Abstract
Models of molecular evolution that incorporate the ratio of nonsynonymous to synonymous polymorphism (dN/dS ratio) as a parameter can be used to identify sites that are under diversifying selection or functional constraint in a sample of gene sequences. However, when there has been recombination in the evolutionary history of the sequences, reconstructing a single phylogenetic tree is not appropriate, and inference based on a single tree can give misleading results. In the presence of high levels of recombination, the identification of sites experiencing diversifying selection can suffer from a false-positive rate as high as 90%. We present a model that uses a population genetics approximation to the coalescent with recombination and use reversible-jump MCMC to perform Bayesian inference on both the dN/dS ratio and the recombination rate, allowing each to vary along the sequence. We demonstrate that the method has the power to detect variation in the dN/dS ratio and the recombination rate and does not suffer from a high false-positive rate. We use the method to analyze the porB gene of Neisseria meningitidis and verify the inferences using prior sensitivity analysis and model criticism techniques.
AS an indicator of the action of natural selection in gene sequences the ratio of nonsynonymous to synonymous substitutions (dN/dS) is versatile and widely used. An excess of nonsynonymous relative to synonymous polymorphism is a clear signal of diversifying selection, whereas a lack of nonsynonymous relative to synonymous polymorphism is indicative of purifying selection imposed by functional constraint.
Nielsen and Yang (1998) proposed a maximum-likelihood phylogenetic approach to estimating the dN/dS ratio that employs a codon-based mutation model (Goldman and Yang 1994) and treats the dN/dS ratio as an unknown parameter ω. This method has subsequently been expanded (Yang et al. 2000; Yang and Swanson 2002; Swanson et al. 2003), adapted into a Bayesian setting (Huelsenbeck and Dyer 2004), and approximated for the purposes of computational efficiency (Massingham and Goldman 2005). Simulation studies have shown that phylogenetic likelihood-based methods can be substantially more powerful than alternative approaches (Anisimova et al. 2001, 2002; Wong et al. 2004; Kosakovsky Pond and Frost 2005).
Estimating the selection parameter ω using these methods has become widespread (e.g., Bishop et al. 2000; Ford 2001; Mondragon-Palomino et al. 2002; Filip and Mundy 2004) and has been applied to many organisms. Analysis of pathogens such as viruses (Twiddy et al. 2002; de Oliveira et al. 2004; Moury 2004) and bacteria (Peek et al. 2001; Urwin et al. 2002) is particularly informative, because they typically have high mutation rates and are consequently genetically diverse, which lends greater statistical power to estimation. The ability to observe these populations evolving in real time makes them especially interesting for the study of evolution (Drummond et al. 2003) and suggests that we may be able to make useful epidemiological inferences from molecular sequence data (Wilson et al. 2005).
However, the use of phylogenetic techniques is questionable in organisms that are highly recombining, because recombination leads to not one, but multiple evolutionary trees along the sequence. If the recombination rate is of the same order as the mutation rate, as has been found in some organisms (McVean et al. 2002; Stumpf and McVean 2003), then there might be a new evolutionary tree for every polymorphic site along the sequence. In such a scenario, which is plausible for many highly recombining microorganisms (Awadalla 2003) and eukaryotic genes containing recombination hotspots (McVean et al. 2004; Winckler et al. 2005), there is little hope of inferring any particular evolutionary tree along the sequence. When a single evolutionary tree is inferred for a sample of gene sequences that have in fact undergone recombination, the resulting tree is likely to have longer terminal branches and total branch length, yet a smaller time to the most recent common ancestor, in a way that superficially resembles the star-shaped topology of an exponentially growing population (Schierup and Hein 2000). The effect on the identification of sites experiencing diversifying selection is to cause a high number of false positives (Anisimova et al. 2003), as high as 90% (Shriner et al. 2003).
In this article we present a new method that coestimates the selection parameter ω and the recombination rate along the sequence. We use a population genetics approximation (Li and Stephens 2003) to the coalescent with recombination (Hudson 1983; Griffiths and Marjoram 1997), rather than using a phylogenetic approach, and we adopt a Bayesian, rather than a maximum-likelihood strategy, to incorporate evolutionary uncertainty. The method uses reversible-jump Markov chain Monte Carlo (MCMC) to obtain the posterior distribution of parameters. We conduct simulation studies, which show that there is good power to detect variation in ω and the recombination rate and that the method has a low false-positive rate. We use the method to analyze the porB gene of the bacterial pathogen Neisseria meningitidis and verify the inferences using prior sensitivity analysis and model criticism techniques.
THEORY
In this article, the parameters of primary interest are the selection parameter ω and the population recombination rate ρ, both of which are allowed to vary along the sequence. The other model parameters are the transition–transversion ratio κ, the rate of synonymous transversion μ, and the insertion/deletion rate φ. Key to maximum-likelihood or Bayesian inference is the likelihood function, 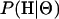 , where H is the data (the haplotypes) and Θ represents our model parameters. Phylogenetic methods typically estimate the maximum-likelihood tree,
, where H is the data (the haplotypes) and Θ represents our model parameters. Phylogenetic methods typically estimate the maximum-likelihood tree, 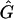 , and then calculate the likelihood conditional on the tree,
, and then calculate the likelihood conditional on the tree, 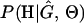 , using the pruning algorithm (Felsenstein 1981). When there is recombination there can be multiple trees along the sequence, and there is typically little power to estimate those trees. Therefore we treat the trees as a nuisance parameter that we wish to average over, so
, using the pruning algorithm (Felsenstein 1981). When there is recombination there can be multiple trees along the sequence, and there is typically little power to estimate those trees. Therefore we treat the trees as a nuisance parameter that we wish to average over, so
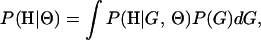 |
(1) |
where 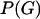 is the probability density of the ancestral tree or trees, including branch lengths. There are various ways to model
is the probability density of the ancestral tree or trees, including branch lengths. There are various ways to model 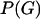 . In the case of no recombination Huelsenbeck and Dyer (2004) used a model in which all unrooted tree topologies were uniformly likely, and branch lengths had an exponential distribution. When the sequences are from a single population a natural choice would be the coalescent (Kingman 1982; Hudson 1983; Griffiths and Marjoram 1997), which models a neutrally evolving, randomly mating population of constant size, with or without recombination. In this article we approximate Equation 1 in the case where
. In the case of no recombination Huelsenbeck and Dyer (2004) used a model in which all unrooted tree topologies were uniformly likely, and branch lengths had an exponential distribution. When the sequences are from a single population a natural choice would be the coalescent (Kingman 1982; Hudson 1983; Griffiths and Marjoram 1997), which models a neutrally evolving, randomly mating population of constant size, with or without recombination. In this article we approximate Equation 1 in the case where 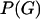 is the coalescent with recombination.
is the coalescent with recombination.
In a coalescent model the expected branch length between a pair of sequences is 2PNe generations (where P is the ploidy and Ne is the effective population size), during which time there are 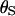 synonymous mutations on average.
synonymous mutations on average. 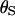 is twice the synonymous mutation rate per PNe generations and, likewise, ρ is twice the recombination rate per PNe generations. We use the codon model of Nielsen and Yang (1998), hereafter NY98, which gives the mutation rate from codon i to j (
is twice the synonymous mutation rate per PNe generations and, likewise, ρ is twice the recombination rate per PNe generations. We use the codon model of Nielsen and Yang (1998), hereafter NY98, which gives the mutation rate from codon i to j ( ) in units of PNe generations as
) in units of PNe generations as
 |
(2) |
where the frequency of codon j is 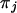 . The diagonal elements of the mutation rate matrix are defined to be
. The diagonal elements of the mutation rate matrix are defined to be 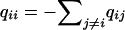 . When there is equal codon usage,
. When there is equal codon usage,
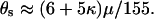 |
(3) |
In appendix a we extend the NY98 model specified by Equation 2 to incorporate an insertion/deletion rate φ.
When there is no recombination, Equation 1 could be computed using importance sampling or MCMC (e.g., Huelsenbeck and Dyer 2004). In the presence of recombination, importance sampling (Fearnhead and Donnelly 2001) and MCMC (Kuhner et al. 2000) have been applied to simpler mutation models. However, these methods are highly computationally intensive. In the context of the NY98 mutation model, such methods are not feasible.
Instead we turn to an approximation to the likelihood in the presence of recombination (Li and Stephens 2003) called the product of approximate conditionals (PAC) likelihood. Their approach relies on rewriting the likelihood as
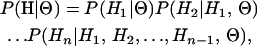 |
(4) |
where 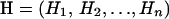 is the sample of n gene sequences (haplotypes). Li and Stephens approximate the
is the sample of n gene sequences (haplotypes). Li and Stephens approximate the 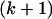 th conditional likelihood:
th conditional likelihood:
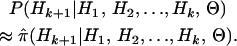 |
The approximate conditional likelihood, 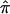 , that they use is a hidden Markov model that is designed to incorporate some key properties of the proper likelihood, notably that (i) the
, that they use is a hidden Markov model that is designed to incorporate some key properties of the proper likelihood, notably that (i) the 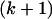 th haplotype is likely to resemble the first k haplotypes but (ii) recombination means that it may be a mosaic of those haplotypes and (iii) mutation means that it may be an imperfect copy. In terms of averaging over possible evolutionary trees, one can think of the hidden Markov model doing so implicitly, but in an approximate way that is highly computationally efficient.
th haplotype is likely to resemble the first k haplotypes but (ii) recombination means that it may be a mosaic of those haplotypes and (iii) mutation means that it may be an imperfect copy. In terms of averaging over possible evolutionary trees, one can think of the hidden Markov model doing so implicitly, but in an approximate way that is highly computationally efficient.
As a result of the approximate nature of the PAC likelihood, the ordering of the n haplotypes can influence the value of the likelihood (were it not for the approximation, the haplotypes would be exchangeable). Therefore, the likelihood is assessed by averaging over multiple orderings of the haplotypes. We use 10 orderings throughout unless otherwise stated.
