Abstract
In yeast, the nonhomologous end joining pathway (NHEJ) mobilizes the DNA polymerase Pol4 to repair DNA double-strand breaks when gap filling is required prior to ligation. Using telomere–telomere fusions caused by loss of the telomeric protein Rap1 and double-strand break repair on transformed DNA as assays for NHEJ between fully uncohesive ends, we show that Pol4 is able to extend a 3′-end whose last bases are mismatched, i.e., mispaired or unpaired, to the template strand.
NONHOMOLOGOUS end joining (NHEJ) is a DNA repair pathway dedicated to double-strand breaks (DSBs) (for review, Wilson et al. 2003; Daley et al. 2005b). NHEJ proceeds by a direct ligation of two ends and is conserved through evolution. The ligation step is performed by an ATP-dependent DNA ligase committed to this pathway, Lig4 and its associated factor Lif1 in Saccharomyces cerevisiae. Repair by NHEJ of DSBs whose ends are perfectly cohesive is essentially a ligation and is a very efficient and accurate process (Lee et al. 1999; Frank-Vaillant and Marcand 2002). NHEJ may also attempt to restore the original sequence at a DSB whose ends are uncohesive due to damaged bases. X rays and γ-rays can release enough energy to produce in a small volume multiple reactive species susceptible to attack simultaneously the two DNA strands and the adjacent bases, creating DSBs whose terminal bases might be lost or damaged (Ward 2000). In higher eukaryotes, the increased sensitivity of NHEJ-deficient cells to ionizing radiations indicates that this pathway can repair some of the induced DSBs (Grawunder et al. 1997). In yeast, an increased sensitivity to ionizing radiations has been observed in some strains defective for NHEJ and only in contexts where DSB repair cannot occur through homologous recombination (Boulton and Jackson 1996; Siede et al. 1996; Schar et al. 1997). Interestingly, Lig4-deficient yeast cells in stationary phase display a reduced level of adaptive mutations (Heidenreich et al. 2003). If the Lig4 pathway is indeed restricted to DSB repair, it suggests that DSB could occur spontaneously in quiescent cells and be repaired by NHEJ. They could be formed by endogenous reactive species and nuclease activities, possibly leaving incompatible DNA ends. It is still unclear how efficient and accurate NHEJ is in these contexts. Repair or removal of modified bases as well as the search for base pairing between the ends implies that processing steps occur prior to ligation. A detailed comprehension of this processing may help to understand how radiation-induced and spontaneous DSBs are repaired by NHEJ.
The processing of imperfectly cohesive ends implies that nuclease and polymerase activities are recruited to correct mismatches, eliminate damaged bases, and fill in gaps. It is still unclear how many nuclease activities can act in the course of NHEJ (Wu et al. 1999; Yu et al. 2004; Pardo and Marcand 2005; Zhang and Paull 2005). In particular, nucleases involved in the processing of mismatches at the 3′-ends remain to be identified. The DNA polymerases mobilized by NHEJ belong to the Pol X family. In S. cerevisiae, the DNA polymerase Pol4 is the only member of this family and is required for NHEJ events, implying gap fill in prior to ligation (Wilson and Lieber 1999; Daley et al. 2005a). In humans, three DNA polymerases from the Pol X family have been implicated in NHEJ: Pol λ, Pol μ, and TdT. (Delarue et al. 2002; Mahajan et al. 2002; Bertocci et al. 2003; Lee et al. 2004; Ma et al. 2004; Nick McElhinny et al. 2005).
In vitro, Pol4 prefers to fill short gaps and lacks an exonuclease proofreading activity (Prasad et al. 1993; Shimizu et al. 1993; Wilson and Lieber 1999; Tseng and Tomkinson 2002; Bebenek et al. 2005). In particular Pol4 can fill in a short gap even if the priming 3′ nucleotide is mispaired to the template (Wilson and Lieber 1999). A similar tolerance to a terminal mismatch was observed with Pol μ in vitro (Zhang et al. 2001; Covo et al. 2004). In these experiments, alignment of the 3′ priming end is accomplished by partial annealing with the template strand. More recently, Pol μ tolerance to a mispaired or even an unpaired priming end was observed in the context of an end-joining reaction carried out in vitro in the presence of Ku and ligase IV-XRCC4 (Nick McElhinny et al. 2005). Here we show that Pol4 tolerance to mismatches at the priming ends can be observed in vivo.
