Abstract
In human genetics a detailed knowledge of linkage disequilibrium (LD) is considered a prerequisite for effective population-based, high-resolution gene mapping and cloning. Similar opportunities exist for plants; however, differences in breeding system and population history need to be considered. Here we report a detailed study of localized LD in different populations of an inbreeding crop species. We measured LD between and within four gene loci within the region surrounding the hardness locus in three different gene pools of barley (Hordeum vulgare). We demonstrate that LD extends to at least 212 kb in elite barley cultivars but is rapidly eroded in related inbreeding ancestral populations. Our results indicate that haplotype-based sequence analysis in multiple populations will provide new opportunities to adjust the resolution of association studies in inbreeding crop species.
LARGE-SCALE investigations of sequence variation within genes and across genomes have only just begun for plant species. Such studies are required to determine the distribution and extent of linkage disequilibrium (LD), the nonrandom association of alleles, since this will determine the resolution power of association-based mapping strategies. In human and mammalian genetics, knowledge of LD is a topic of great current interest and has been successfully used to refine high-resolution mapping studies for complex disease genes and provide new insights into human evolution and the distribution of meiotic crossover events (Ardlie et al. 2002). Similar opportunities exist for plants (Gaut and Long 2003); however, the impact of biological and historical factors influencing the extent of LD need to be assessed before informative mapping strategies can be implemented. Initial studies of LD in maize (outcrosser) showed that the extent of LD decay differs from within a few hundred to 2000 bp, depending on whether landrace or cultivated material is analyzed (Remington et al. 2001; Tenaillon et al. 2001; Palaisa et al. 2003). In contrast, small isolated populations of Arabidopsis (inbreeder) exhibit drastic differences in the extent of LD, ranging from 1 to >50 cM (Nordborg et al. 2002). Garris et al. (2003) studied haplotype diversity and LD around the xa5 locus of rice (Oryza sativa L.), an inbreeder. The data revealed that significant LD was detected between sites 100 kb apart. These studies vividly illustrate the impact different breeding systems can have on LD. The natural decay of LD with distance occurs at a considerably slower rate in inbreeding systems because effective recombination is severely reduced and genetic polymorphisms remain correlated over longer physical distances (Nordborg et al. 2002; Morrell et al. 2005). This extended LD is a potential barrier to the localization of causative polymorphisms of phenotypic effects. Some of the world's most important crops such as rice, soybean, barley, and wheat are inbreeders. The impact of inbreeding on the magnitude and pattern of LD in these plants will also be amplified by human intervention arising from the processes of selection and domestication (Tanksley and McCouch 1997). In addition, the haploid genome size of crop plants such as wheat and barley (5000 Mbp; Arumuganathan and Earle 1991) is 50 times greater than that of Arabidopsis (125 Mb) mainly due to the recent amplification of repetitive DNA (SanMiguel et al. 1998). These factors suggest that studies of LD in inbreeding crop plants are needed to complement information emerging from Arabidopsis to provide a comprehensive picture of the patterns and magnitude of LD in plant genomes. To characterize the extent of LD in an inbreeding crop species, we analyzed inter- and intragenic associations across four gene loci within 215 kb of the barley hardness locus (Ha). The objectives of this study were: (1) to determine the extent and magnitude of LD at a regional level and (2) to relate empirical estimates of LD to genome composition and the varying evolutionary histories of different gene pools. Our data indicate that LD varies dramatically between different inbreeding populations of barley, providing exciting opportunities to pursue a two-tiered LD mapping strategy based on whole-genome scans of cultivated barley germ plasm followed by high-resolution LD mapping in ancestral landrace and Hordeum spontaneum populations.
MATERIALS AND METHODS
Plant material:
Seventy-four cultivated, 23 landrace, and 34 wild barley accessions were used in this study (Table 1). Cultivated material consisted predominantly of European breeding lines. Landrace and wild lines were chosen for their global distribution.
TABLE 1.
