Abstract
The deleterious consequences of inbreeding, especially in the form of inbreeding depression, are well known. However, little is known about how inbreeding affects genome-wide gene expression. Here, we show that inbreeding changes transcription levels for a number of genes. Gene expression profiles of Drosophila melanogaster lines inbred to F ≈ 0.67 at different rates changed relative to those of noninbred lines, but the rate of inbreeding did not significantly affect gene expression patterns. Genes being differentially expressed with inbreeding are disproportionately involved in metabolism and stress responses, suggesting that inbreeding acts like an environmental stress factor.
INBREEDING depression is caused by an increase in the homozygosity of recessive deleterious alleles and loss of overdominance at some loci due to increased homozygosity (Charlesworth and Charlesworth 1987). Empirical studies show the ubiquity of inbreeding depression for traits related to fitness (Charlesworth and Charlesworth 1987; Crnokrak and Roff 1999; Hedrick and Kalinowski 2000; Keller and Waller 2002; Kristensen and Sørensen 2005). However, theory predicts and empirical studies have shown that the level of inbreeding depression is environmental and line specific (Bijlsma et al. 1999; Fowler and Whitlock 1999; Reed et al. 2002; Kristensen et al. 2003; Vermeulen and Bijlsma 2004) and dependent upon the level of genetic load (Ehiobu et al. 1989; Lacy and Ballou 1998; Wang et al. 1999; Day et al. 2003; Reed et al. 2003; Pedersen et al. 2005). Differences between lines in the effects of inbreeding may partly be determined by the rate of inbreeding. If inbreeding is sudden and extreme, the effective population size is strongly reduced, random fixation occurs more often, and selection will have minor impact (Hedrick 1994; Fu et al. 1998; Wang et al. 1999). Consequently, because there are more generations and greater opportunity for selection to act before a given inbreeding level is reached, slower inbreeding is predicted to cause less inbreeding depression than an equivalent level of fast inbreeding (Robertson 1952; Ehiobu et al. 1989; Wang et al. 1999; Day et al. 2003; Reed et al. 2003; Pedersen et al. 2005).
Molecular studies of aging, inbreeding, and environmental stress have been shown to induce similar cellular responses (Kristensen et al. 2002; Pletcher et al. 2002; Sørensen et al. 2003; Girardot et al. 2004; Landis et al. 2004; Pedersen et al. 2005). For instance, the molecular chaperone Hsp70 is found to be upregulated in response to numerous environmental stresses (Feder and Hofmann 1999; Sørensen et al. 2003) and is also upregulated in some inbred lines (Kristensen et al. 2002; Pedersen et al. 2005). However, no studies have investigated how inbreeding affects expression levels of the whole genome. Here, gene-expression profiles of lines inbred to the same level at different rates and noninbred Drosophila melanogaster lines are investigated. On the basis of the obtained results we conclude that inbreeding leads to differential expression of a wide variety of genes with disproportionate representation involved in metabolism and stress resistance.
MATERIALS AND METHODS
Inbreeding procedure and maintenance of the lines:
A genetically diverse mass population of D. melanogaster was founded in August 2002 by mixing 600–700 flies from each of four sets of preexisting populations collected in Denmark, Australia, and The Netherlands. The stocks were all maintained at high population sizes (N > 1000) prior to crossing.
Inbred (fast and slower rate) and “noninbred” control lines were founded from the mass population in December 2002 eight generations after the mass population was founded. Lines with expected equivalent levels of inbreeding (F ≈ 0.67) were obtained by two different rates of inbreeding, either through five generations of full-sib mating (fast rate) or by maintaining a population size of two pairs during nine generations (slower rate). Five independent inbred lines were generated for each of the two breeding regimes. Each inbred line was founded by randomly selecting, respectively, one male and one female (full-sib) and two males and two females (slower inbreeding) from the mass population. Assuming the inbreeding level of the base population to be zero, the expected inbreeding levels were calculated as a measure of coancestry for the full-sib mating (Ft = (1 + Ft-1 − Ft-2)/4) (Falconer and Mackay 1996) and as a measure of genetic drift in the lines inbred by a slower rate of inbreeding (Ft = Ft-1 + (1 − 2Ft-1 + Ft-2)/2Ne) (Crow and Kimura 1970). For the inbreeding procedures, offspring from each line from each consecutive generation were collected as virgins. Four and two pairs were set up per line within the fast inbreeding and the slower inbreeding regimes, respectively, to reduce the extinction of lines throughout the inbreeding procedure. For each breeding regime offspring from one vial were randomly chosen to establish the next generation of inbreeding. However, some lines went extinct through the process of inbreeding; thus to make sure that enough lines reached the expected level of inbreeding, excess lines were set up. Twenty and 15 independent lines were started to make, respectively, the fast and slower inbred lines to compensate for loss of lines as a function of the intensity of inbreeding. Respectively 20 and 10% of the fast and the slower inbred lines went extinct through the inbreeding process. After reaching the desired level of inbreeding, all lines were flushed to minimum sizes of 500 breeding individuals (within two generations) and transferred to bottles. Five “noninbred” control lines, each founded by ∼500 breeding individuals, were established at the time when the inbreeding procedures were initiated. The control lines and the flushed inbred lines were each kept in 10 bottles and within each line flies from all the bottles were mixed in every generation prior to setting up the next generation. The major features of the design used to establish the experimental lines are summarized in Table 1. Throughout and following the inbreeding procedure all flies were maintained in one climate room (25° ± 0.2°, 50% relative humidity, 12/12-hr light/dark cycle).
