Abstract
Inbred mouse strains C57BL/6J (B6) and C3H/HeJ (C3H) differ significantly in atherosclerosis susceptibility and plasma lipid levels on the apolipoprotein E-deficient (apoE−/−) background when fed a Western diet. To determine genetic factors contributing to the variations in these phenotypes, we performed quantitative trait locus (QTL) analysis using an intercross between the two strains carrying the apoE−/− gene. Atherosclerotic lesions at the aortic root and plasma lipid levels of 234 female F2 mice were analyzed after being fed a Western diet for 12 weeks. QTL analysis revealed one significant QTL, named Ath22 (42 cM, LOD 4.1), on chromosome 9 and a suggestive QTL near D11mit236 (20 cM, LOD 2.4) on chromosome 11 that influenced atherosclerotic lesion size. One significant QTL on distal chromosome 1, which accounted for major variations in plasma LDL/VLDL cholesterol and triglyceride levels, coincided with a QTL having strong effects on body weight. Plasma LDL/VLDL cholesterol or triglyceride levels of F2 mice were significantly correlated with body weight, but they were not correlated with atherosclerotic lesion sizes. These data indicate that atherosclerosis susceptibility and plasma cholesterol levels are controlled by separate genetic factors in the B6 and C3H mouse model and that genetic linkages exist between body weight and lipoprotein metabolism.
ATHEROSCLEROTIC cardiovascular disease is a multi-factorial disorder with a strong heritable component. Although environmental factors such as diet, exercise, and smoking play a role in atherosclerosis, genetic factors are a major determinant for the development of the disease (Lusis et al. 2004). Only a small subset of atherosclerosis cases are caused by rare mutants that are observable as Mendelian traits segregating in families. These mutations, which include low-density lipoprotein receptor (LDLR) (Goldstein and Brown 1979), cystathionine β-synthase (Kraus et al. 1999), and ATP-binding cassette A1 (Bodzioch et al. 1999), have been identified. However, the common forms of atherosclerosis are due to a combination of multiple genetic and environmental factors (Lusis et al. 2004). Genes that contribute to the disease remain largely unidentified.
The availability of numerous inbred mouse strains that differ in atherosclerosis susceptibility provides an experimental tool for identifying genetic factors in atherosclerosis. C57BL/6 (B6) and C3H mice are two commonly used inbred strains that exhibit dramatic differences in atherosclerotic lesion formation (Paigen et al. 1985; Qiao et al. 1994). When fed an atherogenic diet containing 15% fat, 1.25% cholesterol, and 0.5% cholic acid (the Paigen diet), B6 mice readily develop fatty-streak lesions at the aortic root, whereas C3H mice are almost totally resistant to lesion formation. On the atherogenic diet, both strains have similarly increased plasma levels of low-density lipoproteins (LDL) and very low-density lipoproteins (VLDL). However, high-density lipoprotein (HDL) cholesterol levels are reduced in B6 mice but not in C3H mice (Paigen et al. 1987). This reduction in HDL levels had been considered to be responsible for the increased susceptibility of B6 mice to atherosclerosis (Paigen et al. 1987; Machleder et al. 1997). Recently, the atherogenic diet has been shown to result in a reduction in serum paraoxonase, an HDL-associated enzyme that protects against LDL oxidation, and chronic inflammation in the liver and other nonvascular tissues of B6 mice but not of C3H mice (Liao et al. 1993, 1994; Shih et al. 1996). To eliminate the influence of the cholate-containing diet, we and other groups constructed a congenic strain of C3H carrying the mutant apoE allele by sequentially backcrossing B6.apoE−/− mice with C3H/HeJ mice (Grimsditch et al. 2000; Shi et al. 2000; Matsushima et al. 2001). B6.apoE−/− mice provide a mouse model in which spontaneous hyperlipidemia and atherosclerosis occur even on a low-fat chow diet (Plump et al. 1992; Zhang et al. 1992). Moreover, these mice develop all phases of atherosclerotic lesions seen in humans, progressing from the early foam-cell stage to the advanced stage with a fibrous cap and necrotic lipid core (Nakashima et al. 1994). Compared to B6.apoE−/− mice, C3H.apoE−/− mice are highly resistant to atherosclerosis, developing much smaller lesions on either chow or Western diets (Shi et al. 2000). Interestingly, both C3H.apoE−/− and B6.apoE−/− mice have comparable plasma HDL cholesterol and paraoxonase levels on a chow diet, indicating that factors other than plasma HDL and paraoxonase are responsible for their differences in atherosclerosis susceptibility.
In pioneering work, using recombinant inbred strains derived from B6 and C3H mice, Paigen et al. (1987) identified a susceptibility locus named Ath-1, which increased fatty-streak lesions at the aortic root and reduced HDL cholesterol levels when the mice were fed the atherogenic diet. However, a more recent study using a large F2 cross (185 female progeny) between the two strains failed to observe any suggestive evidence of linkage to markers on chromosome 1 or elsewhere in the genome (Machleder et al. 1997). The small size and the incomplete penetrance (not all individuals of a cohort have a phenotype) of lesions may explain difficulties in isolating genetic loci controlling atherosclerosis susceptibility using this diet-induced atherosclerosis model. Thus, new approaches are needed with this model to identify responsible genes for atherosclerosis susceptibility. In this study, we report the identification of genetic loci influencing atherosclerotic formation and related traits in an intercross between the two apolipoprotein E-deficient (apoE−/−) strains.
MATERIALS AND METHODS
Mice:
B6.apoE−/− mice at the N10 backcross, constructed from B6;129.apoE−/− mice (Zhang et al. 1992), were purchased from The Jackson Laboratories, and C3H.apoE−/− mice were generated in our laboratory by initially crossing B6.apoE−/− mice with C3H/HeJ mice, followed by 12 sequential backcrossings with C3H/HeJ mice. B6.apoE−/− mice were crossed with C3H.apoE−/− mice to generate F1 hybrids, which were subsequently intercrossed by brother–sister mating to generate 234 female F2 progeny. At 6 weeks of age, F2 mice were started on a Western-type diet containing 42% fat, 0.15% cholesterol, and 19.5% casein without sodium cholate (TD 88137, Teklad, Madison, WI) and maintained on the diet for 12 weeks. All procedures were carried out in accordance with National Institutes of Health guidelines and approved by the University Animal Care and Use Committees.
