540E. High levels of phosphotyrosine residues were observed in cells expressing WT Sti1. Each of the three mutants tested resulted in phosphotyrosine levels that were near the level of control cells that did not express v-src (−) or cells lacking STI1 (sti1). This confirms that the sti1 mutants we isolated affect the activity of Hsp90 client proteins (Figure 4A).
Figure 4.
Hsp90 client activity is disrupted by sti1 mutation. (A) Wild-type or sti1 mutant yeast cultures expressing either the GAL1–v-src (pBv-src) multicopy plasmid or the control plasmid (pB656) were grown overnight in selective media containing raffinose as the carbon source. v-src expression was induced by addition of 20% galactose to a final concentration of 2%. Cells were harvested 6 hr after induction. Upper panel shows immunoblot of yeast lysates using anti-phosphotyrosine antibody 4G10 (Upstate Biologicals). Lower panel shows immunoblot of yeast lysates using control antibody against Tim44 as a protein-loading control. (B) GR activity in sti1 mutant strains. sti1 disruption strain JJ623 containing GR (pRS424GPDGR) and corresponding reporter plasmid (pUCΔ55-26X) was transformed with indicated sti1 mutants expressed on low-copy plasmids. Yeast cultures were grown in selective media overnight at 25°, then diluted into fresh media and grown to mid-log phase. β-Galactosidase assays were performed as described in materials and methods.
Next we examined the effect of sti1 mutation on another Hsp90 client protein, the GR. When expressed in yeast, GR is able to bind hormone and activate transcription in an Hsp90- and hormone concentration-dependent manner. In a prior study, deletion of sti1 reduced the ability of the GR to activate transcription without affecting GR protein levels, and restoring WT STI1 on a plasmid resulted in WT levels of activity (Chang et al. 1997). We transformed sti1 cells expressing WT or mutant STI1 with plasmids that express GR and a glucocorticoid-regulated lacZ reporter gene. GR activity was measured in the absence of hormone and over a range of hormone concentrations. First we examined the effect of the V540E, ΔA304, and N520 mutations (Figure 4B). Over a range of hormone concentrations, each of these mutations produced a level of GR activity similar to that observed in sti1 cells, indicating that these mutations disrupt GR activity.
To compare the relative activity of the GR in the presence of different sti1 mutations, we expressed GR activity in sti1 mutant cells as a percentage of WT activity in each experiment (Figure 5). For simplicity, and because the most dramatic effects of sti1 mutation were observed at the lowest concentration of hormone, 0.1 μM DOC, we present the combined data for this concentration of hormone. As expected on the basis of the predicted structural disturbances caused by each mutation, within each TPR domain successively enhanced defects are observed upon mutation of the basic residue to an alanine residue (R/K to A), to an acidic residue (R/K to E), and upon the more dramatic amino acid deletion mutation. These results indicate that alteration of single conserved basic residues in the EEVD-binding cleft of TPR1 and TPR2A and corresponding residues in TPR2B partially disrupts in vivo activity of Sti1, but the single mutations do not have as dramatic an effect on function as deletion of a conserved amino acid within the region. In addition, mutations in TPR1 did not affect GR activity as much as mutations in TPR2A and TPR2B. The single mutants R79A, R341A, or R469A exhibited 50–60% WT level of GR activity. The combination of R79A + R341A or R341A + R469A resulted in a further loss of GR activity, to about 35–40% of WT levels. However, upon combination of R79A + R469A, the activity dropped to the level of the sti1 strain (10–15%), indicating a specific functional interaction between these TPR domains that is in accordance with observed enhanced growth defects. Consistent with the growth phenotypes, mutations in the DP2 domain had a dramatic effect on GR activity, and in each case the activity was similar to that in the absence of STI1, indicating the importance of this domain in Sti1 function.
Figure 5.
Comparison of GR activity in sti1 mutant strains. WT STI1, vector alone (−) and indicated sti1 mutants were transformed into strain JJ623 containing GR (pRS424GPDGR) and reporter plasmid (pUCΔ55-26X). GR activity was measured as in Figure 4 after 1-hr incubation in the presence of 0.1 μM DOC. This data is combined from different experiments in which activity from each experiment is expressed as a percentage of WT activity.
