Abstract
Small peptides of two to six residues serve as important sources of amino acids and nitrogen required for growth by a variety of organisms. In the yeast Saccharomyces cerevisiae, the membrane transport protein Ptr2p, encoded by PTR2, mediates the uptake of di/tripeptides. To identify genes involved in regulation of dipeptide utilization, we performed a systematic, functional examination of this process in a haploid, nonessential, single-gene deletion mutant library. We have identified 103 candidate genes: 57 genes whose deletion decreased dipeptide utilization and 46 genes whose deletion enhanced dipeptide utilization. On the basis of Ptr2p-GFP expression studies, together with PTR2 expression analysis and dipeptide uptake assays, 42 genes were ascribed to the regulation of PTR2 expression, 37 genes were involved in Ptr2p localization, and 24 genes did not apparently affect Ptr2p-GFP expression or localization. The 103 genes regulating dipeptide utilization were distributed among most of the Gene Ontology functional categories, indicating a very wide regulatory network involved in transport and utilization of dipeptides in yeast. It is anticipated that further characterization of how these genes affect peptide utilization should add new insights into the global mechanisms of regulation of transport systems in general and peptide utilization in particular.
PEPTIDES can serve as a major nutritional source of amino acids and nitrogen for microbial cell growth and for human nutrition as well (Pritchard and Coolbear 1993; Daniel 2004). Peptides are generated by the action of proteases and peptidases in environments where microbes reside (Gonzales and Robert-Baudouy 1996; Daniel 2004). All cellular organisms from bacteria to fungi, plants, and mammals are capable of taking up small peptides (two to six amino acids) through a system mediated by cytoplasmic membrane transporters specific for peptides (Hauser et al. 2001). In mammalian systems, two major peptide transporters, PepT1 and PepT2, have been found in the intestine and kidney, respectively (Fei et al. 1994; Liu et al. 1995). The PepT1 transporter provides the means for absorption of nutritional peptides and peptidomimetic drugs, such as β-lactam antibiotics, angiotensin-converting enzyme inhibitors, renin inhibitors, and antivirals. The substrate specificity of these transporters has provided an impetus to many pharmaceutical companies to develop peptide transporters as drug delivery systems (Lee 2000; Herrera-Ruiz and Knipp 2003).
Two peptide transport systems have been characterized in the model eukaryote Saccharomyces cerevisiae: the peptide transport (PTR) system transports di/tripeptides (Steiner et al. 1995) and the oligopeptide transport (OPT) system highly favors transport of peptides of four to five amino acid residues and glutathione (Lubkowitz et al. 1997; Miyake et al. 2002). While PTR family members are present in all cells studied to date and are in the same family as the mammalian transporters PepT1 and PepT2, OPT family members have been found only in plants and fungi (Hauser et al. 2001). Both the PTR and OPT transport systems are dependent on the proton-motive force, are predicted to have 12 transmembrane domains, and have specific signature sequences distinguishing them from one another and from all other proteins in the database (Hauser et al. 2001). Yet the amino acid sequence of the PTR and OPT members places them in different, unrelated families of transport proteins with presumably totally separate evolutionary origins. The only S. cerevisiae member of the PTR system is Ptr2p, encoded by the PTR2 gene. Di/Tripeptide utilization is regulated by a number of means including PTR2 transcriptional regulation.
In early studies predating the cloning of PTR2, peptide utilization was shown to be upregulated by growing cells on organic nitrogen sources such as allantoin, isoleucine, or proline, defined as poor nitrogen sources in comparison to the rich nitrogen source ammonium sulfate (Island et al. 1991). Peptide utilization was also stimulated markedly by addition of micromolar amounts of certain amino acids, most notably leucine and tryptophan, to the growth medium (Island et al. 1987; Baetz et al. 2004). These environmental conditions were later shown to upregulate the expression of PTR2 (Perry et al. 1994). Although nitrogen source apparently regulates PTR2 expression via nitrogen catabolite repression (Marzluf 1997), amino acids regulate PTR2 expression via the Ssy1p-Ptr3p-Ssy5p (SPS) signal transduction pathway (Barnes et al. 1998; Forsberg and Ljungdahl 2001b; Forsberg et al. 2001a,b).
In the SPS complex, Ssy1p is a transmembrane receptor that senses extracellular amino acids. Ptr3p and Ssy5p interact with the cytoplasmically located N terminus of Ssy1p (Forsberg and Ljungdahl 2001a; Poulsen et al. 2005). Two related transcription factors, Stp1p and Stp2p, are downstream of the SPS complex and regulate the expression of PTR2 and branched-chain amino acid permeases (de Boer et al. 2000). Stp1p and Stp2p are synthesized in an inactive form, and the activation of Stp1p and Stp2p depends on the endoproteolytic processing of their N-terminal domain mediated by Ptr3p and Ssy5p. The truncated forms of Stp1p and Stp2p are translocated into the nucleus to upregulate expression of downstream genes including PTR2 (Andreasson and Ljungdahl 2002, 2004).
In addition to the SPS complex regulation, PTR2 expression is positively regulated by the import of di/tripeptides with basic (Arg, His, or Lys) and bulky (Ile, Leu, Phe, Trp, or Tyr) N-terminal residues that bind to Ubr1p to allosterically activate Ubr1p-mediated degradation of the PTR2 repressor Cup9p (Byrd et al. 1998; Turner et al. 2000; Du et al. 2002). Relief of Cup9p repression of PTR2 results in enhanced PTR2 expression. In a cup9 null mutant strain, PTR2 is overexpressed, resulting in a marked increase in dipeptide uptake (Byrd et al. 1998). Cup9p is degraded by Ptr1p through the ubiquitinylation pathway. In this pathway, Ptr1p acts as a scaffolding protein or ubiquitin ligase (E3) for two other proteins, Ubc2p and Ubc4p, which serve as ubiquitin-conjugating (E2) enzymes in the Cup9p degradation process (Xie and Varshavsky 1999). Deletion of PTR1 results in the stabilization of Cup9p, lack of expression of PTR2, and a null peptide-uptake phenotype (Island et al. 1991; Turner et al. 2000; Du et al. 2002). Other transcription factors, such as Dal81p/Uga35p, have also been reported to be involved in PTR2 expression, in ways apparently different from their involvement in the SPS or Cup9p pathways (Iraqui et al. 1999). Obviously, the systems regulating PTR2 expression are numerous and complex, and there has not been a systematic study to uncover other proteins involved in the regulation of peptide utilization.
Systematic screening of yeast deletion mutant collections has been successfully applied in identifying genes related to drug resistance (Page et al. 2003; Aouida et al. 2004; Baetz et al. 2004), human disease (Steinmetz et al. 2002), telomere function (Askree et al. 2004), centromeric cohesion (Marston et al. 2004), vacuolar protein sorting (Bonangelino et al. 2002), and other biological processes (Scherens and Goffeau 2004). To identify other gene products involved in the regulation of peptide utilization in yeast, we conducted a genomewide screen using the haploid, nonessential, single-gene deletion strain library. We have identified a total of 103 open reading frames (ORFs), accounting for ∼2% of the nonessential deletion mutants as being involved in causing increased or decreased utilization of peptides for growth. We monitored Ptr2p-GFP expression in the identified 103 deletion mutant strains, and genes involved in multiple cellular functions including transcriptional regulation and membrane trafficking were revealed as being involved in dipeptide utilization. The data generated provide a global view of molecular components regulating dipeptide utilization by S. cerevisiae.
MATERIALS AND METHODS
Strains, media, and mutant library growth assay screen:
A yeast haploid, nonessential, single-gene deletion mutant strain library was purchased from Open Biosystems (Huntsville, AL), containing 4827 deletion strains, 4750 in the BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) background and 77 strains in the BY4739 (MATα leu2Δ0 lys2Δ0 ura3Δ0) strain background. The list of strains contained in the library is available at the company's website (http://openbiosystems.com/yeast_knock_outs.php). The deletion mutants were arrayed in 96-well microtiter plates kept at −80° in YPD broth containing G418 and glycerol (1% yeast extract, 2% peptone, 2% dextrose, 200 μg/ml G418, and 15% glycerol). After thawing, the deletion mutants were inoculated into minimal medium supplemented with amino acids to satisfy the auxotrophic requirements, using a 96-well plate replicator. The minimal medium used in this study is referred to as MM + HLKU and contained (per liter) 20 g dextrose, 1.7 g yeast nitrogen base (Difco, Detroit) without (NH4)2SO4 and amino acids, 1 g allantoin as nitrogen source, supplemented with 20 μg/ml His (H), 30 μg/ml Leu (L), 30 μg/ml Lys (K), and 20 μg/ml Ura (U). The plates were incubated at 30° for 2 days, and then the liquid medium growth assay was performed as follows. Five microliters from each well were inoculated into 200 μl of three different liquid media: (1) MM + HLKU medium as described above, (2) MM + His–Leu (H–L)HKU medium, or (3) MM + (H–L)HKU + Trp medium. The MM + (H–L)HKU and MM + (H–L)HKU + Trp media contained 80 μm His–Leu dipeptide in place of leucine with all other components identical to the MM + HLKU medium, except that tryptophan (30 μg/ml) was added to the MM + (H–L)HKU + Trp medium. The purpose of the tryptophan addition was to induce PTR2 expression as shown in previous studies (Island et al. 1987). The efficiency of the utilization of His–Leu was reflected in cell growth. The minicultures in the microtiter plates were grown at 30° with shaking at 200 rpm. The OD620 was measured at incubation times of 0, 12, 24, 48, and 72 hr with a 96-well plate reader (Multiskan MCC/340; Labsystems, Helsinki). In some circumstances, growth at incubation times of 36, 60, 84, and 96 hr was also recorded. The ptr2 deletion mutant was not able to grow in the His–Leu-supplemented medium, although its growth in the medium MM + HKLU was excellent. Strains either defective in peptide utilization (ptr1 deletion) or overexpressing PTR2 (cup9 deletion) were inoculated into wells of each plate as controls showing defective or enhanced dipeptide utilization, respectively. In this manner, deletion strains were identified that grew more, or less, efficiently than wild type in MM + (H–L)HKU or MM + (H–L)HKU + Trp medium.
Solid medium growth assay:
Using the same principle as that employed in the liquid medium growth assay, a solid medium growth assay was used to determine whether the candidate gene deletion strains obtained from the liquid medium growth screen could utilize dipeptide on solid medium as their substrate to satisfy auxotrophic requirements. The deletion mutant candidates from the screen, wild-type BY4742, cup9, and ptr1 deletion mutants were grown overnight in MM + HLKU liquid medium at 30° while shaking at 200 rpm. Cells were harvested by centrifugation and washed three times with sterile, distilled water. Cells were then counted by hemacytometer and adjusted to the cell number of 5 × 106/ml. Five microliters of a suspension containing 2.5 × 104 and 2.5 × 103 yeast cells were spotted onto agar plates containing one of three different media: (1) MM + HLKU, (2) MM + (H–L)HKU, or (3) MM + (H–L)HKU + Trp. The plates were incubated at 30°, and growth was recorded after 2 days. A score was given to the growth of each strain compared to the growth of wild-type, cup9, and ptr1 deletion mutant strains.