We modify the approximation of Li and Stephens to incorporate the NY98 codon-based model with the addition of an insertion/deletion rate φ (see appendix a for details), and we adopt a Bayesian rather than a maximum-likelihood approach. Thus, our object of inference is the posterior distribution of parameters, 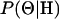 , where
, where
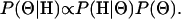 |
(5) |
Here 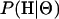 is the likelihood function, described above and in appendix a, and
is the likelihood function, described above and in appendix a, and 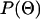 is the prior distribution on the parameters.
is the prior distribution on the parameters.
Our primary aim is to obtain a posterior distribution for ω, allowing ω to vary along the length of the sequence. The information regarding ω at a given position along the sequence is limited by the number of mutations in the underlying evolutionary history. This is a potentially serious limitation, particularly for sequences with low diversity. In an attempt to exploit to the full the available information, we use a prior distribution on ω in which adjacent sites may share a common selection parameter.
For a sequence of length L codons, our prior distribution imposes a “block-like” structure on the variation in ω with two fixed and B 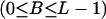 variable transition points,
variable transition points,
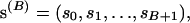 |
where 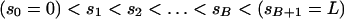 .
.
Block j is delimited by transition points 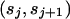 and has a common selection parameter
and has a common selection parameter 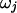 . We model the number of variable transition points in the region as a binomial distribution with parameters
. We model the number of variable transition points in the region as a binomial distribution with parameters 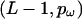 . Given the number of transition points, the selection parameter for each block is independently and identically distributed. For an exponential prior on
. Given the number of transition points, the selection parameter for each block is independently and identically distributed. For an exponential prior on 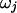 with rate parameter λ, the prior distribution on the transition points and selection parameters can be written
with rate parameter λ, the prior distribution on the transition points and selection parameters can be written
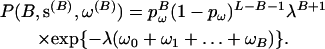 |
(6) |
In this model, the expected length of a block is 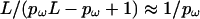 . For
. For 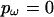 there is a single block, producing a constant model for ω along the sequence, and for
there is a single block, producing a constant model for ω along the sequence, and for 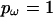 every site has its own independent ω. Therefore the user can choose not to impose a block structure on the variation in ω if desired.
every site has its own independent ω. Therefore the user can choose not to impose a block structure on the variation in ω if desired.
This model for variation in ω is based on the multiple change-point model of Green (1995), which was adopted by McVean et al. (2004) to estimate variable recombination rates along a gene sequence, although the binomial model described here is designed specifically so that transition points must fall between codons at a finite (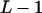 ) number of positions. Multiple change-point models have also been used in the context of detecting parental and recombinant genomes in HIV-1 (Suchard et al. 2002; Minin et al. 2005). We implement a model for the variation in ρ of the same form as that for ω, but the block structure for ρ is independent of the block structure for ω, and the number of variable transition points is binomially distributed with parameters
) number of positions. Multiple change-point models have also been used in the context of detecting parental and recombinant genomes in HIV-1 (Suchard et al. 2002; Minin et al. 2005). We implement a model for the variation in ρ of the same form as that for ω, but the block structure for ρ is independent of the block structure for ω, and the number of variable transition points is binomially distributed with parameters 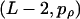 . We assume that recombination occurs only between codons and not within them. In this way we are able to perform inference jointly on variation in ω and ρ along the sequence. We use reversible-jump MCMC to explore the posterior distribution of Θ (see appendix b).
. We assume that recombination occurs only between codons and not within them. In this way we are able to perform inference jointly on variation in ω and ρ along the sequence. We use reversible-jump MCMC to explore the posterior distribution of Θ (see appendix b).
SIMULATIONS
To investigate the performance of the method, we undertook two simulation studies. In the first we simulated data with variation in the selection parameter along the sequence and a constant recombination rate. In the second, we simulated data with variation in the recombination rate along the sequence and a constant selection parameter. Each of these two studies consisted of simulating 100 data sets of 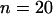 sequences each of length
sequences each of length 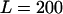 codons, using the coalescent with recombination (Hudson 1983; Griffiths and Marjoram 1997) and the NY98 mutation model.
codons, using the coalescent with recombination (Hudson 1983; Griffiths and Marjoram 1997) and the NY98 mutation model.
To investigate the effect of the block model of variation in ω, a third simulation study was undertaken in which a short sequence of length L = 21 codons was simulated with a single site experiencing diversifying selection in the middle (ω = 5.0) against a background of functionally constrained sites (ω = 0.2). One hundred data sets of n = 20 sequences were simulated and analyzed, using both the block model for variation in ω (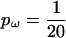 ) and the independent model for variation in ω (pω = 1).
) and the independent model for variation in ω (pω = 1).
In all three simulation studies, the MCMC was run twice for each analysis over 250,000 iterations, with a burn-in of 20,000 iterations. Initial values were chosen randomly from the priors independently for the two runs. The runs were compared for convergence and merged to obtain the posterior distributions.
Simulation study A:
This study was designed to simulate data with variation in ω but not in ρ. We varied ω between 0.1 and 10, as shown by the red line in Figure 1a. The mutation parameters were set at 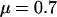 and
and 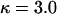 , which gives
, which gives 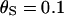 . The recombination rate was set constant at
. The recombination rate was set constant at 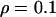 , giving a total recombination distance for the region of
, giving a total recombination distance for the region of 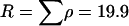 . The mutation and recombination parameters were chosen to mimic those estimated for the housekeeping genes of N. meningitidis (Jolley et al. 2005). Exponential distributions were used for the priors on μ, κ, ω, and ρ, with means 0.7, 3.0, 1.0, and 0.1. A block model of variation in ω and ρ was used with
. The mutation and recombination parameters were chosen to mimic those estimated for the housekeeping genes of N. meningitidis (Jolley et al. 2005). Exponential distributions were used for the priors on μ, κ, ω, and ρ, with means 0.7, 3.0, 1.0, and 0.1. A block model of variation in ω and ρ was used with 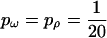 , so that the average length of a block would be ∼10% of the sequence length.
, so that the average length of a block would be ∼10% of the sequence length.
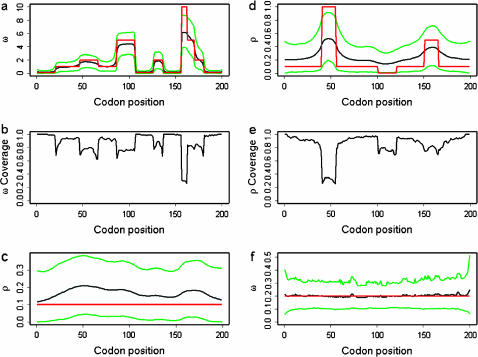
Figure 1.
Results of simulation studies A and B. (a) Average posterior of ω, (b) coverage of ω, and (c) average posterior of ρ in simulation study A. (d) Average posterior of ρ, (e) coverage of ρ, and (f) average posterior of ω in simulation study B. (a, c, d, and f) The red line indicates the truth, the black line indicates the average mean of the posterior, and the green lines indicate the average 95% HPD interval of the posterior. The averages are taken over 100 simulated data sets. (b and e) Coverage is defined as the proportion of the 100 data sets for which the 95% HPD interval encloses the truth.
A permutation test based on the correlation between physical distance and three measures of linkage disequilibrium (LD), 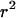 ,
,  , and G4 (see, for example, Meunier and Eyre-Walker 2001; McVean et al. 2002), showed that phylogenetic analysis of these data sets was inappropriate because of the presence of recombination. The numbers of data sets for which the P-value was <0.05 were 99, 93, and 93 for the three test statistics, respectively.
, and G4 (see, for example, Meunier and Eyre-Walker 2001; McVean et al. 2002), showed that phylogenetic analysis of these data sets was inappropriate because of the presence of recombination. The numbers of data sets for which the P-value was <0.05 were 99, 93, and 93 for the three test statistics, respectively.
Figure 1a shows the average over the 100 simulated data sets of the mean and 95% highest posterior density (HPD) interval for the posterior distribution of ω at each site. The average mean posterior density follows the truth closely. Likewise the average 95% HPD interval generally encloses the true value of ω. As expected, the effect of fitting a prior with mean 1 was to cause the posterior to underestimate ω when 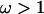 and overestimate ω when
and overestimate ω when 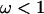 . The effect is not great except for the most extreme values where
. The effect is not great except for the most extreme values where 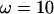 .
.
However, even where the average 95% HPD interval encloses the truth, that does not mean the 95% HPD interval encloses the truth for all simulated data sets. Figure 1b shows the relevant quantity, the coverage of ω, for each site. Coverage is defined here as the proportion of data sets for which the 95% HPD interval encloses the truth. Half of sites have coverage >93%, and 95% of sites have coverage >66%. If a false positive is defined as the lower bound of the 95% HPD interval exceeding 1 when in truth 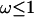 , then the false-positive rate was 0.5%. The estimate of the synonymous transversion rate μ exhibits upward bias (average 0.90), with 63% coverage (Table 1), and the transition–transversion ratio κ is estimated to be 3.1 on average, with 91% coverage.
, then the false-positive rate was 0.5%. The estimate of the synonymous transversion rate μ exhibits upward bias (average 0.90), with 63% coverage (Table 1), and the transition–transversion ratio κ is estimated to be 3.1 on average, with 91% coverage.
TABLE 1.
Summary of posteriors for simulation study A
| Average posterior
|
||||||
|---|---|---|---|---|---|---|
| Parameter | Truth | Prior: mean | Lower 95% HPD | Mean | Upper 95% HPD | Coverage |
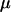 |
0.7 | 0.7 | 0.7 | 0.9 | 1.1 | 0.63 |
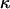 |
3.0 | 3.0 | 2.3 | 3.1 | 3.9 | 0.91 |
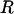 |
19.9 | 19.9 | 22.4 | 33.3 | 44.7 | 0.43 |
Consistent with the findings of Li and Stephens (2003), we observe that the recombination rate estimator has a small upward bias (Figure 1c). The average mean posterior is almost flat, and the average 95% confidence intervals enclose the truth completely, suggesting that the estimator is good notwithstanding its bias. The coverage is almost constant across sites at 95%. Table 1 shows that the estimate of the total recombination distance, R, is also upwardly biased. Coverage of R, however, was only 43%, suggesting that the good coverage for ρ at individual sites may be in part because of poor information. Importantly, Figure 1, a–c, shows that the effect of the selection parameter on the estimate of ρ is negligible, indicating that inference on ρ is not confounded by ω.