Physical interaction of mutant Sti1 with Hsp90:
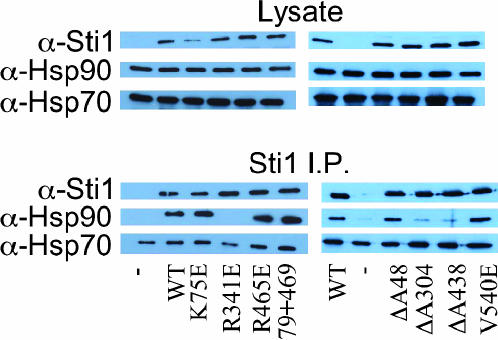
Isolation of Sti1 from yeast extract results in the copurification of yeast Hsp90, which has two isoforms, Hsc82 and Hsp82, and Hsp70s of the Ssa family (Chang et al. 1997). To determine how Sti1 mutation affects interaction with Hsp90 and Hsp70, we immunoprecipitated Sti1 out of yeast lysate using a monoclonal antibody specific for Sti1. Copurifying Hsp90 was detected with a polyclonal antiserum that recognizes Hsc82/Hsp82, and Hsp70 was recognized by an antiserum against Ssa1/2. Sti1 was immunoprecipitated out of a sti1 strain expressing WT or mutant Sti1. In the absence of Sti1, little or no Hsp90 binds the antibody resin, but significant levels of Hsp90 are observed in the presence of Sti1 (Figure 6). Unfortunately, due to high levels of binding of Hsp70 to the antibody-Protein-A Sepharose beads in the absence of STI1, we were unable to assess the effect of these mutations on Ssa interaction using this assay. The interaction of Sti1 with Hsp90 was specifically disrupted by mutations in TPR2A (R341E and ΔA304), but was not disrupted by mutations in TPR1 (K75E, ΔA48), the DP2 region (V540E), or the combination of R79A + R469A. The R465E mutation in TPR2B did not disrupt interaction with Hsp90, but the ΔA438 mutation effectively disrupted binding. We do not yet know if this is due to loss of a specific interaction or a general conformational change caused by the ΔA438 mutation. These results are in accordance with similar studies of binding of mutant Hop to rabbit Hsp90 in which Hsp90 interaction was specifically disrupted by mutation in TPR2A (Carrigan et al. 2004) and support prior studies that indicate that the terminal EEVD residues of Hsp90 specifically interact with TPR2A (Young et al. 1998; Scheufler et al. 2000; Abbas-Terki et al. 2001; Carrigan et al. 2004). Surprisingly, the growth phenotypes caused by Sti1 mutants (Table 1) are unrelated to the pattern of Hsp90 interaction, since the V540E and R79A + R469A mutation cause growth defects but do not appear to affect Hsp90 interaction, and the R341E mutation disrupts Hsp90 interaction without affecting growth in our assays.
Figure 6.
In vivo interaction of Sti1 with Hsc82/Hsp82. Cell lysates were prepared from strain JJ623 expressing WT and mutant Sti1. Sti1 was immunoprecipitated with a monoclonal antibody against Sti1. Resin-bound complexes were separated by SDS–PAGE and immunoblotted with antibodies specific for Sti1, Hsc82/Hsp82 (Hsp90), or Ssa1/2 (Hsp70).
