Abstract
The transition from self-incompatibility (SI) to self-compatibility (SC) is regarded as one of the most prevalent transitions in Angiosperm evolution, having profound impacts on the genetic structure of populations. Yet, the identity and function of mutations that result in the breakdown of SI in nature are not well understood. This work provides the first detailed genetic description of the breakdown of S-RNase-mediated gametophytic self-incompatibility (GSI) in a polyploid species that exhibits genotype-dependent loss of SI. Genetic analyses of six natural sour cherry (Rosaceae, Prunus cerasus) selections identified seven independent, nonfunctional S-haplotypes with disrupted pistil component (stylar-S) and/or pollen component (pollen-S) function. A genetic model demonstrating that the breakdown of SI in sour cherry is due to the accumulation of a minimum of two nonfunctional S-haplotypes within a single individual is developed and validated. Our finding that sour cherry is SI when only one nonfunctional S-haplotype is present has significant evolutionary implications since nonfunctional S-haplotypes would be maintained in the population without causing an abrupt shift to SC. Furthermore, we demonstrate that heteroallelic sour cherry pollen is self-incompatible, which is counter to the well-documented phenomenon in the Solanaceae where SC accompanying polyploidization is frequently due to the SC of heteroallelic pollen.
GAMETOPHYTIC self-incompatibility (GSI) is a common genetic mechanism that promotes outcrossing in flowering plants (de Nettancourt 1977). In GSI, self-incompatibility (SI) is determined by a single, multi-allelic locus, called the S-locus, in which the compatibility of a cross is determined by the haploid genome of the pollen and the diploid genome of the pistil. Pollen tube growth is arrested if the pollen tube has an S-allele in common with one of the two S-alleles in the style. The S-locus contains a minimum of two genes, one controlling stylar specificity and the other controlling pollen specificity of the SI reaction. The stylar-S in three plant families, the Solanaceae, Scrophulariaceae, and Rosaceae is a ribonuclease (S-RNase) (Anderson et al. 1986; McClure et al. 1989; Sassa et al. 1992; Xue et al. 1996), which is expressed in the pistil and specifically degrades RNA of incompatible pollen (McClure et al. 1990). The pollen-S gene is an F-box gene named S-locus F-box (SLF) in Antirrhinum (Lai et al. 2002) and in Prunus mume (Entani et al. 2003), PiSLF in Petunia inflata (Sijacic et al. 2004), and S-haplotype-specific F-box gene (SFB) in Prunus dulcis, Prunus avium, and Prunus cerasus (Ushijima et al. 2003; Yamane et al. 2003; Ikeda et al. 2004). The function of this F-box gene in the SI reaction remains unknown.
Within the Rosaceae, Prunus has emerged as the model GSI genus due to the small physical size of the S-haplotype region that facilitated map-based cloning of the pollen-S (Entani et al. 2003; Ushijima et al. 2003). Four diploid Prunus species, sweet cherry (P. avium), almond (P. dulcis), and apricot (P. mume and Prunus armeniaca) have well-characterized GSI systems with >50 S-RNases and 10 SFBs isolated and sequenced (Ushijima et al. 1998, 2003; Tao et al. 1999; Tamura et al. 2000; Sonneveld et al. 2001, 2003; Yaegaki et al. 2001; Ma and Oliveira 2002; Beppu et al. 2003; Romero et al. 2004; Wünsch and Hormaza 2004; de Cuyper et al. 2005). Within Prunus, cherry represents a natural diploid–tetraploid series with the tetraploid sour cherry arising through hybridization between sweet cherry and the tetraploid ground cherry (Prunus fruticosa) (Olden and Nybom 1968). Like sweet cherry, sour cherry exhibits an S-RNase-based GSI system (Yamane et al. 2001; Hauck et al. 2002; Tobutt et al. 2004); however, in contrast to sweet cherry, natural sour cherry selections include both SI and self-compatible (SC) types (Lansari and Iezzoni 1990).
This genotype-dependent loss of SI in sour cherry indicates that genetic changes, and not polyploidy per se, cause the breakdown of SI. This is in contrast to the Solanaceae where polyploidy can result in the breakdown of SI (Livermore and Johnstone 1940; Stout and Chandler 1942; Pandey 1968). This breakdown of GSI accompanying polyploidy in Solanaceous species is a result of competitive interaction (Lewis 1943; Golz et al. 1999, 2001) in which pollen grains containing two copies of the same pollen-S allele (homoallelic pollen) are arrested if the cognate S-RNase is present in the style, while pollen grains containing two different pollen-S alleles (heteroallelic pollen) are compatible, regardless of the S-RNase composition of the style (Luu et al. 2001).
Competitive interaction describes a specific example of a pollen-part mutation caused by the presence of two different functional pollen-S genes within a single pollen tube. However, numerous other types of mutations have been associated with the occurrence of SC from normally SI populations or individuals. Pollen-part mutations can also result from a structural alteration of the SLF or SFB gene (Ushijima et al. 2004; Sonneveld et al. 2005). Stylar-part mutants can result from a structural alteration of the S-RNase gene or its cis-acting promoter region (Yamane et al. 2003). Finally, SC can be caused by mutations affecting so-called “modifier” genes that are required for pollen rejection but not for the allele specificity of the reaction (McClure et al. 1999).