Simulation study B:
This study was designed to simulate data with variation in ρ but not in ω. Along the sequence we let ρ vary at 0.005, 0.1, 0.5, and 1, for which one would expect 0.018, 0.35, 1.8, and 3.5 recombination events, respectively, per site in the ancestral history under a coalescent model (Griffiths and Marjoram 1997). The total recombination distance was 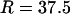 . We let
. We let 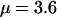 and
and 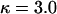 , giving
, giving 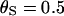 and a constant selection parameter of
and a constant selection parameter of 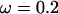 . Exponential distributions were used for the priors on μ, κ, ω and ρ, with means 3.6, 3.0, 1.0, and 0.2. The same model of variation in ω and ρ was used as for simulation study A.
. Exponential distributions were used for the priors on μ, κ, ω and ρ, with means 3.6, 3.0, 1.0, and 0.2. The same model of variation in ω and ρ was used as for simulation study A.
Permutation tests showed that these data sets were not amenable to phylogenetic analysis because of the presence of recombination. All 100 data sets yielded P-values <0.05 for all three measures of LD.
Variation in the recombination rate was detected by the new method, as seen in Figure 1d. The average over the 100 data sets shows that the mean and 95% HPD interval for the posterior distribution of ρ at each site pick up the rate variation, but not to the full extent. As a result, the coverage shown in Figure 1e is generally good, on average 85%, but performs worst for the most extreme peak in rate between sites 41 and 55, where it consistently underestimates the height. The properties of the estimate of the total recombination distance R (Table 2) are similar to those in simulation study A. There is a tendency to overestimate (average 50.9) and as a result coverage is 49%. This bias could be corrected empirically, as in Li and Stephens (2003). Nevertheless, there is power to detect rate variation on such fine scales. The extent to which the posteriors underestimate the deviations from the mean recombination rate reflects the constraining effect of the prior when the signal in the data is weak.
TABLE 2.
Summary of posteriors for simulation study B
| Average posterior
|
||||||
|---|---|---|---|---|---|---|
| Parameter | Truth | Prior: mean | Lower 95% HPD | Mean | Upper 95% HPD | Coverage |
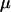 |
3.6 | 3.6 | 3.4 | 4.2 | 5.1 | 0.53 |
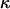 |
3.0 | 3.0 | 2.5 | 3.1 | 3.8 | 0.95 |
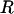 |
37.5 | 39.8 | 37.4 | 50.9 | 65.0 | 0.49 |
Figure 1f shows that on average the estimates of ω are very close to the truth, with the average 95% HPD intervals completely enclosing the true value. Along the sequence, the estimates are flat, with mean 0.21 and coverage 90%. The false-positive rate was zero. Reflecting simulation study A, there was no evidence that variation in the recombination rate confounded inference on the selection parameter. Table 2 shows that there was some upward bias in the mean estimate of 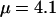 , with 58% coverage, and the transition–transversion ratio was estimated to be 3.2 on average, with 89% coverage. Most importantly, both simulation studies show that when there is variation in ω or ρ it can be detected, when there is no variation none is detected, and there is little or no confounding between ω and ρ.
, with 58% coverage, and the transition–transversion ratio was estimated to be 3.2 on average, with 89% coverage. Most importantly, both simulation studies show that when there is variation in ω or ρ it can be detected, when there is no variation none is detected, and there is little or no confounding between ω and ρ.
Simulation study C:
This study was designed to investigate the smoothing effect of the block-like prior for variation in ω on the detection of diversifying selection and functional constraint. As in simulation study A, the mutation parameters were set at 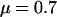 and
and 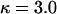 , giving
, giving 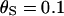 . A single codon in the middle of the L = 21-codon sequence was simulated under diversifying selection (ω = 5.0) whereas all the surrounding sites were functionally constrained (ω = 0.2). As in simulation study A, the recombination rate was set constant at
. A single codon in the middle of the L = 21-codon sequence was simulated under diversifying selection (ω = 5.0) whereas all the surrounding sites were functionally constrained (ω = 0.2). As in simulation study A, the recombination rate was set constant at 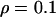 and exponential distributions were used for the priors on μ, κ, ω, and ρ, with means 0.7, 3.0, 1.0, and 0.1. All simulated data sets exhibited nonsynonymous polymorphism at the codon under diversifying selection. Two analyses were conducted for each of the 100 simulated data sets: in one a block model on variation in ω was used (pω =
and exponential distributions were used for the priors on μ, κ, ω, and ρ, with means 0.7, 3.0, 1.0, and 0.1. All simulated data sets exhibited nonsynonymous polymorphism at the codon under diversifying selection. Two analyses were conducted for each of the 100 simulated data sets: in one a block model on variation in ω was used (pω = 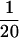 ) and in the other each site had an independent ω (pω = 1). In both a block model on variation in ρ was used (pρ =
) and in the other each site had an independent ω (pω = 1). In both a block model on variation in ρ was used (pρ = 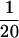 ).
).
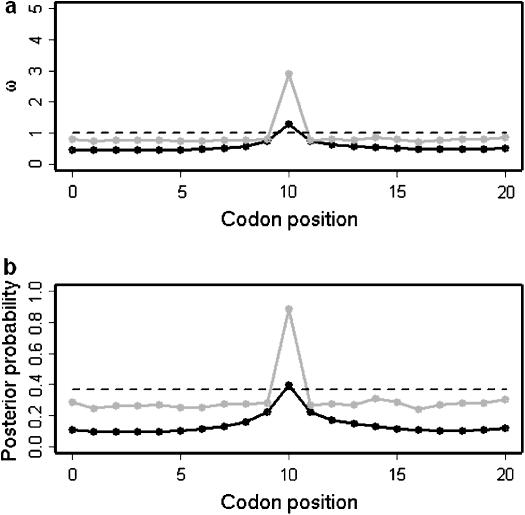
Figure 2a shows the average over the 100 data sets of the mean posterior of ω along the sequence under the block model (solid line) and the independent model (shaded line). The mean of the prior is also shown (dashed line). In both models the mean value of ω is estimated to be >1 for the site under diversifying selection and <1 for the functionally constrained sites. By combining information across functionally constrained sites, the block model has obtained an estimate of ω closer to the truth 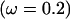 than the independent model. At the site under diversifying selection, the effect of the block model is to smooth the variation in ω, and as a result the estimate is only just >1
than the independent model. At the site under diversifying selection, the effect of the block model is to smooth the variation in ω, and as a result the estimate is only just >1 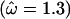 whereas the independent model obtains an estimate closer to the true value of 5.0
whereas the independent model obtains an estimate closer to the true value of 5.0 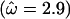 . Both are underestimates, which reflects the effect of the prior when there is little information.
. Both are underestimates, which reflects the effect of the prior when there is little information.
Figure 2.
Results of simulation study C. (a) Average posterior of ω when analyzed using the block model for variation in ω (pω = 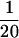 , solid line) and the independent model (pω = 1, shaded line). The average prior of ω is also shown (dashed line). (b) Sitewise posterior probability of diversifying selection (
, solid line) and the independent model (pω = 1, shaded line). The average prior of ω is also shown (dashed line). (b) Sitewise posterior probability of diversifying selection (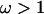 ) when analyzed using the block model (pω =
) when analyzed using the block model (pω = 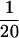 , solid line) and the independent model (pω = 1, shaded line). The prior probability of diversifying selection (
, solid line) and the independent model (pω = 1, shaded line). The prior probability of diversifying selection (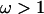 ) is shown (dashed line).
) is shown (dashed line).
For functionally constrained sites, the coverage was 99% for both models, although this partly reflects the wider HPD intervals for the independent model. For the site under diversifying selection, coverage was 16% for the block model and 67% for the independent model. However, for both models there is an appreciable increase in the estimate of ω at the site under diversifying selection, which is seen more clearly in Figure 2b. The sitewise posterior probability of diversifying selection 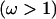 is plotted for the block model (solid line) and the independent model (shaded line). The prior probability of diversifying selection is also indicated (dashed line). The posterior probability increases by a similar amount at the middle site for both models, although the increase is more abrupt for the independent model. By combining information across sites, the block model reports a lower posterior probability of diversifying selection at the functionally constrained sites, but the smoothing effect accordingly produces a lower posterior probability of diversifying selection at the middle site, compared to the independent model.
is plotted for the block model (solid line) and the independent model (shaded line). The prior probability of diversifying selection is also indicated (dashed line). The posterior probability increases by a similar amount at the middle site for both models, although the increase is more abrupt for the independent model. By combining information across sites, the block model reports a lower posterior probability of diversifying selection at the functionally constrained sites, but the smoothing effect accordingly produces a lower posterior probability of diversifying selection at the middle site, compared to the independent model.
As expected, the smoothing effect of a block model for variation in ω improves the estimates for series of sites that share a common selection parameter, but disfavors lone sites with a very different selection parameter, compared to a model in which each site has an independent ω. The decision whether to use a block model or an independent model for variation in ω will depend on the user's prior beliefs as to the nature of variation in the selection parameter and the relative importance of obtaining improved estimates on average at the expense of lone unusual sites. Figure 2b shows that the strength of the block structure (1/pω) should be taken into account when choosing a level above which a site is deemed to undergo diversifying selection; the signal of lone sites experiencing diversifying selection is still apparent for the block model, but the absolute posterior probability is lower.
APPLICATION TO MENINGOCOCCAL PORB
Using the porB locus of N. meningitidis, we demonstrate the application of the Bayesian approach to inference of selection and outline a coherent approach to model-based analysis, from rejection of a model with no recombination through to prior sensitivity analysis and model criticism. Finally, we look at the effect on inference of assuming no recombination.