Genetic interaction of mutant Sti1 with Hsp82–G313S:
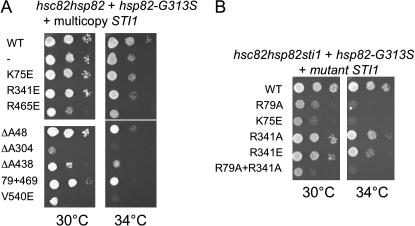
Next we used these mutations to determine whether mutations in TPR2A or other domains disrupt the functional interaction between Sti1 and Hsp90. Deletion of either gene encoding yeast Hsp90 (HSC82 and HSP82) results in only slight growth defects, but deletion of both genes is lethal. A number of mutations in hsp82 that exhibit temperature-sensitive growth when present as the only Hsp90 in the cell have been described (Nathan and Lindquist 1995). Overexpression of WT STI1 had allele-specific effects on Hsp90 mutant alleles, exhibiting no effect, a negative effect, or a positive effect on growth (Chang et al. 1997). In particular, overexpression of STI1 partially rescued the temperature-sensitive growth defects of cells expressing hsp82–G313S. G313 is located in the middle domain of Hsp82, which contains a proposed client binding site and a binding site for the cochaperone Aha1 (Meyer et al. 2003; Meyer et al. 2004). We overexpressed WT and mutant STI1 in cells expressing WT HSP82 or hsp82–G313S as the only Hsp90 in the cell and examined the effect on cell growth (Figure 7A). Overexpression of WT or mutant STI1 did not affect the growth of cells expressing WT HSP82 at 30° or 34° (not shown). As previously reported (Chang et al. 1997), overexpression of WT STI1 was able to slightly suppress the growth defect of hsp82–G313S at 34°. First we found that K75E, R341E, or R465E were not able to suppress the growth of hsp82–G313S at 34°, and overexpression of R465E caused a slight negative growth effect. Overexpression of other sti1 mutants also caused enhanced growth defects. Most dramatically, overexpression of ΔA304 or V540E caused a severe growth defect at 30° and the cells were inviable at 34°. Importantly, overexpression of R79A + R469A did not cause the severe growth defects observed in the presence of the ΔA304 and V540E mutations. In the other assays used to measure Sti1 function, R79A + R469A exhibited the same level of defects as the ΔA304 and V540E mutations. Thus, the function of Hsp82-G313S is specifically inhibited by the overexpression of either the ΔA304 or V540E mutations, even though ΔA304 disrupted Sti1–Hsp90 interaction and V540E did not.
Figure 7.
Genetic interaction between Sti1 and Hsp82-G313S. (A) Strain JJ816 (hsc82hsp82) expressing hsp82-G313S from a low copy plasmid was transformed with WT or mutant STI1 expressed from a multicopy plasmid. Transformants were grown overnight and 10-fold serial dilutions were plated on selective media and grown for 2 days at the indicated temperature. (B) Strain JJ832 (hsc82hsp82sti1/YEp24–HSP82) expressing hsp82–G313S was transformed with WT or mutant STI1 and plated on 5-FOA. Viable strains were grown overnight, serially diluted, plated on rich medium, and grown for 2 days at the indicated temperature.
As another test of the functional interaction of Hsp82–G313S and Sti1, we transformed a hsc82hsp82sti1/Yep24–HSP82 strain (JJ832) with hsp82–G313S to determine if hsp82–G313S is able to support viability in the absence of STI1. We transformed all available sti1 mutants into this strain and tested their ability to support growth in the presence of 5-FOA, which counterselects for the Yep24–HSP82 plasmid. In the absence of STI1, no growth was observed, indicating that STI1 becomes essential when Hsp82–G313S is the only Hsp90 in the cell. In the presence of 5-FOA, colonies appeared only upon expression of WT Sti1, R79A, K75E, R341A, R341E, and R79A + R341A (not shown). We assayed growth of these colonies and found that R341A and R341E exhibited near WT growth, and slow growth was observed in the presence of R79A, K75E, and R79A + R341A (Figure 7B). Thus, the growth of the hsp82–G313S strain is strictly dependent on Sti1, but is specifically unaffected by the R341A/E mutation that disrupts the only identified site of interaction between Hsp90 and Sti1.
The G313 residue is in the middle domain of Hsp90, which is not known to interact with Sti1, and the biochemical defects of this mutant have not been reported. One possible reason that the Hsp82–G313S mutant is unaffected by mutations in TPR2A is that the G313S alteration may directly or indirectly disrupt the interaction of Sti1 with the carboxy-terminal EEVD residues, and thus there is no additional loss of function in the presence of TPR2A mutations. Alternatively, the interaction of Hsp82-G313S with Sti1 may be altered in such a way as to make it resistant to the disruptive effects of TPR2A mutation. To distinguish between these possibilities we immunoprecipitated Sti1 out of an hsc82hsp82 strain expressing WT Hsp82 or Hsp82-G313S (Figure 8, lanes 1 and 2). Similar levels of WT and mutant Hsp82 copurified with Sti1, indicating that the G313S mutation does not disrupt Sti1 interaction. We also immunoprecipitated Sti1 out of the hsc82hsp82sti1 strain expressing Hsp82–G313S in combination with WT Sti1, R341A, or R341E (Figure 8, lanes 4–6). Both the R341A and R341E mutations effectively disrupted the interaction with Hsp82–G313S, suggesting that this mutant does not have altered interaction with the TPR2A domain of Sti1.