Plasma lipid analysis:
Mice were fasted overnight before being sacrificed, and blood was collected by retro-orbital venous plexus puncture with the animals under isoflurane anesthesia. Plasma total cholesterol, high-density lipoprotein (HDL) cholesterol, and triglyceride levels were determined by using the Thermo DMA (Louisville, CO) cholesterol and triglyceride kits, which were adapted for a microplate assay (Tian et al. 2005). Briefly, 6 μl of plasma samples (for cholesterol measurements, plasma was diluted 1:5 in distilled water), lipid standards, and controls were loaded in a 96-well plate and then mixed with 150 μl of cholesterol or triglyceride reagents. After a 5-min incubation at 37°, the absorbance at 500 nm was read on a Molecular Devices (Menlo Park, CA) plate reader. HDL cholesterol levels were determined after precipitation of VLDL and LDL cholesterol fractions with a precipitating reagent provided by the company.
Aortic lesion analysis:
Methods for quantification of atherosclerotic lesions in aortic root were as previously reported by Qiao et al. (1994). Briefly, heart and proximal aorta were excised and embedded in optimal cutting temperature compound. Serial 10-μm thick cryosections from the middle portion of the ventricle to the aortic arch were collected and mounted on pretreated slides. In the region extending from the appearance to the disappearance of the aortic valves, every other section was collected. In all other regions, every fifth section was collected. The total number of sections examined for lesions ranged from 70 to 100 per mouse. Sections were stained with oil red O and hematoxylin and counterstained with fast green. Atherosclerotic lesion areas were quantified throughout the aortic sinus using an ocular with a square-micrometer grid on a light microscope. The lesion areas of five sections with the largest readings were averaged for each mouse and this average was used for statistical analysis.
Genotyping:
Genomic DNA was isolated from the tails of mice by using the standard phenol/chloroform extraction and ethanol precipitation method. A total of 130 microsatellite markers distinguishing strain B6 from strain C3H and covering all 19 autosomes and the X chromosome at an average interval of 13 cM was used to detect simple sequence length polymorphism of all F2 mice to identify which of the two parental strains contributed to alleles at a specific locus of each animal by PCR analysis. Parental and F1 DNA served as controls for each marker.
Statistical analysis:
Linkage analysis was performed to localize quantitative trait loci (QTL) by using Map Manager QTXb20 software (http://mapmgr.roswellpark.org/) (Manly et al. 2001) and the R/qtl program (http://www.biostat.jhsph.edu/∼kbroman/software) (Broman et al. 2003). Lesion areas and LDL/VLDL and HDL cholesterol levels were log transformed before QTL analysis was performed as they were not normally distributed. Body weight and triglyceride level data were directly used for QTL analysis. A likelihood-ratio statistic (LRS) was generated to define the significance of the association of a genetic marker with a trait. Log-of-the-odds-ratio (LOD) scores were calculated by dividing the LRS by 4.6. One thousand permutations of the trait values were used to define the genomewide LOD score threshold required to be significant or suggestive for each specific trait. Loci that exceeded the 95th percentile of the permutation distribution were defined as significant (P < 0.05) and those that exceeded the 37th percentile were suggestive (P < 0.63). The support interval (SI) for each QTL was determined using a 1-LOD drop from the QTL peak. The allele effect of each QTL was determined by calculating the phenotype means for each of the three possible genotypes. ANOVA was used for determining whether the mean phenotype values of progeny with different genotypes at a specific marker were significantly different. Differences were considered statistically significant at P < 0.05.
RESULTS
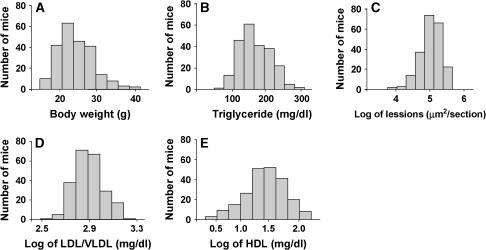
Our recent studies have shown that apoE-deficient mice with a B6 genetic background develop much larger atherosclerotic lesions than C3H mice on either chow or Western diet (Shi et al. 2000). Also, on the Western diet, C3H.apoE−/− mice exhibit significant increases in plasma levels of total and HDL cholesterol and triglyceride compared with B6.apoE−/− mice. To identify genetic factors contributing to the variations in the phenotypes, in this study we performed QTL analysis using an intercross between the two apoE−/− mouse strains. Thus, 234 female F2 mice were fed the Western diet for 12 weeks and typed for genetic markers spanning the genome and for such phenotypes as atherosclerotic lesion size, plasma lipid levels, and body weight. As shown in Figure 1, body weight (Figure 1A) and triglyceride levels (Figure 1B) in the F2 progeny were approximately normally distributed. Log-transformed lesion size (Figure 1C), LDL/VLDL (Figure 1D), and HDL cholesterol levels (Figure 1E) of the F2 population approached normal distributions. These graphs also show a wide range of variations in the phenotypes of F2 mice. These data were analyzed by using the Map Manager and R/qtl programs to identify chromosomal regions segregating with the traits. Those loci exhibiting significant linkage and suggestive linkage are presented.
Figure 1.
Distributions of body weight (A), triglyceride levels (B), log-transformed aortic lesion sizes (C) and LDL/VLDL (D) and HDL cholesterol levels (E) in 234 female F2 mice derived from B6.apoE−/− and C3H.apoE−/− mice. Mice were fed a Western diet for 12 weeks.