Toxic halo assays and osmotic sensitivity assay:
For the toxic dipeptide halo assay, the sensitivity of deletion mutants to the toxic dipeptide Ala–Ethionine (Eth) was measured as previously described (Island et al. 1987). Eth is an analog of methionine, and utilization of Eth causes cell death. This assay is more sensitive than the previously described dipeptide growth assays. Yeast cells having a functional dipeptide transport system will take up the toxic dipeptide and die (indicated by a clear halo of growth inhibition on plates spotted with ethioninyl-containing peptides). Cells with a defective dipeptide uptake system will not take up the dipeptide efficiently and will survive (as indicated by a small halo or no halo). Cells were grown overnight in MM + HLKU medium and then harvested and washed three times with sterile, distilled water. Yeast cells were counted and adjusted to the cell number of 5 × 106/ml. One milliliter of the cell suspension was added to 0.8% noble agar (3 ml) and plated onto solid MM + HLKU medium. Two 6-mm sterile paper disks containing either 0.2 or 0.1 μmol of Ala–Eth were placed on the lawn of cells. The halo size was measured after 2 days of incubation at 30°. The halo size formed by each deletion mutant was compared to the halo size of the wild-type strain by dividing the halo size of the mutant by the halo size of the wild-type strain and multiplied by 100 to get a percentage. A strain that gave a >100% value was considered to be deleted for a gene that was involved in downregulation of peptide utilization, whereas a strain deleted for a gene involved in upregulation would give a <100% value in this test.
For the canavanine toxicity assay, the procedure was the same as that for the toxic dipeptide halo assay. Two 6-mm sterile paper disks containing 1 μg of canavanine were placed on the lawn of cells. The size of halo was measured and the image was taken after 2 days incubation at 30°.
For the osmotic sensitivity assay, the wild-type and deletion mutant strains were grown overnight in MM + HLKU medium and then harvested and washed three times with sterile, distilled water. Yeast cells were counted and adjusted to the cell number of 5 × 106/ml, and then cells were further diluted to 5 × 105/ml, 5 × 104/ml, and 5 × 103/ml. Ten microliters of each cell suspension were spotted into MM + HLKU media containing 0 m, 0.4 m, and 1.0 m NaCl.
Uptake assays:
The strains were grown overnight in MM + HLKU and then subcultured into a fresh medium. Cells were harvested in log phase, washed with 2% glucose, and adjusted to a final concentration to 2 × 108 cells/ml. The uptake assay was initiated by combining equal volumes of prewarmed (30°) cells and uptake assay mixture [2% glucose, 20 mm sodium citrate/potassium phosphate, pH 5.5, 320 μm Leu–Leu (Sigma, St. Louis), and 2 μCi/ml [3H]Leu–Leu (Island et al. 1987)]. After 10 min, portions (100 μl) were removed onto a membrane filter and washed four times by vacuum filtration with 1 ml ice-cold water. The radioactivity retained on the filter was determined by liquid scintillation spectrometry, and results were reported as nmol Leu–Leu uptake/1 × 109 cells/10 min. The accumulation of [3H]Leu–Leu in the ptr2 strain was subtracted from that of the tested strains, and the percentage of the accumulation of [3H]Leu–Leu in each deletion mutant vs. that in the wild type was calculated.
Reporter gene assay:
The centromeric plasmid pRD1, which contained a selectable URA3 marker and the lacZ gene under the control of the PTR2 promoter, was transformed into the tested deletion mutant strains. The reporter gene assay was performed using a protocol adapted from Hoffman (Hoffman et al. 2002). The strains were grown overnight in MM + HLK and subcultured into a fresh medium. Cells were harvested in log phase, washed with sterile water, and adjusted to a final concentration to 1 × 108 cells/ml. Cell suspension (100 μl) was added to a well of a 96-well plate, and then 20 μl fluorescein di-β-d-galactopyranoside (FDG) solution was added. The FDG solution was prepared by mixing solution 1 (0.1 mm FDG diluted in 25 mm PIPES, pH 7.2) and solution 2 (5% Triton X-100 diluted in 250 mm PIPES, pH 7.2) in equal amounts just prior to use. The plate was incubated at 37° for 1.5 hr and then read with a fluorescence multiwell plate reader (Wallac Victor2, 1420 multilabel counter; Perkin-Elmer Life and Analytical Sciences, Wellesley, MA), using 485 and 530 nm as excitation and emission wavelengths, respectively.
Classification of the obtained genes:
The classification of the 103 genes identified through this screen was based on the Gene Ontology (GO) annotation found in the Saccharomyces Genome Database (SGD) (http://db.yeastgenome.org/cgi-bin/GO/goTermMapper) (Christie et al. 2004), where 34 functional categories of biological process were given. Among the 103 genes, some genes fell into several categories due to their multiple known functions. The detailed gene function could also be referenced from the Yeast Proteome Database (YPD) (https://proteome.incyte.com/control/tools/proteome) (Hodges et al. 1999).
Northern analysis of PTR2 mRNA in deletion mutant strains:
Approximately 3 × 108 cells of select candidate deletion mutant strains were harvested after overnight growth in MM + HLKU medium. Yeast total RNA was isolated using an extraction kit (RiboPure-Yeast; Ambion, Austin, TX). Total RNA was quantified by monitoring absorbance at 260 nm, and 20 μg was loaded per lane into a formaldehyde-reducing gel. Gels were run in buffer containing MOPS (pH 7.0) for 5 hr at 75 V and then transferred by capillary action onto a nylon hybridization membrane in 20× SSC. Total RNA was UV crosslinked to the membrane with a Stratalinker UV source. The blot was prehybridized at 65° in Church buffer (7% SDS, 1% BSA, 1 mm EDTA, and 250 mm Na2HPO4, pH 7.2) for 2 hr and hybridized overnight in the same buffer containing radioactive probes at 65°. Radioactive probes were prepared as follows. A 1.7-kb fragment of PTR2 was obtained by PCR amplification using the primers PTR2-F (Northern) and PTR2-R (Northern), and a 0.92-kb fragment of ACT1 was obtained by PCR amplification using the primers ACT1-F (Northern) and ACT1-R (Northern) (primer sequences are represented in supplemental Table S1 at http://www.genetics.org/supplemental/). DNA fragments were labeled with [α-32P]dATP by random-primed probe synthesis. Blots were rinsed with posthybridization buffer (0.1× SSC, 0.1% SDS) at 65°, and the RNA images were developed with a Storm 840 phosphoimager (Molecular Dynamics, Sunnyvale, CA). The expression of PTR2 in each mutant was analyzed at least twice in separate experiments, with similar expression found in each experiment. The quantification of PTR2 and ACT1 mRNA was performed using the software ImageQuant 5.0 (GE Healthcare Technologies, Waukesha, WI).
Real-time RT–PCR:
Approximately 3 × 108 cells of the tested deletion mutant strains were harvested after overnight growth in MM + HLKU medium. Yeast total RNA was isolated using the RiboPure-Yeast extraction kit (Ambion) and then treated with the TURBO DNA-free kit (Ambion) to eliminate the genomic DNA contamination. The amount of total RNA was quantified by monitoring absorbance at 260 nm. To check that the genomic DNA had been eliminated, the obtained total RNA was used for the template to PCR amplify the target genes, and no PCR product was obtained. cDNA was synthesized using M-MLV RT (400 units reverse transcriptase; Invitrogen, San Diego) in a reaction mixture containing 1 μg Oligo(dT), 1 mm deooxynucleoside triphosphates, 14 units anti-RNase, and 1 μg of total RNA at 42° for 45 min, and the reaction was stopped after 5 min by incubation at 72°.
Real-time PCR analysis was performed in the DNA Engine Opticon (MJ Research, Boston, MA). The QuantiTect SYBR Green PCR kit (QIAGEN, Valencia, CA) and the primers (the sequences are presented in supplemental Table S1 at http://www.genetics.org/supplemental/) resulting in ∼100 bp amplicon were applied in the PCR reaction. The reaction contained 12.5 μl of 2× qPCR reaction mix and 15 pmol of primers and was run with a cycle of 50° for 2 min, 95° for 15 min, followed by 40 cycles of 95° for 15 sec, 54° for 30 sec, and 72° for 15 sec. A standard curve for each primer set was performed with 1, 1:10, 1:100, 1:1000, 1:10,000, and 1:100,000 dilutions of the wild-type cDNA. The CT-value, the cycle when sample fluorescence exceeds a chosen threshold above background fluorescence, was determined using the software program. The copy number of PTR2, PMA1, and ACT1 genes in each deletion mutant strain was calculated on the basis of the standard curve. The ratio of the fold change of target genes (PTR2 and PMA1) vs. the fold change of the internal control gene (ACT1) in the tested mutant strains was calculated to show upregulation or downregulation. The ratio of the fold change in the wild type was standardized as 1.0.
Ptr2p localization in deletion mutant strains:
To trace the localization of Ptr2p, a Ptr2p-GFP construct was created as follows. The primers PTR2-FLAG-GFP-F and PTR2-Flag-GFP-R (see supplemental Table S1 at http://www.genetics.org/supplemental/ for the sequence) were used to PCR amplify two tandem copies of the GFP gene using pKW430 [a gift of Mary Miller, Rhodes College, Memphis, TN (Stade et al. 1997)] as the template. The amplified PCR product (1.5 kb) had two copies of GFP sequence with 40 bp of flanking sequence homologous to pMS2, which contained PTR2 with its endogenous promoter and terminator sequences and both FLAG and His tags located at the C terminus. The plasmid was linearized with AgeI at a unique restriction site located between the FLAG and His tags. The centromeric plasmid pMS4, containing Ptr2p-FLAG-GFP2-His6 and the selectable marker URA3, was created by homologous recombination.
To test the localization of Ptr2p-GFP in different yeast strains, the pMS4 construct was transformed into each deletion mutant of interest and transformants were selected by growth in the absence of uracil. Yeast strains carrying pMS4 were pregrown in MM + HLK overnight and then inoculated into fresh MM + HLK at an initial cell concentration of 2 × 106 cells/ml. Cells were concentrated by centrifugation and observed at 10 hr after inoculation by fluorescence microscopy using a 470- to 490-nm excitation wavelength and 515-nm emission filter fitted to an Olympus (Lake Success, NY) microscope. Images were taken with a MicroFire camera (model S99809; Olympus). All mutants were in the log phase at the 10-hr time point as ascertained by the high proportion of cells with buds and the low density of cells under the experimental conditions used. To visualize the amount of GFP expression, each image was captured with the same exposure time. Significantly higher and lower GFP signals could be distinguished from the captured images. For those images with higher GFP signal, the images were taken with a decreased exposure time to be visualized clearly.