N. meningitidis is the bacterium responsible for meningococcal meningitis and septicemia. Despite its notorious pathogenesis, it is commonly found as a commensal organism occupying the nasopharynx of ∼10% of the population (e.g., Jolley et al. 2000). PorB is a porin expressed on the surface of the meningococcus and is thought to be important for both proper cell growth and pathogenesis. Two classes of PorB protein exist, with somewhat different molecular structure and evolutionary ancestry (Smith et al. 1995; Derrick et al. 1999), called PorB2 and PorB3. Urwin et al. (2002) used a maximum-likelihood method (Yang et al. 2000) implemented in the CODEML program of the PAML package (Yang 1997) to infer selection in the porB locus, taking the porB2 and porB3 allelic classes separately. The CODEML method infers a maximum-likelihood phylogenetic tree for the sequences and then makes inference on the selection parameters on the basis of that tree. Therefore it does not take account of recombination that has occurred between those sequences since their most recent common ancestor.
Data:
Here we analyze the 79 porB3 alleles from Urwin et al. (2002), using the new method implemented in the program omegaMap. The 79 alleles do not constitute a random sample of any population in a meaningful sense, thus violating one of the assumptions of the coalescent model. Instead the sequences are a collection taken from an assortment of studies, including 37 isolates from healthy carriers from England and Wales obtained during swabbing programs at a military recruit training camp (see Urwin et al. 2002 for details). Of these 37 isolates, 19 were obtained from 5 of the recruits and the remaining 18 were from 1 each. To account for this sampling bias, we took only 1 isolate from each recruit, yielding a sample size of 23. In the discussion we explain the rationale behind this. We called the sample of 23 the carriage study and the full collection of 79 the global study. Whereas the global study consisted of 77 unique haplotypes, the carriage study consisted of 12 unique haplotypes. R. Urwin kindly provided us with her sequence alignments.
Preliminary analysis:
To test the simpler model of no recombination, we applied the permutation tests described in simulation studies to the carriage and global studies. Table 3 shows the results. For the carriage study, there was a 0.1% probability of observing as extreme a correlation between physical distance and LD under the model of no recombination, regardless of choice of LD statistic. The result was the same for the global study. Therefore these data are not amenable to phylogenetic analysis. In the analyses that follow we specified the codon frequencies using the observed codon frequencies (Nakamura et al. 2000) in the N. meningitidis Z2491 serogroup A genome (Parkhill et al. 2000), excluding the stop codons.
TABLE 3.
Permutation test for recombination
| Carriage study
|
Global study
|
|||
|---|---|---|---|---|
| Correlation | P | Correlation | P | |
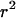 |
−0.18 | 0.001 | −0.15 | 0.001 |
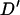 |
−0.24 | 0.001 | −0.16 | 0.001 |
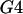 |
−0.23 | 0.001 | −0.15 | 0.001 |
Carriage study:
We chose to use exponential distributions for the priors on μ, κ, φ, ω, and ρ (Table 4, prior A). The mean of the prior on μ was 0.07, and the mean for κ was put at 3. The rate of insertion/deletion was given a mean of 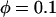 . For ω, the mean of the prior was set to 1, to represent our null model of selective neutrality, and for ρ, the mean was set at 0.1. The prior on the number of blocks for ω and ρ was binomial with pω = pρ =
. For ω, the mean of the prior was set to 1, to represent our null model of selective neutrality, and for ρ, the mean was set at 0.1. The prior on the number of blocks for ω and ρ was binomial with pω = pρ = 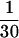 , so that the length of a block would be on average ∼10% of the sequence length (
, so that the length of a block would be on average ∼10% of the sequence length (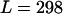 codons). We ran three MCMC chains, each 500,000 iterations in length, with a burn-in of 20,000 iterations. Having compared the chains for convergence, we merged them to obtain the posterior distributions.
codons). We ran three MCMC chains, each 500,000 iterations in length, with a burn-in of 20,000 iterations. Having compared the chains for convergence, we merged them to obtain the posterior distributions.
TABLE 4.
Prior distributions
| Prior A | Prior B | |
|---|---|---|
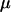 |
Exponential mean 0.07 | Uniform 0–10 |
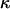 |
Exponential mean 3.0 | Exponential ratio |
| φ | Exponential mean 0.1 | Exponential mean 1.0 |
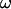 |
Exponential mean 1.0 | Gamma shape 2, scale 0.5 |
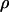 |
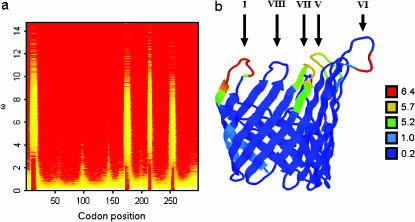
Exponential mean 0.1 | Uniform 0–10 |
Figure 3a shows a fire plot for the posterior distribution of ω at each site. More intense colors (closer to white) represent high posterior probabilities and less intense colors (closer to red) low posterior probabilities. The structure of PorB3 (Urwin et al. 2002) consists of eight putative loop regions that extend out of the cell. Of these, there is clear and strong evidence for diversifying selection at four of the eight loops. In these loop regions the 95% HPD intervals for the peak ω are (3.58, 9.76), (3.01, 8.92), (3.26, 9.68), and (2.58, 7.57) for loops 1, 5, 6, and 7, respectively. Taking the point estimate of ω at a site, 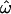 , as the mean of the posterior distribution, then the average
, as the mean of the posterior distribution, then the average 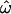 for the sequence is 0.90. Excluding sites for which
for the sequence is 0.90. Excluding sites for which 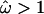 , this drops to 0.16. So the majority of the sequence is under strong functional constraint, but four of the eight loop regions are under strong diversifying selection.
, this drops to 0.16. So the majority of the sequence is under strong functional constraint, but four of the eight loop regions are under strong diversifying selection.
Figure 3.
Posterior distribution of ω in the N. meningitidis porB3 carriage study. (a) Fire plot of the sitewise posterior distribution of ω. More intense colors (closer to white) represent high posterior probabilities and less intense colors (closer to red) low posterior probabilities. (b) Molecular structure of PorB3 color coded according to  , the mean of the posterior distribution of ω. Dark blue indicates strong functional constraint and red indicates strong diversifying selection. This image was produced using protein explorer (Martz 2002; http://proteinexplorer.org). The image is oriented with the surface-exposed regions at the top. Arrows indicate the position of loops I and V–VIII.
, the mean of the posterior distribution of ω. Dark blue indicates strong functional constraint and red indicates strong diversifying selection. This image was produced using protein explorer (Martz 2002; http://proteinexplorer.org). The image is oriented with the surface-exposed regions at the top. Arrows indicate the position of loops I and V–VIII.
Superimposing 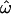 onto the three-dimensional structure of the PorB3 protein (Figure 3b) illustrates the external position of loops 1, 5, 6, and 7. Because PorB3 is a cell surface protein, these outer loops are especially exposed to the immune system and are prime sites for recognition by antibody. It is striking that there is no evidence for diversifying selection outside the loops. Loops 2, 3, and 4 do not appear to be under diversifying selection; the three-dimensional structure suggests that they may be less exposed than the other loops. However, loop 8 is surprising because despite its prominent position (Figure 3b), there is very little support for diversifying selection between codons 280 and 295 (Figure 3a). The light blue shading in Figure 3b occurs at the N and C termini, outside the nucleotide alignment we analyzed. Therefore we have assigned to them the mean of the prior,
onto the three-dimensional structure of the PorB3 protein (Figure 3b) illustrates the external position of loops 1, 5, 6, and 7. Because PorB3 is a cell surface protein, these outer loops are especially exposed to the immune system and are prime sites for recognition by antibody. It is striking that there is no evidence for diversifying selection outside the loops. Loops 2, 3, and 4 do not appear to be under diversifying selection; the three-dimensional structure suggests that they may be less exposed than the other loops. However, loop 8 is surprising because despite its prominent position (Figure 3b), there is very little support for diversifying selection between codons 280 and 295 (Figure 3a). The light blue shading in Figure 3b occurs at the N and C termini, outside the nucleotide alignment we analyzed. Therefore we have assigned to them the mean of the prior, 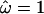 .
.
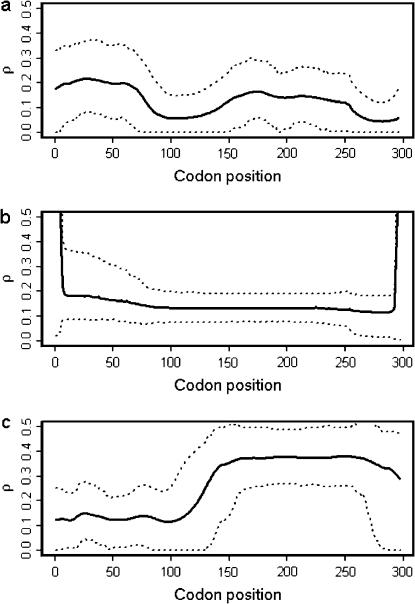
There was some evidence for variation in the recombination rate (Figure 4a). The posterior mean for the total recombination distance, 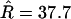 (Table 5), was twice the prior mean of 19.9. The posterior on μ was very different from the prior (
(Table 5), was twice the prior mean of 19.9. The posterior on μ was very different from the prior (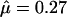 ), while there was little discrepancy for κ and φ (
), while there was little discrepancy for κ and φ (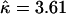 ,
, 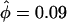 ).
).
Figure 4.
Posterior distribution of ρ in the N. meningitidis porB3 carriage and global studies. The sitewise mean (solid line) and 95% HPD intervals (dotted lines) are shown for (a) the carriage study under prior A, (b) the carriage study under prior B, and (c) the global study under prior A.
TABLE 5.