We used a genetic approach to elucidate the basis for the breakdown of SI in sour cherry. Six diverse sour cherry selections representing the habitat range of the species were used, making it highly unlikely that these selections would contain similar mutations in modifier genes. To determine if the breakdown of SI is due to changes affecting the allele specificity of the SI reaction or to changes affecting the ability to carry out the incompatibility reaction, we took advantage of functional S-haplotypes shared between the sweet and the sour cherry and the full fertility of the reciprocal interspecific crosses. Four functional and seven nonfunctional S-haplotypes present in sour cherry are described. A model was developed and validated, confirming that SC in sour cherry is caused by the presence of two or more nonfunctional S-haplotypes within an individual. Furthermore, the demonstration that heteroallelic pollen is SI in sour cherry suggests that the pollen-S genes of Prunus and the Solanaceae may differ.
MATERIALS AND METHODS
Plant material:
Six SC sour cherry cultivars—Cigány, Érdi Bőtermő (EB), Montmorency (Mont), Rheinische Schattenmorelle (RS), Surefire (Sure), and Újfehértói fűrtős (UF)—and four sweet cherry cultivars—Chelan, Emperor Francis (EF), Gold, and Schmidt—were used (see supplemental Table 1 at http://www.genetics.org/supplemental). The S-alleles for the sweet cherry cultivars have been previously reported (Iezzoni et al. 2005). Initial S-allele characterizations for Cigány, EB, Mont, RS, Sure, and UF have also been previously reported (Yamane et al. 2001). Triploid progeny were generated from reciprocal interspecific crosses between sweet and sour cherry. Tetraploid progeny were generated from self-pollination of each of the sour cherry selections and the following sour cherry crosses: RS × EB, UF × Sure, UF × RS, and UF × Mont. S-haplotype segregation was examined from a total of 1200 progeny from 25 different self- and hybrid populations. For the triploid progeny and a portion of the tetraploid progeny, genotyping was done using DNA extracted from mature seed. All other plant material was grown at the Michigan State University Experimental Stations in Clarksville, Traverse City, or Benton Harbor, Michigan.
TABLE 1.
Segregation of pollen-derived S-haplotypes in interspecific crosses between sour cherry and sweet cherry
| Population size
|
Segregation of paternal S-haplotypes
|
|||
|---|---|---|---|---|
| Parents (S-genotype)a | Observed ratio | Expected ratiob | χ2 (P-value) | |
| RS (S6S13′S26Sa) × Gold (S3S6) | 31 | 31:0 (S3:S6) | 1:1 | 31.0 (<0.0001) |
| Mont (S6 S13′SaSnull) × Gold (S3S6) | 55 | 55:0 (S3:S6) | 1:1 | 55.0 (<0.0001) |
| UF (S1′S4SdSnull) × Schmidt (S2S4) | 66 | 66:0 (S2:S4) | 1:1 | 66.0 (<0.0001) |
| Sure (S4 S13′SaSnull) × EF (S3S4) | 30 | 30:0 (S3:S4) | 1:1 | 30.0 (<0.0001) |
| EB (S4S6mSaSnull) × EF (S3S4) | 18 | 18:0 (S3:S4) | 1:1 | 18.0 (<0.0001) |
| Cigány (S6m2S9S26Sa) × Chelan (S3S9) | 45 | 45:0 (S3:S9) | 1:1 | 45.0 (<0.0001) |
| EB (S4S6m Sa Snull) × Gold (S3S6) | 33 | 22:11 (S3:S6) | 1:1 | 3.67 (0.0555) |
| Cigány (S6m2S9S26Sa) × Gold (S3S6) | 36 | 16:20 (S3:S6) | 1:1 | 0.44 (0.5050) |
The S-haplotypes being tested are underlined.
Observed ratios were tested for fit to the ratio expected if the shared S-haplotype is nonfunctional (1:1). If the shared S-haplotype were functional, it would not be inherited from the paternal parent.
DNA extractions:
From leaves:
DNA extractions were conducted as previously described (Hauck et al. 2002).
From seed:
The testa was removed from the cherry seed and the remaining embryo and cotyledons were ground in liquid nitrogen and mixed in a buffer consisting of 1% CTAB, 150 mm Tris-HCl (pH 8.0), 20 mm EDTA, 800 mm NaCl, 0.25% SDS, and 1% β-mercaptoethanol. The DNA was purified by chloroform extraction and precipitated using isopropanol.
S-RNase genotyping:
The S-RNase gene-specific primer set, composed of Pru-C2 and PCE-R (Yamane et al. 2001), was used for S-haplotype determination for all self- and interspecific seed. This primer pair could differentiate between most S-RNase alleles on the basis of polymorphisms in the length of the second intron in the Prunus S-RNase. However, the S2- and S13′-RNase alleles could not be reliably amplified using this primer pair. Instead, either PaS2-Fnew/PaS2-R (Sonneveld et al. 2003) or newly designed PcS13-F (AGC AAA CCT TCC CAC CAA C)/PcS13-R (AGG AGG GGT GTT CTT CCA GT) was used. In certain crosses, other S-RNase-allele-specific primers were used to verify S-RNase genotypes (Sonneveld et al. 2001, 2003). The Sa- and Sd-haplotypes can be differentiated using RFLP analysis, as described previously (Yamane et al. 2001). We previously aligned the amino acid sequences for the S4-, S6-, S13′-, S26-, and Sa-RNases from sour cherry (Hauck et al. 2002).