RESULTS
Primary screening of deletion mutants to identify strains demonstrating increased or decreased dipeptide utilization:
To identify genes involved in dipeptide utilization, we screened a library of 4826 haploid, single-gene deletion strains for mutants with an altered dipeptide utilization profile. The leucine auxotrophic marker in the BY4742 strain background (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) was used to monitor how well each strain was able to utilize the dipeptide His–Leu. Under the conditions of the screen, cells could grow only if they were able to take up the dipeptide His–Leu from the medium and release leucine from the peptide intracellularly. Numerous previous studies have shown that there is no extracellular peptidase or protease in S. cerevisiae able to catalyze the hydrolysis of dipeptides under the growth conditions used. In this screening, any mutant strain demonstrating either enhanced or decreased growth was considered to have a deletion in a gene involved in dipeptide utilization. Since Ptr2p is the only dipeptide transporter expressed in this yeast under the conditions used for the screen, we expected that genes related to the regulation of PTR2 expression or Ptr2p function would comprise the list of candidates. The yeast deletion mutant library was generated by the Saccharomyces Genome Deletion Project, which involved several laboratories (Winzeler et al. 1999). Only nonessential genes are represented in this deletion mutant collection.
In the primary screening, yeast deletion mutants were grown in a poor nitrogen medium with allantoin as the nitrogen source. These conditions are known to partially upregulate PTR2 expression (Island et al. 1991). All deletion mutant strains not capable of growing in allantoin as the sole nitrogen source were necessarily excluded from this study. One hundred nine strains of the haploid collection were identified in this category. For a full list of the 109 strains see supplemental Table S2 at http://www.genetics.org/supplemental/. The rest of the deletion series, including strains known to be deficient (ptr1) and hyperactive (cup9) for dipeptide utilization, grew similarly in the MM + HLKU medium.
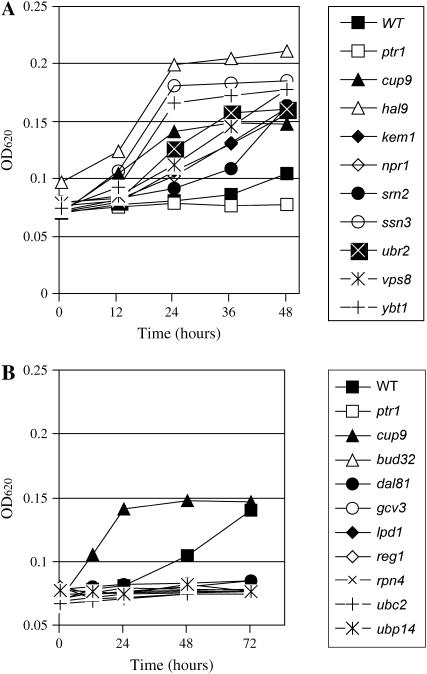
A screen was performed of the mutant collection in medium [MM + (H–L)HKU + Trp] containing His–Leu as the only leucine source. Tryptophan upregulates PTR2 expression via the SPS system over the enhancement induced by growth on MM alone, thus providing the maximal dipeptide utilization phenotype (Island et al. 1987; Forsberg and Ljungdahl 2001b). The screen was also performed without tryptophan addition with identical results observed (data not shown). Two hundred seventy-eight deletion mutants were obtained from the primary screening that showed either enhanced dipeptide utilization (the deleted genes are “negative regulator genes”) or decreased dipeptide utilization (the deleted genes are “positive regulator genes”). As controls for comparison of strains that show increased or decreased dipeptide utilization, cup9 and ptr1 mutants were used. Cup9p is a repressor of PTR2 transcriptional expression; the cup9 deletion mutant strain has a higher expression level of PTR2 in comparison to that of the wild type (Byrd et al. 1998); therefore the hyperactive strain (cup9) grew better than wild type on dipeptide (Figure 1A). Ptr1p is required for degradation of Cup9p; in the ptr1 deletion mutant strain Cup9p is stabilized (Island et al. 1991; Du et al. 2002), and there is an undetectable level of PTR2 expression preventing this strain from growing on dipeptide (Figure 1A). For clarity of presentation and to present data representative of the phenotypes among the mutants identified, we highlight in the results section 18 deletion mutant strains (bud32, cup9, dal81, gcv3, hal9, kem1, lpd1, npr1, ptr1, reg1, rpn4, ssn3, srn2, ubc2, ubp14, ubr2, vps8, and ybt1).
Figure 1.
Growth patterns of various selected strains that demonstrate increased or decreased dipeptide utilization. (A) The growth pattern of wild type (BY4742), ptr1, cup9, and eight other deletion mutant strains in MM + H(H–L)KU + Trp medium. (B) The growth pattern of wild type (BY4742), ptr1, cup9, and nine other deletion mutant strains in MM + H(H–L)KU + Trp medium.
Growth patterns of deletion mutant strains hal9, kem1, npr1, srn2, ssn3, ubr2, vps8, and ybt1 were similar to that of the cup9 deletion mutant in that they had a shorter lag phase and a higher growth rate compared to the wild type (Figure 1A). Therefore, these deletion strains were considered to carry a deletion in a gene involved in decreasing dipeptide utilization. Deletion mutants bud32, dal81, gcv3, lpd1, reg1, rpn4, ubc2, and ubp14 had a similar growth pattern to that of the ptr1 deletion strain, which did not grow in MM + (H–L)HKU + Trp medium (Figure 1B). These deletion mutants were considered to carry a deletion in a gene involved in increasing dipeptide utilization.
Verification of candidate genes—solid media growth assay and toxic dipeptide halo assay:
The candidate mutants were further tested in a solid medium growth assay and a toxic dipeptide halo assay. The deletion mutant strains were grown on solid medium plates containing MM + HLKU, MM + (H–L)HKU, or MM + (H–L)HKU + Trp. Eighteen representative deletion mutant strains with enhanced or decreased dipeptide utilization are shown in Figure 2. All the tested deletion mutant strains grew to a similar extent as compared to that of the wild type in MM + HLKU. The deletion mutants hal9, kem1, npr1, srn2, ssn3, ubr2, vps8, and ybt1 had a similar growth pattern to that of the cup9 deletion mutant strain, which grew better than wild type in both MM + (H–L)HKU + Trp (Figure 2) and MM + (H–L)HKU (data not shown). In contrast, the deletion mutants bud32, dal81, gcv3, lpd1, reg1, rpn4, ubc2, and ubp14 had a similar growth pattern to that of the ptr1 deletion mutant strain, which could not grow in MM + (H–L)HKU + Trp medium (Figure 2) or in MM + (H–L)HKU (data not shown). The growth phenotype found in the liquid growth screening of these chosen candidate mutants with respect to the regulation of dipeptide utilization was therefore confirmed in this solid medium test.
Figure 2.
Growth of selected strains on solid medium. Two cell dilutions, 3 ×104 cells/ml and 3 × 103 cells/ml, were tested on MM + HLKU and MM + (H–L)HKU + Trp media.
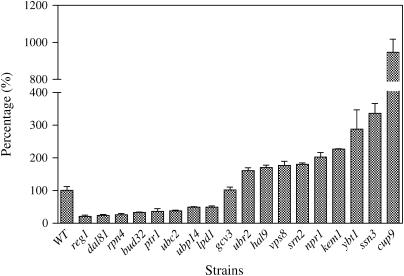
The mutants identified in the initial screen were also subjected to a toxic dipeptide assay for further verification of the peptide transport phenotype. In this assay, cells were grown on MM + HLKU and tested for their sensitivity to Ala–Eth. The halo size of each deletion mutant strain was measured and compared to that of wild type. A larger halo indicated that the deletion mutant strain was more sensitive to Ala–Eth toxic dipeptide than the wild type, while a smaller halo indicated less sensitivity. As expected, cup9 developed a larger halo than that of wild type and ptr1 was not sensitive to the toxic dipeptide. Similar to cup9, deletion strains hal9, kem1, npr1, srn2, ssn3, ubr2, vps8, and ybt1 developed a larger halo than that of wild type (Table 1), indicating that the deleted genes negatively regulated dipeptide utilization. Conversely, similar to ptr1, deletion strains bud32, dal81, gcv3, lpd1, reg1, rpn4, ubc2, and ubp14 were not sensitive to toxic dipeptide (Table 2), indicating that the deleted genes positively regulated dipeptide utilization.
TABLE 1.
The response of deletion mutants with increased dipeptide utilization to Ala-Eth, NaCl, and canavanine
| Gene name | Description of gene product | Toxicity of Ala–Eth (% of wild type)a | NaCl Sensitivityb | Canavanine toxicity (% of the wild type)c |
|---|---|---|---|---|
| Wild type | 100 | |||
| CUP9 | Specific RNA polymerase II transcription factor activity | 120 | — | — |
| HAL9 | Specific RNA polymerase II transcription factor activity | 131 | — | — |
| KEM1 | 5′–3′ exoribonuclease activity | 135 | More sensitive | Less sensitive |
| NPR1 | Serine/threonine protein kinase | 129 | — | Less sensitive |
| SRN2 | Class E vacuolar sorting protein | 109 | More sensitive | More sensitive |
| SSN3 | Cyclin-dependent protein kinase activity | 114 | — | — |
| UBR2 | Ubiquitin-protein ligase activity | 129 | — | — |
| VPS8 | Vacuolar sorting | 121 | More sensitive | More sensitive |
| YBT1 | Bile acid transporter activity | 156 | — | — |
Data are also presented in supplemental Table S3 and Figures S4, and S5 at http://www.genetics.org/supplemental/.
The halo size formed by each deletion mutant was compared to the halo size of the wild-type strain by dividing the halo size of the mutant by the halo size of the wild-type strain and multiplied by 100 to get a percentage.
“—” indicates that the deletion strain had no difference in its sensitivity to NaCl compared to that of the wild type. A strain that was more sensitive exhibited greater growth inhibition to 1.0 m NaCl in comparison to that shown by the wild type.
“—” indicates that the deletion strain had no difference in sensitivity to canavanine compared to that of the wild type.
TABLE 2.
The response of deletion mutants with decreased dipeptide utilization to Ala-Eth, NaCl, and canavanine
| Gene name | Description of gene product | Toxicity of Ala–Eth (% of wild type)a | NaCl sensitivityb | Canavanine toxicity (% of the wild type)c |
|---|---|---|---|---|
| Wild type | 100 | |||
| PTR1 | Ubiquitin–protein ligase activity | 0 | — | — |
| BUD32 | Protein serine/threonine kinase activity | 0 (fuzzy halo) | More sensitive | Less sensitive |
| DAL81 | Specific RNA polymerase II transcription factor activity | 0 | — | More sensitive |
| GCV3 | Glycine dehydrogenase (decarboxylating) activity | 0 | — | More sensitive |
| LPD1 | Dihydrolipoyl dehydrogenase activity | 0 | — | More sensitive |
| REG1 | Protein phosphatase type 1 activity | 0 (fuzzy halo) | — | More sensitive |
| RPN4 | Transcriptional activator activity | 0 | — | — |
| UBC2 | Ubiquitin-conjugating enzyme activity | 0 | — | — |
| UBP14 | Ubiquitin-specific protease activity | 73 | — | — |
These data are also presented in supplemental Table S3 and supplemental Figures S4 and S5 at http://www.genetics.org/supplemental/.