Figure 8.
In vivo interaction of Sti1 with Hsp82-G313S. Sti1 was immunoprecipitated from cell lysates as in Figure 6. Lanes 1 and 2, strain JJ816 (hsc82hsp82) expressing WT Hsp82 (lane 1) or Hsp82–G313S (lane 2); lanes 3–6, strain JJ832 (hsc82hsp82sti1); lane 3, WT Hsp82 with no STI1 present; Lanes 4–6, Hsp82–G313S plus WT Sti1 (lane 4), R341A (lane 5), or R341E (lane 6).
DISCUSSION
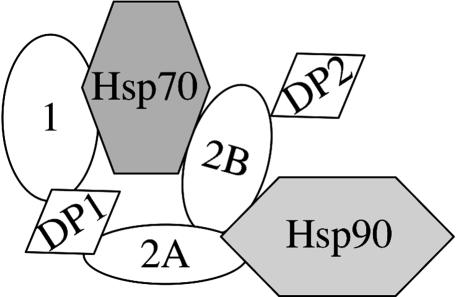
Our study was designed to determine the in vivo importance of the individual TPR domains and the DP2 domain of Sti1. Our results indicate that point mutation of a TPR2A residue disrupts the stable interaction between Sti1 and Hsp90 but only partially disrupts activity. Additionally, mutation of a residue in TPR1 presumed to interact with the terminal EEVD residues of Hsp70 does not disrupt Sti1 function unless combined with mutations in TPR2B, suggesting that TPR1 and TPR2B have overlapping functions. In contrast, Sti1 function was completely disrupted by deletion of an amino acid within the TPR2A domain or mutations in the poorly-understood DP2 region. Our results and data from other laboratories challenge the model that the primary sites of Sti1 interaction with Hsp70 and Hsp90 are mediated through TPR1 and TPR2A, respectively, and are summarized below in a new model of how Sti1 physically and functionally interacts with Hsp70 and Hsp90 (Figure 9).
Figure 9.
Model for the interaction of Sti1/Hop with Hsp70 and Hsp90. See text for details.
Sti1 interaction with Hsp70:
TPR1 of Hop/Sti1 interacts with the conserved terminal EEVD residues of Hsp70 (Demand et al. 1998; Scheufler et al. 2000). The EEVD residues were proposed to have a regulatory effect on Hsp70 activity because deletion or mutation of these residues caused altered ATPase activity and conformation (Freeman et al. 1995). Sti1 was recently shown to stimulate the ATPase activity of Ssa1 (Wegele et al. 2003), but it is unknown if this effect is mediated by TPR1. The Hop–Hsp70 interaction is not strictly dependent upon TRP1–EEVD interaction since Hsp70 mutants lacking the C-terminal 34 amino acids were still able to co-immunoprecipitate with Hop, and mutations outside of TPR1 affect Hsp70 interaction (Odunuga et al. 2003; Carrigan et al. 2004). The interaction of purified Hop with purified Hsp70 was affected by single point mutants targeting basic residues TPR1 and TPR2B, suggesting that both TPR1 and TPR2B are required for direct interaction with Hsp70 (Carrigan et al. 2004). In support of this data, we found that single amino acid substitutions in TPR1 and TPR2B had only mild effects on Sti1 activity but a combination of point mutations in TPR1 and TPR2B (R79A + R469A) completely disrupted the in vivo functions of Sti1. Together these results suggest that TPR1 and TPR2B have redundant or overlapping functions in mediating the interaction of Sti1 with Hsp70.