Significant and suggestive QTL for atherosclerotic lesion size:
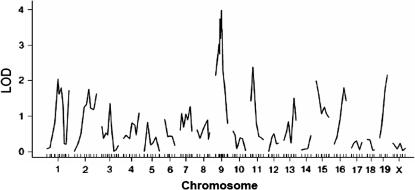
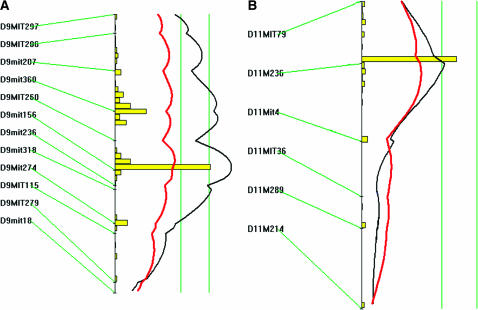
A genomewide scan of the F2 cross revealed loci on chromosomes 9 and 11 influencing atherosclerotic lesion size (Figure 2). Details of the QTL detected, including peak marker locus, LOD score, SI, variance, P-value, and allele conferring the trait, are presented in Table 1. The interval mapping graph for chromosome 9 displayed three peaks with LOD scores exceeding the suggestive LOD score of 2.3 (Figure 3A). The distal peak was near marker D9Mit156 (42 cM from the centromere), which had a significant LOD score of 4.1 and accounted for 34% of the variation in atherosclerotic lesion size of the cross. We designated this locus as Ath22, in keeping with the previous nomenclature for atherosclerosis susceptibility loci in the mouse. This locus exhibited a dominant inheritance pattern, because F2 mice with the heterozygous BC genotype at D9Mit156 had a lesion size comparable to mice with the homozygous BB genotype, while mice homozygous in the CC genotype had significantly smaller lesions (P = 0.000076) (Table 2). The central peak was near marker D9Mit360 (35 cM), which had a significant LOD score of 3.7 and explained 24% of the lesion variation. This locus was not named because it overlapped with Ath22 in the support interval and functioned in the same mode of inheritance as Ath22. The proximal peak, near marker D9Mit206 (20 cM), had a suggestive LOD score of 2.9 and explained 11% of the lesion variation. The locus on chromosome 11 peaked at marker D11Mit236 (20 cM) and had a LOD score of 2.4 (Figure 3B, Table 1). This locus explained 5% of the variation in atherosclerotic lesion size, and it exhibited a dominant effect from the C3H allele on lesion formation since mice homozygous for the C3H allele had a lesion size similar to the heterozygous mice but significantly smaller than mice homozygous for the B6 allele (Table 2).
Figure 2.
A genomewide scan to search for loci influencing atherosclerotic lesion sizes. Chromosomes 1 through X are represented numerically on the x-axis. The relative width of the space allotted for each chromosome reflects the relative length of each chromosome. The y-axis represents the LOD score. Aortic lesion sizes were determined by averaging the lesion sizes of five aortic-root cross sections with the largest readings for each of 234 female F2 mice after being fed the Western diet for 12 weeks.
TABLE 1.
Significant and suggestive QTL for atherosclerotic lesions, plasma LDL/VLDL cholesterol, HDL cholesterol, triglyceride, and body weight identified in the B6.apoE−/− and C3H.apoE−/− intercross
| Chromosome marker(cM)a
|
SIΔ (cM)c
|
Variance (%)d
|
Model of inheritancee
|
Overlapping QTLf
|
||||
|---|---|---|---|---|---|---|---|---|
| Trait | LODb | P-value | Name | Reference | ||||
| D9mit156(42) | Lesion | 4.1 | 37–50 | 34 | 0.00008 | Dominant/B6 | ||
| D11mit236(20) | Lesion | 2.4 | 11–33 | 5 | 0.00361 | Dominant/C3H | ||
| D1mit270(92.3) | LDL/VLDL | 6.3 | 88–97 | 32 | 0.00001 | Additive | Cq2, Hdlq15 | Machleder et al. (1997) |
| D5mit95(68) | LDL/VLDL | 2.2 | 51–81 | 4 | 0.00604 | Dominant/B6 | Unnamed QTL | Machleder et al. (1997) |
| D9mit115(56) | LDL/VLDL | 2.5 | 51–65 | 5 | 0.00323 | Dominant/C3H | Cq4, Cq5 | Suto et al. (2003) |
| D1mit270(92.3) | HDL | 3.0 | 81–105 | 20 | 0.00110 | Additive | Hdlq15 | Machleder et al. (1997) |
| D3mit42(58.8) | HDL | 2.3 | 50–67 | 5 | 0.00282 | Dominant/B6 | Unnamed QTL | Mehrabian et al. (2000) |
| D1mit270(92.3) | Triglyceride | 3.8 | 78–97 | 13 | 0.00642 | Dominant/C3H | Cq2 | Suto et al. (1999) |
| D8mit41(41) | Triglyceride | 3.2 | 33–51 | 10 | 0.00067 | Dominant/B6 | Tgq2 | Shike et al. (2001) |
| D1mit425(81.6) | Weight | 9.0 | 74–87 | 40 | 0.00001 | Dominant/C3H | BW17 | Anunciado et al. (2000) |
| D1mit206(95.8) | Weight | 8.3 | 88–101 | 34 | 0.00060 | Dominant/C3H | BW8q1 | Zhang et al. (2003) |
| D4mit251(66) | Weight | 2.7 | 39–66 | 15 | 0.00217 | Additive | BW7 | Brockmann et al. (2000) |
| D14mit185(54) | Weight | 2.2 | 44–61 | 4 | 0.00726 | Additive | Bwnd2wk7 | Brockmann et al. (2004) |
| D17mit180(29.4) | Weight | 3.4 | 19–35 | 7 | 0.00039 | Dominant/C3H | Wt3q3 | Moody et al. (1999) |
From Mouse Genome Informatics database at http://www.informatics.jax.org.
LOD scores were derived by dividing the LRS by 4.6. Suggestive QTL LOD ≥ 2.3 and significant QTL LOD ≥ 3.3 for lesion size; suggestive QTL LOD ≥ 2.3 and significant QTL LOD ≥ 3.4 for LDL/VLDL; suggestive QTL LOD ≥ 2.3 and significant QTL LOD ≥ 3.3 for HDL-C; suggestive QTL LOD ≥ 2.3 and significant QTL LOD ≥ 3.2 for triglyceride; suggestive QTL LOD ≥ 2.4 and significant QTL LOD ≥ 3.4 for body weight as defined by 1000 permutation tests. The significant loci are underlined to easily distinguish them from suggestive loci.