Accessions and geographical origin of germ plasm sampled and genotyped
| Species | Origin | Accession |
|---|---|---|
| H. vulgare (cv) | Germany | Alexis, Bavaria, Derkado, Haisa, Isaria, Kneifel, Tern, Volla, Wisa |
| H. vulgare (cv) | Netherlands | Aramir, Prisma, Sultan, Vada |
| H. vulgare (cv) | France | Bernice, Natasha |
| H. vulgare (cv) | Denmark | Carlsberg |
| H. vulgare (cv) | Australia | Clipper, Galleon |
| H. vulgare (cv) | Great Britain | Georgie, Golden Promise, Livet, Maris Otter, Optic, Proctor, Tankard |
| H. vulgare (cv) | Sweden | Gotlands, Gull, Ingrid, Kenia, Maja, Opal, Rika |
| H. vulgare (cv) | Canada | Harrington |
| H. vulgare (cv) | India | Monte Cristo |
| H. vulgare (cv) | Abed Binder, B83, Borwina, Carlsberg II, Casino, Chime (AB), Criewener 403, Europa, Fanfare, Franka, Friedrichswerther Berg, Hana, Hatif de Grignon, Heils Franken, Lina, Marinka, Melanie, Morex, Olli, Pallas, Plaisant, Plumage Archer, Puffin, Ragusa, Regina, Romanze, Scarlett, Scots Bere, Sewa, Sonja, Spratt Archer, Static, Svalof Svanhals, Triumph, Trumph, Tschermaks 2 row, Tyra, Union, Valticky, Vogelsanger Gold | |
| H. vulgare (lr) | Cyrrhus | |
| H. vulgare (lr) | SW Syria | NJSS101, NJSS111, NJSS121, NJSS141 |
| H. vulgare (lr) | NW Syria | WS231, WS241, WS281 |
| H. vulgare (lr) | Central Syria | CS301 |
| H. vulgare (lr) | Jordan | SJ31, SJ41 |
| H. vulgare (lr) | NE Syria | NES421 |
| H. vulgare (lr) | Jordan | SJ51, SJ81, SJ91 |
| H. spontaneum | Afghanistan | 180044/HS5700, 220664 |
| H. spontaneum | Iraq | 180046/HS5701, 180049/HS5702 |
| H. spontaneum | Iran | 180052/HS5704, 181164, 181170, 181174, 181319, 2691 |
| H. spontaneum | Greece | 181277/HS5738 |
| H. spontaneum | Uzbekistan | 181498/HS5770 |
| H. spontaneum | Israel | 180994/HS5835, 284738, 296791, 296860, 296874, 296912, 391131, 391132, 391133, 391134, 391135, 466446, 466470 |
| H. spontaneum | Pakistan | 181243/HS5856 |
| H. spontaneum | Jordan | 181436/HS5865 |
| H. spontaneum | Turkey | 181267, 181679, 245739, HIS 34 |
| H. spontaneum | Syria | 181549 |
| H. spontaneum | Siberia, Russia | 39864, 39870 |
SNP discovery and genotyping:
PCR primers were designed to four gene regions of 1.5–3.5 kb in size from the ∼113-kb genic space of the Ha locus (GenBank accessions nos. AY643842–AY643844). Amplicons were sequenced using ABI BigDye Terminators Version 2 on an ABI 3700 automated sequencer and assembled with Sequencer version 4.1.4 (Gene Codes, Ann Arbor, MI) software. All products were sequenced in both directions and the absence of PCR and sequencing errors was confirmed by repeated sequencing of independent amplicons. Sequences were submitted to GenBank under accession nos. AY643845–AY644336.
Data analysis:
Estimates of nucleotide polymorphism (Watterson's estimate, θW; nucleotide diversity, π), neutrality tests (Tajima's D, McDonald–Kreitman, HKA), recombination (Hudson and Kaplan), and linkage disequilibrium (r2) and their statistical significance were calculated using DnaSP Version 3.53 (Lewontin 1964; Lewontin and Kojim 1964; Watterson 1975; Tajima 1983, 1989, 1993; Hudson et al. 1987; Nei 1987; Nei and Miller 1990; McDonald and Kreitman 1991; Rozas and Rozas 1999). All sites with a frequency <0.10 for the rare allele were excluded for LD analysis because r2 has a large variance with rare alleles. Plots of all informative pairwise comparisons and the negative log P-values of significance among associations relative to physical distance (in kilobases), in addition to expected and observed number of significant associations at specific distances, were generated in Microsoft Excel. The median association value for each set of pairwise comparisons between sites located within two different gene regions was calculated and plotted against the corresponding median distance for each set using GenStat for Windows (2002, Release 6.2, 6th Edition, VSN International). Recombination analysis was performed using LDhat (http://www.stats.ox.ac.uk/∼mcvean/LDhat/).
RESULTS
Patterns of LD within candidate gene regions:
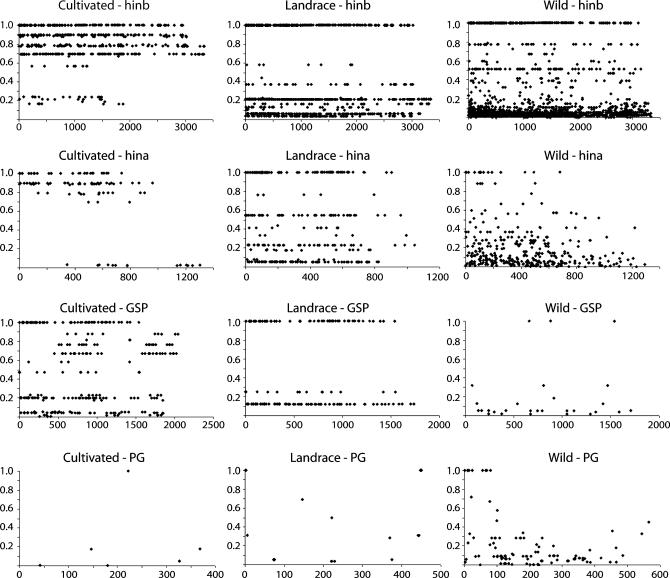
Four gene regions from the previously described (Caldwell et al. 2004) fully sequenced genomic region surrounding the hardness locus in barley, namely hinb, hina, GSP, and a putative gene (PG2), were analyzed to determine the level of LD (r2) between informative sites (rare allele with f > 0.1; Figure 1). High levels of association extended across the entire 3373-bp hinb gene region containing both hinb-1 and hinb-2 in the cultivated sample with relatively few pairwise comparisons demonstrating low association. A scarcity of low association values was also observed at the hina region. However, in contrast to the hinb region, there appeared to be evidence of LD decay after 1000 bp. The plot of LD across the GSP gene revealed a high level of association across the entire 1805-bp region. However, this pattern also differed from that observed at the hinb region as association values demonstrating a range of magnitudes were found between pairs of sites at varying, intermediate distances. An insufficient number of informative sites for accurately assessing LD existed within the PG2 region in the cultivated sample.