Both TPR1 and TPR2B are flanked by a region containing DP repeats, and both the DP1 and DP2 regions contribute to Hop function (Chen and Smith 1998; Carrigan et al. 2004; Carrigan et al. 2005). Our results demonstrate that the DP2 domain of Sti1 is critical for its in vivo functions, and that the single amino acid mutation V540E or truncation of the last 15 amino acids is sufficient to disrupt the function of this domain. Mutations in the DP2 region of human Hop disrupted the interaction of Hsp70 in rabbit reticulocyte lysates (Carrigan et al. 2004), but further studies are needed to determine if the DP2 domain contacts Hsp70 directly.
Sti1 interaction with Hsp90:
The extreme carboxy-terminal sequences of Hsp90, including the terminal MEEVD sequence, compose the only described binding site for Hop/Sti1 (Young et al. 1998; Scheufler et al. 2000). The stable interaction between Sti1/Hop and Hsp90 is disrupted by deletion of the terminal MEEVD residues of Hsp82 (Abbas-Terki et al. 2001) or mutation of a basic residue in TPR2A (Carrigan et al. 2004 and Figure 6). However, the function of this interaction remains unknown. Hsp82 lacking the terminal MEEVD residues was able to support near-wild-type levels of growth when present as the only Hsp90 in the cell (Louvion et al. 1996), and a mutation in TPR2A of Sti1 (R341E) that disrupts stable Hsp90 interaction maintains about 50% of client protein activity and does not affect growth in our assays (Figure 5 and Table 1). In addition, although the growth of an hsp82-G313S strain was highly dependent on STI1, growth of this strain was remarkably unaffected by loss of the TPR2A–MEEVD interaction. Growth of the hsp82–G313S strain was inhibited by overexpression of multiple alleles of STI1, but overexpression of Sti1-R341E had little or no effect. Likewise, STI1 is essential in the hsp82-G313S strain, and the only sti1 mutants that supported WT growth were those that specifically target the TPR2A–MEEVD interaction.
Because additional Hsp90 cochaperones also contain TPR domains and compete with Sti1 for binding to Hsp90 (Smith 2004), interaction of TPR2A with the MEEVD residues of Hsp90 has been proposed to be a mechanism for Sti1 to regulate Hsp90 activity. Sti1 inhibits the ATPase activity of yeast Hsp90 and this inhibition is relieved in the presence of Cpr6 (Prodromou et al. 1999). The G313 residue is located near the binding site for the Aha1 cochaperone, which is able to stimulate the ATPase activity of Hsp90 and may also compete with Sti1 for binding to Hsp90 (Lotz et al. 2003; Meyer et al. 2004). Perhaps conformational changes in Hsp90 or alteration of the Hsp90–Aha1 interaction by the G313S mutation eliminates the requirement for differential TPR protein interaction at the carboxy-terminus of Hsp90, but additional studies will be needed to address this question.
Data from other labs indicate that TPR2A, TRP2B, and the DP2 region may all be critical for Hsp90 regulation and/or interaction (Chen and Smith 1998; Carrigan et al. 2004; Carrigan et al. 2005; Song and Masison 2005). Sti1 is able to inhibit the ATPase activity of Hsp90 and there is evidence that Hop/Sti1 interacts with the amino-terminal ATPase domain of Hsp90 (Chen and Smith 1998; Prodromou et al. 1999; Richter et al. 2003). Our results demonstrated that deletion of an amino acid in TPR2A (ΔA304) or TPR2B (ΔA438) disrupted both Hsp90 interaction and most or all Sti1 activity in vivo. In both cases these deletion mutants had much more dramatic effects than alteration of residues in the peptide binding groove, and it is possible that other interaction sites within these domains are disrupted or that they cause significant conformational changes. Mutations within the DP2 domain severely disrupted Sti1 function and dramatically inhibited the growth of the hsp82-G313S strain, suggesting that the DP2 domain modulates Hsp90 function. A point mutation in DP2 did not disrupt Hsp90 interaction, but further studies will be needed to determine how both the TPR2B and DP2 domains affect Hsp90 interaction.