SI, support intervals, were defined by a 1-unit decrease in LOD score on either side of the peak marker.
Variance (%) indicates the percentage of the total phenotypic variance detected in the F2 cohort with which each marker showing linkage was associated.
Model of inheritance was determined using the MapManager QT program.
Overlapping QTL identified in previous studies.
Figure 3.
Likelihood plots for atherosclerotic lesions on chromosome 9 (A) and chromosome 11 (B). Plots were created using the interval mapping function of Map Manager QTXb20, including a bootstrap test shown as a histogram estimating the confidence interval for the major QTL. Two straight vertical lines on the plot represent significance thresholds for the likelihood-ratio statistic, indicating “suggestive” or “significant” peaks as calculated by permutation analysis (the genomewide significance thresholds of P = 0.63 and P = 0.05, respectively, are shown from left to right). Black plots reflect the likelihood-ratio statistic calculated at 1-cM intervals. The red plot represents the additive regression coefficient, indicating the effect of the B6 allele: if B6 represents the high allele, then the red plot will be to the right of the graph; otherwise, it will be to the left.
TABLE 2.
Effects of C3H (C) and B6 (B) alleles in different QTL on atherosclerosis, plasma lipids, and body weight in the B6.apoE−/− and C3H.apoE−/− intercross
| Trait | Peak marker | BB | BC | CC | P-value |
|---|---|---|---|---|---|
| Lesion | D9mit156 | 178,359 ± 13,867 | 172,756 ± 10,143 | 105,825 ± 9,160 | 7.6 × 10−5 |
| D11mit236 | 196,644 ± 14,310 | 150637 ± 9,138 | 135,065 ± 13,437 | 4.0 × 10−3 | |
| LDL/VLDL | D1mit270 | 691.1 ± 23.9 | 815.5 ± 20.8 | 911.9 ± 36.8 | 1.9 × 10−6 |
| D5mit95 | 767.6 ± 32.5 | 780.3 ± 18.9 | 860.4 ± 37.7 | 4.1 × 10−3 | |
| D9mit115 | 726.3 ± 24.1 | 826.2 ± 22.4 | 832.5 ± 31.7 | 1.8 × 10−3 | |
| HDL | D1mit270 | 24.4 ± 2.1 | 35.0 ± 2.8 | 42.5 ± 4.2 | 1.7 × 10−3 |
| D3mit42 | 31.3 ± 2.6 | 30.6 ± 2.7 | 45.0 ± 4.3 | 3.9 × 10−3 | |
| Triglyceride | D1mit270 | 152.3 ± 4.6 | 178.2 ± 4.5 | 185.5 ± 7.2 | 2.1 × 10−4 |
| D8mit41 | 161.6 ± 5.3 | 167.8 ± 4.1 | 190.5 ± 6.4 | 1.0 × 10−3 | |
| Body weight | D1mit425 | 21.7 ± 0.5 | 26.0 ± 0.4 | 26.0 ± 0.7 | 2.2 × 10−9 |
| D1mit206 | 21.6 ± 0.5 | 25.8 ± 0.4 | 26.0 ± 0.7 | 1.0 × 10−8 | |
| D4mit251 | 23.0 ± 0.6 | 24.7 ± 0.4 | 26.3 ± 0.7 | 2.5 × 10−3 | |
| D14mit185 | 24.3 ± 0.5 | 25.8 ± 0.6 | 23.5 ± 0.5 | 7.8 × 10−3 | |
| D17mit180 | 27.3 ± 0.9 | 24.1 ± 0.4 | 24.2 ± 0.6 | 1.6 × 10−4 |
Measurements are given as means ± SD. The units for these measurements are: Lesion (square micrometers per section), LDL/VLDL cholesterol (milligrams per deciliter), HDL cholesterol (milligrams per deciliter), triglyceride (milligrams per deciliter), and body weight (g). BB, homozygous for C57BL/6-derived alleles of the linked marker; CC, homozygous for C3H alleles of the linked marker; BC, heterozygous for C57BL/6-derived and C3H alleles of the linked marker.
Loci on distal chromosome 1 accounting for major variations in plasma lipid levels and body weight:
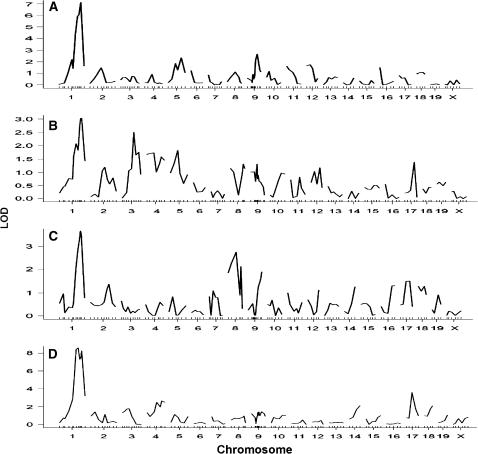
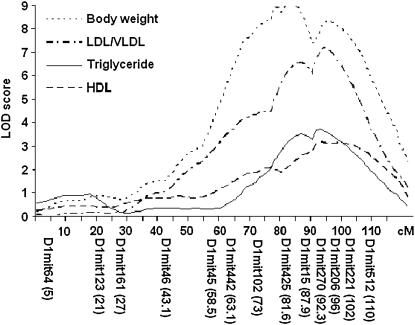
Genomewide scans revealed that loci on chromosome 1 were responsible for the major variations in plasma levels of LDL/VLDL (Figure 4A), HDL cholesterol (Figure 4B), and triglyceride (Figure 4C) and in body weight of the cross (Figure 4D). LOD score plots for chromosome 1 revealed two peaks for either plasma lipids or body weight (Figure 5). For plasma lipids, the distal peak appeared at marker D1Mit270 (92.3 cM), which had LOD scores of 6.3 for LDL/VLDL cholesterol, of 3.0 for HDL cholesterol, and of 3.8 for triglyceride, and the proximal peak appeared at marker D1Mit425 (81.6 cM) with LOD scores of 5.7 for LDL/VLDL cholesterol and 2.5 for triglyceride (Figure 5). The pattern of the LOD score peaks suggests the existence of two loci for plasma lipids in the distal chromosome 1 region. However, because the two peaks overlapped in the support intervals, they were considered as a single QTL. This locus modulated LDL/VLDL cholesterol in a codominant manner but influenced HDL cholesterol and triglyceride in a dominant manner from a C3H allele (Table 2). For body weight, the distal peak with a LOD score of 8.3 was observed at marker D1Mit206 (95.8 cM), and the proximal peak with a LOD score of 9.0 was observed at marker D1Mit425. Because the two peaks were not overlapping in the support intervals, they were considered as two individual QTL. Both loci exhibited a dominant effect from a C3H allele on body weight (Table 2). The proximal peak overlaps with the QTL Bw17, which was mapped to distal chromosome 1 at 75 cM with the use of recombinant inbred strains derived from SM/J and A/J mice (Anunciado et al. 2000). The distal peak overlaps with QTL Bw8q1, which was mapped to distal chromosome 1 at 100 cM in a B6 × A/J F2 cross (Zhang and Gershenfeld 2003).