Figure 1.
Plots of LD (r2) as a function of distance (in base pairs) between informative (f > 0.1) polymorphic sites in four different gene regions and three different gene pools.
The overall extent of LD observed within gene regions in the landrace sample was similar to that observed within the cultivated material (Figure 1); high levels of association stretched across each gene region. However, a substantial number of pairwise comparisons within the hinb and hina gene regions gave moderate to low association values that were not detected in the cultivated material. Furthermore, there was a complete absence of intermediate association values in the GSP region. This bimodal distribution could be attributed to an elevated level of high association values as a result of small sample size. Although more informative SNPs were available for association analysis in the PG2 region relative to the cultivated sample, the paucity of these events still prevented accurate assessment of LD.
High levels of association extended across the entire hinb-1 and hinb-2 gene region in the wild sample (Figure 1). However, in contrast to the cultivated data, a substantial number of intermediate and low association values were observed at a range of distances within the gene region. Likewise, a more distinct pattern of LD decay within the hina region was observed in the wild sample relative to the cultivated material with complete decay to <0.2 by 1100 bp. An even greater rate of decay was observed in the PG2 region where association values dropped to <0.2 by 400 bp. A general trend of low association values is apparent within the GSP region despite the small number of informative SNPs.
Patterns of LD across a contiguous 212-kb region:
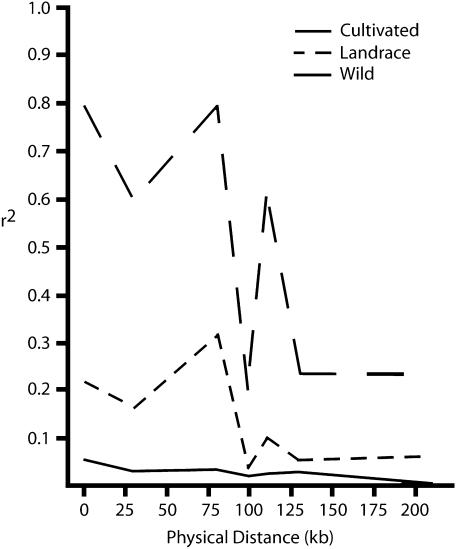
Estimates of association were also determined among the different gene regions, which are nonuniformly distributed across the 212 kb contiguous sequence (Figure 2). Estimates of LD between all pairs of informative sites are summarized in Figures 3 and 4; supplemental Figure 1 at http://www.genetics.org/supplemental/. High levels of association were found to stretch across the entire region in the cultivated sample. Although a gradual decay of LD was observed with distance, highly significant (P < 0.0001) LD values extended across the entire 212-kb region and the median level for each distance group never decayed below 0.2.
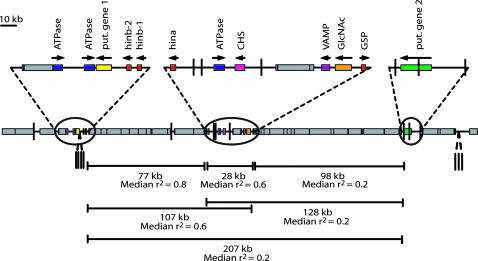
Figure 2.
Representation of the intergenic space between 12 genes located in a contiguous 303-kb region surrounding the Ha locus. Median LD values across these regions for the cultivated sample are indicated. Coding sequence is represented by colored boxes and arrows designate gene orientation. Location of repetitive sequence is indicated by shaded boxes. Mini-inverted repetitive element insertions are represented by a vertical bar.
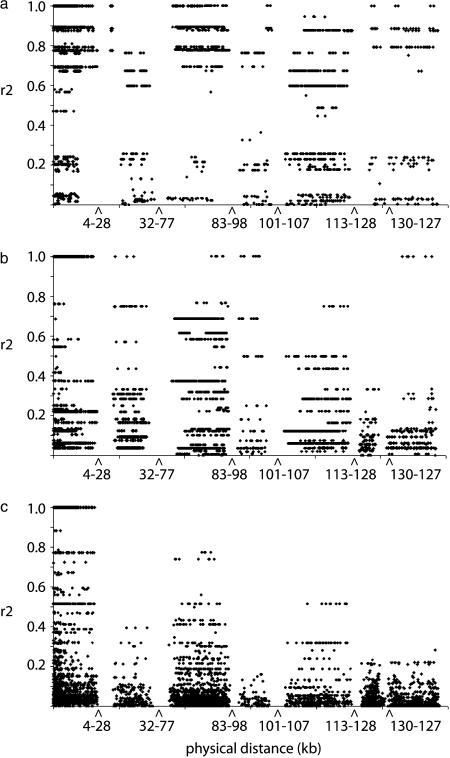
Figure 3.
Plots of LD (r2) as a function of distance (in kilobases) for the (a) cultivated, (b) landrace, and (c) wild (H. spontaneum) samples.