Pol4 requirement for fusions between telomeres exposed to NHEJ:
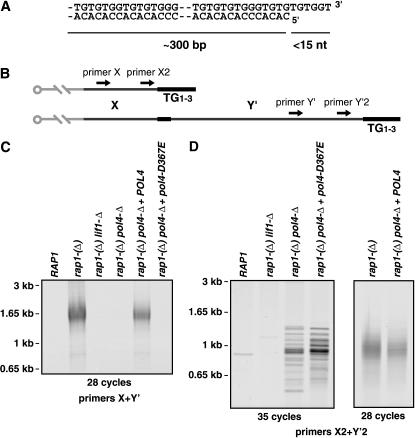
Fusions between telomeres offer an in vivo situation where NHEJ might involve Pol4. In S. cerevisiae, the sequence of telomeric DNA consists of a tandem array of TG1–3 repeats (Figure 1A). It is an oriented structure with the G-rich strand running 5′ to 3′ toward the distal end of the chromosome. The last base at the 3′-end of a telomere is randomly a T or a G (Forstemann et al. 2000). The length of TG1–3 telomeric repeats is kept within a narrow size distribution around a mean value of ∼300 bp (Forstemann et al. 2000). Yeast telomeres for the most part are double stranded and end with a short single-stranded 3′ overhang. The length of this single-stranded DNA is dynamic during replication and remains <15 bases outside of S phase (Larrivee et al. 2004).
Figure 1.
Telomere fusions induced by Rap1 loss involve Pol4. (A) Schematic of a yeast telomere. (B) Relative positions of the primers used to detect telomere fusions. Primers X, X2, Y′, and Y′2 anneal at a distance from the TG1–3 repeats of ∼520, ∼340, 480, and 120 bp, respectively. (C) Pol4 activity is required for most telomere fusions. Yeast strains ZMY60 (wild type), Lev391 [rap1-(Δ)], Lev396 [rap1-(Δ) lif1-Δ], and Ybp25 [rap1-(Δ) pol4-Δ] transformed with plasmid pRS314 and strain Ybp25 transformed with plasmid pRS314-POL4 and plasmid pRS314-pol4-D367E were grown to saturation in synthetic medium lacking tryptophan for 5 days (Table 1). Telomere fusions were amplified by PCR with primers X and Y′. (D) Increased number of PCR cycles detects a low level of Lif1-dependent fusions in the absence of Pol4 activity. Telomere fusions were amplified by PCR with primers X2 and Y′2. The weak signals sometimes observed from the wild-type and rap-(Δ) lif1-Δ strains seem to be due to nonspecific amplifications although rare fusion events cannot be excluded. The methods used were the following: the POL4 gene (including 399 bp upstream of the start codon and 275 bp downstream of the stop codon) was amplified by PCR and inserted into pRS314 (CEN, TRP1), creating plasmid pRS314-POL4. The pol4-D367E allele was obtained from plasmid pTW305 (Wilson and Lieber 1999) and introduced into pRS314-POL4 by gap repair into yeast cell. The new plasmid, pRS314-pol4-D367E, was transformed and amplified in E. coli. The presence of the mutation was confirmed by sequencing. Telomere fusions were amplified by PCR with primer X and Y′ as described previously (Pardo and Marcand 2005). Genomic DNA was prepared by phenol–chloroform extraction and resuspended in TE, pH 8.0 buffer. Primer X2 (TGTGGTGGTGGGATTAGAGTGGTAG) has a sequence from X elements (e.g., coordinates 417–441 of chromosome XV). Primer Y′2 (TTAGGGCTATGTAGAAGTGCTG) has a sequence from Y′ elements (e.g., coordinates 174–195 of chromosome XII). PCR reactions (30 μl) contained genomic DNA ∼10 ng, Hot Start buffer 1× supplemented with MgSO4 0.83 mm, dNTP 0.3 mm each, primers 1 μm each, 1.2 units of HotStarTaq (QIAGEN, Chatsworth, CA), and 0.12 unit of ProofStart (QIAGEN). The conditions were: 95° for 15 min and then 28 or 35 cycles of 94° for 30 sec, 68° for 30 sec, 72° for 1 min 15 sec, followed by 72° for 3 min. The products were run through a 1% agarose gel and visualized by ethidium bromide staining. Amplified fusions were cloned by HindIII–EcoRI digestion into pUC18 using primers with added restriction sites. The clones were amplified in XL1-blue cells grown at 25°, analyzed by restriction, and sequenced.