Complex interaction of Hop/Sti1 with Hsp70 and Hsp90:
We have evidence that mutations in TPR2A and TPR2B affect Hsp90 interaction in vivo and are conducting additional studies with purified Sti1, Hsp90, and Hsp70 to determine how these mutations affect Sti1 interaction with those proteins. Sti1 functions as a dimer (Prodromou et al. 1999), but the dimerization site and the in vivo importance of this function are unknown. In addition, Sti1 interaction with proteins other than Hsp70 and Hsp90 may also be critical for its function, since Drosophila Hop is able to bind yeast Hsp70 and Hsp90 but is unable to functionally replace Sti1 in assays of GR function (Carrigan et al. 2005). Two candidates are the Hsp90 cochaperone Cdc37 and the molecular chaperone Hsp104, both of which directly interact with Sti1 (Abbas-Terki et al. 2001; Abbas-Terki et al. 2002).
Recently the proposed function of Hop/Sti1 has expanded from acting merely as a bridge to bring Hsp70 and Hsp90 together in a ternary complex (Chen and Smith 1998) to one that also includes a role for Hop/Sti1 in regulating the ATPase activity of both Hsp70 and Hsp90 (Prodromou et al. 1999; Richter et al. 2003; Wegele et al. 2003). During the same time period, there has been increasing evidence that Hop/Sti1 has physical contacts with Hsp70 and Hsp90 in addition to the TPR–EEVD interactions. There are also unresolved questions about how Hop/Sti1 mediates formation of the ternary complex between Hsp70, Hop/Sti1, and Hsp90, particularly since it was demonstrated that the stoichiometry and affinity of the Hop–Hsp70 interaction was influenced by the presence of Hsp90 (Hernandez et al. 2002). The mutants we isolated are valuable tools that will help clarify the cellular functions of Sti1 during assembly of Hsp90-client protein complexes and will provide novel information about how Hsp90 and cochaperones cooperate to mediate the folding of diverse cellular proteins.
Acknowledgments
We thank Elizabeth Craig, David Toft, David Smith, Frank Boschelli, Didier Picard, Susan Lindquist, and Kevin Morano for reagents. We also thank David Smith and Elizabeth Craig for helpful comments on this manuscript. This work was supported by the National Institutes of Health P20 RR-15587 and was funded in part by P20 RR-16454 from the BRIN/INBRE Program of the National Center for Research Resources, and an American Cancer Society grant through Washington State University, IRG-77-003-26.
References
- Abbas-Terki, T., O. Donze, P. A. Briand and D. Picard, 2001. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 21: 7569–7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas-Terki, T., P. A. Briand, O. Donze and D. Picard, 2002. The Hsp90 co-chaperones Cdc37 and Sti1 interact physically and genetically. Biol. Chem. 383: 1335–1342. [DOI] [PubMed] [Google Scholar]
- Caplan, A. J., and M. G. Douglas, 1991. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 114: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrigan, P. E., G. M. Nelson, P. J. Roberts, J. Stoffer, D. L. Riggs et al., 2004. Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J Biol. Chem. 279: 16185–16193. [DOI] [PubMed] [Google Scholar]
- Carrigan, P. E., D. L. Riggs, M. Chinkers and D. F. Smith, 2005. Functional comparison of human and Drosophila Hop reveals novel role in steroid receptor maturation. J. Biol. Chem. 280: 8906–8911. [DOI] [PubMed] [Google Scholar]
- Chang, H. C., D. F. Nathan and S. Lindquist, 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17: 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., and D. F. Smith, 1998. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 273: 35194–35200. [DOI] [PubMed] [Google Scholar]
- Demand, J., J. Luders and J. Hohfeld, 1998. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol. Cell. Biol. 18: 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, B., A. J. Caplan and F. Boschelli, 1996. The Ydj1 molecular chaperone facilitates formation of active p60v-src in yeast. Mol. Biol. Cell 7: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom, G., J. Weekes and J. L. Johnson, 2005. Novel interaction of the Hsp90 chaperone machine with Ssl2, an essential DNA helicase in Saccharomyces cerevisiae. Curr. Genet. 47: 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman, B. C., M. P. Myers, R. Schumacher and R. I. Morimoto, 1995. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14: 2281–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., R. H. Schiestl, A. R. Willems and R. A. Woods, 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360. [DOI] [PubMed] [Google Scholar]
