Abstract
Maize (Zea mays) pollen tubes grow in the styles at a rate of >1 μm/sec. We describe here a gene required to attain that striking rate. The aberrant pollen transmission 1 (apt1) gene of maize was identified by an Ac-tagged mutation that displayed a severe pollen transmission deficit in heterozygotes. Rare apt1 homozygotes can be recovered, aided by phenotypic selection for Ac homozygotes. Half of the pollen in heterozygotes and most of the pollen in homozygotes germinate short and twisted pollen tubes. The apt1 gene is 26 kb long, makes an 8.6-kb pollen-specific transcript spliced from 22 exons, and encodes a protein of 2607 amino acids. The APT1 protein is homologous to SABRE and KIP, Arabidopsis proteins of unknown function involved in the elongation of root cortex cells and pollen tubes, respectively. Subcellular localization analysis demonstrates that APT1 colocalizes with a Golgi protein marker in growing tobacco pollen tubes. We hypothesize that the APT1 protein is involved in membrane trafficking and is required for the high secretory demands of tip growth in pollen tubes. The apt1-m1(Ac) mutable allele is an excellent tool for selecting Ac transpositions because of the strong negative selection pressure operating against the parental Ac site.
IN higher plants, pollination is initiated with the transfer of pollen from the anther to the stigma. Once recognized by the stigma, the pollen hydrates and germinates a pollen tube. The pollen tube elongates along the transmitting tract in the style and delivers its two sperm cells to the female gametophyte to undergo double fertilization (McCormick 2004). Elongation of the pollen tube can be over a distance of a few hundred micrometers to several centimeters and constitutes the most rapid growth of any cell known (Hepler et al. 2001). This elongation is particularly remarkable in the polystigmatic styles of maize (Zea mays), where the pollen tube grows at the astounding rate of 1 cm/hr (Bedinger 1992). Evidence from a large collection of maize aneuploid stocks, in which development of the deficient microspores was slowed down or arrested, suggests that genes required for pollen development are expressed gametophytically and dispersed throughout the genome (Kindiger et al. 1991). By implication, mutations that affect male gametophytic development will be recovered rarely through the pollen. They can, however, be identified by the aberrant segregation of linked markers.
Several molecules that control pollen germination and the elongation and guidance of pollen tubes have been identified recently. Lipophilic molecules in the exine wall of the pollen (Lord 2000; Lord and Russell 2002), lipids (Preuss et al. 1993; Hulskamp et al. 1995a), pollen-coat proteins (Mayfield and Preuss 2000; Mayfield et al. 2001), and a calmodulin-binding protein (Golovkin and Reddy 2003) play important roles in pollen germination. Extracellular cues in the pistil and proteins produced by the embryo sac are likely involved in guiding the pollen tube toward the ovules and female gametophytes (Cheung et al. 1995; Hulskamp et al. 1995b; Ray et al. 1997; Shimizu and Okada 2000; Huck et al. 2003; Marton et al. 2005). Even the arrest of pollen tube growth by the embryo sac is under genetic control, as demonstrated by the feronia mutation in Arabidopsis, in which synergid degeneration fails to occur and pollen tubes continue to grow, leading to impaired fertility (Huck et al. 2003).
Substantial molecular, genetic, and cellular biological data have clearly demonstrated that pollen tube growth is tightly regulated by a Ca2+ gradient (Pierson et al. 1994; Holdaway-Clarke et al. 1997), a K+ channel of the Shaker family (Mouline et al. 2002), a Ca2+-dependent protein kinase (Estruch et al. 1994; Moutinho et al. 1998), an intact actin cytoskeleton (Chen et al. 2002, 2003), F-actin level (Fu et al. 2001), Rop GTPase (Li et al. 1999; Arthur et al. 2003), and Rab GTPase (Cheung et al. 2002). The protein products of two homologs, SABRE and the SABRE-like KIP (KINKY POLLEN), are required for the normal growth of Arabidopsis roots and pollen tubes, respectively (Aeschbacher et al. 1995; Procissi et al. 2003). SABRE affects root cortex cell expansion and appears to be expressed only in the sporophyte, whereas the kip mutation arrests the elongation of both pollen tubes and root hairs. The mechanism by which SABRE and KIP regulate cell expansion and tip growth, respectively, is not known.
We describe here a mutation with striking effects on pollen tube growth in maize. The mutation was uncovered by its aberrant pollen transmission in heterozygotes and named aberrant pollen transmission 1 (apt1) (Xu and Dooner 2003). apt1 is tagged by Activator (Ac), making it possible to use Ac's trans-acting phenotype, the spotted seed color produced by a reporter allele, to follow its pollen transmission in maize. We show that pollen tube development and growth rate are severely arrested in the apt1 mutant. The apt1 gene, a homolog of SABRE and KIP, is extremely large for a plant gene: it extends over 26 kb, contains 22 exons, and makes an 8.6-kb transcript. Data from fluorescent APT1 fusion proteins point to a possible Golgi localization, suggesting a role for APT1 in vesicular trafficking during pollen tube elongation.
MATERIALS AND METHODS
Plant genetic stocks:
All genetic stocks used in this study are in a W22 background. The apt1 mutant was recovered in family 929.20 from an Ac insertion library derived from the wx-m7(Ac) mutable allele (Cowperthwaite et al. 2002). The bz-m2(D1) stock, which carries a Ds element in the second exon of the bz gene, was used throughout as Ac reporter and as the wild-type sibling control for the apt1 mutant.
In vitro maize pollen germination:
Full, freshly dehisced anthers were collected from apt1 heterozygous, homozygous, and wild-type plants. The pollen germination medium is made up of 12% sucrose, 0.03% calcium chloride, 0.01% boric acid, 0.4% DMSO, 0.6% Seakem LE agarose (Cambrex Bio Science, Rockland, ME). Pollen was gently squeezed from the anther and evenly dusted on the surface of a plate with germination medium. Plates were incubated in the dark at 25°. Pollen tubes were photographed at serial time points of 15 min, 30 min, 1 hr, 2 hr, 3 hr, and 4 hr on a dissecting microscope using bright field illumination. Pictures were saved as TIFF files in Adobe Photoshop and opened by an NIH imaging program (http://rsb.info.nih.gov/ij/docs/intro.html). Pollen tube length was then measured by tracing along the length of the tube in the picture. Pollen tubes were numbered to compare growth rates of individual tubes at different time points. Measurements were taken for 158 pollen tubes from three wild-type plants and for 176 pollen tubes from two apt1 homozygous plants.
Chromosome mapping:
Recombinant inbred lines from a cross between T232 and CM37 were used to the map the genetic location of apt1 (Burr et al. 1988). Gels were blotted onto membranes and hybridized with a radiolabeled tac929.20 fragment (apt1 exons 14 and 15). Several restriction enzymes were tested on the parental genomic DNA and EcoRI gave the most distinct RFLPs. The segregation of RFLPs in recombinant inbred (RI) populations was scored and compared with the RI database from Brookhaven National Laboratory (http://www.maizegdb.org/cgi-bin/displayposrecord.cgi?id=41579).
Nucleic acid extraction and hybridization:
Genomic DNA from seedlings and mature leaves was prepared by a urea extraction procedure (Greene et al. 1994). DNA blots and RNA blots were performed as previously described (Cowperthwaite et al. 2002).