In wild-type cells, NHEJ is suppressed at telomeres, ensuring that chromosome end-to-end fusions do not occur (Ferreira et al. 2004). Recently, we and others showed that in yeasts the telomere-binding protein Rap1 is required to establish NHEJ suppression at telomeres (Miller et al. 2005; Pardo and Marcand 2005). We used a conditional allele of RAP1, called rap1-(Δ), which causes the Rap1 protein level to drop in cells progressing toward stationary phase, resulting in telomere fusions (Pardo and Marcand 2005). A PCR strategy was used to detect fusions between telomeres (Mieczkowski et al. 2003). In S. cerevisiae, a conserved element, X, is located adjacent to every telomere. About half of the chromosome ends display a second element, Y′, inserted between X and the telomere (Figure 1B). First, two primers annealing with X and Y′, respectively, were used to amplify fusions occurring between a X telomere and a Y′ telomere. In rap1-(Δ) cells having reached stationary phase, fusions between X and Y′ telomeres can be amplified (Figure 1C). Since telomere length distribution is heterogeneous, telomere fusions are detected as a smearing PCR signal. Their frequency was estimated at about one fusion per genome (Pardo and Marcand 2005). As previously reported, fusions could not be detected in rap1-(Δ) lif1-Δ cells defective for NHEJ, indicating that they are primarily produced by this pathway. In the absence of Pol4, the fusions are not detected (Figure 1C). Complementation of the pol4 disruption by a centromeric plasmid encoding a wild-type Pol4 restores the appearance of fusions in rap1-(Δ) cells. An allele encoding a catalytically inactive form of Pol4 fails to restore the fusions, indicating that the activity of Pol4 is required for the high level of fusions observed in cells defective for Rap1.
To determine if the loss of fusions in a pol4-Δ mutant is as severe as that in a lif1-Δ mutant, we used an improved set of primers, X2 and Y′2, that generate less nonspecific products at higher numbers of PCR cycles (Figure 1B). X2 and Y′2 anneal at ∼340 and 120 bp, respectively, from the beginning of the telomeric repeats. A telomere fusion should give a PCR product of ∼460 bp plus the length of TG1–3 repeats at the junction. As shown in Figure 1D, increased numbers of PCR cycles allow the detection of bands from rap1-(Δ) pol4-Δ cells and from rap1-(Δ) pol4-Δ cells with the plasmid encoding a catalytically inactive form of Pol4. In the same conditions, no such signals are amplified from wild-type cells and from rap1-(Δ) lif1-Δ cells defective for NHEJ. The size range of the bands is similar to the smears observed in rap1-(Δ) cells with a wild-type Pol4 activity. The discrete band pattern agrees with a low abundance of telomere fusions. PCR products were cloned, amplified in Escherichia coli, and sequenced, revealing an X element end, a Y′ element end, and TG1–3 repeats pointing at each other (data not shown). When genomic DNA from rap1-(Δ) cells is diluted by 100- to 1000-fold, the PCR produces a signal with a discrete band pattern similar in intensity to the one observed with undiluted DNA from rap1-(Δ) pol4-Δ cells (data not shown). These results suggest that, in the absence of Pol4, telomere fusions by NHEJ can still occur although at a frequency reduced by two to three orders of magnitude.
Since most of the telomere fusions induced by Rap1 loss require the catalytic activity of Pol4, we infer that NHEJ between telomeres involves gap filling between the telomeric overhangs. The 3′-ends of S. cerevisiae telomeres are made of only G and T bases, ruling out normal base pairing between telomeric ends exposed to NHEJ. This leaves two possibilities for Pol4. If a 3′ overhang remains on both telomere ends, Pol4 could extend a 3′-end whose last bases are unpaired or mispaired with the aligned template strand. Or, following the degradation of the 3′ overhang on one telomere, Pol4 could extend a blunt end over the remaining 3′ overhang; i.e., Pol4 could polymerize across a nick in the template strand.