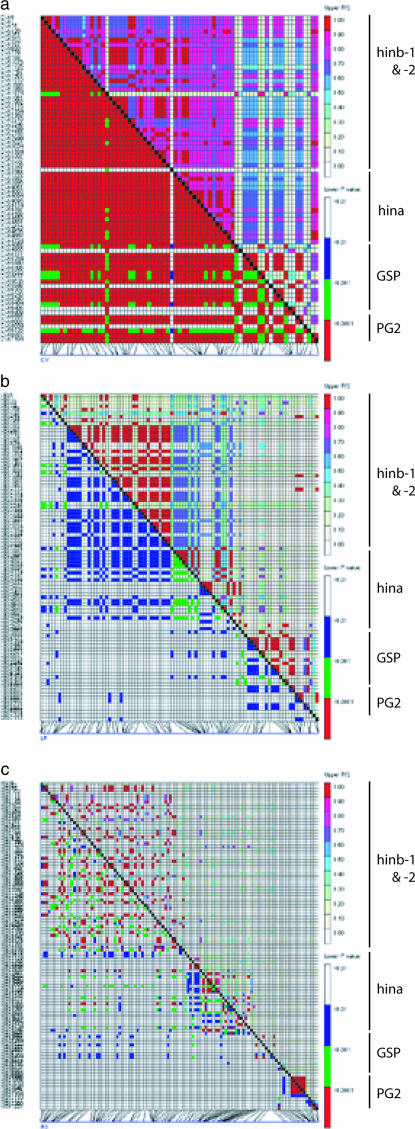
Figure 4.
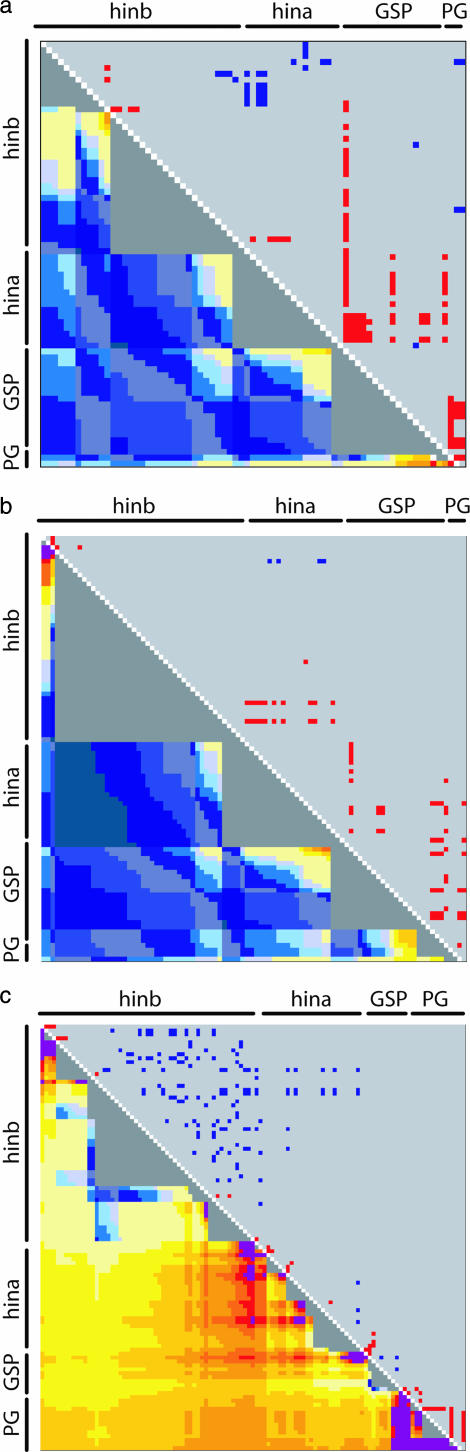
Matrix of pairwise association between sites for the (a) cultivated, (b) landrace, and (c) wild (H. spontaneum) samples. Above the diagonal are r2 values; below the diagonal are P-values.
To determine if this pattern and the level of LD were maintained in ancestral gene pools, the landrace and wild samples were assessed across the same region. LD and its significance decreased as a function of increasing distance in both ancestral samples (Figures 3 and 4; supplemental Figure 1 at http://www.genetics.org/supplemental). However, this pattern was particularly distinct in the wild material (Figures 3 and 4). Significant median LD values at a level >0.1 extended as far as 83 kb in the landrace sample and complete decay was seen by 98 kb (Figure 5). In contrast, complete equilibrium outside intragenic associations was observed in the wild sample with no median values at a level >0.1 (Figure 5). Although a few pairwise comparisons at distances >100 kb demonstrated perfect association (r2 = 1) in the landrace sample, it is likely that these were spurious associations detected as a consequence of the small sample size and inability to recognize and remove rare polymorphisms (Figure 3).
Figure 5.
Plots of the median association value for each group of pairwise comparisons against the corresponding median distance. Groups are: within-gene comparisons (1–4 kb), comparisons between markers within hina and GSP (28–32 kb), comparisons between markers within hinb and hina (77–83 kb), comparisons between markers within GSP and PG2 (98–101 kb), comparisons between markers within hinb and GSP (107–113 kb), comparisons between markers within hina and PG2 (128–131 kb), and comparisons between markers within hinb and PG2 (207–212 kb).