PCR, BAC screening, and sequencing:
PCR was performed according to the protocol of QiaTaq (QIAGEN, Valencia, CA). PCR products were cloned into pGEM-T Easy vector (Promega, Madison, WI) and transformed into DH5α competent cells. Plasmids were purified with a QIAGEN spin miniprep kit. DNA sequencing was carried out in an ABI 377 sequencer (Perkin-Elmer, Torrance, CA) following the manufacturer's instructions. The tac929.20 fragment was isolated by inverse (I)PCR, which was optimized for isolation of Ac tac sites in the Dooner laboratory (Cowperthwaite et al. 2002). Two pairs of primers, which were designated as Ac2578R and Ac4473 and Ac2494R and Ac4542, were used for IPCR and a subsequent nested PCR. The sequences of the four oligonucleotide primers were as follows: Ac2578R, 5′-GACAAACATACC-TGCGAGGATCAC-3′; Ac4473, 5′-TCCCGTTTTCGTTTCCGTC-3′; Ac2494R, 5′-ACGCATCCCTCAACATCAAATAGG-3′; and Ac4542, 5′-CGTTACCGACCGTTTTCATCCCTA-3′. BAC filters harboring CHORI MboI B73 BAC clones were screened with a tac929.20 probe and a series of positive clones were obtained. The B73 BAC clone ZMMBBc0301K11 was selected for sequencing and was confirmed to contain the central and 3′ part of the apt1 gene. Another B73 BAC clone, ZMMBBc0235P22, containing the 5′ and central part of the apt1 gene and overlapping with the previous BAC clone, was identified among the maize FPC BAC contigs (http://www.genome.arizona.edu/fpc/maize/WebAGCoL/WebFPC/). By sequencing subclones from these two BAC clones, a complete apt1 genomic sequence was obtained. The apt1 genomic sequence has been deposited in GenBank under accession no. DQ020097.
Amplification of apt1 cDNA:
A partial apt1 cDNA was initially amplified by RT–PCR. RT–PCR was performed according to the protocol of QiaTaq (QIAGEN, Valencia, CA). Total RNA extracted from W22 mature pollen was used as a RT–PCR template. Six overlapping pairs of primers were used to cover the full length of the cDNA: primers 5′-UTR-1 and apt 6423R, from the 5′-UTR to exon 8 (1.5 kb); primers apt 6218F and apt 14054R, from exon 8 to exon 9 (2.5 kb); primers xbaI-7F and xbaI-12R, from exon 9 to exon 10 (2.0 kb); primers xbaI-11F and apt 18109R, from exon 10 to exon 12 (1.0 kb); primers con7-5′ and apt 23888R, from exon 12 to exon 19 (1.0 kb); and primers con4-5′ and BACEcorI3′-1R, from exon 18 to exon 22 and the 3′-UTR (1.5 kb). The sequences of the 12 oligonucleotide primers are provided in supplemental Table 1 (http://www.genetics.org/supplemental/). RT–PCR products were cloned into pGEM-T Easy (Promega) and then sequenced on an ABI 377 sequencer (Perkin-Elmer). An apt1 full-length cDNA was constructed and fused to the GFP N terminus (see below). The apt1 transcript sequence has been deposited in GenBank under accession no. DQ020098.
GFP and DsRed fusion protein construction:
A pLAT52–GUS vector was kindly provided by Sheila McCormick. The fragment containing the LAT52 promoter was released by ApaI and NcoI digestion from pLAT52–GUS and then cloned into pGEM-T Easy (Promega). This pLAT52–GEM-T Easy vector contains an ATG translation start codon at the NcoI site. An alanine linker with KpnI and NcoI adapter sites was cloned into the multiple cloning site of a plant GFP expression vector, pFF–GFP, kindly provided by Gregorio Segal. The GFP–35s terminator segment of pFF–GFP was released by NcoI and EcoRI digestion and used to replace the GUS–NOS terminator segment of pLAT52–GUS, yielding pLAT52–GFP–35sT. The APT1 S4 fragment, which covers the apt1 transcript from site 7055 to 7533, was amplified with primers SacII 7055F and KpnI 7533R. The S4 fragment was digested with SacII and KpnI and cloned, together with the pFF–GFP fragment alanine linker–GFP–35s terminator (released by KpnI and EcoRI), into SacII–EcoRI double-digested LAT52–pGEM-T Easy in a three-way ligation. We call the resulting construct pLAT52–S4–Ala–GFP. This construct was used as the backbone for the following GFP fusion constructs: R1R6, R2, CHO1, CHO2, CHO3, CHO4, S2, S5, and N6, as described in the supplemental Materials and Methods (http://www.genetics.org/supplemental/).
The DsRed coding sequence was amplified from the pDsRed2–C1 vector (CLONTECH, Palo Alto, CA) by PCR with a high-fidelity polymerase rTth XL. A pair of SacII- and SphI-adapted primers, RedC1-5′SacII and RedC1-3′SphI, was used for the amplification. The DsRed fragment was digested with SacII and SphI and then cloned into the pLAT52–S4–Ala–GFP backbone by replacing the S4–Ala–GFP fragment. A pLAT52–DsRed–35sT vector was produced, with the multiple cloning sites at the C terminus of DsRed. The c1BK1, c1Bk2, and c1BK3 fusions to DsRed were constructed as described in supplemental Materials and Methods (http://www.genetics.org/supplemental/).
The GmManI coding region was amplified from the GmManI∷GFP fusion vector pBP30, kindly provided by Andreas Nebenführ. A pair of BglII- and KpnI-adapted primers, designated as Man-BglII5′F and Man-KpnI3′R, was used for this amplification. The amplified GmManI fragment was digested with BglII and KpnI and then cloned into the pLAT52–DsRed–35sT vector at the multiple cloning site, producing pLAT52–DsRed–GmManI. The sequences of the alanine linker primers are: top, 5′-CAAGCTTGCTGCCGCACCTGTCGCGGCAGCCGCTGCGGC-3′; bottom, 5′-CATGGCCGCAGCGGCTGCCGCGACAGGTGCGGCAGCAAGCTTGGTAC-3′. T4 PNKase-treated oligos were denatured and then annealed to form a short fragment for the cloning described above. Sequences of the other primers are provided in supplemental Table 1 (http://www.genetics.org/supplemental/).
Microprojectile bombardment of tobacco pollen:
The biolistic particle delivery system PDS-1000 (Bio-Rad Laboratories, Hercules, CA) was used for transient expression of GFP and DsRed marker in tobacco pollen. Gold particle preparation and bombardment procedure followed a previously reported protocol in Z. Yang's laboratory (Fu et al. 2001). Colocalization analysis of GFP and DsRed markers was accomplished with mixing both GFP and DsRed constructs in one bombardment shooting.
RESULTS
The apt1 mutation is defective in Ac pollen transmission:
Ac segregates aberrantly in the self progeny of plants carrying Ac929.20, one of a collection of transposed Ac (trAc) elements derived from wx-m7(Ac) (Cowperthwaite et al. 2002). The bz mutable allele bz-m2(D1), which harbors a Dissociation (Ds) element in the bz gene, was used as a reporter of Ac activity. In the presence of Ac, this reporter produces an unstable aleurone color phenotype consisting of purple spots on a bronze background (bz-m); in its absence, it produces a solid bronze phenotype (bz). Because Ac generally has a negative dosage effect, the degree of spotting is inversely correlated with the dosage of Ac in the genome. One dose of Ac produces a coarse spotting pattern (bz-mC) and additional doses lead to increasingly finer spotting. Thus, it is possible to score the activity and dosage of Ac in each individual kernel of a segregating ear. In the self progeny of transposant 929.20 the segregation ratio of Ac to no Ac was 1:1, instead of 3:1, and very fine spotted kernels, indicative of Ac homozygotes, were rare. Yet, the mutant ear had a normal seed set, suggesting that the trAc had caused a mutation defective in pollen, but not egg, viability. The genetic analysis described below confirmed that the mutation had been caused by an Ac insertion, so we designated the affected gene apt1 for aberrant pollen transmission 1 and the new mutant allele apt1-m1(Ac).