Figure 4.
Genomewide QTL scans for loci affecting plasma levels of LDL/VLDL cholesterol (A), HDL cholesterol (B), triglyceride (C), and body weight (D) in the F2 population. Chromosomes 1 through X are represented numerically on the x-axis and the y-axis represents the LOD score.
Figure 5.
Detailed LOD score plots for LDL/VLDL cholesterol, HDL cholesterol, triglyceride, and body weight on chromosome 1. The x-axis depicts the marker positions in centimorgans, and the y-axis depicts the LOD score. The microsatellite markers typed are listed below the x-axis, corresponding to their map locations on the chromosome.
Loci for plasma lipids and body weight on other chromosomes:
For LDL/VLDL cholesterol, we found two suggestive loci at marker D5Mit95 (LOD 2.2, 68 cM) and marker D9Mit115 (LOD 2.5, 56 cM) (Table 1). The locus on chromosome 5 colocalized with a locus identified using a MRL/lpr × BALB/c cross (Gu et al. 1999). A suggestive locus for HDL cholesterol was detected on chromosome 3 near marker D3Mit42 (LOD 2.3, 58.8 cM). For triglyceride, we identified a suggestive loci near marker D8Mit41 (LOD 3.2, 41 cM), which overlapped with QTL identified previously in different crosses (Shike et al. 2001; Suto and Sekikawa 2003). A locus on chromosome 17 near marker D17Mit180 (29.4 cM) gave a significant LOD score of 3.4 for body weight. A QTL, Wt2q3, for body weight has been observed in this region in a previous study (Moody et al. 1999). We also found two suggestive loci for body weight near marker D4Mit251 (LOD 2.7, 66 cM) and marker D14Mit185 (LOD 2.2, 54 cM). These two loci colocalized with previously reported QTL Bw7 and Bwnd2wk7, respectively (Brockmann et al. 2000, 2004).
Relationships of plasma lipids with atherosclerotic lesions and body weight:
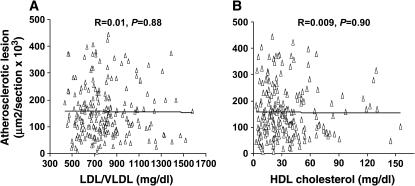
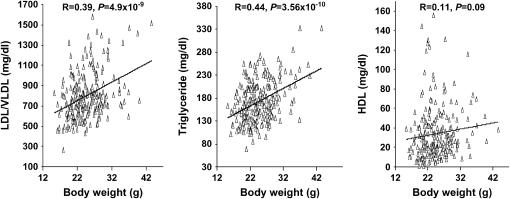
The relationships of plasma lipids with atherosclerotic lesions and body weight were analyzed using the F2 population. No significant correlations were observed between lesion sizes and LDL/VLDL cholesterol levels (R = 0.01, P = 0.88) or between lesion sizes and HDL cholesterol levels (R = 0.016 and P = 0.97) (Figure 6). Plasma LDL/VLDL cholesterol levels of the F2 mice were significantly correlated with body weight (R = 0.39, P = 4.9 × 10−9), as were the triglyceride levels (R = 0.44, P = 3.56 × 10−10) (Figure 7). However, plasma HDL cholesterol levels were poorly correlated with body weight (R = 0.11, P = 0.09).
Figure 6.
Scatterplots showing relationships between atherosclerotic lesion formation and plasma levels of LDL/VLDL (A) or HDL (B) cholesterol in the F2 cross. Each point represents an individual value of a F2 mouse. The correlation coefficient (R) and significance (P) are shown. There were no correlations between the sizes of atherosclerotic lesion sizes and the plasma levels of LDL/VLDL or HDL cholesterol.
Figure 7.
Scatterplots showing correlations of plasma lipids (in milligrams per deciliter) with body weight (in grams) in the F2 cross. Each point represents an individual value of a F2 mouse. The correlation coefficient (R) and significance (P) are shown. Plasma levels of LDL/VLDL cholesterol and triglyceride but not HDL cholesterol are significantly correlated with body weight in the cross.
DISCUSSION
In this study, we used the apoE−/− mouse model to search for QTL that contribute to the differences between strains B6 and C3H in atherosclerosis and plasma lipid levels. We found that atherosclerosis susceptibility and plasma lipid levels were controlled by separate genetic loci: a QTL on chromosome 9 explained the major variation in atherosclerotic lesion size whereas a QTL on chromosome 1 accounted for the major variation in plasma LDL/VLDL and HDL cholesterol and triglyceride levels. Moreover, we found that the QTL for plasma lipids coincided with the loci for body weight on distal chromosome 1.