Posterior distributions
| Carriage study
|
|||||
|---|---|---|---|---|---|
| Prior A | Prior B | Prior A: 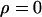
|
Global study: prior A | ||
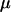 |
Mean | 0.27 | 0.35 | 0.45 | 0.31 |
| 95% HPD | (0.18, 0.36) | (0.23, 0.48) | (0.33, 0.58) | (0.22, 0.40) | |
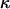 |
Mean | 3.61 | 3.09 | 3.69 | 3.34 |
| 95% HPD | (2.38, 5.00) | (1.94, 4.24) | (2.69, 4.83) | (2.41, 4.33) | |
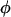 |
Mean | 0.09 | 0.17 | 0.29 | 0.08 |
| 95% HPD | (0.02, 0.19) | (0.03, 0.37) | (0.08, 0.56) | (0.02, 0.16) | |
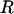 |
Mean | 37.7 | 46.8 | — | 78.0 |
| 95% HPD | (27.2, 49.0) | (26.2, 75.0) | — | (61.6, 94.5) | |
Prior sensitivity analysis:
To determine the influence of our choice of priors on the posteriors, we repeated the analyses with alternative priors (Table 4, prior B). For μ and ρ we fit a uniform prior between 0 and 10 (10 being the highest value we considered plausible for either parameter). Following Huelsenbeck and Dyer (2004) we fit a prior distribution on κ describing the ratio of two independent and identically distributed exponential random variables. The moments, including the mean, for this distribution are undefined, but the median equals 1. For φ we changed the mean of the exponential prior from 0.1 to 1. Finally, for ω we used a gamma distribution still with a mean of 1, but with shape parameter 2, which gives the distribution a mode at 0.5. This distribution retains the case of selective neutrality for its mean, but it tails off toward zero rather than increasing. We ran three MCMC chains, each 250,000 iterations in length, with a burn-in of 20,000 iterations. The chains were merged to obtain the posteriors.
Ninety-five percent HPD intervals for the peak ω in loops 1, 5, 6, and 7 show that the magnitude of the estimates has been reduced by the gamma prior to (2.76, 6.80), (2.16, 5.79), (2.31, 6.70), and (2.16, 5.66), respectively. Despite this, the relative height of the peaks is conserved. The average 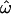 for the sequence is 0.68, reflecting the more conservative effect of the gamma prior. Excluding sites for which
for the sequence is 0.68, reflecting the more conservative effect of the gamma prior. Excluding sites for which 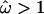 , this drops to 0.17, which is almost identical to the inference based on prior A. This suggests that information about the absolute magnitude of sites under functional constraint is less influenced by the prior. Despite differences concerning the magnitude of ω, the priors strongly agree on which sites are under diversifying selection (Figure 5). The posterior probability of diversifying selection at a given site is
, this drops to 0.17, which is almost identical to the inference based on prior A. This suggests that information about the absolute magnitude of sites under functional constraint is less influenced by the prior. Despite differences concerning the magnitude of ω, the priors strongly agree on which sites are under diversifying selection (Figure 5). The posterior probability of diversifying selection at a given site is
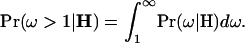 |
(7) |
Prior A is represented in Figure 4 by the shaded line and prior B by the dashed line. The two lines are virtually indistinguishable from one another at every site, indicating that our inference on diversifying selected sites in porB3 is robust to the choice of prior.
Figure 5.
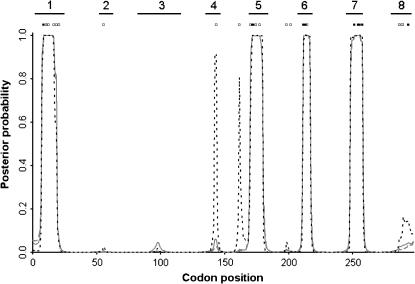
Sitewise posterior probability of diversifying selection (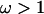 ) for the N. meningitidis porB3 carriage study, under prior A (shaded solid line), prior B (shaded dashed line), and prior A with the recombination rate forced to equal zero (dotted line). The loop regions are numbered above. Those sites identified as under weak (open squares) and strong (solid squares) diversifying selection by Urwin et al. (2002) are shown.
) for the N. meningitidis porB3 carriage study, under prior A (shaded solid line), prior B (shaded dashed line), and prior A with the recombination rate forced to equal zero (dotted line). The loop regions are numbered above. Those sites identified as under weak (open squares) and strong (solid squares) diversifying selection by Urwin et al. (2002) are shown.
Figure 4, a and b, compares the posterior probability of ρ given priors A and B. Under prior B, the posterior on ρ is somewhat flatter, with tighter credible intervals. The average 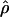 is largely the same for most of the sequence, except at the far ends, where
is largely the same for most of the sequence, except at the far ends, where 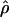 increases sharply. This is an edge effect where, in the lack of information about the recombination rate, the posterior has been overwhelmed by the prior. The uniform prior on ρ has mean 5, explaining the rapid increase. The effect is reflected in the posterior on R (Table 5), which has a similar lower bound, but a much increased upper bound. This striking sensitivity to the prior at the edges suggests that we should be cautious in interpreting the rates at the extremes of the sequence.
increases sharply. This is an edge effect where, in the lack of information about the recombination rate, the posterior has been overwhelmed by the prior. The uniform prior on ρ has mean 5, explaining the rapid increase. The effect is reflected in the posterior on R (Table 5), which has a similar lower bound, but a much increased upper bound. This striking sensitivity to the prior at the edges suggests that we should be cautious in interpreting the rates at the extremes of the sequence.
The posterior on μ is influenced by the high mean of the uniform prior (Table 5), to the extent that 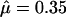 under prior B, which is only just inside the upper bound of the credible interval under prior A. In contrast, κ is not particularly sensitive to the prior, with largely overlapping credible intervals. φ shows a similar sensitivity to μ in responding to a considerable increase in the prior mean. The lower bound is almost unaffected, but the mean and upper bound show a marked increase.
under prior B, which is only just inside the upper bound of the credible interval under prior A. In contrast, κ is not particularly sensitive to the prior, with largely overlapping credible intervals. φ shows a similar sensitivity to μ in responding to a considerable increase in the prior mean. The lower bound is almost unaffected, but the mean and upper bound show a marked increase.
Model criticism:
An essential property of any statistical model is that it should be falsifiable. A useful approach in Bayesian inference, and one that we use here, is that of posterior predictive P-values (Rubin 1984; see also, e.g., Bollback 2002, 2005; Nielsen and Huelsenbeck 2002). Here we take model to mean the probability model together with the posterior distribution of the model parameters. In essence, if the model is a good description of the data, then further data sets simulated under that model ought to “look like” the real data. If they do not, then the model is failing in some important way. By look like we mean that with respect to some statistic D, the observed value of that statistic, 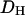 should fall well within the range of values for the simulated data sets,
should fall well within the range of values for the simulated data sets, 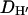 , where we use
, where we use 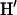 to denote a simulated data set.
to denote a simulated data set.
The posterior predictive P-value is defined as the probability under the model of observing a discrepancy statistic D as large as that observed,
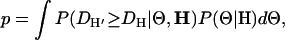 |
(8) |
where the integration is approximated by
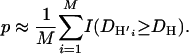 |
(9) |
In this equation, M is a large number (we used 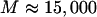 ),
), 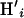 is simulated from the posterior distribution
is simulated from the posterior distribution 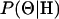 , and I is the indicator function. It is important to note that we simulated under the exact probability model specified by the PAC likelihood and used in inference, which is not the coalescent but an approximation to it.
, and I is the indicator function. It is important to note that we simulated under the exact probability model specified by the PAC likelihood and used in inference, which is not the coalescent but an approximation to it.
Discrepancy statistics have to be chosen that describe some aspect of the data that should be fit well by the model. This is important because it is unlikely that a model will fit all aspects of the data well. Statistics that are sensitive to mutation are S, the number of segregating sites and 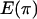 , the average number of pairwise differences. For recombination, the variance in the number of pairwise differences
, the average number of pairwise differences. For recombination, the variance in the number of pairwise differences  and the minimum number of recombination events
and the minimum number of recombination events 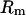 (Hudson and Kaplan 1985) are useful statistics. We also used the correlation between physical distance and LD (
(Hudson and Kaplan 1985) are useful statistics. We also used the correlation between physical distance and LD (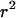 ,
, 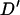 , and G4) that we used previously in the permutation tests. For selection we introduce U, which is sensitive to any tendency for the simulated data to have too many or too few nonsynonymous changes on average,
, and G4) that we used previously in the permutation tests. For selection we introduce U, which is sensitive to any tendency for the simulated data to have too many or too few nonsynonymous changes on average,
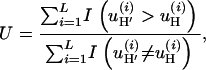 |
(10) |
where 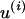 is the number of nonsynonymous pairwise differences minus the number of synonymous pairwise differences at site i. U should be centered around 0.5.
is the number of nonsynonymous pairwise differences minus the number of synonymous pairwise differences at site i. U should be centered around 0.5. 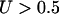 indicates a bias toward diversifying selection and
indicates a bias toward diversifying selection and 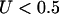 a bias toward functional constraint. Finally, we use Tajima's (1989) D, which is sensitive to directional selection, balancing selection, and demography, not forces that we modeled explicitly.
a bias toward functional constraint. Finally, we use Tajima's (1989) D, which is sensitive to directional selection, balancing selection, and demography, not forces that we modeled explicitly.
As with a classical P-value, if P is very small then the model does not fit the data well. Table 6 shows the observed values of all the discrepancy statistics and the two-tailed posterior predictive P-values for the carriage study under priors A and B. Of all the discrepancy statistics, the only posterior predictive P-value in the first two columns <0.05 is S for prior B. To obtain a single posterior predictive P-value for each model we combined information from one each of the mutation-sensitive, recombination-sensitive, and selection-sensitive statistics (S, 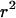 , and U). Accounting for the multiplicity of P-values using Bonferroni would be too conservative because the statistics are not independent. Instead we use the procedure in appendix c. Table 6 shows that the combined posterior predictive P-values for the carriage study under priors A and B are
, and U). Accounting for the multiplicity of P-values using Bonferroni would be too conservative because the statistics are not independent. Instead we use the procedure in appendix c. Table 6 shows that the combined posterior predictive P-values for the carriage study under priors A and B are 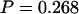 and
and 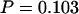 , respectively. Neither one is in the 5% tail of the distribution, suggesting that the model fit is adequate with respect to mutation, recombination, and selection insofar as the dN/dS ratio is concerned. Tajima's D was positive (
, respectively. Neither one is in the 5% tail of the distribution, suggesting that the model fit is adequate with respect to mutation, recombination, and selection insofar as the dN/dS ratio is concerned. Tajima's D was positive (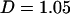 ), which may indicate balancing selection or population structure, but the P-value for each prior was not in the 5% tail. So while we have not attempted to model these forces, the model fit appears to be adequate.