Table 1 shows the analysis of Ac transmission in apt1-m1(Ac)/+ heterozygotes. Self progenies of apt1-m1(Ac)/+; bz-m2(D1) segregated 353 bz-m:364 bz, which means that the ratio of kernels with and without Ac was 1:1, instead of the expected 3:1. Testcross progenies of apt1-m1(Ac)/+; bz-m2(D1) as the maternal parent produced 1295 bz-m and 1277 bz, i.e., the expected 1:1 ratio of kernels with and without Ac. Therefore, the female transmission of Ac is normal. The reciprocal cross using apt1-m1(Ac)/+; bz-m2(D1) as the paternal parent produced a total of 305 bz-m and 3797 bz, i.e., a 1:12 ratio of kernels with and without Ac. Therefore, the male transmission of Ac is defective. Representative ears from these reciprocal crosses are shown in Figure 1: In the ear from the cross using the apt1-m1(Ac)/+ parent as female and the bz-m2(D1) reporter as male (Figure 1A), half of the kernels are spotted, as expected, whereas in the ear from the reciprocal cross (Figure 1B), very few kernels are spotted, indicating that Ac is transmitted poorly by the pollen parent.
TABLE 1.
Ac transmission analysis in apt1(Ac)/+ heterozygotes
| Seed phenotypesa
|
||||
|---|---|---|---|---|
| Type of mating | Genotype of parents in mating | bz-m | bz | Ac:no Ac ratio |
| Self | apt1(Ac)/+; bz-m2(D1) × same | 353 | 364 | 1:1 |
| Ac as female | apt1(Ac)/+; bz-m2(D1) × + ; bz-m2(D1) | 1295 | 1277 | 1:1 |
| Ac as male | + ; bz-m2(D1) × apt1(Ac)/+; bz-m2(D1) | 305 | 3797 | 1:12 |
bz-m, spotted bronze kernel phenotype resulting from the presence of Ac in the genome; bz, uniformly bronze kernel phenotype resulting from the absence of Ac.
Figure 1.
Ears from reciprocal crosses between apt1-m1(Ac)/+; bz-m2(D1) × +; bz-m2(D1) individuals, showing Ac transmission in the female and male gametophytes. (A) An ear using the apt1-m1(Ac)/+ parent as female. The bz-m and bz kernel phenotypes segregate in a 1:1 ratio, indicating that Ac transmission by the female gametophyte is normal. (B) An ear of the reciprocal cross using the apt1-m1(Ac)/+ parent as male. The bz-m and bz kernel phenotypes segregate in a 1:12 ratio, indicating that Ac transmission by the male gametophyte is defective.
The apt1 mutation is caused by an Ac insertion:
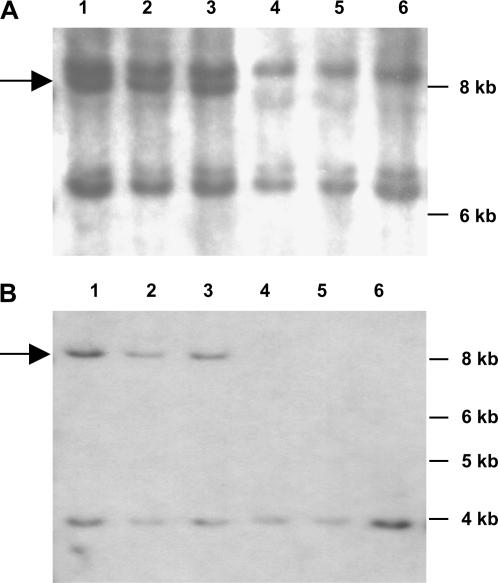
To verify that apt1 cosegregated with Ac, DNA blots of spotted and nonspotted segregants from the testcross progeny of the apt1-m1(Ac)/+ female parents were hybridized with the central part of the Ac element. Figure 2 shows the result of hybridizing a blot containing SacI-digested DNA from representative individuals. A unique band, indicated with an arrow, is present only in DNA from spotted seeds (Figure 2A). A similar result was obtained with EcoRI-digested DNA (not shown). The unique band was present in the 60 spotted segregants and absent from the 20 nonspotted segregants analyzed. These results confirm that apt1 is linked with an Ac-hybridizing band. The fragment adjacent to the Ac insertion site was isolated by IPCR, using as a template EcoRI-digested, size-fractionated DNA from apt1-m1(Ac)/+ heterozygotes and primers based on Ac (Cowperthwaite et al. 2002). The amplified products contained a sequence with a perfect match to the 3′ end of Ac plus 1.2 kb of putative tac929.20 sequence. The original DNA blots were rehybridized with the putative tac929.20 site and the results for the SacI digest are shown in Figure 2B. The apt1-m1(Ac)/+ heterozygous plants are also heterozygous for a band that is 4.6 kb larger than the single band found in wild type, thus validating the tac929.20 site and showing that it is present in single copy in the maize genome. The tac929.20 site was mapped with RI lines (Burr et al. 1988) to position 78 in the long arm of chromosome 9 (bin 9.04), 23 cM away from wx. The RI mapping result is consistent with the placement of Ac to a location linked to wx in the F2 progeny of a Wx apt1-m1(Ac)/ wx + heterozygote (Cowperthwaite et al. 2002).
Figure 2.
DNA blot showing that the apt1 mutation cosegregates with Ac. Genomic DNA from spotted (lanes 1–3) and nonspotted (lanes 4–6) testcross segregants was digested with SacI. (A) Filter hybridized with an Ac internal probe. (B) The same filter hybridized with a tac929.20 probe. The arrow shows the unique band common to both panels.