The halo size formed by each deletion mutant was compared to the halo size of the wild-type strain by dividing the halo size of the mutant by the halo size of the wild-type strain and multiplied by 100 to get a percentage.
“—” indicates that the deletion strain had no difference in its sensitivity to NaCl compared to that of the wild type. A strain that was more sensitive exhibited greater growth inhibition to 1.0 m NaCl in comparison to that shown by the wild type.
“—” indicates that the deletion strain had no difference in sensitivity to canavanine compared to that of the wild type.
Of the 278 strains identified in the initial liquid growth screen, 103 strains showed increased or decreased peptide utilization in all three assays: liquid and solid media growth and the toxic dipeptide halo assay [supplemental Tables S3 and S4 (http://www.genetics.org/supplemental/) listed in order of biological process according to Gene Ontology annotation]. These 103 genes were considered to be the set of genes involved as either negative or positive regulators of peptide utilization as determined by the three experimental tests. As expected, 5 genes (PTR1, CUP9, UBC2, DAL81/UGA35, and STP2) previously known to be involved in dipeptide utilization were identified among the 103 genes, which validated the utility of the screening method. Three other genes (SSY1, PTR3, and SSY5), known to affect dipeptide utilization via the regulation of PTR2 mRNA expression, were not found in the screen since they are not included in the deletion mutant library. The number of deletion mutant strains found by our methods to modulate dipeptide utilization accounted for ∼2% of the entire collection. Of the 46 genes whose deletion mutant strains enhanced dipeptide utilization (supplemental Table S3), 20 encoded proteins with more than a 30% protein sequence identity to a human protein (supplemental Table S3, footnote a). Additionally, of the 57 genes whose deletion mutant strain decreased dipeptide utilization (supplemental Table S4), 33 encoded proteins with more than a 30% sequence identity to a human protein (supplemental Table S4, footnote a). According to GO annotation in the SGD, the 103 genes covered 29 of the 34 given Biological Processes GO categories. The categories of each gene are indicated in supplemental Tables S3 and S4.
Ptr2p-GFP expression and localization in the identified deletion strains:
To further explore the impact of the identified 103 genes on dipeptide utilization, we measured Ptr2p expression and localization using a Ptr2p-GFP chimera. This construct (pMS4), which encoded Ptr2p-GFP under the control of its native promoter, was transformed into the 103 deletion strains, and Ptr2p-GFP in log phase cells was observed by fluorescence microscopy. The addition of the GFP tag did not affect the function of Ptr2p as demonstrated by halo and uptake assays (data not shown). Ptr2p-GFP was primarily localized to the plasma membrane in wild-type log-phase cells (Figure 3). The amount of the Ptr2p-GFP expression signal and its localization in the 103 deletion mutant strains revealed seven phenotypic patterns (Table 3).
Figure 3.
Ptr2p-GFP expression and localization in the wild type (WT) and nine deletion strains that demonstrated increased dipeptide utilization. The right-hand side of each strain shows phase microscopy of the same field as the left-hand side, which shows the fluorescently labeled Ptr2p-GFP. Deletion strain ssn3 showed a very high amount of GFP signal under the same exposure time as that of the wild type.
TABLE 3.
Ptr2p-GFP expression and localization in 103 identified strains with decreased or increased dipeptide utilization
| Phenotypic category | Increased dipeptide utilization (46 strains) | Decreased dipeptide utilization (57 strains) |
|---|---|---|
| Strains with enhanced Ptr2p-GFP expression | arp5, arp8, cup9, csm1, dbr1, eaf3, hal9, ies6, kem1, mrps9, pat1, sfp1, srb8, ssd1, ssn2, ssn3, ssn8, tom72, ubr2, yor322c, ynl123w, yfr044c, ylr114c | asm4, ydr015c, ydr290w |
| Strains with altered Ptr2p-GFP localization | bni1, def1, eaf7, lst4, npr1, sac6, snf7, srn2, tpm1, vam10, vps8, vps36, ypl073c, ynl295w | bem4, bud32, csf1, etr1, gcv2, gcv3, hfm1, ilm1, isa1, isa2, mck1, mre11, npl3, shp1, reg1, rmd12, rps9b, tsa1, vbm1, ydr433w, yjl046w, ydr157w, yjl175w |
| Strains with no apparent effect on Ptr2p-GFP localization or expression | cik1, kti12, mlh1, mrp17, pho2, rim101, tif3, ybt1, ydr417c | bud28, bye1, elf1, hfa1, ira2, lat1, lpd1, mct1, oar1, pdb1, pdx1, prd1, rad23, spo21, ypl098c |
| Strains with decreased Ptr2p-GFP expression | No mutant strains were found in this category. | dal81, hof1, ipk1, nfu1, pho85, ptr1, rpl21a, rpn4, shm2, stp2, taf14, thp1, uba3, ubp14, ubc2, ypr174c |
Ptr2p-GFP expression and localization in the underlined strains are shown in Figures 3 and 4. The underlined strains were the representatives chosen to highlight in the results section. Ptr2p-GFP expression and localization of all the listed strains are shown in supplemental Figures S1 and S3 at http://www.genetics.org/supplemental/. The strains in boldface type (cup9, dal81, ptr1, stp2, and ubc2) contain the deleted genes previously identified as being involved in regulation of PTR2 transcription.
Ptr2p-GFP expression and localization in deletion strains with increased dipeptide utilization:
The Ptr2p-GFP expression signal and localization in 46 deletion strains with increased dipeptide utilization are shown in supplemental Figure S1 at http://www.genetics.org/supplemental/ (9 deletion strains and wild type are shown in Figure 3 as representative of the full set).
Strains with enhanced Ptr2p-GFP expression signal:
A higher level of Ptr2p-GFP expression signal in the plasma membrane was observed in the cup9 deletion strain as compared to the wild type (Figure 3, Table 3). The enhanced Ptr2p-GFP expression level corresponded with the phenotype of increased dipeptide utilization as reflected in growth on dipeptide in liquid and solid medium and toxicity to Ala–Eth (Figures 1 and 2, and Table 1). In addition, Ptr2p-GFP was also observed in the vacuole, possibly due to the overexpressed PTR2 in the cup9 strain. Similar to the cup9 strain, the deletion of HAL9, KEM1, SSN3, and UBR2 showed an increased Ptr2p-GFP expression signal in both the plasma membrane and the vacuole. Enhanced Ptr2p-GFP expression signal was also observed in 18 other deletion strains including four unknown gene deletion mutants, yfr044c, ylr114c, ynl123w, and yor322c (Table 3 and supplemental Figure S1 at http://www.genetics.org/supplemental/). Consistently, the increased Ptr2p-GFP expression agreed with increased sensitivity to toxic dipeptide and better growth on dipeptide substrate (supplemental Table S3 at http://www.genetics.org/supplemental/).
Strains with altered Ptr2p-GFP localization:
Fourteen deletion strains with increased dipeptide utilization showed altered localization of Ptr2p-GFP (Table 3 and supplemental Figure S1 at http://www.genetics.org/supplemental/). An example of three deletion mutants, npr1, srn2, and vps8 is shown in Figure 3. These gene products had GO annotations of transport, vesicle-mediated transport, cytokinesis, or related to protein modification. Ptr2p-GFP signal in the srn2 and vps8 deletion mutants was localized mainly to the plasma membrane and the endosome. In addition, Ptr2p-GFP remained in the endosome in stationary phase cells and targeting of Ptr2p-GFP to the vacuole was defective (data not shown). Similar to srn2 and vps8 strains, the deletion strains eaf7, sac6, snf7, vps36, and ypl073c exhibited Ptr2p-GFP localization to the plasma membrane and to the endosome (supplemental Figure S1 at http://www.genetics.org/supplemental/), suggesting that these gene products are involved in a similar cellular process regulating dipeptide utilization. Snf7p, Srn2p, and Vps36p are three key proteins involved in ESCRT (endosomal sorting complex required for transport) protein complexes required for endocytotic degradation of membrane proteins (Bowers et al. 2004).
To further characterize the ESCRT protein complexes involved in Ptr2p endocytotic degradation, deletion mutant strains of other components of the ESCRT protein complexes (vps2, vps20, vps22, vps23, vps24, vps25, and vps28) were also transformed with the pMS4 construct. The localization of Ptr2p-GFP in these strains was similar to that in snf7, srn2, and vps36 strains (supplemental Figure S2 at http://www.genetics.org/supplemental/). Additionally, vps2, vps20, vps22, vps23, vps24, vps25, and vps28 strains were more sensitive to toxic dipeptide as compared to the wild type (supplemental Table S5 at http://www.genetics.org/supplemental/), confirming the role of the ESCRT protein complex in dipeptide utilization.
The deletion strain npr1 showed an accumulation of Ptr2p-GFP signal in both endosomal vesicles and the vacuole in addition to the plasma membrane localization (Figure 3). The Ptr2p sorting process is likely regulated by Npr1p, a protein kinase involved in the regulation of vesicle transport systems (supplemental Table S3 at http://www.genetics.org/supplemental/). Similar to Ptr2p-GFP localization in the npr1 strain, deletion strains bni1, def1, lst4, and tpm1 showed Ptr2p-GFP localization to the plasma membrane and the vacuole (supplemental Figure S1 at http://www.genetics.org/supplemental/), implicating them as well as being involved in the protein trafficking of Ptr2p.
Strains with no apparent effect on Ptr2p-GFP expression or localization:
The deletion strain ybt1 showed no apparent difference in the expression level of Ptr2p-GFP signal in the cytoplasmic membrane (Figure 3), although there was an increase in the Ptr2p-GFP signal in the vacuole. In this strain dipeptide utilization was increased (Figures 1 and 2 and Table 1). In addition, seven other deletion strains, cik1, kti12, mlh1, mrp17, pho2, rim101, tif3, and ydr417c demonstrated increased dipeptide utilization with no apparent effect on Ptr2p-GFP expression in the cytoplasmic membrane (Table 3 and supplemental Figure S1 at http://www.genetics.org/supplemental/). These gene products might impact dipeptide utilization independently of Ptr2p function. For example, Ybt1p, known as an ABC transporter and localized on the vacuolar membrane, may be involved in the uptake of dipeptides into the vacuole. The deletion of this gene resulted in increased growth response and dipeptide toxicity.