), which may indicate balancing selection or population structure, but the P-value for each prior was not in the 5% tail. So while we have not attempted to model these forces, the model fit appears to be adequate.
TABLE 6.
Posterior predictive P-values
| Carriage study
|
Global study | |||||
|---|---|---|---|---|---|---|
| Observed | Prior A | Prior B | Prior A: 
|
Observed | Prior A | |
 |
67 | 0.236 | 0.039 | 0.008 | 92 | 0.391 |
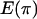 |
25.3 | 0.340 | 0.179 | 0.003 | 26.9 | 0.068 |
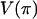 |
94.0 | 0.268 | 0.391 | 0.000 | 98.2 | 0.118 |
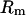 |
15 | 0.293 | 0.658 | 0.070 | 12 | 0.036 |
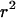 |
−0.13 | 0.247 | 0.265 | 0.002 | −0.07 | 0.002 |
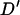 |
−0.24 | 0.440 | 0.353 | 0.000 | −0.10 | 0.059 |
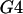 |
−0.22 | 0.443 | 0.332 | 0.000 | −0.09 | 0.144 |
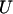 |
0.5 | 0.543 | 0.878 | 0.711 | 0.5 | 0.621 |
 |
1.05 | 0.121 | 0.058 | 0.567 | 0.97 | 0.398 |
| Combined | 0.268 | 0.103 | 0.001 | 0.013 | ||
Global study:
We analyzed the 79-sequence PorB3 data of Urwin et al. (2002) to investigate how the violation of the coalescent model would affect inference, using prior A. For computational tractability we used one randomly chosen ordering of the haplotypes. We ran three MCMC chains, each 500,000 iterations in length, with a burn-in of 20,000 iterations. The chains were merged to obtain the posteriors. Table 5 shows that 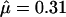 was barely larger than that for the carriage study, and the credible intervals overlapped almost entirely. The rate of insertion/deletion, φ was not greatly affected (
was barely larger than that for the carriage study, and the credible intervals overlapped almost entirely. The rate of insertion/deletion, φ was not greatly affected (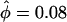 ), nor was the transition–transversion ratio (
), nor was the transition–transversion ratio (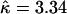 ). But the total recombination rate doubled to
). But the total recombination rate doubled to 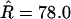 with no overlap in the credible intervals. Across the sites, the recombination map (Figure 4c) does not differ greatly in the left half of the sequence (cf. Figure 4a), but thereafter rises rapidly to ∼
with no overlap in the credible intervals. Across the sites, the recombination map (Figure 4c) does not differ greatly in the left half of the sequence (cf. Figure 4a), but thereafter rises rapidly to ∼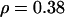 . The low posterior predictive P-values for the recombination-sensitive discrepancy statistics (Table 6) suggest caution in the interpretation of
. The low posterior predictive P-values for the recombination-sensitive discrepancy statistics (Table 6) suggest caution in the interpretation of 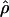 .
.
However, inference on ω was hardly affected. Loops 1, 5, 6, and 7 still have very high posterior probabilities of diversifying selection. The magnitude of ω inferred for each loop is comparable, with the 95% HPD intervals for the four loops (2.89, 7.28), (3.47, 8.17), (3.22, 8.79), and (3.10, 7.60). The only substantive difference is in loop 8, which now also has high posterior probability of 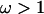 . The 95% HPD interval for the peak ω in loop 8 is (0.66, 2.87) and
. The 95% HPD interval for the peak ω in loop 8 is (0.66, 2.87) and 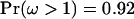 . This difference can be explained by sites in loop 8 that exhibit amino acid variation in the global study but not in the carriage study. The average
. This difference can be explained by sites in loop 8 that exhibit amino acid variation in the global study but not in the carriage study. The average 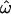 for the whole sequence is 0.91, and excluding sites for which
for the whole sequence is 0.91, and excluding sites for which 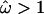 , it drops to 0.22, both values comparable to those of the carriage study.
, it drops to 0.22, both values comparable to those of the carriage study.
Effect of recombination on inference:
Ancestral recombination can cause false positives in phylogenetic methods (Anisimova et al. 2003; Shriner et al. 2003). If this has had an important effect on the analysis of meningococcal PorB3 then we should expect to see those false positives when we compare the results of the CODEML analysis (Urwin et al. 2002) to those presented here. Those sites identified as under weak (open squares) and strong (solid squares) diversifying selection by CODEML are illustrated in Figure 5. All of the strongly selected sites and all but five of the weakly selected sites fall within loops 1 and 5–8. With the exception of loop 8 all these sites had high posterior probability of diversifying selection for the carriage study (Figure 5). When the global study is analyzed, loop 8 also has high posterior probability of diversifying selection. Therefore there are just five sites where CODEML inferred diversifying selection but omegaMap did not. These are candidates for false positives.
There are a number of possible explanations for discrepancies of this kind, including the following:
The approximation in omegaMap has given rise to false negatives. The PAC likelihood does not explicitly model the genealogy and this might have unexpected effects.
The block-like prior in omegaMap caused false negatives. Imposing a model in which adjacent sites share a common selection parameter might disfavor isolated sites under diversifying selection.
Recombination has caused CODEML to give false positives.
In an attempt to distinguish between the explanations, we performed an analysis of the carriage study in which we forced the recombination rate to equal zero. Using prior A, we ran three chains for 500,000 iterations each. After a burn-in of 20,000 iterations the chains were compared for convergence and merged to give the posterior.
In Figure 5 the sitewise posterior probability of diversifying selection is plotted (dotted line) for comparison with the other analyses. The false-positive candidates are located at sites 55, 143, 161, 198, and 201. Of these, the first two are located in loops 2 and 4, respectively. The remaining three are not in loops. Comparison of our Figure 3b to Urwin et al.'s (2002) Figure 2b shows that these latter three disputed sites are located in a cytoplasmic region of the protein. The sitewise posterior probability of diversifying selection is very similar to our other analyses (Figure 5), except at two positions. These two positions correspond to two of the five false-positive candidates: sites 143 and 161. Although we cannot be certain that these sites are false positives, the results are suggestive.
The posterior predictive P-values (Table 6) show that the deleterious effect of assuming no recombination is not confined to recombination-sensitive discrepancy statistics. The mutation-sensitive parameters also have extremely low P-values [0.008 and 0.003 for S and 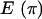 , respectively]. The combined test shows that the model as a whole is a very poor description of the data (
, respectively]. The combined test shows that the model as a whole is a very poor description of the data (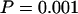 ). Although the selection-sensitive parameters do not have significant P-values, the consequence of the model inadequacy is to cast doubt on all inferences made from it.
). Although the selection-sensitive parameters do not have significant P-values, the consequence of the model inadequacy is to cast doubt on all inferences made from it.
The PAC model in the absence of recombination does not default to the coalescent with no recombination because the tree is still not modeled explicitly. Therefore it is unlikely that the assumption of no recombination will affect a PAC model and a phylogenetic model in an exactly equivalent fashion. Nevertheless, when we assume there is no recombination, sites that otherwise had low posterior probability of diversifying selection attained high posterior probabilities. This outcome is exactly what is predicted by the work of Shriner et al. (2003) and Anisimova et al. (2003).
DISCUSSION
In this article we have presented a new method for estimating the selection parameter ω and the recombination rate ρ from a sample of gene sequences. Uncertainty in the evolutionary history was taken into account using a coalescent-based approximate (PAC) likelihood. Variation in ω and ρ was modeled as a block-like structure with a variable number of blocks. We averaged over the number and position of the blocks using reversible-jump MCMC to obtain the posterior distribution of the parameters. Using simulations, we showed that the new method has good power to detect variation in ω and ρ, and that the two do not appear to be confounded. The method has a low false-positive rate for detecting sites under diversifying selection. We applied the method to the porB locus of N. meningitidis and performed prior sensitivity analysis and model criticism to verify the results.
In addition to the ability to coestimate ω and ρ, there are several advantages to the new method. Some of these are a consequence of the Bayesian approach, and all of them rely on the computational tractability of the PAC model. First among these is that our posterior probabilities of diversifying selection are fully Bayesian, so they incorporate uncertainty about the evolutionary history, as well as uncertainty in the other parameters. In the presence of recombination, there is likely to be a great deal of uncertainty in the evolutionary history. The computationally efficient PAC likelihood means that in the posterior, ω can take on any positive value, rather than having to constrain it to a discrete number of points or approximate a continuous distribution in a similar manner.
In any Bayesian approach it is necessary to specify a prior distribution on all parameters. It is possible to represent a lack of prior knowledge with relatively flat priors, although we note that in reversible-jump MCMC it is not possible to use an improper prior (Green 1995). However, in this article we have taken a different approach, that of prior sensitivity analysis. Prior sensitivity analysis reveals which aspects of the posterior distribution, if any, are unduly influenced by the choice of prior. This in turn reveals which aspects of the model the data are uninformative about. For example, Figure 4b shows that the data contained very little information about recombination rates at the extremes of the sequence. In contrast, inference about diversifying selection in porB3 (Figure 5) was robust to the prior.