SFB genotyping:
Thirty-three progeny from the cross RS × EB, 17 from UF × Mont, and 22 from UF × RS were genotyped using allele-specific primers for each of the functional SFB alleles to verify cosegregation of the S-RNase and SFB alleles. Allele-specific primers for SFB4 (PaSFB4-F/PaSFB4-R) and SFB6 (PaSFB6-F/PaSFB6-R) were used as previously described (Ikeda et al. 2005). The newly designed PcSFB26-F (GATTTGCTTGCTTTTTAAATGTTACGG)/PcSFB26-R (CTTAATTCTTGTGTCAAGAACTTGCC) were used for SFB26 genotyping.
Model testing:
The S-RNase genotypes for 92 mature seedlings with known pedigrees were determined using RFLP analyses following digestion with either HindIII or DraI as previously described (Yamane et al. 2001). Predictions of the SI or SC phenotype for seedlings were made on the basis of our developed hypothesis of the genetic control of SI and SC in sour cherry. The growth of self-pollen in each of the 92 seedlings was observed by aniline blue staining and UV microscopy (Hauck et al. 2002).
RESULTS
Sour cherry styles reject sweet cherry pollen in an S-allele-specific manner:
The ability of sour cherry styles to arrest pollen in an S-haplotype-specific manner was tested by crossing sour cherry and sweet cherry cultivars that have common S-haplotypes. When the sour cherry cultivar RS (S6S13′ S26Sa) was pollinated with pollen from the sweet cherry cultivar Gold (S3S6), all the progeny contained the S3-haplotype (Table 1). This indicates that Gold S3 pollen was compatible in RS styles, whereas the Gold S6 pollen was arrested by the presence of a functional S6-RNase (Figure 1A). Likewise, S6 and not S3 pollen was selectively inhibited in Mont (S6S13′ SaSnull) styles, S4 and not S2 pollen was selectively inhibited in UF (S1′ S4SdSnull) styles, S4 and not S3 pollen was selectively arrested in Sure (S4S13′ SaSnull) and EB (S4S6mSaSnull) styles, and S9 and not S3 pollen was selectively inhibited in Cigány (S6m2S9S26Sa) styles (Table 1). These results demonstrate that sour cherry retains the ability to reject pollen in an S-haplotype-specific manner; therefore, SC must be caused by genetic changes affecting the specificity of the GSI reaction. See supplemental Table 2 (http://www.genetics.org/supplemental) for complete segregation of the S-genotypes in the triploid progeny from the interspecific reciprocal crosses between sweet and sour cherry.
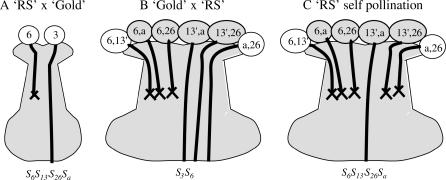
Figure 1.
Schematics of the interspecific crosses between RS (S6S13′ S26Sa) and Gold (S3S6) and the self-pollination of RS. (A) Pollination of RS styles with Gold pollen results in the rejection of all pollen containing the S6-haplotype. Pollen containing the S3-haplotype is successful. (B) Pollination of Gold styles with RS pollen results in the rejection of all pollen containing the S6-haplotype. Any pollen that does not contain the S6-haplotype is successful. Because sour cherry exhibits homologous and occasional nonhomologous pairing (Beaver and Iezzoni 1993), all possible chromosome-pairing configurations are considered. Pollen types formed by homologous pairing are shaded. (C) Self-pollination of RS results in rejection of all pollen containing either S6 or S26 or both. The only successful pollen is S13′Sa.
TABLE 2.
S-haplotypes of successful pollen types from interspecific crosses between sweet cherry and sour cherry selections
| Possible sour cherry pollen types
|
|||
|---|---|---|---|
| Parents (S-genotype)a | No. of progeny | Successful | Not detected |
| Gold (S3S6) × RS (S6S13′S26Sa) | 13 | S13′ S26, S13′Sa |
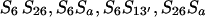 b b
|
| Gold (S3S6) × EB (S4S6mSaSnull) | 14 | S4Sa, S4Snull, SaSnull | S6mSa, S6mSnull, S4S6m |
| Gold (S3S6) × Cigány (S6m2S9S26Sa) | 40 | S9Sa, S9S26, S26Sa | S6m2Sa, S6m2S26, S6m2S9 |
| Gold (S3S6) × Mont (S6S13′SaSnull) | 15 | S13′Snull, S13′Sa, SaSnull | S6Snull, S6Sa, S6S1′ |
| EF (S3S4) × Sure (S4S13′SaSnull) | 37 | S13′Snull, S13′Sa, SaSnull | S4Snull, S4Sa, S4S13′ |
| EF (S3S4) × UF (S1′S4SdSnull) | 40 | S1′Snull, S1′Sd, SdSnull | S4Snull, S4Sd, S1′S4 |
| EF (S3S4) × EB (S4S6mSaSnull) | 20 | S6mSnull, S6mSa, SaSnull | S4Snull, S4Sa, S4S6m |
The S-haplotypes being tested are underlined.
The S26Sa gamete type is rare, resulting in only 3% of the progeny in a fully compatible cross (see supplemental Figure 1 at http://www.genetics.org/supplemental/).