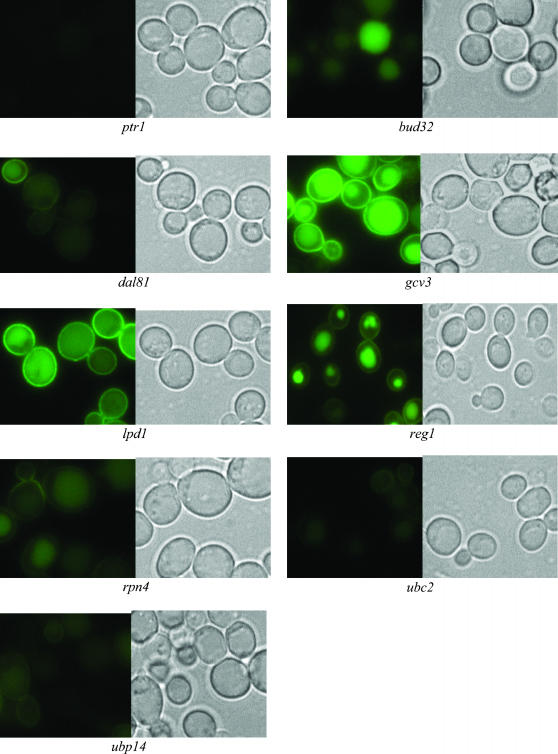
Ptr2p-GFP expression and localization in deletion strains with decreased dipeptide utilization:
The expression level of Ptr2p-GFP signal and localization of 9 representative strains are shown in Figure 4, and all 57 deletion strains listed in Table 3 with decreased dipeptide utilization are shown in supplemental Figure S3 at http://www.genetics.org/supplemental/.
Figure 4.
Ptr2p-GFP expression and localization in nine selected deletion strains that demonstrated decreased dipeptide utilization. The right-hand side of each strain shows phase microscopy of the same field as the left-hand side, which shows the fluorescently labeled Ptr2p-GFP.
Gene deletion enhanced the expression level of Ptr2p-GFP signal:
The deletion strains asm4, ydr015c, and ydr290w showed an enhanced expression level of Ptr2p-GFP at the cytoplasmic membrane and the vacuole (supplemental Figure S4 at http://www.genetics.org/supplemental/). These strains showed a decreased sensitivity to toxic dipeptide, however, with no growth change on dipeptide (supplemental Table S4 at http://www.genetics.org/supplemental/). In the asm4 strain, a fuzzy halo was observed in the toxic dipeptide halo assay, indicating a transient growth inhibition. ASM4 encodes a component of the karyopherin docking complex of the nuclear pore (Marelli et al. 1998), indicating that some cellular protein involved in ethionine toxicity requires Asm4p for its full expression.
Strains with altered Ptr2p-GFP localization:
Compared to the wild type, deletion strains bud32 and reg1 showed higher expression level of Ptr2p-GFP signal at the vacuole and less expression in the plasma membrane (reg1) or no visible plasma membrane expression (bud32) (Figure 4). In addition, deletion strain gcv3 showed Ptr2p-GFP localization both at the vacuole and at the plasma membrane (Figure 4). Similar to the Ptr2p-GFP localization at the vacuole in bud32, reg1, or gcv3 strains, the deletion strains bem4, csf1, etr1, gcv2, hfm1, ilm1, isa1, isa2, mck1, mre11, npl3, rps9b, rmd12, shp1, tsa1, vbm1 and four unknown gene deletion strains (ydr157w, ydr433w, yjl046w, and yjl175w) resulted in Ptr2p-GFP localization both at the vacuole and at the plasma membrane (supplemental Table S4 and supplemental Figure S3 at http://www.genetics.org/supplemental/).
Strains with no apparent effect on the expression or localization of Ptr2p-GFP:
No apparent change in the expression or localization of Ptr2p-GFP was observed in the lpd1 deletion strain (Figure 4). Similarly, Ptr2p-GFP localization and expression level was not different in the deletion strains bye1, bud28, elf1, hfa1, ira2, lat1, mct1, oar1, pdb1, pdx1, rad23, and ypl098c as compared to that in the wild type (supplemental Table S4 and supplemental Figure S3 at http://www.genetics.org/supplemental/).
Strains with decreased expression of Ptr2p-GFP:
The expression level of Ptr2p-GFP signal was lower in the ptr1, dal81, rpn4, ubc2, and ubp14 deletion strain as compared to that in the wild type (Figure 4). A decreased Ptr2p-GFP expression signal was also observed in the deletion strains hof1, ipk1, nfu1, pho85, rpl21a, shm2, stp2, taf14, thp1, uba3, and ypr174c (supplemental Table S4 and supplemental Figure S3 at http://www.genetics.org/supplemental/). The decreased Ptr2p-GFP expression signal in these strains was consistent with a decrease of dipeptide utilization that was previously documented (supplemental Table S4) and is consistent with the observation that Ptr1p, Stp2p, Ubc2p, and Dal81p positively regulate PTR2 transcription (Bernard and Andre 2001; Andreasson and Ljungdahl 2002; Du et al. 2002).
Transcriptional regulation of PTR2:
To explore whether strains with enhanced or reduced expression level of Ptr2p-GFP showed a change in the transcriptional regulation of PTR2, the fold change of PTR2 mRNA compared to that of the wild type was measured by real-time reverse transcription PCR in the 18 representative deletion strains. In control experiments, PTR2 mRNA expression level in the cup9 strain was upregulated more than nine times that of the wild type (Table 4). This result also agreed with the increased PTR2 mRNA level in Northern analysis and lacZ activity as compared to the wild type (data not shown). The increase in PTR2 mRNA expression in the cup9 strain is consistent with the increased expression level of Ptr2p-GFP signal (Figure 3). Similar to that of the cup9 strain, PTR2 mRNA expression level was highly upregulated in ssn3 and kem1. PTR2 mRNA expression was upregulated in ubr2, hal9, ybt1, vpc8, and gcv3 less than twofold. In contrast to the cup9, hal9, kem1, ssn3, ubr2, vps8, and ybt1 strains, which showed increased dipeptide utilization, the gcv3 strain exhibited a decrease in peptide utilization (supplemental Table S4 at http://www.genetics.org/supplemental/). The apparent discrepancy between decrease in utilization and robust expression in the gcv3 strain could result from a post-translational alteration of Ptr2p function or the alteration of the metabolic pathway(s) of dipeptide utilization.
TABLE 4.
The expression of PTR2 and PMA1 in various strains
| Strains | PTR2 expressiona | PMA1 expressiona |
|---|---|---|
| ubc2 | 0.06 | 0.94 |
| ptr1 | 0.07 | 1.08 |
| ubp14 | 0.08 | 1.09 |
| rpn4 | 0.21 | 1.23 |
| npr1 | 0.48 | 1.33 |
| dal81 | 0.79 | 1.70 |
| reg1 | 0.88 | 1.03 |
| lpd1 | 0.82 | 2.30 |
| srn2 | 0.92 | 0.87 |
| bud32 | 0.94 | 1.44 |
| ubr2 | 1.23 | 2.95 |
| hal9 | 1.37 | 1.93 |
| ybt1 | 1.51 | 2.37 |
| vps8 | 1.66 | 2.09 |
| gcv3 | 1.93 | 1.22 |
| ssn3 | 5.00 | 2.41 |
| cup9 | 9.36 | 0.98 |
| kem1 | 12.12 | 3.06 |
Expression level measured by real-time RT–PCR analysis is shown as a ratio calculated by the fold change of the target gene (PTR2 or PMA1) in the mutant compared to that of the wild type divided by the fold change of ACT1 in the mutant compared to that of the wild type.
PTR2 mRNA expression level in the ptr1 strain was only 7% that of the wild type (Table 4) in accordance with the known requirement of Ptr1p for PTR2 transcription (Byrd et al. 1998; Turner et al. 2000). This result also agreed with the decreased PTR2 mRNA level in Northern analysis and lacZ activity as compared to that in the wild type (data not shown). The decrease in PTR2 mRNA expression in the ptr1 strain is consistent with the decreased expression level of Ptr2p-GFP signal (Figure 4). Similar to that in the ptr1 strain, PTR2 mRNA was highly downregulated in ubc2, ubp14, and rpn4 strains. The expression of PTR2 mRNA in npr1, dal81, reg1, lpd1, srn2, and bud32 strains was not more than twofold less than that of the wild type.
Dipeptide uptake capability:
To explore whether altered dipeptide utilization was correlated directly to flux of dipeptide, the accumulation of [3H]Leu–Leu dipeptide was measured in the 18 representative strains (Figure 5). In a control experiment, the accumulation of [3H]Leu–Leu increased remarkably in the cup9 strain as compared to that in the wild type (Figure 5). Similar to that in the cup9 strain, the accumulation of [3H]Leu–Leu was higher in the hal9, kem1, npr1, srn2, ssn3, ubr2, vps8, and ybt1 strains as compared to that in the wild type. The increased uptake in these strains was reflected in their increased ability to utilize dipeptide. In contrast to the increased accumulation of dipeptide in hal9, kem1, npr1, srn2, ssn3, ubr2, vps8, and ybt1 strains, the accumulation of [3H]Leu–Leu in gcv3 was not significantly different as compared to that in the wild type, suggesting that deletion of this gene did not impact the import of dipeptide even though dipeptide utilization was decreased.
Figure 5.
The uptake of [3H]Leu–Leu dipeptide in log phase cells of the wild type, cup9, and ptr1 and the 16 deletion strains bud32, dal81, gcv3, hal9, kem1, lpd1, npr1, reg1, rpn4, srn2, ssn3, ubc2, ubr2, ubp14, vps8, and ybt1. The percentage of the accumulation of [3H]Leu–Leu for each deletion mutant was calculated as [(the accumulation of [3H]Leu–Leu in each mutant strain)/(the accumulation of [3H]Leu–Leu in the wild type) × 100].
The accumulation of [3H]Leu–Leu in the ptr1 strain was lower than that in the wild type. The reduced accumulation of dipeptide was consistent with the decreased expression level of PTR2 mRNA and Ptr2p-GFP in the ptr1 strain. Similar to that in the ptr1 strain, the accumulation of [3H]Leu–Leu was lower in the bud32, dal81, lpd1, reg1, rpn4, ubc2, and ubp14 strains as compared to that in the wild type (Figure 5). The decreased uptake in these strains was reflected in their decreased expression of PTR2 in dal81, rpn4, ubc2, and ubp14 strains (Table 4), decreased amount of Ptr2p-GFP in the cytoplasmic membrane (Figure 4), and the alteration localization of Ptr2p in bud32 and reg1 strains (Figure 4). In contrast, although the transport of dipeptide was reduced in the lpd1 strain, the expression level of Ptr2p-GFP remained similar to that in the wild type (Figure 4).