Although strains B6 and C3H are prototype mouse models for genetic studies of atherosclerosis, the genetic factors that influence the trait remain undefined. In pioneering studies of recombinant inbred strains derived from the two strains, as well as from B6 and BALB/c, Paigen et al. (1987) identified the first atherosclerosis susceptibility locus, termed Ath-1. However, there are several limitations in that study: first, the number rather than the size of atherosclerotic lesions was measured. However, the number of the lesion was not as relevant to the disease as the lesion area. Second, the mapping was performed using only 10 recombinant inbred strains. Mapping using recombinant inbred strains has low power to detect QTL, mainly owing to the small number of available strains in each set (Silver 1995). And third, there were fewer polymorphic markers available at the time when the study was conducted. In a more recent study of a large F2 cross (185 female progeny) derived from the strains B6 and C3H, Machleder et al. (1997) failed to observe any significant or suggestive loci for atherosclerotic lesions on chromosome 1 or elsewhere in the genome, largely owing to the smaller size of atherosclerotic lesions. Foam cells start to develop 6 weeks after initiation of the atherogenic diet in susceptible B6 mice (Paigen et al. 1987), but the F2 mice were fed the diet for only 8 weeks.
In this study, we identified a significant QTL and a suggestive QTL that contributed to the development of atherosclerotic lesions using a F2 cross between B6.apoE−/− and C3H apoE−/− mice. These loci acted independently of plasma lipids because there were no correlations between plasma lipids and lesion sizes nor did they share any chromosomal regions that affected plasma lipid levels. The QTL on chromosome 9 accounted for 34% of the variance in lesion sizes of the cross. We have designated this QTL as Ath22 because it had not been reported in previous studies in the mouse. The LOD score plot for chromosome 9 shows three peaks with significant or suggestive LOD scores, suggesting a possibility of existence of three linked loci for atherosclerosis on the chromosome. However, because these loci overlapped in the support intervals and all exhibited a dominant effect from a B6 allele effect, we considered them as a single QTL. Nevertheless, there is a possibility that one or three of the peaks harbor more than one responsible gene. Fine mapping is needed to determine if there is only one or three linked loci in the chromosomal region. We also identified a locus with a suggestive LOD score of 2.4 on chromosome 11 influencing atherosclerotic lesion size. In a recent study of a B6x129S1/SvImJ cross fed an atherogenetic diet, Ishimori et al. (2004) also identified an atherosclerosis susceptibility locus, Ath19, on chromosome 11. However, these two QTL appear to be different loci because Ath19 is identified as a susceptibility locus only through putative interaction with a QTL on another chromosome while the locus identified in this study exhibited a dominant effect from a C3H allele on lesion formation.
The QTL for plasma lipids on distal chromosome 1 have been reported previously in several crosses, including a B × H cross (Machleder et al. 1997; Dansky et al. 2002; Lyons et al. 2003), but previous studies have not examined the association of plasma lipids with body weight. In this study, we have observed that the QTL for plasma lipids on distal chromosome 1 colocalized with the loci for body weight. The colocalization of QTL peaks for plasma lipids and body weight suggests a possibility that a single gene within the region impacts these two types of traits. Indeed, Apoa2 (92.6 cM) appears to be the causal gene responsible for the variations in both plasma lipids and body weight. Sequence analysis of Apoa2 cDNA from several inbred strains, including B6 and C3H mice, has revealed a number of nucleotide substitutions (Doolittle et al. 1990). In a set of recombinant inbred strains derived from B6 and C3H mice, HDL cholesterol levels segregated with the Apoa2 locus on chromosome 1 (Leboeuf et al. 1990). Moreover, transgenic overexpression of Apoa2 elevates plasma HDL, LDL/VLDL, and triglyceride levels (Hedrick et al. 1993) and Apoa2 deficiency reduces plasma HDL, LDL/VLDL, and triglyceride levels in mice (Weng and Breslow 1996). Interestingly, Apoa2 transgenic mice exhibit increases in adipose mass compared with B6 control mice (Castellani et al. 2001). Soat1 (81.6 cM), encoding acyl-CoA:cholesterol acyltransferase (ACAT)-1, is another positional candidate gene on chromosome 1 that could contribute to the variations in plasma lipids and body weight. Sulfonylureas, including glibenclamide, have been used clinically to treat noninsulin-dependent diabetes mellitus because these ACAT inhibitors have been shown to cause decreases in chylomicron, VLDL, and LDL levels and in body weight (Chan et al. 2004). Our finding that HDL cholesterol, triglycerides, and body weight all show a dominant C3H allele effect for increased levels also supports the possibility that these traits are controlled by the same locus. However, this is just speculation until these traits can be separated using recombinant congenic strains.
Increases in plasma LDL/VLDL levels or decreases in HDL levels have been considered to be major risk factors for the development of atherosclerosis. However, in the B6 and C3H mouse model, atherosclerosis susceptibility cannot be attributed to variations in plasma lipids. Indeed, on the chow diet, C3H.apoE−/− mice develop 140-fold larger aortic lesions than B6.apoE−/− mice despite their comparable levels of plasma LDL/LDL and HDL cholesterol (Shi et al. 2000). On the Western diet, C3H.apoE−/− mice exhibit 6-fold increases in HDL cholesterol and 10-fold decreases in aortic lesions compared with B6.apoE−/− mice. However, it is difficult to assess the contribution of HDL increases to the reduction in atherosclerotic lesions in C3H.apoE−/− mice using the parental strains. This study of F2 mice has clearly demonstrated that atherosclerosis susceptibility is independent of plasma lipids because the size of atherosclerotic lesions is not correlated with plasma HDL cholesterol or LDL/VLDL cholesterol. The recent studies of other genetic crosses carrying the Ldlr−/− (Welch et al. 2001) or apoE−/− gene (Dansky et al. 2002) also demonstrated no significant associations between atherosclerosis susceptibility and plasma lipoprotein levels.
In humans, dyslipidemia is an integral component of the metabolic perturbations in type II diabetes, obesity, and the metabolic syndrome and is featured by moderate-to-marked elevation of triglyceride and LDL and reduction of HDL cholesterol levels. Our study has revealed coincident loci for plasma lipids and body weight. Furthermore, we have demonstrated the strong correlation of plasma triglyceride and LDL/VLDL cholesterol with body weight. Thus, our findings suggest a genetic linkage between the phenotypes of dyslipidemia and obesity.