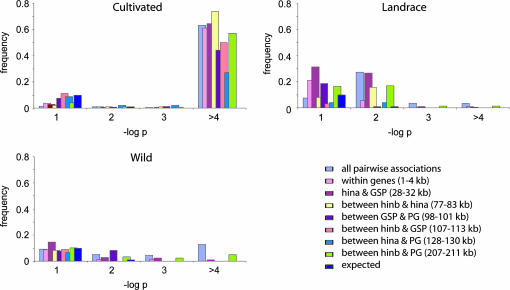
The stark contrast in LD between the cultivated and ancestral gene pools is further exemplified by the distribution of −log P-values for pairwise associations (Figure 6). Although all three sample sets demonstrated higher levels of significant association than would be expected in the absence of LD, the majority (>57%) of pairwise associations in the cultivated material demonstrated −log P-values >4% compared to ≤5% in the other two gene pools.
Figure 6.
Distribution of −log P-values of pairwise association.
LD and its relation to genome organization:
Although the overall trend reveals a gradual decrease of LD with physical distance, there is also an undulating pattern in the levels of association among the different between-gene comparison groups in all three sample sets studied (Figure 3). This pattern is even more striking when the median LD value plots are considered (Figure 5). In an effort to explain this pattern, focus was turned to the genome composition and organization of the region. The sequence spanning the candidate grain texture genes includes several different patterns of genome organization that are typical of small-grained cereals, including both solitary genes and gene clusters separated by stretches of nested repetitive element insertions. The presence of either singular or nested transposable elements has previously been implicated as a mechanism for recombination suppression in several eukaryotic genomes and, therefore, could have an impact on local levels of LD (Charlesworth et al. 1994; Arabidopsis Genome Initiative 2000; Fu et al. 2002; Yao et al. 2002).
Two groups of pairwise comparisons involved sites from genes separated by large expanses of nested repetitive sequence: hinb and hina and GSP and PG2. The hinb and hina gene regions are separated by 77 kb, 93% of which is composed of transposable elements (Figure 2). This region occupies a 2.5-fold greater intergenic interval than that observed between hina and GSP, which contains only one small ∼5-kb retrotransposon (Figure 2). Nevertheless, association values between pairwise sites flanking the 77-kb transposable element cluster were higher and more significant than those between the 28-kb intergenic region composed primarily of low-copy genic sequence (Figures 4 and 5). This result suggests recombination suppression in the former region. Plots showing local pairwise deviations from the assumption of a constant rate of recombination across the region indicated that recombination is indeed reduced between hinb and hina within the cultivated sample (Figure 7). This localized region of suppression is even more evident in the wild sample.
Figure 7.
Plots indicating pairs of sites that demonstrate deviation from the assumption of a constant recombination rate across the region for the (a) cultivated, (b) landrace, and (c) wild (H. spontaneum) samples. Above the diagonal, marginal likelihood ratios >2.0 are blue and red for pairs with more and less LD than expected, respectively. Below the diagonal are the minimum number of detectable recombination events normalized by physical distance between pairs of sites. Recombination rates of a magnitude of 10e−4 are indicated by blue as recombination rates increase. Recombination rates of a magnitude of 10e−3 are indicated by light yellow (1 × 10e−3), yellow (2 × 10e−3), gold (3 × 10e−3), light orange (4 × 10e−3), orange (5 × 10e−3), and red (6 × 10e−3). Recombination rates of a magnitude >7.0 × 10e−3 are purple. Gray boxes indicate the absence of detectable recombination.
The intergenic space between GSP and PG2 was also primarily composed of nested transposable elements (96%) and spanned a region even larger than that observed between hina and hinb (96 kb; Figure 2). However, in contrast to the hina and hinb region, the GSP and PG2 region demonstrated one of the lowest median association values of all groups considered (Figures 4 and 5). This suggests that the extensive regional expansion caused by element insertion has had negligible suppression on the recombination between these two genes. On the contrary, GSP and PG appear to be recombination hotspots compared to the rest of the region analyzed (Figure 7). Therefore, the presence/absence of repetitive DNA cannot solely account for the punctuated pattern of LD observed across the entire sequenced region.
Impact of selection on local levels of LD:
Contrasting gene histories for the different regions could also contribute to the undulating pattern of LD. In all three sample sets, the hinb-1 gene demonstrated the strongest evidence for selection (McDonald–Krietman test, P < 0.05; HKA test, P < 0.05 with Triticum tauschii sequence as an outgroup). It is striking, therefore, that both prominent peaks present on the plots of median LD values correspond to pairwise comparisons involving sites located within the hinb-1 gene region (Figure 5). Although median LD values are too low to observe this pattern in the wild germ plasm, plots of the 95th percentile are consistent with this observation (data not shown). In contrast, no evidence for selection was observed for the PG2 gene region regardless of the sample set analyzed or the test statistic employed. Median values of pairwise comparison groups involving sites located within the PG2 gene demonstrate the lowest LD values observed.
DISCUSSION
Contrasting evolutionary histories of different germ-plasm samples as a tool for association mapping strategies:
Our observations in barley provide an example of significantly different patterns of LD detected across a relatively short physical distance among different samples of an autogamous species. These contrasting patterns exist despite similar levels of inbreeding and most likely reflect different population histories associated with the occurrence of bottlenecks and selection within the domesticated germ plasm. Similar observations have recently been reported for 25 accessions of H. spontaneum, with intralocus LD decaying rapidly at a rate similar to that observed in the outbreeding species, Zea mays (Morrell et al. 2005). The observations have crucial implications for the design and execution of association studies, suggesting a two-tiered strategy for LD mapping. To take advantage of large LD in elite germ plasm, low-resolution whole-genome scans could be deployed to identify candidate gene regions. This would be complemented by fine-scale, high-resolution LD mapping utilizing landrace and wild germ plasm to identify candidate genes. The possibility of using a two-tiered approach has previously been reported in human and maize LD studies (Reich et al. 2001; Nordborg et al. 2002) Recently, a haplotype-based approach, rather than individual SNP associations, was used to identify and elucidate natural allelic variation at the CRY2 flowering-time gene in Arabidopsis (Olsen et al. 2004). This study, together with our own observations of barley, suggest that haplotype tagging coupled with LD mapping in different populations of barley will provide new opportunities to connect sequence diversity to complex phenotypes in crop plants.