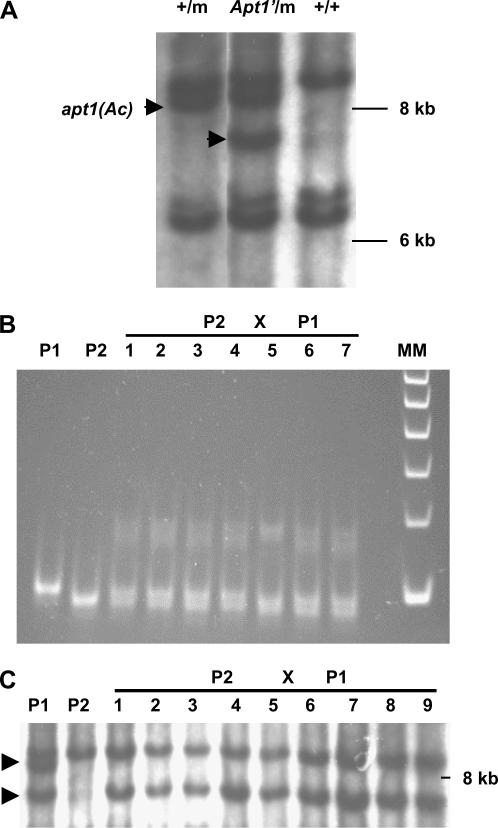
To confirm that apt1 was tagged by Ac, the spotted seed progeny of apt1-m1(Ac)/+ heterozygotes was screened for restoration of apt1 gene function following transposition of Ac to a new location in the genome. Apt1′ revertants were sought first without selection among the spotted F2 progeny of an apt1-m1(Ac)/+ heterozygote. Of 36 individuals tested for their ability to transmit Ac as male parents, 35 resembled the apt1-m1(Ac)/+ parent in transmitting Ac poorly, but one transmitted Ac normally. DNA blot analysis of this putative revertant showed that it had two Acs, one at the original apt1-m1(Ac) location and another one at a new location (Figure 3A). Subsequent genetic analysis revealed that this individual carried a trAc 6 cM away from a revertant Apt1′ allele containing a typical 8-bp footprint (data not shown). As is discussed later, the extra 8 bases do not cause a mutant phenotype because Ac is inserted in an intron of the apt1 gene. The presence of the 8-bp footprint allows one to distinguish the Apt1′ revertant allele from the wild-type allele. Primers flanking the Ac insertion site were used to analyze the genotypes of the progeny from a cross between a wild-type female and the Apt1′/apt1-m1(Ac), trAc/+ heterozygous revertant as male. PCR with these primers will amplify the wild-type and Apt1′ revertant alleles, but not the apt1-m1(Ac) mutant allele. Figure 3B shows that the revertant (lane P1) produces a larger band than wild type (lane P2). The progeny of the cross using the revertant as male are all heterozygous for an Apt1′ revertant allele and a wild-type allele. So, loss of Ac from the apt1-m1(Ac) allele appears to restore normal male transmission. DNA blot analysis (Figure 3C) showed that all the bz-m (i.e., Ac-carrying) progeny of the cross had the trAc band, but not the apt1-m1(Ac) band. Therefore, the trAc, which is closely linked to the Apt1′ revertant allele, is male transmitted, but the Ac in the apt1-m1(Ac) mutant allele is not. From the above reversion analysis we conclude that an Ac insertion caused the apt1 mutation.
Figure 3.
Ac excision restores the pollen transmission of Apt1′ revertant alleles. (A) DNA blot analysis of an Apt1′ revertant. The revertant has two Acs (indicated with arrowheads): One is the Ac element in apt1, and the second one is a trAc. Genomic DNA was digested with SacI and hybridized with an Ac internal probe. Lanes: +/m, +/apt1-m1(Ac) heterozygote; Apt1′/m, Apt′/apt1-m1(Ac) heterozygous revertant; +/+, wild type. (B) Pollen transmission of the Apt1′ excision allele. Wild-type and revertant alleles were PCR amplified using apt1 primers flanking the Ac insertion site and the PCR products were separated in an 8% PAGE gel. P1, Apt1′/apt1-m1(Ac) heterozygous revertant parent (the mutant allele fails to amplify because of the 4.6-kb Ac insertion); P2, wild type; lanes 1–7, randomly selected progenies from the cross between a P2 female and a P1 male (all are heterozygous for the fragments amplified in the parents); MM, 100-bp DNA ladder. The band in P1 is 8 bp larger than that in P2, as determined by sequencing. (C) DNA blot analysis of the pollen transmission of the two Acs present in the Apt1′ revertant. Genomic DNA was digested with SacI and the blot was hybridized with an Ac probe. The Ac in apt1 (top band) is not male transmitted, but the trAc (bottom band) is. P1, apt1-m1(Ac)/Apt1′ +/trAc double heterozygote; P2, wild type; lanes 1–9, bz-m progenies of the cross between a P2 female and a P1 male.
The apt1-m1(Ac) allele can be occasionally transmitted by the pollen:
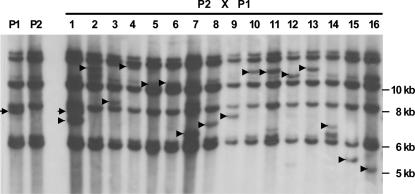
Given the selective disadvantage of apt1-m1(Ac) pollen in apt1-m1(Ac)/+ heterozygotes, it is likely that many of the Ac progeny of an apt1-m1(Ac)/+ pollen parent will carry a trAc element rather than the one originally found at the apt1 locus. To test this prediction, 16 bz-m progeny seeds of the cross between a +; bz-m2(D1) tester female and an apt1-m1(Ac) /+; bz-m2(D1) heterozygous male were analyzed. Figure 4 shows the result of hybridizing a blot containing SacI-digested DNA with an Ac probe. An apt1-m1(Ac) unique band (confirmed with the tac929.20 probe, data not shown) is present in the male parent (lane P1). All 16 bz-m progeny carry a trAc band (marked by an arrowhead), but only the individual in lane 1 carries a parental apt1-m1(Ac) band. Therefore, only 1 of 16 Ac selections (∼6%) carries the apt1-m1(Ac) allele. The other selected individuals carry trAcs. An additional 120 bz-m selections from the same cross were planted, and the selections were either selfed or crossed as males to wild type. All of these progenies showed normal transmission of Ac, indicating that Ac had transposed from apt1. We conclude that apt1-m1(Ac) is only rarely male transmitted. On the basis of the Ac male transmission data in Table 1 and the results of the above DNA blot analysis, the male transmission ratio of the apt1-m1(Ac) chromosome relative to its normal homolog is estimated to be ∼5:1000 [(305/3797) × (1/16)].
Figure 4.
DNA blot analysis of pollen-transmitted Ac elements from apt1-m1(Ac)/+. Genomic DNA from apt1-m1(Ac)/+ heterozygotes (lane P1), wild type (lane P2), and randomly selected spotted seed (i.e., carrying Ac) of the cross between a P2 female and a P1 male (lanes 1–16) was digested with SacI and hybridized with an Ac internal probe. The arrowhead in each lane indicates a unique trAc band; the full arrow indicates the parental apt1-m1(Ac) band.
apt1 homozygous mutants exhibit short and twisted pollen tubes and strong selection for reversion events:
Because apt1-m1(Ac) can be occasionally male transmitted, we set out to recover apt1-m1(Ac) homozygotes, which would enable us to examine the sporophytic effects of the mutation. From self matings of 150 apt1-m1(Ac)/+; bz-m2(D1) plants, 120 fine-spotted (bz-mF) seeds were obtained among 22,500 seeds. These bz-mF seeds should carry more than one copy of Ac and are, therefore, homozygous apt1-m1(Ac) candidates. Seventy-eight of these 120 bz-mF individuals were genotyped by DNA blotting and 4 of them turned out to be homozygous apt1-m1(Ac) mutants.
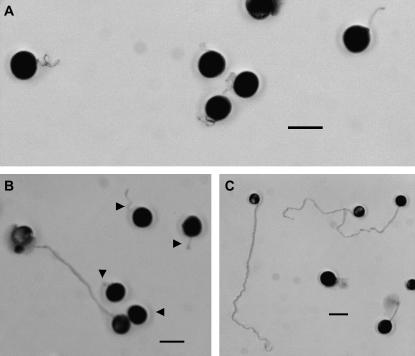
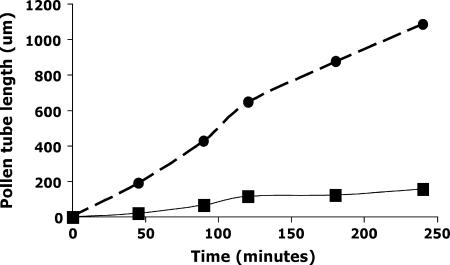
Mature pollen from apt1-m1(Ac) homozygous, heterozygous, and wild-type plants was collected and germinated in vitro. Compared with wild type, the apt1-m1(Ac) homozygous mutant shows mostly short and twisted pollen tubes (Figure 5, A and C). Sporadically, however, the mutant also shows some long and normal pollen tubes, which are most likely Apt1′ revertants arising from Ac excision (see previous section). For example, in the plating of pollen from one anther, 4 out of 77 germinated pollen tubes appeared normal. This is a manifestation of the mutable phenotype of the apt1-m1(Ac) homozygous mutant. The heterozygous apt1-m1(Ac)/+ plants show a 1:1 segregation of wild-type and mutant pollen tube phenotypes (Figure 5B). The gross appearance of mature pollen grains was examined with a portable 10× field microscope, used routinely to score pollen abortion in deficiency and translocation heterozygotes, but no difference could be seen among the pollen grains of an apt1-m1(Ac)/+ heterozygous plant. The growth rates of wild-type and mutant pollen grains were compared by in vitro germination tests. As shown in Figure 6, wild-type pollen tubes grow dramatically faster than mutant pollen tubes (255 μm/hr vs. 34 μm/hr), suggesting that the mutant pollen tube's competitive ability in heterozygotes is severely compromised.