Two stylar-part mutants are identified in sour cherry:
Sweet cherry S4 and S9 pollen was selectively inhibited in EB (S4S6mSaSnull) and Cigány (S6m2S9S26Sa) styles, respectively, indicating that these sour cherry cultivars are able to carry out an SI reaction (Table 1). In contrast, S6 pollen from the sweet cherry cultivar Gold successfully grew down the styles of these two selections, indicating that the S6-RNases in these two cultivars are nonfunctional (Table 1). These nonfunctional stylar-part mutations, which can be distinguished on the basis of RFLP patterns (Yamane et al. 2001) and PCR amplification products (Yamane et al. 2003), are termed S6m and S6m2 in EB and Cigány, respectively. We previously have shown that S6m consists of a functional S6-SFB but a nonfunctional S6-RNase due to a 2600-bp insertion upstream from the S6-RNase (Yamane et al. 2003).
Sweet cherry styles reject sour cherry pollen in an allele-specific manner:
When RS (S6S13′ S26Sa) pollen was placed on Gold (S3S6) styles, the absence of progeny containing both the S3- and S6-haplotypes indicated that S6-containing pollen from RS was selectively rejected by the S6-RNase in Gold styles, regardless of what other S-haplotype was in the pollen (Table 2; Figure 1B). This establishes that the RS S6-haplotype also exhibits S6-pollen-specific rejection and is, therefore, fully functional. S6-containing pollen of EB and Cigány was selectively rejected in Gold styles, indicating that the S6m- and S6m2-haplotypes in these selections have a functional pollen-S (Table 2). Likewise, S6-containing pollen from Mont was selectively rejected in Gold styles, and S4-containing pollen of Sure, UF, and EB was selectively rejected in EF (S3S4) styles (Table 2). This demonstrates that sour cherry pollen containing a functional pollen-S from an S-haplotype that is identical to the one in sweet cherry is always rejected. This allele-specific pollen rejection occurred regardless of the other S-haplotype present in the diploid pollen.
Self-pollinated progeny of sour cherry segregate for functional and nonfunctional S-haplotypes:
All of the progeny from the self-pollination of RS (S6S13′ S26Sa) inherited the S13′- and Sa-haplotypes, whereas the S6- and S26-haplotypes segregated 1:1 (present:absent) (Table 3). This can be explained by the arrest of pollen containing either the S6- or S26-haplotype, or both, and the self-compatibility of S13′Sa-containing pollen (Figure 1C). Therefore, we conclude that both the S6- and the S26-haplotypes are fully functional, as pollen containing either of these S-haplotypes was incapable of self-fertilization, whereas the S13′- and Sa-haplotypes were nonfunctional. S13′ was also determined to be a nonfunctional S-haplotype on the basis of self-pollinations of Sure and Mont (Table 3). S13′ was previously shown to have a functional stylar component in crosses with sweet cherry containing an S13-allele (Tobutt et al. 2004); therefore, we predict that the mutation affects the pollen component. Sa was also confirmed to be a nonfunctional S-haplotype from self-pollinations of Cigány, EB, Sure, and Mont (Table 3). Finally, S26 was also confirmed to be a fully functional S-haplotype on the basis of the self-pollination of Cigány (Table 3).
TABLE 3.
Segregation of S-haplotypes following self-pollination of six sour cherry selections to determine the functionality of each S-haplotype
| Parent (S-genotype)
|
No. of progeny observed
|
Segregation of S-haplotypes
|
|||
|---|---|---|---|---|---|
| S-haplotype | Observed ratio | Expected ratioa | χ2 (P-value) | ||
| RS | 54 | S6 | 28:26 | 1:1 | 0.07 (0.7855) |
| (S6S13′S26Sa) | S13′ | 54:0 | 1:1 | 54.0 (<0.0001) | |
| S26 | 23:31 | 1:1 | 1.19 (0.2763) | ||
| Sa | 54:0 | 1:1 | 54.0 (<0.0001) | ||
| Cigány | 59 | S6m2 | 59:0 | 1:1 | 59.0 (<0.0001) |
| (S6m2S9S26Sa) | S9 | 24:35 | 1:1 | 2.05 (0.1521) | |
| S26 | 36:23 | 1:1 | 2.86 (0.0906) | ||
| Sa | 59:0 | 1:1 | 59.0 (<0.0001) | ||
| EB | 25 | S4 | 9:16 | 1:1 | 1.96 (0.1615) |
| (S4S6mSaSnull) | S6m | 25:0 | 1:1 | 25.0 (<0.0001) | |
| Sa | 20:5 | 1:1 | 9.00 (0.0027) | ||
| Sure | 64 | S4 | 35:29 | 1:1 | 0.56 (0.4533) |
| (S4S13′SaSnull) | S13′ | 64:0 | 1:1 | 64.0 (<0.0001) | |
| Sa | 63:1 | 1:1 | 60.1 (<0.0001) | ||
| UF | 102 | S1′ | 102:0 | 1:1 | 102.0 (<0.0001) |
| (S1′S4SdSnull) | S4 | 60:42 | 1:1 | 3.18 (0.0747) | |
| Sd | 98:4 | 1:1 | 86.6 (<0.0001) | ||
| Mont | 135 | S6 | 72:63 | 1:1 | 0.60 (0.4386) |
| (S6S13′SaSnull) | S13′ | 131:4 | 1:1 | 119.4 (<0.0001) | |
| Sa | 131:4 | 1:1 | 119.4 (<0.0001) | ||
A 1:1 ratio is expected if the shared S-haplotype is fully functional, resulting in pollen rejection. A shared nonfunctional S-haplotype would not result in pollen rejection; therefore, the shared S-haplotype would be transmitted to the progeny at a higher frequency than expected.