Pol4 can act on mismatched 3′-ends:
In the previous experiment, the sizes of the single-stranded 3′ overhang are variable among the telomeres and cannot be deduced from the fusions. To address this limitation and to further test how Pol4 can act in the absence of correct base pairing, we used a plasmid transformation assay. This approach is based on the observation that, following transformation into cells, NHEJ can repair a DSB created in vitro on a plasmid by enzymatic restriction (Orr-Weaver and Szostak 1983; Boulton and Jackson 1996). Positive selection for recircularized plasmid and sequencing of the junctions provides a simple assay for NHEJ. We took advantage of the BstXI restriction enzyme to generate TGTG-3′ single-stranded extensions on a plasmid prior to transformation into wild type, pol4-Δ, and lif1-Δ strains (Figure 2A). Transformation efficiency was reduced compared to that obtained with circular plasmid DNA (Table 2) or KpnI-digested plasmid DNA displaying cohesive ends (data not shown), suggesting that uncohesive ends are not efficiently repaired by NHEJ in this assay. Joints created after repair were amplified by PCR and analyzed by restriction (Table 2). Some clones have conserved the linker between the two BstXI sites, an outcome of undigested or partially digested DNA. Others reproducibly fail to generate a PCR product or have lost one of the two restriction sites adjacent to the ends, indicating extensive processing on at least one side. A few clones display insertions. All those clones were not investigated further. The remaining clones were repaired with limited processing and could be potentially meaningful for the purpose of this analysis: 77/200 in the wild type, slightly less in the pol4-Δ mutant (53/200, P = 0.01; t-test), and none in the lif1-Δ mutant (0/100), indicating that they are produced by NHEJ. These joints with limited loss from the ends were sequenced.
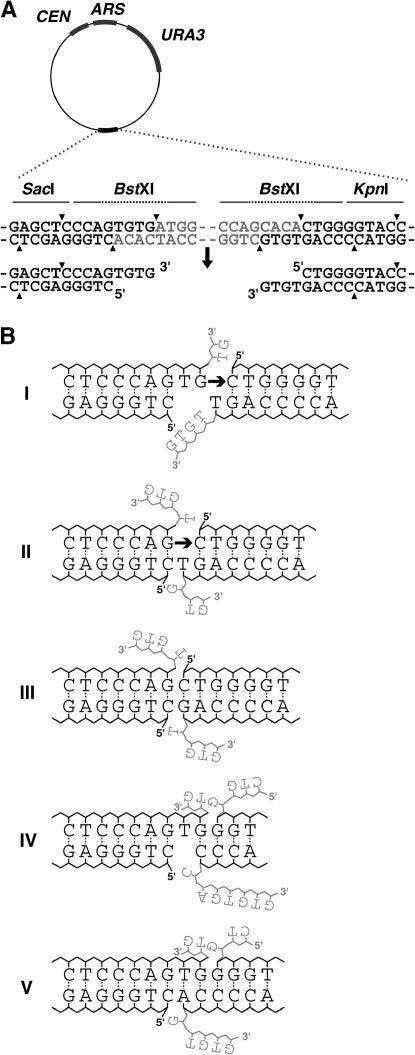
Figure 2.
Plasmid transformation assay with two noncohesive TGTG 3′ overhangs. (A) Schematic of plasmid sp469. Arrowheads indicate the cleavage sites for SacI, BstXI, and KpnI. (B) Possible end alignments for each type of joints observed in the plasmid transformation assay with two noncohesive TGTG 3′ overhangs. Deleted bases are shaded. The following methods were used: the polylinker of pRS316 (CEN, URA3) was replaced from SacI to KpnI by the sequence CCAGTGTGATGGGAGCAACTCATCTCTATTCACCAGCACACTGG, which includes two BstXI sites separated by a 20-bp linker, creating plasmid sp469. About 50 ng of plasmid sp469, intact or digested with BstXI (New England Biolabs, Beverly, MA), was transformed into yeast strains Lev488, Ybp50, and Ybp52 by the lithium acetate transformation method (Table 1). The transformed cells were plated on synthetic media lacking uracil. Colonies were counted after incubation at 30° for 3 days. Individual colonies were subjected to PCR using primers framing the junction (pRS#1 CACACCCGCCGCGCTTAATG, 232 bp upstream of SacI; pRS#2 CAATACGCAAACCGCCTCTCCC, 266 bp downstream of KpnI). The PCR products, purified on a Qiaquick column (QIAGEN), were digested by KpnI and SacI and analyzed on a 1.5% agarose gel. Clones that had conserved the two restriction sites were sequenced. Numerical data were compared by the Student's t-test.