Figure 5.
Phenotypes of pollen tubes germinated in vitro. (A) Pollen from an apt1 homozygous mutant grown for 4 hr. (B) Pollen from an apt1/+ heterozygote grown for 4 hr. Arrowheads show the pollen tubes originating from mutant pollen grains. Compared to wild type, the apt1 pollen tubes are short and twisted. (C) Wild-type pollen grown for 4 hr. Some wild-type pollen tubes have burst over this time period. Bars, 150 μm.
Figure 6.
Growth rate comparison of wild type (•) and apt1 mutant (▪) pollen tubes germinated in vitro. See materials and methods for experimental details.
Analysis of the self progenies of apt1-m1(Ac) homozygous mutants reveals a strong selection for Ac excision products. In one self-pollinated ear from an apt1-m1(Ac) bz-m2(D1) homozygote, roughly equal numbers of bz-mF and bz-mC seeds were obtained, rather than the bz-mF majority expected from an Ac homozygote. The coarse spotted phenotype suggested that the bz-mC progeny were Ac heterozygotes and, in fact, all 23 individuals tested showed a heterozygous, i.e., revertant, pollen tube phenotype in in vitro germination tests (1:1 ratio of long and short pollen tubes). Twenty-four bz-mF progenies were genotyped by DNA blots, using an apt1 probe: 22 of them were Apt′/apt1-m1(Ac) heterozygous revertants and only 2 were apt1-m1(Ac) homozygotes (data not shown). The latter 2 progenies showed a homozygous mutant pollen tube phenotype and, again, evidence for strong selection of male gametes carrying Ac excisions. Maintaining the homozygous apt1-m1(Ac) mutant stock is not easy. Homozygous apt1-m1(Ac) individuals must first be screened on the basis of Ac's dosage effect on the spotted-seed phenotype and, then, genotyped by DNA blots. Only a small fraction of apt1-m1(Ac) homozygotes are obtained. The strong tendency of the apt1-m1(Ac) mutant to revert provides further confirmation that the mutation is Ac tagged and indicates that the revertant pollen easily outcompetes the mutant pollen in the styles, thus preferentially transmitting its sperms to the next generation.
The apt1 mutation does not appear to affect the sporophyte. The aerial parts of homozygous apt1-m1(Ac) plants show no difference from wild type, even in fast tip-growing tissues like the silks. We did not see differences between the roots of mutant and wild-type seedlings, but did not attempt to examine the roots of mature plants after the homozygotes had been transferred to soil.
The protein encoded by apt1 is homologous to the SABRE and KIP proteins in Arabidopsis:
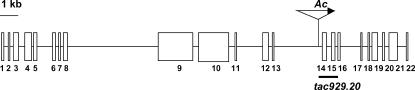
As reported earlier (Cowperthwaite et al. 2002), the amino acid sequence encoded by the tac929.20 site shows homology to SABRE in Arabidopsis, which has been annotated as a very large protein (∼2600 aa). Because isolation of the complete progenitor wild-type allele of the apt1-m1(Ac) mutation from the W22 inbred would have been difficult and time consuming, we decided to take advantage of a maize public B73 BAC library to clone the Apt1-B73 allele, instead. Two overlapping BACs were identified that contained the entire apt1 gene. First, BAC clone ZMMBBc0301K11 was isolated using a tac929.20 probe and, then, the contiguous BAC ZMMBBc0235P22 clone was identified from the maize FPC map (http://www.genome.arizona.edu/fpc/maize/WebAGCoL/WebFPC/). Subclones covering 31 kb across these two BACs, and expected to contain the entire apt1 gene, were made and sequenced. As anticipated, the apt1 gene is very large, extending over 26 kb from one end to the other. A comparison of the sequences of apt1 and tac929.20 with those in the GenBank database revealed that Ac had inserted in a gene encoding a protein highly similar to SABRE and KIP from Arabidopsis (Aeschbacher et al. 1995; Procissi et al. 2003) and to related, but not annotated, proteins from rice. Figure 7 diagrams the structure of the apt1 gene and the site of insertion of Ac in the mutant. The apt1 gene has 22 predicted exons and is bordered by a retrotransposon sequence ∼1.5 kb upstream of the putative start codon and by another gene 2 kb downstream of the putative stop codon (not shown).
Figure 7.
Structure of the apt1 gene. A diagram of the apt1 genomic sequence is shown. The boxes stand for exons and the intervening lines for introns. In the mutable allele apt1-m1, Ac is inserted in intron 13 in the same transcriptional orientation as apt1 (arrow). The location of the tac929.20 probe (Cowperthwaite et al. 2002) is indicated with a bar.
The apt1 cDNA was amplified in overlapping segments by RT–PCR, using total RNA of W22 mature pollen as template. A full-length cDNA sequence was then constructed from the overlapping RT–PCR amplification products and verified by sequencing. The mature transcript measures 8.6 kb and is made up of 22 exons. Its coding region is 7821 nucleotides long and encodes a 2607-amino-acid protein that is homologous to the SABRE and KIP proteins from Arabidopsis and to two related proteins in rice (supplemental Figure 1 at http://www.genetics.org/supplemental/). The five proteins are very conserved throughout their lengths and share homology with other unknown yeast and animal proteins in the central and C-terminal segments. The C terminus of these proteins is the most highly conserved and may define a novel class of proteins, possibly targeted to the Golgi (see below).
The apt1 gene is expressed in pollen only:
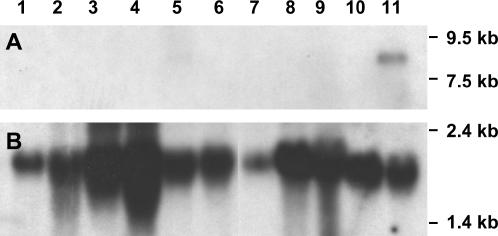
To characterize the expression pattern of apt1, total RNA from different wild-type tissues was analyzed by RNA blots, using tac 929.20 as probe (tac929.20 carries exons 14 and 15; see Figure 7). The apt1 gene appears to be expressed only in mature pollen (Figure 8A, lane 11; the weak signal detected in the mature tassel sample in lane 5 most likely originates from the pollen). On the basis of the RNA markers in the blot, the transcript size is estimated to be ∼8 kb, in agreement with the length of the apt1 cDNA. No signal could be detected in root, stem, leaf, immature tassel, immature ear, endosperm, or silk, even though the RNA from some of these tissues was overloaded relative to the pollen (Figure 8B).