Contrasting gene histories generate a punctuated pattern of LD:
A smooth progression of decreasing association with increasing distance was not observed among the different barley samples (Figures 3 and 5). Instead, an undulating pattern of LD was observed with several regions of notable increase in LD at intermediate distances (Figures 3 and 5). This observation is similar to that described for humans for which the “haplotype-block” model of LD has gained prominence (Jeffreys et al. 2001) and has also been described in Arabidopsis (Haubold et al. 2002). One plausible explanation for this punctuated pattern of LD is the presence of contrasting gene histories within the same local chromosomal region. The different patterns of nucleotide diversity observed among the genes analyzed suggested varying intensities of selection (Table 2). Furthermore, individual comparisons of association within genes demonstrated differences in the magnitude and extent of LD (Figure 1). In all three sample sets, the region harboring the two hinb gene copies demonstrated the highest levels of association with negligible signs of LD decay (Figure 1). This is consistent with evidence that suggests that the hinb-1 region was subjected to past directional selection (K. S. Caldwell, P. Langridge and W. Powell, unpublished results). In contrast, LD was found to decay within only a few hundred base pairs at the PG2 gene region in the wild germ plasm (Figure 1). Limited LD maintenance within this gene region is supported by the lack of evidence to suggest any past selection (K. S. Caldwell, P. Langridge and W. Powell, unpublished results). These local signatures of selection may explain the undulating pattern of LD across the entire region; peaks of high LD corresponded perfectly to associations involving the putatively selected hinb-1 gene region (Figures 3 and 4). Likewise, depressions of low LD corresponded to associations involving the assumed neutral PG2 gene region (Figures 3 and 4).
TABLE 2.
Estimates of nucleotide polymorphism within different germ-plasm samples across the genes
| No. of haplotypes
|
No. of mutations (η)
|
θ/bp based on:
|
|||||
|---|---|---|---|---|---|---|---|
| Gene | Region | Length (bp) | Sna | π | Tajima's D | ||
| Cultivated sample (n = 74) | |||||||
| GSP | Overall | 1788 (1802) | 5 | 21 | 0.00240 | 0.00324 | 1.05816 |
| Silent | 1414.057 | 17 | 0.00247 | 0.00331 | |||
| Nonsyn | 379.941 | 4 | 0.00216 | 0.00298 | |||
| hina | Overall | 1428 (1475) | 6 | 24 | 0.00341 | 0.00297 | −0.39626 |
| Silent | 1102.963 | 18 | 0.00335 | 0.00256 | |||
| Nonsyn | 342.036 | 6 | 0.00360 | 0.00428 | |||
| hinb-1 | Overall | 1583 (1596) | 6 | 29 | 0.00376 | 0.00309 | −0.55869 |
| Silent | 1243.722 | 26 | 0.00429 | 0.00352 | |||
| Nonsyn | 339.277 | 3 | 0.00181 | 0.00153 | |||
| hinb-2 | Overall | 1760 (1776) | 8 | 35 | 0.00408 | 0.00235 | −1.35707 |
| Silent | 1423.274 | 32 | 0.00461 | 0.00263 | |||
| Nonsyn | 335.725 | 4 | 0.00244 | 0.00177 | |||
| PG | Overall | 591 (594) | 6 | 15 | 0.00521 | 0.00336 | −1.02410 |
| Silent | 143.455 | 14 | 0.02002 | 0.01162 | |||
| Nonsyn | 441.545 | 1 | 0.00046 | 0.00073 | |||
| Landrace sample (n = 15) | |||||||
| GSP | Overall | 1788 (1802) | 5 | 26 | 0.00446 | 0.00463 | 0.16105 |
| Silent | 1414.099 | 21 | 0.00457 | 0.00477 | |||
| Nonsyn | 379.900 | 5 | 0.00405 | 0.00411 | |||
| hina | Overall | 1428 (1475) | 6 | 28 | 0.00596 | 0.00695 | 0.69458 |
| Silent | 1103.399 | 21 | 0.00585 | 0.00670 | |||
| Nonsyn | 341.600 | 7 | 0.00630 | 0.00775 | |||
| hinb-1 | Overall | 1583 (1596) | 5 | 21 | 0.00408 | 0.00460 | 0.52060 |
| Silent | 1243.932 | 19 | 0.00470 | 0.00530 | |||
| Nonsyn | 339.067 | 2 | 0.00181 | 0.00202 | |||
| hinb-2 | Overall | 1760 (1776) | 7 | 25 | 0.00441 | 0.00530 | 0.84917 |
| Silent | 1408.266 | 22 | .00480 | .00598 | |||
| Nonsyn | 337.733 | 4 | 0.00364 | 0.00407 | |||
| PG | Overall | 591 (594) | 4 | 12 | 0.00622 | 0.00488 | −0.83954 |
| Silent | 145.211 | 9 | 0.01906 | 0.01296 | |||
| Nonsyn | 442.789 | 3 | 0.00208 | 0.00229 | |||
| Wild sample (n = 34) | |||||||
| GSP | Overall | 1788 (1802) | 26 | 147 | 0.02011 | 0.00754 | −2.36210** |
| Silent | 1407.945 | 128 | 0.02223 | 0.00837 | |||
| Nonsyn | 380.054 | 19 | 0.01223 | 0.00449 | |||
| hina | Overall | 1428 (1475) | 22 | 63 | 0.01062 | 0.01000 | −0.27063 |
| Silent | 1096.774 | 44 | 0.00981 | 0.00918 | |||
| Nonsyn | 331.225 | 19 | 0.01403 | 0.01270 | |||
| hinb-1 | Overall | 1583 (1596) | 22 | 86 | 0.01329 | 0.00916 | −1.15921 |
| Silent | 1243.76 | 77 | 0.01514 | 0.01042 | |||