TABLE 2.
Junctions from two TGTG 3′ overhangs in a plasmid transformation assay
Relative transformation efficiencies for each genotype are expressed as the ratio of the colony count obtained with BstXI-digested plasmid over the colony count obtained with undigested plasmid. Mean and standard deviation were calculated from three independent transformations.
Insertions of 100–300 bp that might come from the carrier DNA (Decottignies 2005).
Boldface type indicates bases stemming from the 3′ overhangs; underlining indicates possible base pairing involved in the junction.
The observed joints are listed in Table 2 and are clustered into five types (Figure 2B). On one hand, we observed joints that did not involve base pairing and have either preserved a piece of the two overhangs (type I), only one overhang (type II), or lost the two overhangs (type III). On the other hand, we observed joints that could have involved a single (type IV) or two (type V) base pairings.
Type V is the most frequent type (Table 2). Its occurrence is not affected by the loss of Pol4, as expected for a joint that does not require gap filling (Figure 2B). In contrast, type I joints entail gap filling on both strands and are missing in cells lacking Pol4 (0/200 compared to 10/200 in wild-type cells; P = 0.0013). Type II joints require gap filling on only one strand and are still observed in pol4-Δ cells, although at a lesser frequency (2/200 compared to 9/200 in wild-type cells; P = 0.032). This would suggest that, following ligation by Lig4 of the template strand, general repair factors unrelated to NHEJ (e.g., Pol δ and Cdc9) could replace Pol4 and Lig4 to fill and ligate the second strand. The absence of Pol4 does not seem to influence the occurrence of type III and IV joints, which either do not require gap filling or do so on only one strand.
Joints that preserved a piece of the two overhangs (type I) imply that, following partial degradation of a 3′ overhang, a 3′-end that is unpaired or mispaired with the template strand is extended prior to ligation (Figure 2B). Since these events require Pol4, they demonstrate that Pol4 is able to extend a 3′-end whose last bases are mismatched. The observed joints did not reveal any random nucleotide addition, suggesting that elongation by Pol4 remains template dependent in this context. However, we cannot rule out a putative terminal transferase activity of Pol4 proofread by a nuclease.
Joints of type I, II, and III that have not involved base pairing resemble fusions between telomeres but their occurrence in the plasmid assay seems much lower. One possibility is that the stability of telomeres allows multiple attempts at a fusion whereas a transformed plasmid is probably rapidly degraded if repair is not immediately successful. It is also possible that the plasmid transformation assay is unable to fully reconstitute the NHEJ pathway. In support of the latter, we note that, even with perfectly cohesive ends, a marked difference is observed between repair of transformed DNA ends and repair of a break generated in vivo: the absolute repair efficiency is lower and the error rate higher with the plasmid transformation assay (Lee et al. 1999; Frank-Vaillant and Marcand 2002; Karathanasis and Wilson 2002).
Could Pol4 mismatch-tolerant priming contribute to an accurate DSB repair by NHEJ?
Mismatch-tolerant primer extension by DNA polymerase Pol4 allows the NHEJ machinery to repair ends for which conventional base pairing is unavailable. This property may not be needed for repair per se in most situations: further degradation of terminal bases can usually expose one or two bases for pairing between the two ends to facilitate end joining, as observed in the plasmid transformation assay described here. A possibility is that mismatch tolerance is important only for very specific situations where two ends cannot base pair over a long stretch. Fusions between yeast telomeres would mimic these rare situations. Such a model implies that, in the absence of base pairing between ends, NHEJ does not attempt to restore the original sequence and usually proceeds to repair with a small deletion.