Effects of gene deletions on osmotic and canavanine sensitivity and PMA1 expression:
To determine whether gene deletions also affected general membrane properties, additional assays were performed. Osmotic sensitivity, toxicity of canavanine, and the expression level of PMA1 encoding a proton transporter localized in the cytoplasmic membrane were examined. NaCl at 1.0 m inhibits the growth of wild-type S. cerevisiae by causing osmotic destabilization of the cell membrane (Hohmann 2002). NaCl at 1.0 m significantly inhibited the growth of all strains on MM + HLKU plates. Fourteen deletion mutant strains, cup9, dal81, gcv3, hal9, lpd1, npr1, ptr1, reg1, rpn4, ssn3, ubc2, ubp14, ubr2, and ybt1, showed similar effects to NaCl as the wild type (Tables 1 and 2 and supplemental Figure S4 at http://www.genetics.org/supplemental/), suggesting that the deletion of those genes did not change the osmotic sensitivity of the deletion mutant strains. In contrast, four deletion mutant strains, bud32, kem1, srn2, and vps8, showed greater sensitivity to 1.0 m NaCl than the wild type (Tables 1 and 2 and supplemental Figure S4).
Canavanine is transported by the arginine permease and changes in canavanine toxicity have been correlated directly to arginine permease function (Hampsey 1997). In the canavanine toxicity assay, nine deletion mutant strains, cup9, hal9, ptr1, rpn4, ssn3, ubc2, ubp14, ubr2, and ybt1, did not show significant alteration in the canavanine sensitivity as compared with that of the wild type. In contrast, six deletion mutants, dal81, gcv3, lpd1, reg1, srn2, and vps8, were more sensitive to canavanine, and three deletion mutant strains, bud32, kem1, and npr1, were less sensitive than the wild type (Tables 1 and 2 and supplemental Figure S5 at http://www.genetics.org/supplemental/).
To examine whether the gene representative deletion mutants were also affected in the expression of other transport proteins, the expression level of PMA1 encoding a proton transporter was measured. PMA1 mRNA expression level in cup9 and ptr1 strains was similar to that of the wild type (Table 4). In contrast, PMA1 mRNA level was upregulated more than twofold in kem1, lpd1, ssn3, ubr2, vps8, and ybt1 strains and less than twofold in bud32, dal81, gcv3, hal9, npr1, reg1, rpn4, srn2, ubc2, and ubp14 strains (Table 4). Overall, these changes in expression levels of PMA1 in the various mutant strains did not correlate with the changes in expression in PTR2 except for upregulation of both genes in the ssn3 and kem1 strains.
DISCUSSION
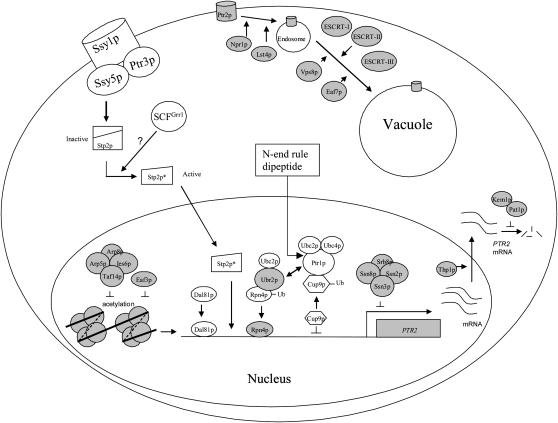
We have systematically screened a haploid, single-gene, deletion mutant library and identified 103 genes involved in dipeptide utilization. To our knowledge, this is the first such screen for a membrane transport system. PTR2 expression is known to be regulated by amino acids via the SPS protein complex and by dipeptides themselves (Figure 6). Import of di/tripeptides containing basic or bulky hydrophobic N-terminal residues (N-end rule peptides) induces PTR2 by reducing cellular levels of the PTR2 repressor Cup9p (Byrd et al. 1998; Turner et al. 2000; Du et al. 2002). In the SPS complex regulatory pathway a few downstream proteins, such as STP2 and components of the SCFGrr1 ubiquitin ligase complex (Skp1p, Hrt1p, Cdc34p, and Cdc53p), are involved in the Ssy1p-induced signal transduction pathway via proteolytic processing of Stp2p (Bernard and Andre 2001; Andreasson and Ljungdahl 2002). Due to the complex nature of signal transduction pathways, such as the well-studied pheromone-mediated mitogen-activated protein (MAP) kinase pathway in yeast (Gustin et al. 1998), we expected that other regulatory proteins would be identified in this screen.
Figure 6.
A hypothetical model for the regulation of dipeptide utilization in S. cerevisiae. The proteins with clear background represent those that have been reported previously to be involved in PTR2 regulation. The shaded background represents those proteins that have been determined in this study. The SPS protein complex (Ssy1p-Ptr3p-Ssy5p) is an amino acid sensor, which leads to a cleavage of the N-terminal of Stp2p. The activated form of Stp2p is localized to the nucleus and regulates PTR2 expression. In addition, Dal81p synergistically regulates PTR2 expression together with Stp2p. Other proteins, Rpn4p, a positive regulator, and Cup9p, a negative regulator, bind at the PTR2 promoter region and are also involved in PTR2 transcription. The stability of Rpn4p is modulated by Ubr2p and Ubc2p and the stability of Cup9p by Ptr1p, Ubc2p, and Ubc4p via the ubiquitination pathway. Binding of N-end rule dipeptides to Ptr1p/Ubr1p results in ubiquitin-mediated degradation of Cup9p. Ubr2p competes with Ptr1p for the degradation of Cup9p. Arp5p, Arp8p, Ies6p, and Taf14p (components of the INO80 complex) together with Eaf3p possibly negatively regulate PTR2 transcription via repressing the acetylation of histones in the PTR2 coding region. The transcription of PTR2 is negatively regulated by a RNA polymerase mediator protein complex, which includes Ssn2p, Ssn3p, Ssn8p, and Srb8p. Thp1p might be involved in the translocation of PTR2 mRNA from the nucleus to the cytosol. The interaction between Kem1p and Pat1p is potentially involved in the degradation of PTR2 mRNA. Dipeptide utilization is also modulated by the Ptr2p trafficking system; Srn2p, Vps36p, and Snf7p represent components of ESCRT-I, -II, and -III, together with Vps8p and Eaf7p, which appear to regulate Ptr2p internalization from the endosome to the vacuole. In addition, Npr1 and Lst4p are also involved in Ptr2p trafficking and affect the retention of Ptr2p in the plasma membrane.
The 103 genes identified in the screen cover a number of different biological processes based on the Gene Ontology annotation. It is clear from the broad range of mutants identified that genes involved in both direct and indirect regulation of peptide utilization were discovered. Such a global array of genes supports the interconnectivity of networks involved in regulating biological processes (Lee et al. 2002; Chen et al. 2003), but such a heterogeneous collection of interacting genes has not been identified previously as specifically regulating membrane transport systems.
Genes that regulate PTR2 transcription:
In this study we have considerably expanded the identification of genes involved in PTR2 transcriptional regulation. The deletion of these genes results in the alteration of dipeptide utilization.
First, we have shown that dipeptide utilization can be affected by gene products involved in the modification of the nucleosome, namely Arp5p, Arp8p, Ies6p, and Taf14p. These proteins belong to the components of the chromatin remodeling INO80 protein complex that carries ATPase activity, DNA binding, and nucleosome mobilization (Sanders et al. 2002) and that preferentially interacts with histones H3 and H4 (Shen et al. 2003). Another histone- modification-related gene product, Eaf3p, was also identified in our screen. Eaf3p is a component of the NuA4 histone acetyltransferase complex that maintains the acetylation of histones H3 and H4 (Reid et al. 2004). It is possible that these chromatin remodeling proteins negatively regulate PTR2 transcription via the modification of histones. A model representing the potential involvement of these proteins and others is shown (Figure 6).
Second, four gene products with transcription factor activity, Cup9p, Dal81p, Stp2p, and Rpn4p were shown to regulate PTR2 expression and affected dipeptide utilization. Cup9p binds the region between −488 and −897 upstream of the PTR2 start codon (Byrd et al. 1998), while Rpn4p may bind between −610 and −603 due to the existence of the Rpn4p binding consensus sequence (5′-GGTGGAAA-3′) present at this location (Mannhaupt et al. 1999). Cup9p is a repressor of PTR2, whereas Rpn4p is a “positive regulator” as reflected in the observation that the cup9 deletion strain showed an increase in peptide utilization and the rpn4 strain has decreased peptide utilization. Subsequent to amino acid induction via the SPS system and its conversion into the active form after proteolytically processing and translocation into the nucleus (Andreasson and Ljungdahl 2002, 2004), the active form of Stp2p (truncated at the N terminus) binds to the upstream activation sequence (UAS) of the promoter region of BAP2 and BAP3 (de Boer et al. 2000; Nielsen et al. 2001). Because BAP2, BAP3, and PTR2 are simultaneously regulated in response to amino acid induction via the SPS-Stp2p system, the active form of Stp2p likely also binds to the PTR2 promoter region and regulates its transcription. Dal81p is a transcriptional activator essential for PTR2 expression under the induction by the SPS signal pathway as well (Bernard and Andre 2001). Dal81p acts synergistically with Stp1p to regulate the transcription of AGP1 in response to the amino acid induction, and the sequence 5′-CGGC-3′ of a UAS element is important for AGP1 transcriptional regulation (Abdel-Sater et al. 2004). Since the PTR2 promoter region has this consensus sequence and because Dal81p was identified as important for PTR2 transcription and dipeptide utilization in our screening, we speculate that Dal81p and Stp2p act synergistically in PTR2 transcription in a similar manner.
Third, other gene products identified in this screen such as Ubr2p, Ptr1p, and Ubc2p regulate PTR2 expression by modulating the stability of Rpn4p and Cup9p (Figure 6). Cup9p stability is regulated by Ptr1p, Ubc2p, and Ubc4p (Du et al. 2002). Ubr2p is associated with Rpn4p to regulate proteasome gene expression through the ubiquitination pathway (Wang et al. 2004). In addition, Ubr2p has high amino acid sequence similarity to Ptr1p, presumably allowing a competition with Ptr1p for the stabilization of Cup9p (Bartel et al. 1990). Therefore, Ubr2p probably regulates PTR2 transcription by impacting the stability of both Rpn4p and Cup9p (Figure 6).
Fourth, we identified one other gene product (SSN3) involved in PTR2 transcription that is known to be general transcription factor, which also affected PMA1 expression. Ssn3p/Srb10p, Ssn8p/Srb11p, Ssn2p/Srb9p, and Srb8p have been identified as a cyclin-dependent serine/threonine protein kinase complex that functions as a mediator of RNA polymerase (Kornberg 2005). This complex is also involved in the glucose signaling pathway (Balciunas and Ronne 1995; Kuchin et al. 1995). Deletion of these genes leads to derepression of a wide variety of downstream genes, including GAL and SUC, which are important for galactose and raffinose metabolism, respectively (Myer and Young 1998; Kuchin et al. 2000). The deletion of SSN3 resulted in a change of PMA1 and PTR2 expression level.