Figure 8.
RNA blot analysis of apt1 expression in various tissues. (A) Blot hybridized with the tac929.20 probe. (B) Same filter as in A rehybridized with an F-actin probe after removing the first probe. Lane 1, mature root; lane 2, stem; lane 3, young leaf; lane 4, mature leaf; lane 5, mature tassel; lane 6, immature tassel; lane 7, mature ear; lane 8, 14-day-old endosperm; lane 9, 35-day-old endosperm; lane 10, unpollinated silk; lane 11, mature pollen.
The APT1 protein colocalizes with Golgi-targeted proteins in tobacco pollen tubes:
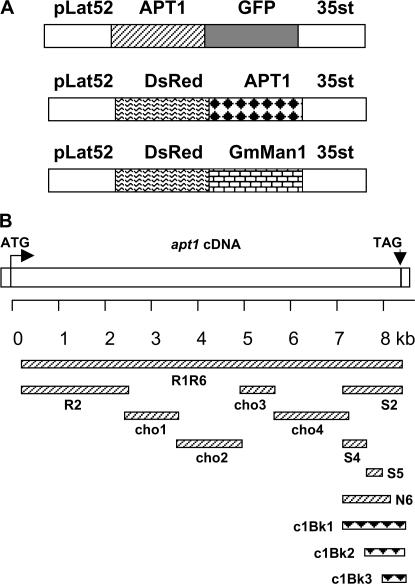
To investigate the subcellular localization of the APT1 protein and, thus, obtain some indication of its possible function, several translational fusions of APT1 to a fluorescent protein were transformed into tobacco pollen tubes by particle bombardment. All the constructs are driven by the tomato LAT52 pollen promoter (Twell et al. 1989) and carry either the green fluorescent protein GFP or the red fluorescent protein DsRed (Figure 9A). Figure 9B shows a schematic diagram of the different parts of the APT1 protein that were fused to the fluorescent protein markers. Details of the constructs are provided in materials and methods.
Figure 9.
Diagram of constructs used in subcellular localization experiments. (A) General organization of the fusion constructs pLAT52–APT1–GFP (top), pLAT52–DsRed–APT1 (middle), and pLat52–DsRed–GmManI (bottom). pLAT52 is a tomato promoter expressed exclusively in the anthers. 35sT indicates a fragment containing the polyadenylation signal from the cauliflower mosaic virus 35S terminator. (B) Specific APT1–GFP and DsRed–APT1 fusion constructs. The kilobase scale refers to locations in the apt1 cDNA. APT1–GFP fusions, upward diagonal pattern; DsRed–APT1 fusions, solid triangle pattern.
GFP was fused to the C terminus of either the entire APT1 protein (R1R6) or a part thereof (R2, CHO1, CHO2, CHO3, CHO4, S2, S4, S5, and N6). In control experiments with GFP alone, GFP was usually distributed throughout the whole pollen tube (data not shown). In the bombardment experiments with APT1–GFP fusion constructs, R2, CHO1, and S4 produced diffusible signals like the GFP control. S2, S5, and N6 generated targeted signals in growing tobacco pollen tubes (Figure 10, A–C, and data not shown). The rest failed to produce a GFP image, possibly because of degradation of the fusion protein. To confirm the protein targeting of N6, S2, and S5, another type of construct was made, in which the N6 region was either extended or shortened and fused with DsRed at the N terminus (Figure 9B: c1Bk1–c1Bk3). In pollen bombardment experiments, the c1Bk1 and c1Bk2 constructs produced clear DsRed signals with the same distribution as the GFP signal of N6, S2, and S5 (data not shown). The c1Bk1 construct is the DsRed counterpart of S2, whereas the c1Bk2 construct shares the right end of c1Bk1, but is shorter than N6 at the left (N) end. Taken together, these results indicate that the C terminus of the APT1 protein contains signals for subcellular targeting. Specifically, the results with the S5 construct show that the region can be narrowed down to a 108-amino-acid stretch in the C terminus.
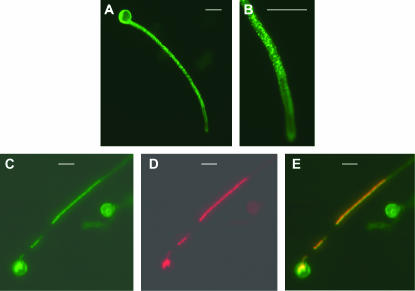
Figure 10.
Localization of the APT1(N6)∷GFP fusion protein in tobacco pollen tubes. Tobacco pollen was cobombarded with the APT1(N6)∷GFP and the DsRed∷GmManI constructs. (A) Pollen tube showing a punctate pattern of GFP signals. (B) Enlargement (2×) of the tip of the pollen tube shown in A. (C–E) Images of a different pollen tube. (C) GFP signal; (D) DsRed signal from the same pollen tube as in C; (E) merged images of C and D. Bars, 30 μm.
The distribution of the green fluorescent signal in germinating pollen tubes (Figure 10, A and B) resembled the distribution of proteins, such as Rab2 GTPase, which have been shown to localize to Golgi bodies in elongating pollen tubes (Cheung et al. 2002). To test whether APT1 also localized to Golgi bodies, a construct was made with the Golgi-targeted GmManI α-1,2 mannosidase I protein (Nebenfuhr et al. 1999) fused to DsRed:LAT52–DsRed–GmManI (Figure 9A). Cobombardment of pollen tubes with LAT52–APT1(N6)–GFP and LAT52–DsRed–GmManI showed that APT1(N6) and GmManI are expressed together in the same pollen tube (Figure 10, C and D) and the merged images clearly establish that APT1(N6) and GmManI colocalize to the same organelle (Figure 10E).
The apt1-m1(Ac) mutation is a powerful tool for selecting Ac transpositions:
As reported above, the male transmission ratio of Ac to wild type in apt1-m1(Ac)/+ heterozygotes is 1:12. All 16 Ac progeny from an apt1-m1(Ac)/+ pollen parent had a trAc element (Figure 4). Therefore, the frequency of Ac progeny that carry a trAc element is much higher in apt1-m1(Ac) than the 1–3% frequency seen in most mutable alleles. The position of the trAcs relative to the apt1-m1(Ac) donor locus was characterized as follows. First, 57 plants carrying trAc elements of independent origin [bz-m2(D1); trAc/+] were crossed as males to bz-m2(D1); apt1-m1(Ac)/+ females. From each of the above crosses, 5 plants derived from bz-mF seeds, which are putative apt1-m1(Ac)/+ heterozygotes carrying either a linked or an unlinked trAc, were pollinated with a bz-m2(D1) tester. This testcross generated a segregating population of bz-mF, bz-mC, and bz seed. The genetic distance between the trAc and the apt1-m1(Ac) donor locus was estimated on the basis of the frequency of bz-mF seeds, which should carry two copies of Ac and should arise mostly by recombination between the trAc and the apt1-m1(Ac) allele. As with other loci, about half of the trAcs were found to be linked to the donor locus (supplemental Figure 2 at http://www.genetics.org/supplemental/).