Another possibility is that mismatch tolerance favors the conservation of the original sequence and thus the overall fidelity of NHEJ. In the plasmid transformation assay with TGTG 3′ overhangs, although the events involving Pol4 cannot be described as being accurate, they are the ones that preserved most of the original sequence. In cells in stationary phase, the absence of Pol4 increases the rate of −1 frameshift adaptive mutations, i.e., of sequence deletions (Heidenreich and Eisler 2004). This increase is dependent upon Lig4, suggesting that Pol4 favors sequence conservation during NHEJ on DSBs occurring spontaneously (Heidenreich and Eisler 2004). In a model where the NHEJ machinery attempts an accurate repair when confronted with a DSB with damaged and uncohesive 3′ overhangs, the impossibility of elongating a 5′-end leaves mismatch tolerance at the priming 3′-end as the sole option to restore the original sequence.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| ZMY60 | MATaura3-52 trp1-Δ1 ade2-101 pACE1-UBR1 pACE1-ROX1 |
| Lev391 | ZMY60 rap1-(Δ)∷KanR |
| Lev396 | Lev391 lif1-Δ∷klURA3 |
| Ybp25 | Lev391 pol4-Δ∷klURA3 |
| W303-1a | MATaade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 rad5-535 |
| Lev488 | W303-1a RAD5 lys2∷pGAL-ISCEI bar1-Δ |
| Ybp50 | Lev488 pol4-Δ∷spHIS5 |
| Ybp52 | Lev488 lif1-Δ∷spHIS5 |
In strain ZMY60, UBR1 and ROX1 are under the control of the ACE1 promoter (Moqtaderi et al. 1996). In strain Lev391, rap1 is under the control of a promoter repressible by Rox1 and expresses a protein with a N-terminal tag that makes it a target for Ubr1 and degradation by the N-end rule (Pardo and Marcand 2005). Gene deletions were made by PCR-mediated transformation.
Acknowledgments
We thank Thomas Wilson for the gift of the pol4-D367E allele and Emmanuelle Martini, Ariane Gratias, Serge Boiteux, Stéphanie Marsin, Madalena Tarsounas, Francis Fabre, Serge Gangloff, and Xavier Veaute for suggestions and comments. This work is supported by grants from the Association pour la Recherche sur le Cancer, the Fondation de France (programme Tumeurs), the Fondation pour la Recherche Médicale, and the Ministère délégué à la Recherche (Action Concertée Incitative jeunes chercheurs).
References
- Bebenek, K., M. Garcia-Diaz, S. R. Patishall and T. A. Kunkel, 2005. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J. Biol. Chem. 280: 20051–20058. [DOI] [PubMed] [Google Scholar]
- Bertocci, B., A. De Smet, C. Berek, J. C. Weill and C. A. Reynaud, 2003. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity 19: 203–211. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15: 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Covo, S., L. Blanco and Z. Livneh, 2004. Lesion bypass by human DNA polymerase mu reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J. Biol. Chem. 279: 859–865. [DOI] [PubMed] [Google Scholar]
- Daley, J. M., R. L. Laan, A. Suresh and T. E. Wilson, 2005. a DNA joint dependence of pol × family polymerase action in nonhomologous end joining. J. Biol. Chem. 280: 29030–29037. [DOI] [PubMed] [Google Scholar]
- Daley, J. M., P. L. Palmbos, D. Wu and T. E. Wilson, 2005. b Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451. [DOI] [PubMed] [Google Scholar]
- Decottignies, A., 2005. Capture of extranuclear DNA at fission yeast double-strand breaks. Genetics 171: 1535–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue, M., J. B. Boule, J. Lescar, N. Expert-Bezancon, N. Jourdan et al., 2002. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J. 21: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, M. G., K. M. Miller and J. P. Cooper, 2004. Indecent exposure: when telomeres become uncapped. Mol. Cell 13: 7–18. [DOI] [PubMed] [Google Scholar]
- Forstemann, K., M. Hoss and J. Lingner, 2000. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 28: 2690–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant, M., and S. Marcand, 2002. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell 10: 1189–1199. [DOI] [PubMed] [Google Scholar]
- Grawunder, U., M. Wilm, X. Wu, P. Kulesza, T. E. Wilson et al., 1997. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388: 492–495. [DOI] [PubMed] [Google Scholar]
- Heidenreich, E., and H. Eisler, 2004. Non-homologous end joining dependency of gamma-irradiation-induced adaptive frameshift mutation formation in cell cycle-arrested yeast cells. Mutat. Res. 556: 201–208. [DOI] [PubMed] [Google Scholar]