Finally, we identified gene products involved in PTR2 transcription by means that are not clear. For example, UBP14 was important for PTR2 expression since deletion of these genes significantly decreased PTR2 mRNA level (Table 4), and the strain carrying deletion of this gene exhibited decreased peptide utilization. Ubp14p has ubiquitin-specific protease activity (supplemental Table S4 at http://www.genetics.org/supplemental/). It is probably modulating PTR2 transcription via the ubiquitination pathway. In addition, the deletion of UBP14 had no effect on the expression of PMA1 or on sensitivity to NaCl and canavanine. Further experiments are needed to provide some insight into whether the gene product acts directly or indirectly to control PTR2 expression.
Genes involved in PTR2 mRNA maturation:
Gene products (Kem1p, Pat1p, and Thp1p) involved in post-transcriptional regulation impacting the stability and transport of PTR2 mRNA also affected dipeptide utilization (Figure 6). Both Kem1p and Pat1p belong to an mRNA decay protein complex (Bonnerot et al. 2000; Bouveret et al. 2000). PTR2 mRNA was highly upregulated in the kem1 strain (Table 4). It is likely that PTR2 mRNA was more stable in kem1 and pat1 strains, which resulted in more Ptr2p being synthesized, leading to enhanced Ptr2p-GFP expression (Figure 3) and increased dipeptide utilization (Table 1 and Figures 1 and 2). The deletion of KEM1 resulted in also an increased expression level of PMA1 and changes in sensitivity to NaCl and canavanine (Table 1). It is not surprising that the deletion of this gene affected several phenotypes related to membrane function, since a deletion in this gene would impact gene expression of many different genes. Another gene shown to impact dipeptide utilization was Thp1p, which is involved in mRNA export from the nucleus to the cytoplasm (Fischer et al. 2002). The thp1 strain probably has a defect in transporting PTR2 mRNA into the cytoplasm for protein synthesis. Therefore this mutant demonstrated a decrease in peptide utilization.
Genes that regulate Ptr2p trafficking:
This screen revealed that dipeptide utilization is regulated by the trafficking system. Deletion of components of ESCRT-I, ESCRT-II, and ESCRT-III protein complexes showed a defect in Ptr2p-GFP trafficking from the endosome to the vacuole (Figure 6 and supplemental Figure S2 at http://www.genetics.org/supplemental/). These protein complexes are essential for sorting ubiquitinylated membrane proteins from the plasma membrane to the multivesicular body and to the vacuole for further degradation (Raiborg et al. 2003). Ptr2p has been shown to be ubiquitinylated (Hitchcock et al. 2003). Strains srn2, vps36, and snf7 demonstrated increased dipeptide utilization perhaps due to an increased residence time of Ptr2p at the cytoplasmic membrane. In addition, the eaf7 and vps8 strains showed Ptr2p localization similar to that of the ESCRT protein mutants. Surprisingly, Eaf7p is annotated as a component of the NuA4 HAT complex, which is an essential histone acetyltransferase complex that acetylates the N-terminal tails of histones H4 and H2A (Krogan et al. 2004). It is possible that Eaf7p has dual functions or indirectly affects Ptr2p localization through other proteins. Both srn2 and vps8 strains showed an altered sensitivity to NaCl and canavanine. It has been reported that the osmotic sensitivity changes if vacuolar development is affected (Hampsey 1997). Thus a disruption of the protein-sorting process that affects the vacuole could lead to alteration of osmotic sensitivity.
Deletion mutants (npr1, lst4) with increased dipeptide utilization and mutants (bud32, reg1) with decreased utilization showed an accumulation of Ptr2p-GFP in vesicles. NPR1 encodes a Ser/Thr protein kinase, involved in post-translational control of Gap1p. Npr1p is required for Gap1p to be targeted to the plasma membrane, and an npr1 deletion mutant loses Gap1p function by sorting Gap1p from the Golgi into the vacuole, bypassing the plasma membrane (De Craene et al. 2001). Similarly, mutations in LST4 reduced the function of Galp1p and other amino acid permeases by the interruption of sorting of these transporters to the cell surface (Roberg et al. 1997). Our observations indicated that NPR1 and LST4 regulate Ptr2p differently from the way they regulate Gap1p. It is not clear how Lst4p and Npr1p regulate Ptr2p in a manner opposite to that of their regulation of Gap1p. Bud32p is a Ser/Thr protein kinase involved in polar bud site selection and changing the vacuolar morphology (Bonangelino et al. 2002). Reg1p is a regulatory subunit for protein phosphatase Glc7p involved in the repression of many glucose-regulated genes and vesicular trafficking (Cui et al. 2004). The involvement of Bud32p and Reg1p in dipeptide utilization reflects their roles in Ptr2p trafficking.
Genes that regulate dipeptide utilization independent of Ptr2p:
The alteration of dipeptide utilization also results from Ptr2p-independent metabolic processes as indicated by the observation of no apparent phenotypic change in expression or localization of Ptr2p-GFP in a number of mutants (Table 3). Mutation in genes encoding a variety of cellular metabolic processes (supplemental Figure S6 at http://www.genetics.org/supplemental/) (Nagarajan and Storms 1997; Schneider et al. 1997; Stoops et al. 1997), such as the conversion of pyruvate into acetyl-CoA by pyruvate dehydrogenase complex (Pdx1p, Lpd1, Pdb1p, and Lat1p), the conversion of acetyl-CoA to malonyl-CoA by Hfa1p, fatty acid synthesis by fatty acid synthase complex (Oar1p, Mct1p, and Etr1p), glycine degradation (Gcv2p, Gcv3p, and Lpd1p), and a peptidase activity (Prd1p), resulted in a decrease of dipeptide utilization (Table 3). In these mutants the impairment of the dipeptide utilization appeared to be independent of Ptr2p transport activity since Ptr2p-GFP could be observed at the plasma membrane (supplemental Figure S3 at http://www.genetics.org/supplemental/). The assay of [3H]Leu–Leu uptake in gcv3 strains suggested that the import of dipeptide was still maintained at a wild-type level, which showed that Ptr2p was functional (Figure 5), indicating that the decrease of dipeptide utilization resulted from the inability of the mutant to release amino acid from accumulated dipeptide or from a metabolic block in using the released amino acid for protein biosynthesis. Prd1p is a metalloendoproteinase (Buchler et al. 1994). The deletion mutant might be impaired in the degradation of intracellularly transported dipeptide, which would result in a decrease of dipeptide utilization.
Deletion strains cik1, kti12, mlh1, mrp17, pho2, rim101, tif3, ybt1, and ydr417c showed no apparent effect on Ptr2p-GFP localization or expression but demonstrated increased dipeptide utilization. The utilization of dipeptide in these strains appears to involve Ptr2p-independent cellular processes as well. For example, Ybt1p belongs to the ABC transporter family, exhibiting ATP-dependent bile acid transport (Ortiz et al. 1997; Paulsen et al. 1998). As expected for an ABC transporter, the ybt1 mutant was not only more sensitive to the toxic dipeptide Ala–Eth, but also this strain was more sensitive to ethionine and Lys–Ala–Eth (data not shown). We speculate that this protein might be involved in the transport of dipeptide into the vacuole. In the deletion mutant the defect in the uptake of dipeptide into the vacuole would lead to an increase of dipeptide availability in the cytoplasm. It is also possible that the deletion of a gene might result in a derepression or change in function of other transport systems that may mediate dipeptide utilization in the mutant without alteration of PTR2 expression or Ptr2p function. We are currently working on unraveling the involvement of these metabolic genes in dipeptide utilization.
Unknown genes that regulate dipeptide utilization:
Fifteen unknown gene deletion mutant strains were identified in this screen, including 7 strains exhibiting increased and 8 strains showing decreased dipeptide utilization (Table 3). Deletion mutants yor322c, ynl123w, yfr044c, and ylr114c showed enhanced expression level of Ptr2p-GFP signal compared to that of wild type, and ypr174c had reduced expression of Ptr2p-GFP; these results were consistent with dipeptide utilization in these strains, suggesting that these unknown genes products are involved in the regulation of PTR2 expression. Deletion mutants ynl295w, ypl073c, yjl175w, ydr157w, and ydr433w showed accumulation of Ptr2p-GFP signal in the vesicles or the vacuole, suggesting that these unknown gene products are involved in Ptr2p trafficking and localization. Similar to the expression level of Ptr2p-GFP in the asm4 strain, enhanced Ptr2p-GFP expression was observed in the deletion mutant strains of unknown function, ydr015c and ydr290w. These two mutants, however, showed a decreased sensitivity to toxic dipeptide and no growth change on dipeptide; it is not clear how these genes regulate dipeptide utilization.
Overall, this investigation has provided a list of genes involved directly and indirectly in the utilization of dipeptides in yeast. In our screen, 53 genes encoding proteins with >30% identity to human genes have been identified (see supplemental Tables S3 and S4, footnote a, at http://www.genetics.org/supplemental/). To date no genes have been reported in the regulation of the human PTR2 homologs PEPT1 and PEPT2 (Daniel 2004). Further investigation of these human genes may promote understanding of the regulation of dipeptide utilization in mammalian systems. This study represents the first such global analysis of a membrane transport process and as such provides a rich starting ground for future investigations.
Acknowledgments
We thank Melinda Hauser, Amy Wiles, and Tom Masi for helpful discussions and critical review of the manuscript.