For four of the sour cherry selections (EB, Sure, Mont, and UF), only three different S-haplotypes could be identified (Yamane et al. 2001). Segregation data presented in this study indicate that each S-haplotype was present in a single copy (Table 3). Therefore the fourth S-haplotype is hypothesized to be Snull, containing a deletion of the S-locus since no RFLP fragment associated with Snull was visualized with either an S-RNase or an SFB probe (Yamane et al. 2001).
In UF, S4 is the only fully functional S-haplotype, whereas S1′ and Sd are nonfunctional S-haplotypes (Table 3). Preliminary sequence and genetic analyses indicate that S1′ is a pollen-part mutant (N. R. Hauck, unpublished results). The nonfunctional Sa- and Sd-haplotypes likely represent different mutations of a common S-haplotype, since partial S-RNase and SFB sequences of the Sa- and Sd-haplotypes are identical (N. R. Hauck, unpublished results). These two S-haplotypes can be differentiated on the basis of HindIII S-RNase fragments (Sa, 6.4 kb; Sd, 6.2 kb) (Yamane et al. 2001).
Heteroallelic sour cherry pollen is SI:
The presence of two fully functional S-haplotypes (S6 and S26) in RS allowed us to test whether heteroallelic pollen is SI or SC. Evidence that RS S6S26 pollen is viable is provided by the fully compatible cross UF × RS where 11 of 59 progeny inherited S6S26 pollen from RS (see supplemental Figure 1 at http://www.genetics.org/supplemental/). RS S6S26 pollen was always rejected by Gold (S3S6) styles and self-styles, presumably due to the presence of the S6-RNase in the Gold and the S6- and S26-RNases in RS (Tables 2 and 3). Rejection of the RS S6S26 pollen containing two functional pollen-S alleles in both Gold and RS styles indicates that heteroallelic pollen is SI in sour cherry.
Additional evidence that the breakdown of GSI in sour cherry is not caused by the SC heteroallelic pollen is provided by the self-pollinations of Cigány, which contains two fully functional S-haplotypes (S9 and S26), and EB, which contains at least two functional pollen-S genes (S4 and S6m) (Table 3). Similar to RS, self-pollination of Cigány and EB resulted in the rejection of pollen containing these S-haplotypes. Cigány and EB pollen containing the S4-, S6m-, or S6m2-haplotypes was also arrested in styles of sweet cherry cultivars containing the S4- or S6-haplotypes, respectively (Table 2 and see supplemental Table 2 at http://www.genetics.org/supplemental/). Additionally, previous work with the SI sour cherry cultivar Crisana (S-RNase phenotype: S1S4Sd) demonstrated that it contains two fully functional S-haplotypes (S1 and S4) and all Crisana pollen was rejected in sweet cherry styles known to contain functional S1- and S4-RNases (Hauck et al. 2002).
The SI of heteroallelic pollen in sour cherry could be due to either the absence of competitive interaction or the presence of genetic dominance/recessive relationships between pollen-S alleles similar to that exhibited by the sporophytic SI system in Brassica (Thompson and Taylor 1966). Although the crosses made cannot conclusively distinguish between these two possibilities, we obtained no data consistent with dominant/recessive relationships among the six functional pollen-S alleles identified (S4, S6, S6m, S6m2, S9, and S26) as allele-specific pollen rejection occurred regardless of the other S-haplotype present in the diploid pollen (Table 2).
Model development and testing:
Taken together, our data indicate that the breakdown of GSI in sour cherry is caused by the accumulation of stylar-part and pollen-part mutants affecting multiple S-haplotypes (Figure 2). In sour cherry, four functional (S4, S6, S9, and S26) and seven nonfunctional S-haplotypes (S1′, S6m, S6m2, S13′, Sa, Sd, and Snull) (Hauck et al. 2002; Yamane et al. 2003; Tobutt et al. 2004; this work) have been identified. A comparison of the SI and SC selections revealed that the SI selections contained only one nonfunctional S-haplotype, whereas the SC selections contained two to four nonfunctional S-haplotypes. From this, we developed the “one-allele-match” model, in which a match between a functional pollen-S gene product in the pollen and its cognate functional S-RNase in the style would result in an incompatible reaction. A similar reaction would occur regardless of whether the pollen contained a single functional pollen-S gene or two different functional pollen-S genes. The absence of a functional match would result in a compatible reaction; thus, for successful self-fertilization, pollen must contain two nonfunctional S-haplotypes.
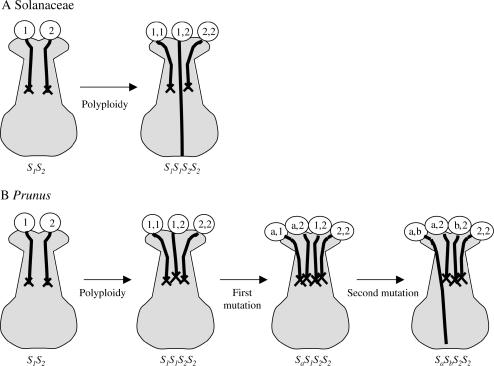
Figure 2.