In a Bayesian setting it is entirely natural to impose a block-like structure on the joint distribution of ω across sites. At sites where the data are compatible with a block structure this allows information about ω to be combined across sites, but when the signal in the data is strong enough it will overwhelm the block model. The sensitivity to the signal is controlled by 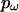 . The user can also specify an independent ω for every site
. The user can also specify an independent ω for every site 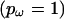 or a single ω for the whole sequence
or a single ω for the whole sequence 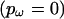 . Imposing a block-like structure is biologically justifiable insofar as adjacent sites in the primary sequence will be closely juxtaposed in the tertiary structure and, as such, are more likely to perform similar functional duties. If anything, the model is overly simplistic because the tertiary structure could in principle be used to impose longer-range dependencies on the prior. In a maximum-likelihood setting, implementing the block structure described here would be computationally unfeasible.
. Imposing a block-like structure is biologically justifiable insofar as adjacent sites in the primary sequence will be closely juxtaposed in the tertiary structure and, as such, are more likely to perform similar functional duties. If anything, the model is overly simplistic because the tertiary structure could in principle be used to impose longer-range dependencies on the prior. In a maximum-likelihood setting, implementing the block structure described here would be computationally unfeasible.
On the basis of previous work (Schierup and Hein 2000; Anisimova et al. 2003; Shriner et al. 2003) and because of clear model misspecification we have claimed that it is inappropriate to analyze data that show evidence for recombination using phylogenetic methods. Yet neither the coalescent nor the approximation to the coalescent we use here inevitably fits data from a recombining population. That is why we have advocated the use of goodness-of-fit testing. Posterior predictive P-values allow for goodness-of-fit testing in a Bayesian setting when no explicit alternative model is specified. The posterior predictive P-values in Table 6 showed that the model with no recombination is a very poor fit to the data, and Figure 5 showed that in the PAC model the assumption of no recombination leads to an increase in the number of sites experiencing diversifying selection, which would be expected if this assumption increases the false-positive rate. Posterior predictive P-values have been criticized for being conservative in the sense that the true discrepancy between the model and the data is suppressed by using the same data for both fitting the model and evaluating its goodness-of-fit (see, e.g., Meng 1994). However, in the absence of truly independent subsets of the data, caused by shared evolutionary ancestry in the gene sequences, posterior predictive P-values are a pragmatic choice for the important task of goodness-of-fit testing.
Posterior predictive P-values (Table 6) suggested that the coalescent approximation was not a good fit to the N. meningitidis global study. This was not unexpected because the global study did not represent a random sample from any population in a meaningful sense. In constructing the carriage study we were careful not to include more than one haplotype from any one host. The idea was to envisage the bacterial population as a metapopulation in which each deme corresponds to a host; colonization and extinction correspond to infection and clearing of infection. Wakeley and Aliacar (2001) have shown that a metapopulation model with many demes converges to a coalescent model when migration (transmission) events are random and each deme is represented by no more than a single haplotype. Consistent with this model, the posterior predictive P-values showed that the coalescent approximation did provide an adequate fit to the carriage study (Table 6). There is more work to be done on formalizing the relationship between genetic models, such as the coalescent, and epidemiological models, but it may be possible in the future to use models such as the one presented here to estimate parameters of epidemiological relevance.
Acknowledgments
We thank Rachel Urwin for her help and advice and providing the sequence alignments for the meningococcal data; Jeremy Derrick, who provided the molecular structure of PorB3; Ziheng Yang for kindly offering part of his C code for this work; and Stephen Leslie, Jonathan Marchini, and Bob Griffiths for useful ideas and advice. This work was conducted on a multinode AMD compute cluster that was bought with a grant awarded by the Wolfson Foundation to Peter Donnelly, without which it could not have been completed. D.J.W. is funded by the Biotechnology and Biological Sciences Research Council. The program omegaMap can be downloaded from www.danielwilson.me.uk.
APPENDIX A: MUTATION MODEL
Li and Stephens (2003) use a hidden Markov model (HMM) to model the sampling distribution of haplotypes in the presence of recombination. Under the model, the 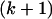 th haplotype is a mosaic copy of the first k haplotypes. The latent variable of the HMM records which of the first k haplotypes the
th haplotype is a mosaic copy of the first k haplotypes. The latent variable of the HMM records which of the first k haplotypes the 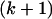 th is a copy of at a given site. Conditional on the latent variable, the emission probability gives the probability of observing state
th is a copy of at a given site. Conditional on the latent variable, the emission probability gives the probability of observing state 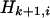 at site i.
at site i.
Informally, we think of the latent variable as recording the haplotype that is the least distant in the evolutionary tree at that site (call this haplotype x, 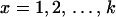 ). Under a coalescent model (Kingman 1982; Hudson 1983), the time (in units of PNe generations) to the common ancestor of haplotypes x and
). Under a coalescent model (Kingman 1982; Hudson 1983), the time (in units of PNe generations) to the common ancestor of haplotypes x and 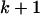 is known (R. C. Griffiths, unpublished data) and to the order of the approximation is exponentially distributed with rate k.
is known (R. C. Griffiths, unpublished data) and to the order of the approximation is exponentially distributed with rate k.
Let 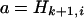 and
and 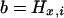 . The probability of observing a pair of states
. The probability of observing a pair of states 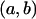 given the time t to their common ancestor for a reversible mutation rate matrix is
given the time t to their common ancestor for a reversible mutation rate matrix is
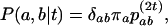 |
(A1) |
where
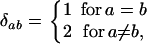 |
(A2) |
and 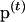 is the transition probability matrix. The transition probability can be solved numerically (e.g., Grimmett and Stirzaker 2001), so
is the transition probability matrix. The transition probability can be solved numerically (e.g., Grimmett and Stirzaker 2001), so
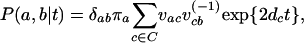 |
(A3) |
where C represents the possible states (in our case 61 codons), v is a matrix of eigenvectors of the mutation rate matrix, v−1 is its inverse, and d is a vector of the corresponding eigenvalues. Thus using the coalescent model for the time t, we can obtain an expression for the HMM emission probability under any reversible mutation model:
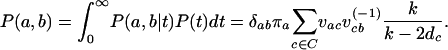 |
(A4) |
To be able to handle indels, we use a very simple extension of NY98 in which there is an extra indel state. This model is applied only to sites in the alignment that are segregating for an indel. Codons mutate to the indel state at rate 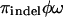 and back at rate
and back at rate 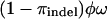 . Here πindel is the equilibrium frequency of indels (in sites segregating for indels),
. Here πindel is the equilibrium frequency of indels (in sites segregating for indels), 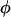 is the rate of insertion/deletion, and ω is the selection parameter for the block containing that site. The motivation for using this model is to capture the information regarding the underlying tree structure and mode of selection at sites segregating for indels, in the simplest possible way.
is the rate of insertion/deletion, and ω is the selection parameter for the block containing that site. The motivation for using this model is to capture the information regarding the underlying tree structure and mode of selection at sites segregating for indels, in the simplest possible way.
APPENDIX B: MCMC MOVES
In the MCMC scheme we use standard Metropolis–Hastings moves to change μ and κ, which are of the same form as move A below. To explore the block structure for the variation in the selection parameter we have four moves. Moves A and B are Metropolis–Hastings moves, while moves C and D are complementary reversible-jump moves (Green 1995). The moves for exploring the recombination rate are of the same form as those described here. For the purpose of illustration, we use an exponential prior for ω with rate parameter λ.
Move A—change ω within a block:
A new value ω′ is chosen so that 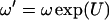 , where U ∼ Uniform(−1, 1). The acceptance probability is
, where U ∼ Uniform(−1, 1). The acceptance probability is
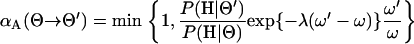 |
(B1) |
Move B—extend a block 5′ or 3′:
The block to extend is chosen uniformly at random, and for each block the direction is chosen with equal probability. If the 5′-most or the 3′-most block is chosen to be extended 5′ or 3′, respectively, the move is rejected. The number of sites to extend the block, 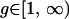 is chosen from a geometric distribution with some parameter. If extending the block g sites in the chosen direction would cause it to merge with the adjacent block, the move is rejected. The acceptance probability is
is chosen from a geometric distribution with some parameter. If extending the block g sites in the chosen direction would cause it to merge with the adjacent block, the move is rejected. The acceptance probability is
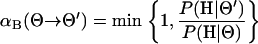 |
(B2) |
Following Green (1995), when there are B transition points, moves C and D are proposed with relative probabilities 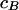 and
and 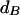 , where
, where
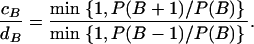 |
Move C—split a block:
A position s* is chosen uniformly at random to create a new transition point. The block spanning position s*, which we denote block j and has parameter 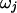 , is split and the two new blocks are assigned parameters
, is split and the two new blocks are assigned parameters 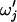 and
and 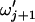 , respectively, such that
, respectively, such that
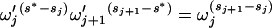 |
and
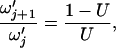 |
where U ∼ Uniform(0, 1). The acceptance probability is
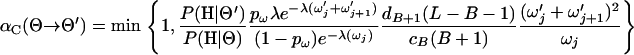 |
(B3) |
Move D—merge a block:
One of the 5′-most B blocks is chosen uniformly at random to merge with its 3′ neighbor. The parameter for the merged block 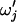 is chosen from the current blocks' parameters
is chosen from the current blocks' parameters 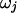 and
and 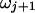 so that
so that
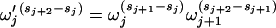 |
and the acceptance probability is
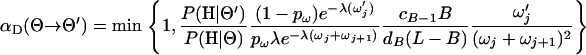 |
(B4) |
APPENDIX C: COMBINING P-VALUES
From the posterior distribution of parameters we simulate a large number of data sets, M. For any particular discrepancy statistic we can calculate a marginal posterior predictive P-value using Equation 9. The P-value is made two-tailed in the usual way. To combine two-tailed P-values for N different discrepancy statistics, denote the vector of discrepancy statistics for data set j:
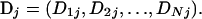 |
(C1) |
Transform the marginal distribution of each discrepancy statistic i (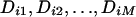 ) into a standard normal distribution, so that
) into a standard normal distribution, so that
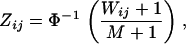 |
(C2) |
where 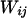 is the marginal rank (with respect to j) of discrepancy statistic
is the marginal rank (with respect to j) of discrepancy statistic 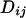 , and
, and 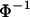 is the quantile function (inverse cumulative distribution function) for the standard normal distribution. We then assume that the joint distribution of
is the quantile function (inverse cumulative distribution function) for the standard normal distribution. We then assume that the joint distribution of 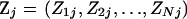 is multivariate normal with zero mean and variance–covariance matrix
is multivariate normal with zero mean and variance–covariance matrix 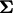 , where
, where
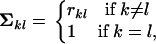 |
(C3) |
where 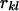 is the correlation coefficient between the transformed discrepancy statistics k and l (
is the correlation coefficient between the transformed discrepancy statistics k and l (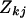 and
and 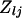 ) over data sets j. Next transform
) over data sets j. Next transform 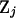 to remove the correlation structure
to remove the correlation structure
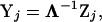 |
(C4) |
where 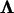 is obtained from the matrix factorization
is obtained from the matrix factorization
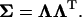 |
(C5) |
Include the observed values of the discrepancy statistics 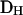 in the above procedure to obtain
in the above procedure to obtain 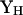 . Assuming that the uncorrelated transformed discrepancy statistics are independent, then
. Assuming that the uncorrelated transformed discrepancy statistics are independent, then
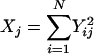 |
(C6) |
has a chi-squared distribution with N degrees of freedom. This can be verified by a histogram of the Xj's. A one-tailed chi-square test of 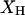 combines the two-tailed posterior predictive P-values.
combines the two-tailed posterior predictive P-values.