References
- Abdel-Sater, F., I. Iraqui, A. Urrestarazu and B. Andre, 2004. The external amino acid signaling pathway promotes activation of Stp1 and Uga35/Dal81 transcription factors for induction of the AGP1 gene in Saccharomyces cerevisiae. Genetics 166: 1727–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson, C., and P. O. Ljungdahl, 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16: 3158–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson, C., and P. O. Ljungdahl, 2004. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol. Cell. Biol. 24: 7503–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouida, M., N. Page, A. Leduc, M. Peter and D. Ramotar, 2004. A genome-wide screen in Saccharomyces cerevisiae reveals altered transport as a mechanism of resistance to the anticancer drug bleomycin. Cancer Res. 64: 1102–1109. [DOI] [PubMed] [Google Scholar]
- Askree, S. H., T. Yehuda, S. Smolikov, R. Gurevich, J. Hawk et al., 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz, K., L. McHardy, K. Gable, T. Tarling, D. Reberioux et al., 2004. Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. Proc. Natl. Acad. Sci. USA 101: 4525–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas, D., and H. Ronne, 1995. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 23: 4421–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, D., W. Lai, M. Breslav, F. Naider and J. M. Becker, 1998. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol. Microbiol. 29: 297–310. [DOI] [PubMed] [Google Scholar]
- Bartel, B., I. Wunning and A. Varshavsky, 1990. The recognition component of the N-end rule pathway. EMBO J. 9: 3179–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, F., and B. Andre, 2001. Ubiquitin and the SCF(Grr1) ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 496: 81–85. [DOI] [PubMed] [Google Scholar]
- Bonangelino, C. J., E. M. Chavez and J. S. Bonifacino, 2002. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot, C., R. Boeck and B. Lapeyre, 2000. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 20: 5939–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret, E., G. Rigaut, A. Shevchenko, M. Wilm and B. Seraphin, 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19: 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K., J. Lottridge, S. B. Helliwell, L. M. Goldthwaite, J. P. Luzio et al., 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5: 194–210. [DOI] [PubMed] [Google Scholar]
- Buchler, M., U. Tisljar and D. H. Wolf, 1994. Proteinase yscD (oligopeptidase yscD). Structure, function and relationship of the yeast enzyme with mammalian thimet oligopeptidase (metalloendopeptidase, EP 24.15). Eur. J. Biochem. 219: 627–639. [DOI] [PubMed] [Google Scholar]
- Byrd, C., G. C. Turner and A. Varshavsky, 1998. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 17: 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Y. Liu, K. M. Goldstein, J. M. Becker, Y. Xu et al., 2003. A computational study on the signal transduction pathway for amino acid and peptide transport in yeast: bridging the gap between high-throughput data and traditional biology. Appl. Genomics Proteomics 2: 43–50. [Google Scholar]
- Christie, K. R., S. Weng, R. Balakrishnan, M. C. Costanzo, K. Dolinski et al., 2004. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32: D311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, D. Y., C. R. Brown and H. L. Chiang, 2004. The type 1 phosphatase Reg1p-Glc7p is required for the glucose-induced degradation of fructose-1,6-bisphosphatase in the vacuole. J. Biol. Chem. 279: 9713–9724. [DOI] [PubMed] [Google Scholar]
- Daniel, H., 2004. Molecular and integrative physiology of intestinal peptide transport. Annu. Rev. Physiol. 66: 361–384. [DOI] [PubMed] [Google Scholar]
- de Boer, M., P. S. Nielsen, J. P. Bebelman, H. Heerikhuizen, H. A. Andersen et al., 2000. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 28: 974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene, J. O., O. Soetens and B. Andre, 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276: 43939–43948. [DOI] [PubMed] [Google Scholar]
- Du, F., F. Navarro-Garcia, Z. Xia, T. Tasaki and A. Varshavsky, 2002. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. USA 99: 14110–14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Y. J., Y. Kanai, S. Nussberger, V. Ganapathy, F. H. Leibach et al., 1994. Expression cloning of a mammalian proton-coupled oligopeptide transporter. Nature 368: 563–566. [DOI] [PubMed] [Google Scholar]
- Fischer, T., K. Strasser, A. Racz, S. Rodriguez-Navarro, M. Oppizzi et al., 2002. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21: 5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, H., and P. O. Ljungdahl, 2001. a Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21: 814–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, H., and P. O. Ljungdahl, 2001. b Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40: 91–109. [DOI] [PubMed] [Google Scholar]
- Forsberg, H., C. F. Gilstring, A. Zargari, P. Martinez and P. O. Ljungdahl, 2001. a The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol. 42: 215–228. [DOI] [PubMed] [Google Scholar]
- Forsberg, H., M. Hammar, C. Andreasson, A. Moliner and P. O. Ljungdahl, 2001. b Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158: 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, T., and J. Robert-Baudouy, 1996. Bacterial aminopeptidases: properties and functions. FEMS Microbiol. Rev. 18: 319–344. [DOI] [PubMed] [Google Scholar]
- Gustin, M. C., J. Albertyn, M. Alexander and K. Davenport, 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey, M., 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133. [DOI] [PubMed] [Google Scholar]
- Hauser, M., V. Narita, A. M. Donhardt, F. Naider and J. M. Becker, 2001. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol. Membr. Biol. 18: 105–112. [PubMed] [Google Scholar]
- Herrera-Ruiz, D., and G. T. Knipp, 2003. Current perspectives on established and putative mammalian oligopeptide transporters. J. Pharm. Sci. 92: 691–714. [DOI] [PubMed] [Google Scholar]
- Hitchcock, A. L., K. Auld, S. P. Gygi and P. A. Silver, 2003. A subset of membrane-associated proteins is ubiquitinated in response to mutations in the endoplasmic reticulum degradation machinery. Proc. Natl. Acad. Sci. USA 100: 12735–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges, P. E., A. H. McKee, B. P. Davis, W. E. Payne and J. I. Garrels, 1999. The Yeast Proteome Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 27: 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, G. A., T. R. Garrison and H. G. Dohlman, 2002. Analysis of RGS proteins in Saccharomyces cerevisiae. Methods Enzymol. 344: 617–631. [DOI] [PubMed] [Google Scholar]
- Hohmann, S., 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66: 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui, I., S. Vissers, F. Bernard, J. O. de Craene, E. Boles et al., 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19: 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Island, M. D., F. Naider and J. M. Becker, 1987. Regulation of dipeptide transport in Saccharomyces cerevisiae by micromolar amino acid concentrations. J. Bacteriol. 169: 2132–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Island, M. D., J. R. Perry, F. Naider and J. M. Becker, 1991. Isolation and characterization of S. cerevisiae mutants deficient in amino acid-inducible peptide transport. Curr. Genet. 20: 457–463. [DOI] [PubMed] [Google Scholar]
- Kornberg, R. D., 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30: 235–239. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., K. Baetz, M. C. Keogh, N. Datta, C. Sawa et al., 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101: 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin, S., P. Yeghiayan and M. Carlson, 1995. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 92: 4006–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin, S., I. Treich and M. Carlson, 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97: 7916–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph et al., 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804. [DOI] [PubMed] [Google Scholar]
- Lee, V. H., 2000. Membrane transporters. Eur. J. Pharm. Sci. 11(Suppl 2): S41–S50. [DOI] [PubMed] [Google Scholar]
- Liu, W., R. Liang, S. Ramamoorthy, Y. J. Fei, M. E. Ganapathy et al., 1995. Molecular cloning of PEPT 2, a new member of the H+/peptide cotransporter family, from human kidney. Biochim. Biophys. Acta 1235: 461–466. [DOI] [PubMed] [Google Scholar]
- Lubkowitz, M. A., L. Hauser, M. Breslav, F. Naider and J. M. Becker, 1997. An oligopeptide transport gene from Candida albicans. Microbiology 143(2): 387–396. [DOI] [PubMed] [Google Scholar]
- Mannhaupt, G., R. Schnall, V. Karpov, I. Vetter and H. Feldmann, 1999. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450: 27–34. [DOI] [PubMed] [Google Scholar]
- Marelli, M., J. D. Aitchison and R. W. Wozniak, 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143: 1813–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, A. L., W. H. Tham, H. Shah and A. Amon, 2004. A genome-wide screen identifies genes required for centromeric cohesion. Science 303: 1367–1370. [DOI] [PubMed] [Google Scholar]
- Marzluf, G. A., 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61: 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake, T., M. Kanayama, H. Sammoto and B. Ono, 2002. A novel cis-acting cysteine-responsive regulatory element of the gene for the high-affinity glutathione transporter of Saccharomyces cerevisiae. Mol. Genet. Genomics 266: 1004–1011. [DOI] [PubMed] [Google Scholar]
- Myer, V. E., and R. A. Young, 1998. RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem. 273: 27757–27760. [DOI] [PubMed] [Google Scholar]
- Nagarajan, L., and R. K. Storms, 1997. Molecular characterization of GCV3, the Saccharomyces cerevisiae gene coding for the glycine cleavage system hydrogen carrier protein. J. Biol. Chem. 272: 4444–4450. [DOI] [PubMed] [Google Scholar]
- Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jorgensen et al., 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264: 613–622. [DOI] [PubMed] [Google Scholar]
- Ortiz, D. F., M. V. St. Pierre, A. Abdulmessih and I. M. Arias, 1997. A yeast ATP-binding cassette-type protein mediating ATP-dependent bile acid transport. J. Biol. Chem. 272: 15358–15365. [DOI] [PubMed] [Google Scholar]
- Page, N., M. Gerard-Vincent, P. Menard, M. Beaulieu, M. Azuma et al., 2003. A Saccharomyces cerevisiae genomewide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163: 875–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, I. T., M. K. Sliwinski, B. Nelissen, A. Goffeau and M. H. Saier, Jr., 1998. Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 430: 116–125. [DOI] [PubMed] [Google Scholar]
- Perry, J. R., M. A. Basrai, H. Y. Steiner, F. Naider and J. M. Becker, 1994. Isolation and characterization of a Saccharomyces cerevisiae peptide transport gene. Mol. Cell. Biol. 14: 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen, P., B. Wu, R. F. Gaber, K. Ottow, H. A. Andersen et al., 2005. Amino acid sensing by Ssy1. Biochem. Soc. Trans. 33: 261–264. [DOI] [PubMed] [Google Scholar]
- Pritchard, G. G., and T. Coolbear, 1993. The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiol. Rev. 12: 179–206. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., T. E. Rusten and H. Stenmark, 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15: 446–455. [DOI] [PubMed] [Google Scholar]
- Reid, J. L., Z. Moqtaderi and K. Struhl, 2004. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg, K. J., S. Bickel, N. Rowley and C. A. Kaiser, 1997. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics 147: 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link and P. A. Weil, 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22: 4723–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherens, B., and A. Goffeau, 2004. The uses of genome-wide yeast mutant collections. Genome Biol. 5: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, R., B. Brors, F. Burger, S. Camrath and H. Weiss, 1997. Two genes of the putative mitochondrial fatty acid synthase in the genome of Saccharomyces cerevisiae. Curr. Genet. 32: 384–388. [DOI] [PubMed] [Google Scholar]
- Shen, X., R. Ranallo, E. Choi and C. Wu, 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12: 147–155. [DOI] [PubMed] [Google Scholar]
- Stade, K., C. S. Ford, C. Guthrie and K. Weis, 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050. [DOI] [PubMed] [Google Scholar]
- Steiner, H. Y., F. Naider and J. M. Becker, 1995. The PTR family: a new group of peptide transporters. Mol. Microbiol. 16: 825–834. [DOI] [PubMed] [Google Scholar]
- Steinmetz, L. M., C. Scharfe, A. M. Deutschbauer, D. Mokranjac, Z. S. Herman et al., 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31: 400–404. [DOI] [PubMed] [Google Scholar]
- Stoops, J. K., R. H. Cheng, M. A. Yazdi, C. Y. Maeng, J. P. Schroeter et al., 1997. On the unique structural organization of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. J. Biol. Chem. 272: 5757–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, G. C., F. Du and A. Varshavsky, 2000. Peptides accelerate their uptake by activating a ubiquitin-dependent proteolytic pathway. Nature 405: 579–583. [DOI] [PubMed] [Google Scholar]
- Wang, L., X. Mao, D. Ju and Y. Xie, 2004. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 279: 55218–55223. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Xie, Y., and A. Varshavsky, 1999. The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 18: 6832–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]