Schematic of the affects of polyploidy on GSI in (A) the Solanaceae and (B) Prunus. In the Solanaceae, polyploidy directly causes the conversion from SI to SC due to the compatibility of heteroallelic pollen. In Prunus, polyploidy does not directly result in a breakdown of SI. Rather, SC requires the loss-of-function for a minimum of two S-haplotype-specificity components. Polyploidization creating tetraploid sour cherry presumably resulted from the mating of a 2n gamete from sweet cherry and an n gamete from tetraploid ground cherry (Iezzoni and Hancock 1984).
To test this model, we genotyped 92 seedlings from four crosses among five sour cherry selections. All seedlings that contained only one nonfunctional S-haplotype (n = 17) were SI and all seedlings that contained two or more nonfunctional and noncomplementary S-haplotypes (n = 75) were SC (Table 4 and see supplemental Table 3 at http://www.genetics.org/supplemental/). Since the nonfunctional Sa- and Sd-haplotypes likely represent different mutations of a common S-haplotype, we hypothesize that Sa and Sd have complementary pistil-S and pollen-S mutations, resulting in a functional S-haplotype. Therefore, these results validate the one-allele-match model for the genetic control of SC and SI in sour cherry.
TABLE 4.
Number of nonfunctional S-haplotypes and the SI or SC phenotypes for 92 sour cherry seedlings
| No. of nonfunctional S-haplotypes in each Seedling
|
No. of seedlings analyzed
|
Phenotype of seedlings
|
|
|---|---|---|---|
| No. SI | No. SC | ||
| 1 | 17 | 17 | 0 |
| 2 | 17 | 3a | 14 |
| 3 | 37 | 0 | 37 |
| 4 | 21 | 0 | 21 |
Three progeny with S-genotype S4S6SaSd were determined to be SI, despite having two nonfunctional S-haplotypes. Partial S-RNase and SFB sequences from the Sa- and Sd- haplotypes are identical (N. R. Hauck, unpublished results), suggesting that the Sa and Sd represent different mutations of a common S-haplotype. We are currently testing the possibility that Sa has a functional S-RNase and a nonfunctional SFB, whereas Sd has a nonfunctional S-RNase and a functional SFB. In this case, S4S6SaSd individuals would be predicted to be SI under the one-allele-match model since SaSd pollen would be rejected due to a match between a functional Sa-RNase and SFBd.
DISCUSSION
The transition from SI to SC has occurred repeatedly and has had a profound impact on angiosperm evolution, yet the genetic and molecular basis of this transition is not well understood. This study provides the first detailed genetic analysis of GSI breakdown in a diploid–polyploidy series involving multiple independent S-haplotype mutations. In each case the mutations were not in “modifier” genes that would cause a disruption in the ability to carry out the SI reaction as four S-haplotypes were found to be fully functional (S4, S6, S9, and S26). Instead, all mutations affected the allele specificity of the reaction by disrupting pistil-S and/or pollen-S function.
The one-allele-match model suggests a fundamental difference in the effect of polyploidy on SI between the Solanaceae and Prunus. In the Solanaceae, polyploidy is a direct cause of SC as a result of competitive interaction (Figure 2A), whereas in Prunus, polyploidy does not directly cause SC since heteroallelic pollen retains its SI phenotype. Rather, in sour cherry, mutations of the stylar- and pollen-specificity components have occurred and then accumulated to result in SC (Figure 2B). Our finding that sour cherry is SI despite the presence of one nonfunctional S-haplotype also has significant evolutionary implications in that nonfunctional S-haplotypes could be maintained in the population without causing an abrupt shift to SC.
Molecular characterization of five of the seven nonfunctional S-haplotypes that are completed or in progress, reveal structural changes of the S-haplotype (Yamane et al. 2003; N. R. Hauck, unpublished results). The S6m-haplotype has a transposon-like element insertion in the putative promoter region of the S6-RNase (Yamane et al. 2003). The coding sequence of the pollen-part mutant SFB1′ contains a 615-bp Ds-like element, while the coding sequence of the pollen-part mutant SFB13′ contains a nonsense mutation (N. R. Hauck, unpublished results). The Snull presumably resulted from a deletion that encompasses the S-RNase and SFB genes. The molecular characterizations of the S6m2-, Sa-, and Sd-haplotypes are not yet complete. However, we predict that at a minimum the Sd-haplotype will have an ∼2-kb deletion within the S-haplotype region that is not present in the Sa-haplotype.
Phylogenetic analyses of S-RNases from the Solanaceae, Scrophulariaceae, and Rosaceae support the conclusion of a common evolutionary origin for S-RNase-mediated GSI (Igic and Kohn 2001; Steinbachs and Holsinger 2002). The finding that the pollen-S in these three families is an F-box protein implicates ubiquitination as a common mechanism for S-RNase degradation (Kao and Tsukamoto 2004). Yet, the SI of heteroallelic pollen in sour cherry suggests that the pollen-S differs between Prunus (Rosaceae) and the Solanaceae. Two other lines of evidence support this contention. First, sweet cherry pollen carrying the mutated SFB3′, characterized by the complete deletion of a functional SFB3, is viable and SC (Sonneveld et al. 2005). However, in the Solanaceae, loss of the pollen-S gene is predicted to be lethal to the pollen (Golz et al. 1999, 2001). Second, the pollen-S allele in Prunus, SFB, exhibits a higher degree of sequence diversity than the pollen-S allele, SLF, in Antirrhinum and Petunia (Ikeda et al. 2004; Kao and Tsukamoto 2004). Further insight will require an understanding of the biochemical interactions involving the pollen-S and stylar-S genes in both the Solanaceae and Prunus.