- Hernandez, M. P., W. P. Sullivan and D. O. Toft, 2002. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J. Biol. Chem. 277: 38294–38304. [DOI] [PubMed] [Google Scholar]
- Johnson, J. L., and E. A. Craig, 2000. A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol. Cell. Biol. 20: 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., I. Yahara and S. Lindquist, 1995. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science 268: 1362–1365. [DOI] [PubMed] [Google Scholar]
- Lee, P., A. Shabbir, C. Cardozo and A. J. Caplan, 2004. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol. Biol. Cell 15: 1785–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. D., K. A. Morano and D. J. Thiele, 1999. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 274: 26654–26660. [DOI] [PubMed] [Google Scholar]
- Lotz, G. P., H. Lin, A. Harst and W. M. Obermann, 2003. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 278: 17228–17235. [DOI] [PubMed] [Google Scholar]
- Louvion, J. F., R. Warth and D. Picard, 1996. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc. Natl. Acad. Sci. USA 93: 13937–13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, P., C. Prodromou, B. Hu, C. Vaughan, S. M. Roe et al., 2003. Structural and functional analysis of the middle segment of hsp90. Implications for ATP hydrolysis and client protein and cochaperone interactions. Mol. Cell 11: 647–658. [DOI] [PubMed] [Google Scholar]
- Meyer, P., C. Prodromou, C. Liao, B. Hu, S. Mark Roe et al., 2004. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 23: 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, D. F., and S. Lindquist, 1995. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15: 3917–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet, C. M., and E. A. Craig, 1989. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 3638–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odunuga, O. O., J. A. Hornby, C. Bies, R. Zimmermann, D. J. Pugh et al., 2003. Tetratricopeptide repeat motif-mediated Hsc70-mSTI1 interaction. Molecular characterization of the critical contacts for successful binding and specificity. J. Biol. Chem. 278: 6896–6904. [DOI] [PubMed] [Google Scholar]
- Odunuga, O. O., V. M. Longshaw and G. L. Blatch, 2004. Hop: more than an Hsp70/Hsp90 adaptor protein. BioEssays 26: 1058–1068. [DOI] [PubMed] [Google Scholar]
- Piper, P. W., S. H. Millson, M. Mollapour, B. Panaretou, G. Siligardi et al., 2003. Sensitivity to Hsp90-targeting drugs can arise with mutation to the Hsp90 chaperone, cochaperones and plasma membrane ATP binding cassette transporters of yeast. Eur. J. Biochem. 270: 4689–4695. [DOI] [PubMed] [Google Scholar]
- Pratt, W. B., and D. O. Toft, 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228: 111–133. [DOI] [PubMed] [Google Scholar]
- Prodromou, C., and L. H. Pearl, 2003. Structure and functional relationships of Hsp90. Curr. Cancer Drug Targets 3: 301–323. [DOI] [PubMed] [Google Scholar]
- Prodromou, C., G. Siligardi, R. O'Brien, D. N. Woolfson, L. Regan et al., 1999. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 18: 754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, K., P. Muschler, O. Hainzl, J. Reinstein and J. Buchner, 2003. Sti1 is a non-competitive inhibitor of the Hsp90 ATPase. Binding prevents the n-terminal dimerization reaction during the ATPase cycle. J. Biol. Chem. 278: 10328–10333. [DOI] [PubMed] [Google Scholar]
- Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder et al., 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101: 199–210. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. F., 2004. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones 9: 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., and D. C. Masison, 2005. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/90 organizing protein Sti1 (Hop1). J. Biol. Chem. 280: 34178–34185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele, H., M. Haslbeck, J. Reinstein and J. Buchner, 2003. Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 278: 25970–25976. [DOI] [PubMed] [Google Scholar]
- Workman, P., 2003. Overview: translating Hsp90 biology into Hsp90 drugs. Curr. Cancer Drug Targets. 3(5): 297–300. [DOI] [PubMed] [Google Scholar]
- Young, J. C., W. M. Obermann and F. U. Hartl, 1998. Specific binding of tetratricopeptide repeat proteins to the C-terminal 12-kDa domain of hsp90. J. Biol. Chem. 273: 18007–18010. [DOI] [PubMed] [Google Scholar]