DISCUSSION
The apt1 mutation is defective in Ac male transmission and pollen tube elongation:
In this study, we took advantage of two well-known properties of the maize transposon Ac to identify and characterize apt1, a mutation affecting male gametophytic development. An Ac trans-acting phenotype—spotted seed color—was used to follow Ac pollen transmission and the negative dosage effect of Ac on spot size and number was used to screen for homozygous mutant plants. The Ac-linked apt1 mutation was discovered by the aberrant pollen transmission of Ac in apt1-m1(Ac)/+ heterozygotes. The transmission ratio of Ac and non-Ac gametes by the pollen is 5:1000, so Ac is rarely male transmitted. Heterozygous apt1 mutant plants show no obvious phenotype other than the defective pollen tube elongation of half of their pollen grains. Rare homozygous apt1 mutant plants, also apparently normal, were recovered by screening the self progeny of apt1/+ heterozygotes on the basis of Ac's negative dosage effect and DNA blot genotyping. The apt1 mutant pollen germinates short and twisted tubes that elongate very slowly. Gametes carrying Ac excisions are recovered frequently from apt1 homozygotes because the revertant pollen tubes elongate much faster than the mutant ones and outcompete them to the embryo sacs.
APT1 is a homolog of the SABRE and KIP proteins required for cell elongation:
The apt1 gene was sequenced from two overlapping BACs from the inbred B73. At 26 kb, apt1 is one of the longest plant genes known. A full-length apt1 cDNA was cloned from maize mature pollen. The apt1 transcript measures 8.6 kb and is made up of 22 exons, which encode a predicted protein of 2607 aa and 292 kDa. apt1 expression occurs only in mature pollen and depends on the genotype of the gametophyte, making the apt1 promoter an excellent candidate for a maize pollen-specific promoter.
The APT1 protein is homologous across its entire length to SABRE and KIP, two novel proteins involved in the elongation of root cortex cells and pollen tubes in Arabidopsis. Hereafter, we refer to this plant gene family as SAK (for SABRE, APT1, KIP). T-BLASTN analysis of the rice sequences in GenBank using APT1 as query disclosed two high-scoring, but misannotated, homologs in chromosome 3 (AP008209.1). The exon–intron junctions of the rice SAK genes were determined with the Softberry sequence analysis program (http://www.softberry.com/berry.phtml), aided by the maize apt1 full-length cDNA sequence. The rice homologs are highly similar to SABRE, APT1, and KIP (supplemental Figure 1): One of them is closer to APT1/KIP and the other one to SABRE. Maize is likely to have at least one other SAK homolog because BLAST searches of the maize Genome Survey Sequence database uncovered related sequences with 80% identity to apt1. Thus, monocots and dicots appear to have two closely related SAK family members, but the extent of conservation of function is unclear at this time as, unlike KIP, apt1 is not expressed in roots. APT1 homologs were also detected among pollen or floral RNAs from maize, rice, sorghum, and sugar cane by T-BLASTN translation of the GenBank EST database. Therefore, APT1 is a member of a conserved protein family most likely required for cell elongation in higher plants.
Curiously, different members of the SAK family differ slightly in their intron–exon structure. SABRE and KIP have 23 exons and completely conserved intron locations relative to each other, whereas apt1 has 1 exon less. Exon 19 in apt1 is split into two in SABRE and KIP by an 87-bp intron, which, although defined experimentally, neither removes a stop codon in the spliced sequence nor shifts the reading frame. The two SAK genes in rice appear to have the same genomic organization as apt1. A caveat in the preceding discussion is that, except for apt1, the complete exon–intron structure has not been determined experimentally for any SAK gene.
APT1 appears to be a Golgi-localized protein required for pollen tube elongation:
The pollen tube elongates by rapid apical extension (Franklin-Tong 1999). A highly active inverse fountain cytoplasmic streaming leads to a large accumulation of Golgi-derived vesicles in what is known as the “clear zone” at the extreme tip of the growing pollen tube. There, membrane and cell wall components and secretory proteins are produced to support the pollen tube's rapid elongation (Hepler et al. 2001). Recent pharmacological and genetic data point to a major role of the Golgi body in this elongation. The importance of an intact Golgi network for pollen tube elongation was first demonstrated with the organelle movement inhibitor brefeldin A. Treatment of pollen tubes with this drug alters cytoplasmic streaming, causes the disappearance of Golgi bodies, and arrests pollen tube growth (Rutten and Knuiman 1993; Cheung et al. 2002; Parton et al. 2003). Recent genetic data confirm the importance of the Golgi for pollen tube growth. Mutations in genes encoding Golgi-localized proteins known to regulate vesicle trafficking in animal and yeast cells, such as a tobacco Rab2 GTPase (Cheung et al. 2002) and an Arabidopsis homolog of yeast Vps52p (Lobstein et al. 2004), are impaired in pollen tube growth. However, the mechanisms by which these proteins regulate pollen tube elongation are not yet understood.
In this study, we have shown that the APT1 protein, which is required for normal pollen tube elongation in maize, may also be targeted to the Golgi body. Different parts of APT1 were fused to GFP or DsRed and bombarded into tobacco pollen. We found that, although the GFP fusion construct containing the entire APT1 was not expressed, possibly because of the unusual length of the APT1 protein, all the constructs containing the carboxy terminus of APT1 produced targeted signals in germinating pollen tubes. A fusion protein of GFP with the APT1 C terminus (residues 2351–2458) colocalized with the Golgi body marker GmManI, indicating that that part of the protein contains a Golgi localization signal. Interestingly, the same part of APT1 shows conservation with animal and yeast proteins of unknown function. We hypothesize that APT1 is a Golgi-localized protein that most likely regulates membrane trafficking and is required for rapid vesicle accumulation at the tip of the pollen tube. Since APT1, KIP, and SABRE are members of a novel protein family and appear to have similar effects on cell elongation, study of the subcellular localization of other SAK proteins could provide clues to their possible mode of action.
The apt1 mutation constitutes a useful transposition selection tool:
We demonstrate here that the apt1-m1(Ac) mutation is a very efficient system for selecting Ac transpositions in maize. Utilizing the apt1 mutation as male, a large number of Ac transpositions can be recovered in one generation because each ear yields an average of ∼10 trAcs. Among the transpositions selected, approximately equal numbers are linked and unlinked to the donor apt1 locus, similar to what has been observed at other loci (Greenblatt 1984; Dooner and Belachew 1989). In the original apt1-m1(Ac) mutation, Ac is inserted in intron 13. In apt1-m2(Ac), a second apt1(Ac) mutant allele isolated recently, Ac is inserted in intron 8 (data not shown). This allele shows the same Ac male transmission defect as apt1-m1(Ac). Because in these two mutants Ac is inserted in an intron, where excision footprints tend to be tolerated, most Ac excisions restore gene function and the standard distribution of linked and unlinked trAcs is not altered. Therefore, apt1-m1(Ac) and apt1-m2(Ac) are particularly useful for selecting Ac transpositions to closely linked sites. Recovery of an intragenic transposition of Ac into an apt1 exon would increase the versatility of the apt1 mutation as a paternal donor of trAcs because it would provide a tool for selecting Ac transpositions to unlinked sites. Most Ac excisions from such an allele would not restore gene function and would be subject to the same negative selection as the parental apt1 mutant allele. Therefore, Acs that transpose to unlinked sites and cosegregate with the wild-type Apt1 allele would be recovered preferentially, the probability of recovery of a trAc being inversely proportional to its linkage to the apt1 mutation.