Acknowledgments
We thank A. Sebolt for technical assistance and H. Sassa, N. Jiang, and S. van Nocker for their critical reviews. This work was partially supported by a grant from the U.S. Department of Agriculture, Cooperative State Research, Education, and Extension Service–National Research Initiative, Plant Genome, Bioinformatics and Genetics Resources Program.
References
- Anderson, M. A., E. C. Cornish, S.-l. Mau, E. G. Williams, R. Hoggart et al., 1986. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321: 38–44. [Google Scholar]
- Beaver, J. A., and A. F. Iezzoni, 1993. Allozyme inheritance in tetraploid sour cherry (Prunus-cerasus L.). J. Am. Soc. Hortic. Sci. 118: 873–877. [Google Scholar]
- Beppu, K., Y. Takemoto, H. Yamane, H. Yaegaki, M. Yamaguchi et al., 2003. Determination of S-haplotypes of Japanese plum (Prunus salicina Lindl.) cultivars by PCR and cross-pollination tests. J. Hort. Sci. Biotechnol. 78: 315–318. [Google Scholar]
- de Cuyper, B., T. Sonneveld and K. R. Tobutt, 2005. Determining self-incompatibility genotypes in Belgian wild cherries. Mol. Ecol. 14: 945–955. [DOI] [PubMed] [Google Scholar]
- de Nettancourt, D., 1977. Incompatibility in Angiosperms. Springer-Verlag, Berlin.
- Entani, T., M. Iwano, H. Shiba, F.-S. Che, A. Isogai et al., 2003. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8: 203–213. [DOI] [PubMed] [Google Scholar]
- Golz, J. F., V. Su, A. E. Clarke and E. Newbigin, 1999. A molecular description of mutations affecting the pollen component of the Nicotiana alata S locus. Genetics 152: 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J. F., H.-Y. Oh, V. Su, M. Kusaba and E. Newbigin, 2001. Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc. Natl. Acad. Sci. USA 98: 15372–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, N. R., H. Yamane, R. Tao and A. F. Iezzoni, 2002. Self-compatibility and incompatibility in tetraploid sour cherry (Prunus cerasus L.). Sex. Plant Reprod. 15: 39–46. [Google Scholar]
- Iezzoni, A. F., and A. M. Hancock, 1984. A comparison of pollen size in sweet and sour cherry. HortScience 19: 560–562. [Google Scholar]
- Iezzoni, A. F., R. L. Anderson, H. Schmidt, R. Tao, K. R. Tobutt et al., 2005. Proceedings of the S-allele workshop at the 2001 International Cherry Symposium. Acta Hortic. 667: 25–35. [Google Scholar]
- Igic, B., and J. R. Kohn, 2001. Evolutionary relationships among self-incompatibility RNases. Proc. Natl. Acad. Sci. USA 98: 13167–13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, K., B. Igic, K. Ushijima, H. Yamane, N. R. Hauck et al., 2004. Primary structure futures of the S haplotype-specific F-box protein, SFB, in Prunus. Sex. Plant Reprod. 16: 235–243. [Google Scholar]
- Ikeda, K., K. Ushijima, H. Yamane, R. Tao, N. R. Hauck et al., 2005. Linkage and physical distances between the S-haplotype S-RNase and SFB genes in sweet cherry. Sex. Plant Reprod. 17: 289–296. [Google Scholar]
- Kao, T.-H., and T. Tsukamoto, 2004. The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16(Suppl.): S72–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., W. Ma, B. Han, L. Liang, Y. Zhang et al., 2002. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 50: 29–42. [DOI] [PubMed] [Google Scholar]
- Lansari, A., and A. Iezzoni, 1990. A preliminary analysis of self-incompatibility in sour cherry. HortScience 25: 1636–1638. [Google Scholar]
- Lewis, D., 1943. Physiology of incompatibility in plants. III. Autotetraploids. J. Genet. 45: 171–185. [Google Scholar]
- Livermore, J. R., and F. E. Johnstone, 1940. The effect of chromosome doubling on the crossability of Solanum chacoense, S. jamesii and S. bulbocastanum with S. tuberosum. Am. Potato J. 17: 170–173. [Google Scholar]
- Luu, D. T., X. K. Qin, G. Laublin, Q. Yang, D. Morse et al., 2001. Rejection of S-heteroallelic pollen by a dual-specific S-RNase in Solanum chacoense predicts a multimeric SI pollen component. Genetics 159: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, R. C., and M. M. Oliveira, 2002. Evolutionary analysis of S-RNase genes from Rosaceae species. Mol. Genet. Genomics 267: 71–78. [DOI] [PubMed] [Google Scholar]
- McClure, B. A., V. Haring, P. R. Ebert, M. A. Anderson, R. J. Simpson et al., 1989. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342: 955–957. [DOI] [PubMed] [Google Scholar]
- McClure, B. A., J. E. Gray, M. A. Anderson and A. E. Clarke, 1990. Self-incompatibility in Nicotiana alata involves degradation of pollen ribosomal RNA. Nature 347: 757–760. [Google Scholar]