The demonstration that the differences in atherosclerosis susceptibility between inbred mouse strains B6 and C3H are not due to genetic differences in lipid metabolism provides an explanation for the clinical observation that a large fraction of patients develop atherosclerosis despite normolipidemia (McGill et al. 2000). Also, we have identified new QTL for atherosclerosis, although further study is needed to identify QTL genes. Once QTL genes are identified in mice, they can be tested by association studies for their relevance in human atherosclerotic disease.
Acknowledgments
This work was supported by National Institutes of Health grant HL71844, the Partners' Fund Award from the Cardiovascular Research Center, and the Dean's Research and Development award from the University of Virginia, School of Medicine.
References
- Anunciado, R. V., T. Ohno, M. Mori, A. Ishikawa, S. Tanaka et al., 2000. Distribution of body weight, blood insulin and lipid levels in the SMXA recombinant inbred strains and the QTL analysis. Exp. Anim. 49: 217–224. [DOI] [PubMed] [Google Scholar]
- Bodzioch, M., E. Orso, J. Klucken, T. Langmann, A. Bottcher et al., 1999. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 22: 347–351. [DOI] [PubMed] [Google Scholar]
- Brockmann, G. A., J. Kratzsch, C. S. Haley, U. Renne, M. Schwerin et al., 2000. Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F(2) variance of growth and obesity in DU6i × DBA/2 mice. Genome Res. 10: 1941–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann, G. A., E. Karatayli, C. S. Haley, U. Renne, O. J. Rottmann et al., 2004. QTLs for pre- and postweaning body weight and body composition in selected mice. Mamm. Genome 15: 593–609. [DOI] [PubMed] [Google Scholar]
- Broman, K. W., H. Wu, S. Sen and G. A. Churchill, 2003. QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Castellani, L. W., A. M. Goto and A. J. Lusis, 2001. Studies with apolipoprotein A-II transgenic mice indicate a role for HDLs in adiposity and insulin resistance. Diabetes 50: 643–651. [DOI] [PubMed] [Google Scholar]
- Chan, D. C., H. P. Barrett and G. F. Watts, 2004. Dyslipidemia in visceral obesity: mechanisms, implications, and therapy. Am. J. Cardiovasc. Drugs 4: 227–246. [DOI] [PubMed] [Google Scholar]
- Dansky, H. M., P. Shu, M. Donavan, J. Montagno, D. L. Nagle et al., 2002. A phenotype-sensitizing Apoe-deficient genetic background reveals novel atherosclerosis predisposition loci in the mouse. Genetics 160: 1599–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle, M. H., R. C. LeBoeuf, C. H. Warden, L. M. Bee and A. J. Lusis, 1990. A polymorphism affecting apolipoprotein A-II translational efficiency determines high density lipoprotein size and composition. J. Biol. Chem. 265: 16380–16388. [PubMed] [Google Scholar]
- Goldstein, J. L., and M. S. Brown, 1979. The LDL receptor locus and the genetics of familial hypercholesterolemia. Annu. Rev. Genet. 13: 259–289. [DOI] [PubMed] [Google Scholar]
- Grimsditch, D. C., S. Penfold, J. Latcham, M. Vidgeon-Hart, P. H. Groot et al., 2000. C3H apoE(−/−) mice have less atherosclerosis than C57BL apoE(−/−) mice despite having a more atherogenic serum lipid profile. Atherosclerosis 151: 389–397. [DOI] [PubMed] [Google Scholar]
- Gu, L., M. W. Johnson and A. J. Lusis, 1999. Quantitative trait locus analysis of plasma lipoprotein levels in an autoimmune mouse model: interactions between lipoprotein metabolism, autoimmune disease, and atherogenesis. Arterioscler. Thromb. Vasc. Biol. 19: 442–453. [DOI] [PubMed] [Google Scholar]
- Hedrick, C. C., L. W. Castellani, C. H. Warden, D. L. Puppione and A. J. Lusis, 1993. Influence of mouse apolipoprotein A-II on plasma lipoproteins in transgenic mice. J. Biol. Chem. 268: 20676–20682. [PubMed] [Google Scholar]
- Ishimori, N., R. Li, P. M. Kelmenson, R. Korstanje, K. A. Walsh et al., 2004. Quantitative trait loci analysis for plasma HDL-cholesterol concentrations and atherosclerosis susceptibility between inbred mouse strains C57BL/6J and 129S1/SvImJ. Arterioscler. Thromb.Vasc. Biol. 24: 161–166. [DOI] [PubMed] [Google Scholar]
- Kraus, J. P., M. Janosik, V. Kozich, R. Mandell, V. Shih et al., 1999. Cystathionine beta-synthase mutations in homocystinuria. Hum Mutat. 13: 362–375. [DOI] [PubMed] [Google Scholar]
- LeBoeuf, R. C., M. H. Doolittle, A. Montcalm, D. C. Martin, K. Reue et al., 1990. Phenotypic characterization of the Ath-1 gene controlling high density lipoprotein levels and susceptibility to atherosclerosis. J. Lipid Res. 31: 91–101. [PubMed] [Google Scholar]
- Liao, F., A. Andalibi, F. C. deBeer, A. M. Fogelman and A. J. Lusis, 1993. Genetic control of inflammatory gene induction and NF-kappa B-like transcription factor activation in response to an atherogenic diet in mice. J. Clin. Invest. 91: 2572–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, F., A. Andalibi, J. H. Qiao, H. Allayee, A. M. Fogelman et al., 1994. Genetic evidence for a common pathway mediating oxidative stress, inflammatory gene induction, and aortic fatty streak formation in mice. J. Clin. Invest. 94: 877–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis, A. J., R. Mar and P. Pajukanta, 2004. Genetics of atherosclerosis. Annu. Rev. Genomics Hum. Genet. 5: 189–218. [DOI] [PubMed] [Google Scholar]