Acknowledgments
We thank Gregorio Segal for his help and advice with the biolistics experiments, Jun Huang and Gregorio Segal for assistance with sequencing, Wonkeun Park for RNA samples, Joachim Messing for the use of the B73 BAC filters, Xuemei Chen for the use of microscope facilities, Junjie Li for help with photography, and members of the Dooner lab for comments on the manuscript. We also acknowledge the following people for providing constructs: Sheila McCormick, for pLAT52–GUS; Andreas Nebenführ, for GmManI–GFP; Gregorio Segal, for pFF–GFP; and Howard Chang, for pDsRed2–C1. Z.X. acknowledges Waksman and Busch predoctoral fellowships from Rutgers University. This work is from a thesis submitted by Z.X. to the Rutgers University Graduate School in partial fulfillment of the requirements for a Ph.D. The apt1 mutant was isolated in a National Science Foundation Plant Genome Program project supported by grant DBI-9813364 to H.K.D.
References
- Aeschbacher, R. A., M. T. Hauser, K. A. Feldmann and P. N. Benfey, 1995. The SABRE gene is required for normal cell expansion in Arabidopsis. Genes Dev. 9: 330–340. [DOI] [PubMed] [Google Scholar]
- Arthur, K. M., Z. Vejlupkova, R. B. Meeley and J. E. Fowler, 2003. Maize ROP2 GTPase provides a competitive advantage to the male gametophyte. Genetics 165: 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger, P., 1992. The remarkable biology of pollen. Plant Cell 4: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr, B., F. A. Burr, K. H. Thompson, M. C. Albertson and C. W. Stuber, 1988. Gene mapping with recombinant inbreds in maize. Genetics 118: 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. Y., E. I. Wong, L. Vidali, A. Estavillo, P. K. Hepler et al., 2002. The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. Y., A. Y. Cheung and H. M. Wu, 2003. Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 15: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A. Y., H. Wang and H. M. Wu, 1995. A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82: 383–393. [DOI] [PubMed] [Google Scholar]
- Cheung, A. Y., C. Y. Chen, R. H. Glaven, B. H. de Graaf, L. Vidali et al., 2002. Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14: 945–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite, M., W. Park, Z. Xu, X. Yan, S. C. Maurais et al., 2002. Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H. K., and A. Belachew, 1989. Transposition pattern of the maize element Ac from the bz-m2(Ac) allele. Genetics 122: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch, J. J., S. Kadwell, E. Merlin and L. Crossland, 1994. Cloning and characterization of a maize pollen-specific calcium-dependent calmodulin-independent protein kinase. Proc. Natl. Acad. Sci. USA 91: 8837–8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin-Tong, V. E., 1999. Signaling and the modulation of pollen tube growth. Plant Cell 11: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., G. Wu and Z. Yang, 2001. Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152: 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin, M., and A. S. Reddy, 2003. A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc. Natl. Acad. Sci. USA 100: 10558–10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt, I. M., 1984. A chromosome replication pattern deduced from pericarp phenotypes resulting from movement of the transposable element Modulator in maize. Genetics 108: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, B., R. Walko and S. Hake, 1994. Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics 138: 1275–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler, P. K., L. Vidali and A. Y. Cheung, 2001. Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187. [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke, T. L., J. A. Feijo, G. R. Hackett, J. G. Kunkel and P. K. Hepler, 1997. Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9: 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck, N., J. M. Moore, M. Federer and U. Grossniklaus, 2003. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130: 2149–2159. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., S. D. Kopczak, T. F. Horejsi, B. K. Kihl and R. E. Pruitt, 1995. a Identification of genes required for pollen-stigma recognition in Arabidopsis thaliana. Plant J. 8: 703–714. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., K. Schneitz and R. E. Pruitt, 1995. b Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindiger, B., J. B. Beckett and E. H. Coe, Jr., 1991. Differential effects of specific chromosomal deficiencies on the development of the maize pollen grain. Genome 34: 579–594. [Google Scholar]
- Li, H., Y. Lin, R. M. Heath, M. X. Zhu and Z. Yang, 1999. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11: 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobstein, E., A. Guyon, M. Ferault, D. Twell, G. Pelletier et al., 2004. The putative Arabidopsis homolog of yeast vps52p is required for pollen tube elongation, localizes to Golgi, and might be involved in vesicle trafficking. Plant Physiol. 135: 1480–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, E., 2000. Adhesion and cell movement during pollination: cherchez la femme. Trends Plant Sci. 5: 368–373. [DOI] [PubMed] [Google Scholar]
- Lord, E. M., and S. D. Russell, 2002. The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 18: 81–105. [DOI] [PubMed] [Google Scholar]
- Marton, M. L., S. Cordts, J. Broadhvest and T. Dresselhaus, 2005. Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576. [DOI] [PubMed] [Google Scholar]
- Mayfield, J. A., and D. Preuss, 2000. Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat. Cell Biol. 2: 128–130. [DOI] [PubMed] [Google Scholar]
- Mayfield, J. A., A. Fiebig, S. E. Johnstone and D. Preuss, 2001. Gene families from the Arabidopsis thaliana pollen coat proteome. Science 292: 2482–2485. [DOI] [PubMed] [Google Scholar]
- McCormick, S., 2004. Control of male gametophyte development. Plant Cell 16(Suppl): S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouline, K., A. A. Very, F. Gaymard, J. Boucherez, G. Pilot et al., 2002. Pollen tube development and competitive ability are impaired by disruption of a Shaker K(+) channel in Arabidopsis. Genes Dev. 16: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho, A., A. J. Trewavas and R. Malho, 1998. Relocation of a Ca2+-dependent protein kinase activity during pollen tube reorientation. Plant Cell 10: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenfuhr, A., L. A. Gallagher, T. G. Dunahay, J. A. Frohlick, A. M. Mazurkiewicz et al., 1999. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121: 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton, R. M., S. Fischer-Parton, A. J. Trewavas and M. K. Watahiki, 2003. Pollen tubes exhibit regular periodic membrane trafficking events in the absence of apical extension. J. Cell Sci. 116: 2707–2719. [DOI] [PubMed] [Google Scholar]
- Pierson, E. S., D. D. Miller, D. A. Callaham, A. M. Shipley, B. A. Rivers et al., 1994. Pollen tube growth is coupled to the extracellular calcium ion flux and the intracellular calcium gradient: effect of BAPTA-type buffers and hypertonic media. Plant Cell 6: 1815–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, D., B. Lemieux, G. Yen and R. W. Davis, 1993. A conditional sterile mutation eliminates surface components from Arabidopsis pollen and disrupts cell signaling during fertilization. Genes Dev. 7: 974–985. [DOI] [PubMed] [Google Scholar]
- Procissi, A., A. Guyon, E. S. Pierson, A. Giritch, B. Knuiman et al., 2003. KINKY POLLEN encodes a SABRE-like protein required for tip growth in Arabidopsis and conserved among eukaryotes. Plant J. 36: 894–904. [DOI] [PubMed] [Google Scholar]
- Ray, S. M., S. S. Park and A. Ray, 1997. Pollen tube guidance by the female gametophyte. Development 124: 2489–2498. [DOI] [PubMed] [Google Scholar]
- Rutten, T. L., and B. Knuiman, 1993. Brefeldin A effects on tobacco pollen tubes. Eur. J. Cell Biol. 61: 247–255. [PubMed] [Google Scholar]
- Shimizu, K. K., and K. Okada, 2000. Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127: 4511–4518. [DOI] [PubMed] [Google Scholar]
- Twell, D., R. Wing, J. Yamaguchi and S. McCormick, 1989. Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 217: 240–245. [DOI] [PubMed] [Google Scholar]
- Xu, Z., and H. K. Dooner, 2003. The Ac-tagged aberrant pollen transmission 1 (apt1) is a homologue of the Arabidopsis gene SABRE required for root cell expansion. Abstracts of 45th Annual Maize Genetics Conference, Lake Geneva, WI, p. 112.