- McClure, B., B. Mou, S. Canevacini and R. Bernatzky, 1999. A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc. Natl. Acad. Sci. USA 96: 13548–13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olden, E. J., and N. Nybom, 1968. On origin of Prunus cerasus L. Hereditas 59: 327–345. [Google Scholar]
- Pandey, K. K., 1968. Colchicine induced changes in the self-incompatibility behaviour of Nicotiana. Genetica 39: 257–271. [Google Scholar]
- Romero, C., S. Vilanova, L. Burgos, J. Martínez-Calvo, M. Vicente et al., 2004. Analysis of the S-locus structure in Prunus armeniaca L. Identification of S-haplotype specific S-RNase and F-box genes. Plant Mol. Biol. 56: 145–157. [DOI] [PubMed] [Google Scholar]
- Sassa, H., H. Hirano and H. Ikehashi, 1992. Self-incompatibility-related RNases in styles of Japanese pear (Pyrus serotina Rehd). Plant Cell Physiol. 33: 811–814. [Google Scholar]
- Sijacic, P., X. Wang, A. L. Skirpan, L. Wang, P. E. Dowd et al., 2004. Identification of the pollen determinant of S-RNase-mediated self–incompatibility. Nature 429: 302–305. [DOI] [PubMed] [Google Scholar]
- Sonneveld, T., T. P. Robbins, R. Boskovic and K. R. Tobutt, 2001. Cloning of six cherry self-incompatibility alleles and development of allele-specific PCR detection. Theor. Appl. Genet. 102: 1046–1055. [Google Scholar]
- Sonneveld, T., K. R. Tobutt and T. P. Robbins, 2003. Allele specific PCR detection of sweet cherry SI (S) alleles S1 to S16 using consensus and allele specific primers. Theor. Appl. Genet. 107: 1059–1070. [DOI] [PubMed] [Google Scholar]
- Sonneveld, T., K. R. Tobutt, S. P. Vaughan and T. P. Robbins, 2005. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17: 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbachs, J. E., and K. E. Holsinger, 2002. S-RNase-mediated gametophytic self-incompatibility is ancestral in eudicots. Mol. Biol. Evol. 19: 825–829. [DOI] [PubMed] [Google Scholar]
- Stout, A. B., and C. Chandler, 1942. Hereditary transmission of induced tetraploidy in fertilization. Science 96: 257. [DOI] [PubMed] [Google Scholar]
- Tamura, M., K. Ushijima, H. Sassa, H. Hirano, R. Tao et al., 2000. Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor. Appl. Genet. 101: 344–349. [Google Scholar]
- Tao, R., H. Yamane, A. Sugiura, H. Murayama, H. Sassa et al., 1999. Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J. Am. Soc. Hortic. Sci. 124: 224–233. [Google Scholar]
- Thompson, K. F., and J. P. Taylor, 1966. Non-linear dominance relationships between S alleles. Heredity 21: 345–362. [Google Scholar]
- Tobutt, K. R., R. Boskovic, R. Cerovic, T. Sonneveld and D. Ruzic, 2004. Identification of incompatibility alleles in the tetraploid species sour cherry. Theor. Appl. Genet 108: 775–785. [DOI] [PubMed] [Google Scholar]
- Ushijima, K., H. Sassa, R. Tao, H. Yamane, A. M. Dandekar et al., 1998. Cloning and characterization of cDNAs encoding S-RNases from almond (Prunus dulcis): primary structural features and sequence diversity of the S-RNases in Rosaceae. Mol. Gen. Genet. 260: 261–268. [DOI] [PubMed] [Google Scholar]
- Ushijima, K., H. Sassa, A. M. Dandekar, T. M. Gradziel, R. Tao et al., 2003. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., H. Yamane, A. Watari, E. Kakehi, K. Ikeda et al., 2004. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 39: 573–586. [DOI] [PubMed] [Google Scholar]
- Wünsch, A., and J. I. Hormaza, 2004. Cloning and characterization of genomic DNA sequences of four self-incompatibility alleles in sweet cherry (Prunus avium L.). Theor. Appl. Genet. 108: 299–305. [DOI] [PubMed] [Google Scholar]
- Xue, Y., R. Carpenter, H. G. Dickinson and E. S. Coen, 1996. Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8: 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaegaki, H., T. Shimada, T. Moriguchi, H. Hayama, T. Haji et al., 2001. Molecular characterization of S-RNase genes and S-genotypes in the Japanese apricot (Prunus mume Sieb. et Zucc.). Sex. Plant Reprod. 13: 251–257. [Google Scholar]
- Yamane, H., R. Tao, A. Sugiura, N. R. Hauck and A. F. Iezzoni, 2001. Identification and characterization of S-RNases in tetraploid sour cherry (Prunus cerasus). J. Am. Soc. Hortic. Sci. 126: 661–667. [Google Scholar]
- Yamane, H., K. Ikeda, N. R. Hauck, A. F. Iezzoni and R. Tao, 2003. Self-incompatibility (S) locus region of the mutated S6-haplotype of sour cherry (Prunus cerasus) contains a functional pollen S allele and a non-functional pistil S allele. J. Exp. Bot. 54: 2431–2434. [DOI] [PubMed] [Google Scholar]