- Heidenreich, E., R. Novotny, B. Kneidinger, V. Holzmann and U. Wintersberger, 2003. Non-homologous end joining as an important mutagenic process in cell cycle-arrested cells. EMBO J. 22: 2274–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanasis, E., and T. E. Wilson, 2002. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161: 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee, M., C. Lebel and R. J. Wellinger, 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18: 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. W., L. Blanco, T. Zhou, M. Garcia-Diaz, K. Bebenek et al., 2004. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J. Biol. Chem. 279: 805–811. [DOI] [PubMed] [Google Scholar]
- Lee, S. E., F. Paques, J. Sylvan and J. E. Haber, 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9: 767–770. [DOI] [PubMed] [Google Scholar]
- Ma, Y., H. Lu, B. Tippin, M. F. Goodman, N. Shimazaki et al., 2004. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell 16: 701–713. [DOI] [PubMed] [Google Scholar]
- Mahajan, K. N., S. A. Nick McElhinny, B. S. Mitchell and D. A. Ramsden, 2002. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol. 22: 5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski, P. A., J. O. Mieczkowska, M. Dominska and T. D. Petes, 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc. Natl. Acad. Sci. USA 100: 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. M., M. G. Ferreira and J. P. Cooper, 2005. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 24: 3128–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil and K. Struhl, 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383: 188–191. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny, S. A., J. M. Havener, M. Garcia-Diaz, R. Juarez, K. Bebenek et al., 2005. A gradient of template dependence defines distinct biological roles for family × polymerases in nonhomologous end joining. Mol. Cell 19: 357–366. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., and J. W. Szostak, 1983. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA 80: 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo, B., and S. Marcand, 2005. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 24: 3117–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, R., S. G. Widen, R. K. Singhal, J. Watkins, L. Prakash et al., 1993. Yeast open reading frame YCR14C encodes a DNA beta-polymerase-like enzyme. Nucleic Acids Res. 21: 5301–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schar, P., G. Herrmann, G. Daly and T. Lindahl, 1997. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 11: 1912–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K., C. Santocanale, P. A. Ropp, M. P. Longhese, P. Plevani et al., 1993. Purification and characterization of a new DNA polymerase from budding yeast Saccharomyces cerevisiae. A probable homolog of mammalian DNA polymerase beta. J. Biol. Chem. 268: 27148–27153. [PubMed] [Google Scholar]
- Siede, W., A. A. Friedl, I. Dianova, F. Eckardt-Schupp and E. C. Friedberg, 1996. The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics 142: 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, H. M., and A. E. Tomkinson, 2002. A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J. Biol. Chem. 277: 45630–45637. [DOI] [PubMed] [Google Scholar]
- Ward, J. F., 2000. Complexity of damage produced by ionizing radiation. Cold Spring Harbor Symp. Quant. Biol. 65: 377–382. [DOI] [PubMed] [Google Scholar]
- Wilson, T. E., and M. R. Lieber, 1999. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J. Biol. Chem. 274: 23599–23609. [DOI] [PubMed] [Google Scholar]
- Wilson, T. E., L. M. Topper and P. L. Palmbos, 2003. Non-homologous end-joining: bacteria join the chromosome breakdance. Trends Biochem. Sci. 28: 62–66. [DOI] [PubMed] [Google Scholar]
- Wu, X., T. E. Wilson and M. R. Lieber, 1999. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA 96: 1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., K. Marshall, M. Yamaguchi, J. E. Haber and C. F. Weil, 2004. Microhomology-dependent end joining and repair of transposon-induced DNA hairpins by host factors in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., and T. T. Paull, 2005. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Rep. 4: 1281–1294. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., X. Wu, F. Yuan, Z. Xie and Z. Wang, 2001. Highly frequent frameshift DNA synthesis by human DNA polymerase mu. Mol. Cell. Biol. 21: 7995–8006. [DOI] [PMC free article] [PubMed] [Google Scholar]