References
- Anisimova, M., J. P. Bielawski and Z. Yang, 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18: 1585–1592. [DOI] [PubMed] [Google Scholar]
- Anisimova, M., J. P. Bielawski and Z. Yang, 2002. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol. Biol. Evol. 19: 950–958. [DOI] [PubMed] [Google Scholar]
- Anisimova, M., R. Nielsen and Z. Yang, 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164: 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla, P., 2003. The evolutionary genomics of pathogen recombination. Nat. Rev. Genet. 4: 50–60. [DOI] [PubMed] [Google Scholar]
- Bishop, J. G., A. M. Dean and T. Mitchell-Olds, 2000. Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA 97: 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollback, J. P., 2002. Bayesian model adequacy and choice in phylogenetics. Mol. Biol. Evol. 19: 1171–1180. [DOI] [PubMed] [Google Scholar]
- Bollback, J. P., 2005. Posterior mapping and posterior predictive distributions, pp. 439–462 in Statistical Methods in Molecular Evolution, edited by R. Nielsen. Springer-Verlag, New York.
- de Oliveira, T., M. Salemi, M. Gordon, A. Vandamme, E. J. van Rensburg et al., 2004. Mapping sites of positive selection and amino acid diversification in the HIV genome. Genetics 167: 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrick, J. P., R. Urwin, J. Suker, I. M. Feavers and M. C. J. Maiden, 1999. Structural and evolutionary inference from molecular variation in Neisseria porins. Infect. Immun. 67: 2406–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A. J., O. G. Pybus, A. Rambaut, R. Forsberg and A. G. Rodrigo, 2003. Measurably evolving populations. Trends Ecol. Evol. 18: 481–488. [Google Scholar]
- Fearnhead, P., and P. Donnelly, 2001. Estimating recombination rates from population genetic data. Genetics 159: 1299–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17: 368–376. [DOI] [PubMed] [Google Scholar]
- Filip, L. C., and N. I. Mundy, 2004. Rapid evolution by positive Darwinian selection in the extracellular domain of the abundant lymphocyte protein CD45 in primates. Mol. Biol. Evol. 21: 1504–1511. [DOI] [PubMed] [Google Scholar]
- Ford, M. J., 2001. Molecular evolution of transferrin: evidence for positive selection in salmonids. Mol. Biol. Evol. 18: 639–647. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Green, P. J., 1995. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82: 711–732. [Google Scholar]
- Griffiths, R. C., and P. Marjoram, 1997. An ancestral recombination graph, pp. 257–270 in Progress in Population Genetics and Human Evolution, edited by P. Donnelly and S. Tavare. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Grimmett, G., and D. Stirzaker, 2001. Probability and Random Processes, Ed. 3. Oxford University Press, London/New York/Oxford.
- Hudson, R. R., 1983. Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23: 183–201. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA-sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and K. A. Dyer, 2004. Bayesian estimation of positively selected sites. J. Mol. Evol. 58: 661–672. [DOI] [PubMed] [Google Scholar]
- Jolley, K. A., J. Kalmusova, E. J. Feil, S. Gupta, M. Musilek et al., 2000. Carried meningococci in the Czech Republic: a diverse recombining population. J. Clin. Microbiol. 38: 4492–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley, K. A., D. J. Wilson, P. Kriz, G. McVean and M. C. J. Maiden, 2005. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 22: 562–569. [DOI] [PubMed] [Google Scholar]
- Kingman, J. F. C., 1982. On the genealogy of large populations. J. Appl. Probab. 19A: 27–43. [Google Scholar]
- Kosakovsky Pond, S. L., and S. D. Frost, 2005. Not so different after all: a comparison of methods for detecting amino-acid sites under selection. Mol. Biol. Evol. 22: 1208–1222. [DOI] [PubMed] [Google Scholar]
- Kuhner, M. K., J. Yamato and J. Felsenstein, 2000. Maximum likelihood estimation of recombination rates from population data. Genetics 156: 1393–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N., and M. Stephens, 2003. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165: 2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martz, E., 2002. Protein explorer: easy yet powerful macromolecular visualization. Trends Biochem. Sci. 27: 107–109. [DOI] [PubMed] [Google Scholar]
- Massingham, T., and N. Goldman, 2005. Detecting amino acid sites under positive selection and purifying selection. Genetics 169: 1753–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean, G., P. Awadalla and P. Fearnhead, 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean, G. A. T., S. R. Myers, S. Hunt, P. Deloukas, D. R. Bentley et al., 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Meng, X.-L., 1994. Posterior predictive P-values. Ann. Stat. 22: 1142–1160. [Google Scholar]
- Meunier, J., and A. Eyre-Walker, 2001. The correlation between linkage disequilibrium and distance: implications for recombination in hominid mitochondria. Mol. Biol. Evol. 18: 2132–2135. [DOI] [PubMed] [Google Scholar]
- Minin, V. N., K. S. Dorman, F. Fang and M. A. Suchard, 2005. Dual multiple change-point model leads to more accurate recombination detection. Bioinformatics 21: 3034–3042. [DOI] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., B. C. Meyers, R. W. Michelmore and B. S. Gaut, 2002. Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12: 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury, B., 2004. Differential selection of genes of cucumber mosaic virus subgroups. Mol. Biol. Evol. 21: 1602–1611. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., T. Gojobori and T. Ikemura, 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R., and J. P. Huelsenbeck, 2002. Detecting positively selected amino acid sites using posterior predictive p-values, pp. 576–588 in Pacific Symposium on Biocomputing, Proceedings, edited by R. B. Altman, A. K. Dunker, L. Hunter, K. Lauderdale and T. E. Klein. World Scientific, Singapore. [DOI] [PubMed]
- Nielsen, R., and Z. Yang, 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher et al., 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404: 502–506. [DOI] [PubMed] [Google Scholar]
- Peek, A. S., V. Souza, L. E. Eguiarte and B. S. Gaut, 2001. The interaction of protein structure, selection, and recombination on the evolution of the type-1 fimbrial major subunit (fimA) from Escherichia coli. J. Mol. Evol. 52: 193–204. [DOI] [PubMed] [Google Scholar]
- Rubin, D. B., 1984. Bayesianly justifiable and relevant frequency calculations for the applied statistician. Ann. Stat. 12: 1151–1172. [Google Scholar]
- Schierup, M. H., and J. Hein, 2000. Consequences of recombination on traditional phylogenetic analysis. Genetics 156: 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner, D., D. C. Nickle, M. A. Jensen and J. I. Mullins, 2003. Potential impact of recombination on sitewise approaches for detecting positive natural selection. Genet. Res. 81: 115–121. [DOI] [PubMed] [Google Scholar]
- Smith, N. H., J. M. Smith and B. G. Spratt, 1995. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis—evidence of positive Darwinian selection. Mol. Biol. Evol. 12: 363–370. [DOI] [PubMed] [Google Scholar]
- Stumpf, M. P. H., and G. A. T. McVean, 2003. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 4: 959–968. [DOI] [PubMed] [Google Scholar]
- Suchard, M. A., R. E. Weiss, K. S. Dorman and J. S. Sinsheimer, 2002. Oh brother, where art thou? A Bayes factor test for recombination with uncertain heritage. Syst. Biol. 51: 715–728. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., R. Nielsen and Q. Yang, 2003. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 20: 18–20. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiddy, S. S., C. H. Woelk and E. C. Holmes, 2002. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J. Gen. Virol. 83: 1679–1689. [DOI] [PubMed] [Google Scholar]
- Urwin, R., E. C. Holmes, A. J. Fox, J. P. Derrick and M. C. J. Maiden, 2002. Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol. Biol. Evol. 19: 1686–1694. [DOI] [PubMed] [Google Scholar]
- Wakeley, J., and N. Aliacar, 2001. Gene genealogies in a metapopulation. Genetics 159: 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, D. J., D. Falush and G. Mcvean, 2005. Germs, genomes and genealogies. Trends Ecol. Evol. 20: 39–45. [DOI] [PubMed] [Google Scholar]
- Winckler, W., S. R. Myers, D. J. Richter, R. C. Onofrio, G. J. Mcdonald et al., 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308: 107–111. [DOI] [PubMed] [Google Scholar]
- Wong, W. S., Z. Yang, N. Goldman and R. Nielsen, 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and W. J. Swanson, 2002. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol. Biol. Evol. 19: 49–57. [DOI] [PubMed] [Google Scholar]
- Yang, Z., R. Nielsen, N. Goldman and A. M. Pedersen, 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]