| Nonsyn | 339.123 | 9 | 0.00649 | 0.00454 | |||
| hinb-2 | Overall | 1760 (1776) | 23 | 104 | 0.01446 | 0.00917 | −1.37269 |
| Silent | 1422.078 | 93 | .01599 | .01055 | |||
| Nonsyn | 337.922 | 11 | 0.00796 | 0.00335 | |||
| PG | Overall | 591 (594) | 20 | 33 | 0.01359 | 0.01389 | 0.08042 |
| Silent | 145.181 | 26 | 0.04380 | 0.04514 | |||
| Nonsyn | 445.818 | 7 | 0.00384 | 0.00382 | |||
Number of haplotypes, number of mutations (η), and gene length were calculated after the omission of indels. Length of regions including indels is denoted in parentheses. Nonsyn, nonsynonymous. Tajima's D statistic significance levels: *P < 0.5 and **P < 0.1.
Number of segregating sites.
Evidence of changes in local levels of LD as a result of contrasting gene history has been previously reported at the different adh loci in H. spontaneum (Lin et al. 2001, 2002). However, such examples are not limited to Hordeum species. The FRI gene in Arabidopsis may have been subjected to local adaptation, which could account for the extended range of LD (250 kb) surrounding the region (Johanson et al. 2000; Nordborg et al. 2002). In comparison, regions in Arabidopsis demonstrating evidence of balancing selection, CLV2 and RPS5 loci, showed a reduction in the extent of LD by as much as a factor of 10 (Tian et al. 2002; Shepard and Purugganan 2003). Breeding selection can also leave a mark in the patterns of LD within a plant system. Despite the close relationship between Y1 and PSY2 in maize, the two loci demonstrate drastically different nucleotide diversity levels with Y1 displaying a >10-fold increase in the extent of LD relative to PSY2 (Remington et al. 2001). This is predicted to be a result of human selection (Palaisa et al. 2003). These results indicate that association analysis could be exploited as a valuable tool for understanding the genetic basis of adaptation to new environments and the phenotypic diversity arising from the crop domestication process (Kraakman et al. 2004).
The role of transposable elements in observed LD patterns:
Several studies in plant species indicate that recombination is predominantly active in gene-rich chromosomal regions (Gill et al. 1996a,b; Schnable et al. 1998; Faris et al. 2000; Kunzel et al. 2000). Furthermore, single transposable elements and clusters of nested repetitive elements within plant species are believed to be either recombinantly inert or suppressors of recombination (Fu et al. 2002; Yao et al. 2002). As a consequence, successful fine-scale mapping of causative mutations based on association mapping may be largely dependent upon the genome composition surrounding candidate genes.
Pairwise associations between two different sets of genes each separated by >77 kb of nested repetitive sequence (Figure 1) revealed substantially different levels of association. High levels of association extended across the intergenic region between hinb-1 and hina with a median value of 0.8 (Figure 1). The region between GSP and PG2, however, showed minimal evidence of strong association and a median value of 0.2 (Figure 1). This suggests that the presence of large segments of transposable sequence was not sufficient to suppress recombination between the genes characterized in this region of the barley genome. However, this assumes that all the lines represented in this study maintain the same or have a very similar gene content and genome organization, which may not be the case. A comparison of the bronze locus in two maize inbred lines revealed that the location and extent of element insertion as well as gene content were highly variable (Fu and Dooner 2002). Furthermore, the results reported here cannot distinguish between recombination events occurring within repetitive or low-copy regions in the intergenic space. Only exon 3 of PG2 was included in the nucleotide diversity and association analysis. Therefore, the 4 kb of low-copy sequence, including the latter portion of the gene and the 3′ flanking region, could harbor a recombination hotspot accounting for the low levels of association involving the PG2 region. This would be consistent with the observation that exon 3 of PG2 contained the highest level of recombination observed within all regions analyzed (Figure 7). Similarly, although the transposable elements in the intergenic region between hinb-1 and hina could have an effect on the local recombination rate, it would be difficult to distinguish this effect from that of regional selection on the basis of information available here.