- Lyons, M. A., H. Wittenburg, R. Li, K. A. Walsh, M. R. Leonard et al., 2003. Quantitative trait loci that determine lipoprotein cholesterol levels in DBA/2J and CAST/Ei inbred mice. J. Lipid Res. 44: 953–967. [DOI] [PubMed] [Google Scholar]
- Machleder, D., B. Ivandic, C. Welch, L. Castellani, K. Reue et al., 1997. Complex genetic control of HDL levels in mice in response to an atherogenic diet. Coordinate regulation of HDL levels and bile acid metabolism. J. Clin. Invest. 99: 1406–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly, K. F., Jr., R. H. Cudmore and J. M. Meer, 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930–932. [DOI] [PubMed] [Google Scholar]
- Matsushima, Y., T. Sakurai, A. Ohoka, T. Ohnuki, N. Tada et al., 2001. Four strains of spontaneously hyperlipidemic (SHL) mice: phenotypic distinctions determined by genetic backgrounds. J. Atheroscler. Thromb. 8: 71–79. [DOI] [PubMed] [Google Scholar]
- McGill, H. C., Jr., C. A. McMahan, A. W. Zieske, G. D. Sloop, J. V. Walcott et al., 2000. Associations of coronary heart disease risk factors with the intermediate lesion of atherosclerosis in youth: the Pathobiological Determinants of Atherosclerosis in Youth (PDAY) Research Group. Arterioscler. Thromb. Vasc. Biol. 20: 1998–2004. [DOI] [PubMed] [Google Scholar]
- Mehrabian, M., L. W. Castellani, P. Z. Wen, J. Wong, T. Rithaporn et al., 2000. Genetic control of HDL levels and composition in an interspecific mouse cross (CAST/Ei × C57BL/6J). J. Lipid Res. 41: 1936–1946. [PubMed] [Google Scholar]
- Moody, D. E., D. Pomp, M. K. Nielsen and L. D. Van Vleck, 1999. Identification of quantitative trait loci influencing traits related to energy balance in selection and inbred lines of mice. Genetics 152: 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, Y., A. S. Plump, E. W. Raines, J. L. Breslow and R. Ross, 1994. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler. Thromb. 14: 133–140. [DOI] [PubMed] [Google Scholar]
- Paigen, B., A. Morrow, C. Brandon, D. Mitchell and P. Holmes, 1985. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis 57: 65–73. [DOI] [PubMed] [Google Scholar]
- Paigen, B., D. Mitchell, K. Reue, A. Morrow, A. J. Lusis et al., 1987. Ath-1, a gene determining atherosclerosis susceptibility and high density lipoprotein levels in mice. Proc. Natl. Acad. Sci. USA 84: 3763–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plump, A. S., J. D. Smith, Y. Hayek, K. Aalto-Setala, A. Walsh et al., 1992. Severe hyper-cholesterolemia and atherosclerosis in apoliprotein E-deficient mice created by homologus recombination in ES cells. Cell 71: 343–353. [DOI] [PubMed] [Google Scholar]
- Qiao, J. H., P. Z. Xie, M. C. Fishbein, J. Kreuzer, T. A. Drake et al., 1994. Pathology of atheromatous lesions in inbred and genetically engineered mice. Genetic determination of arterial calcification. Arterioscler. Thromb. 14: 1480–1497. [DOI] [PubMed] [Google Scholar]
- Shi, W., N. J. Wang, D. M. Shih, V. Z. Sun, X. Wang et al., 2000. Determinants of atherosclerosis susceptibility in the C3H and C57BL/6 mouse model: evidence for involvement of endothelial cells but not blood cells or cholesterol metabolism. Circ. Res. 86: 1078–1084. [DOI] [PubMed] [Google Scholar]
- Shih, D. M., L. Gu, S. Hama, Y. R. Xia, M. Navab et al., 1996. Genetic-dietary regulation of serum paraoxonase expression and its role in atherogenesis in a mouse model. J. Clin. Invest. 97: 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shike, T., S. Hirose, M. Kobayashi, K. Funabiki, T. Shirai et al., 2001. Susceptibility and negative epistatic loci contributing to type 2 diabetes and related phenotypes in a KK/Ta mouse model. Diabetes 50: 1943–1948. [DOI] [PubMed] [Google Scholar]
- Silver, L. M., 1995. Mouse Genetics. Oxford University Press, New York.
- Suto, J., and K. Sekikawa, 2003. Quantitative trait locus analysis of plasma cholesterol and triglyceride levels in KK × RR F2 mice. Biochem. Genet. 41: 325–341. [DOI] [PubMed] [Google Scholar]
- Suto, J., S. Matsuura, H. Yamanaka and K. Sekikawa, 1999. Quantitative trait loci that regulate plasma lipid concentration in hereditary obese KK and KK-Ay mice. Biochim. Biophys. Acta 1453: 385–395. [DOI] [PubMed] [Google Scholar]
- Tian, J., H. Pei, J. C. James, Y. Li, A. H. Matsumoto et al., 2005. Circulating adhesion molecules in apoE-deficient mouse strains with different atherosclerosis susceptibility. Biochem. Biophys. Res. Commun. 329: 1102–1107. [DOI] [PubMed] [Google Scholar]
- Welch, C. L., S. Bretschger, M. Bezouevski, Y. Guo, N. Pleskac et al., 2001. Localization of atherosclerosis susceptibility loci to chromosomes 4 and 6 using the Ldlr knockout mouse model. Proc. Natl. Acad. Sci. USA 98: 7946–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, W., and J. L. Breslow, 1996. Dramatically decreased high density lipoprotein cholesterol, increased remnant clearance, and insulin hypersensitivity in apolipoprotein A-II knockout mice suggest a complex role for apolipoprotein A-II in atherosclerosis susceptibility. Proc. Natl. Acad. Sci. USA 93: 14788–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and H. K. Gershenfeld, 2003. Genetic contributions to body weight in mice: relationship of exploratory behavior to weight. Obes. Res. 11: 828–838. [DOI] [PubMed] [Google Scholar]
- Zhang, S. H., R. L. Reddick, J. A. Piedrahita and N. Maeda, 1992. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apoliprotein E. Science 258: 468–471. [DOI] [PubMed] [Google Scholar]