Conclusions:
This work represents a detailed study into the levels and patterns of local or short-range LD within an inbreeding crop species and suggests that LD-based approaches will be a powerful tool for identifying the allelic variants that contribute to complex traits in crop plants. The contrasting evolutionary histories of crop gene pools represent a unique biological resource that will allow the scale and resolution of association-based studies to be modulated in a highly flexible manner. Furthermore, the punctuated pattern of LD generated by different gene histories within the 212-kb region indicate that association analysis could also be a valuable tool for locating genes involved in local adaptation and the domestication process.
Acknowledgments
We thank J. McNicol for statistical support, D. Charlesworth for helpful discussions, and A. Rafalski for critical review of the manuscript. We also thank the Scottish Executive Environment and Rural Affairs Department for their financial support.
References
- Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Ardlie, K. G., L. Kruglyak and M. Seielstad, 2002. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3: 299–309. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 211–215. [Google Scholar]
- Caldwell, K. S., P. Langridge and W. Powell, 2004. Comparative sequence analysis of the region harboring the hardness locus in barley and its collinear region in rice. Plant Physiol. 136: 3177–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., P. Sniegowski and W. Stephan, 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Faris, J. D., K. M. Haen and B. S. Gill, 2000. Saturation mapping of a gene-rich recombination hot spot region in wheat. Genetics 154: 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H. H., and H. K. Dooner, 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H. H., Z. W. Zheng and H. K. Dooner, 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris, A. J., S. R. McCouch and S. Kresovich, 2003. Population structure and its effect on haplotype diversity and linkage disequilbrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and A. D. Long, 2003. The lowdown on linkage disequilibrium. Plant Cell 15: 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, K. S., B. S. Gill, T. R. Endo and E. V. Boyko, 1996. a Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143: 1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, K. S., B. S. Gill, T. R. Endo and T. Taylor, 1996. b Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 144: 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubold, B., J. Kroymann, A. Ratzka, T. Mitchell-Olds and T. Wiehe, 2002. Recombination and gene conversion in a 170-kb genomic region of Arabidopsis thaliana. Genetics 161: 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A. J., L. Kauppi and R. Neumann, 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29: 217–222. [DOI] [PubMed] [Google Scholar]
- Johanson, U., J. West, C. Lister, S. Michaels, R. Amasino et al., 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347. [DOI] [PubMed] [Google Scholar]
- Kraakman, A. T., R. E. Niks, P. M. Van Den Berg, P. Stam and F. A. Van Eeuwijk, 2004. Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 168: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzel, G., L. Korzun and A. Meister, 2000. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin, R. C., 1964. The interaction of selection and linkage. I. General considerations: heterotic models. Genetics 49: 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin, R. C., and K. Kojima, 1964. The evolutionary dynamics of complex polymorphisms. Evolution 14: 458–472. [Google Scholar]
- Lin, J. Z., A. H. D. Brown and M. T. Clegg, 2001. Heterogeneous geographic patterns of nucleotide sequence diversity between two alcohol dehydrogenase genes in wild barley (Hordeum vulgare subspecies spontaneum). Proc. Natl. Acad. Sci. USA 98: 531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. Z., P. L. Morrell and M. T. Clegg, 2002. The influence of linkage and inbreeding on patterns of nucleotide sequence diversity at duplicate alcohol dehydrogenase loci in wild barley (Hordeum vulgare ssp. spontaneum). Genetics 162: 2007–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- Morrell, P. L., D. M. Toleno, K. E. Lundy and M. T. Clegg, 2005. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp spontaneum) despite high rates of self-fertilization. Proc. Natl. Acad. Sci. USA 102: 2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nei, M., and J. C. Miller, 1990. A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics 125: 873–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory et al., 2002. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193. [DOI] [PubMed] [Google Scholar]
- Olsen, K. M., S. S. Halldorsdottir, J. R. Stinchcombe, C. Weinig, J. Schmitt et al., 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa, K. A., M. Morgante, M. Williams and A. Rafalski, 2003. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15: 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, D. E., M. Cargill, S. Bolk, J. Ireland, P. C. Sabeti et al., 2001. Linkage disequilibrium in the human genome. Nature 411: 199–204. [DOI] [PubMed] [Google Scholar]
- Remington, D. L., J. M. Thornsberry, Y. Matsuoka, L. M. Wilson, S. R. Whitt et al., 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., B. S. Gaut, A. Tikhonov, Y. Nakajima and J. L. Bennetzen, 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43–45. [DOI] [PubMed] [Google Scholar]
- Schnable, P. S., A. P. Hsia and B. J. Nikolau, 1998. Genetic recombination in plants. Curr. Opin. Plant Biol. 1: 123–129. [DOI] [PubMed] [Google Scholar]
- Shepard, K. A., and M. D. Purugganan, 2003. Molecular population genetics of the Arabidopsis CLAVATA2 region: the genomic scale of variation and selection in a selfing species. Genetics 163: 1083–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1993. Measurement of DNA polymorphism, pp. 37–59 in Mechanisms of Molecular Evolution, edited by N. Takahata and A. G. Clark. Sinauer Associates, Sunderland, MA.
- Tanksley, S. D., and S. R. McCouch, 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, A. D. Long, R. L. Gaut, J. F. Doebley et al., 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D. C., H. Araki, E. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Yao, H., Q. Zhou, J. Li, H. Smith, M. Yandeau et al., 2002. Molecular characterization of meiotic recombination across the 140-kb multigenic a1-sh2 interval of maize. Proc. Natl. Acad. Sci. USA 99: 6157–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]