TABLE 1.
Expected effective population sizes (Ne) in each of the 15 lines being either inbred or control
| Ne | t(Ne) | t(Ne ≈ 500) | E(Ft) | |
|---|---|---|---|---|
| Treatment | ||||
| Control | — | — | 14 | ≈0 |
| Slow inbreeding | 4 | 9 | 5 | ≈0.67 |
| Fast inbreeding | 2 | 5 | 9 | ≈0.67 |
t(Ne) is the number of generations populations are held at the expected Ne specified in the Ne column. t(Ne ≈ 500) specifies the number of generations where all populations were held at Ne ≈ 500 prior to the experiment. E(Ft) is the expected inbreeding coefficient within the three treatments following the bottleneck.
Sampling of flies and replication:
The inbred flies had a lower productivity, and the density within bottles was therefore lower. To get around this problem, the number of flies was controlled in all generations so that 20, 25, and 30 parental pairs were set up for egg laying before being discarded 24 hr later, within the control, slower inbred, and fast inbred lines, respectively. The numbers of flies emerging from the bottles were not significantly different across the three treatments (control, 356 ± 6, fast inbreeding, 387 ± 11; slower inbreeding, 357 ± 28). Flies were never exposed to strong crowding.
Twenty virgin male flies were collected from each line by sampling four males (<8 hr old) from each of five randomly chosen bottles. Sampling was done by four people in the afternoon within 3 hr. Flies from the different treatments were sampled in rotating order, so that the time of collection and the person collecting were randomized between the treatments. In each bottle the first flies emerging were used. After sampling, flies were immediately frozen in liquid nitrogen (flies <11 hr old). This procedure was followed for all 15 lines (5 controls, 5 fast inbred, and 5 slower inbred lines). A pool of RNA from 20 flies from each replicate line was hybridized to Affymetrix chips.
RNA purification:
For RNA purification, 20 virgin male flies from each line were collected and frozen in liquid nitrogen and stored at −80°. Flies were homogenized with a FP-120 Fast Prep bead beater according to manufacturer protocols (Bio-101, Carlsbad, CA) in 1.5 ml Trizol reagent (Invitrogen, San Diego) and 150 μl chloroform. Labeling, hybridization, and staining were performed essentially as described by Dyrskjot et al. (2003). Briefly, double-strand cDNA was prepared from 5 μg of total RNA using the SuperScript Choice system (Life Technologies) according to the manufacturer's instructions except using an oligo(dT) primer containing a T7 RNA polymerase promoter site. Biotin-labeled cRNA was prepared using the BioArray High Yield RNA transcript labeling kit (Enzo). Following the IVT reaction, the unincorporated nucleotides were removed using RNeasy columns (QIAGEN, Valencia, CA).
Array hybridization and scanning:
Fifteen micrograms of cRNA was fragmented at 94° for 35 min in a final volume of 40 μl in a buffer containing 40 mm Tris-acetate pH 8.1, 100 mm KOAc, and 30 mm MgOAc. Next, 260 μl of 6× SSPE-T hybridization buffer (1 m NaCl, 10 mm Tris pH 7.6, 0.005% Triton) was added and the cRNA was denatured by heating to 95° for 5 min. The hybridization mixture was loaded onto the Affymetrix probe array cartridge (Drosophila Genome Array Version 1) and incubated for 16 hr at 45° at constant rotation (60 rpm). The washing and staining procedure was performed in the Affymetrix Fluidics Station. The probe array was exposed to 10 washes in 6× SSPE-T at 25° followed by 4 washes in 0.5× SSPE-T at 50°. The biotinylated cRNA was stained with a streptavidin-phycoerythrin conjugate, final concentration 2 μg/μl (Molecular Probes, Eugene, OR) in 6× SSPE-T for 30 min at 25° followed by 10 washes in 6× SSPE-T at 25°. An antibody amplification step followed, using normal goat IgG as blocking reagent, final concentration 0.1 mg/ml (Sigma, St. Louis), and biotinylated anti-streptavidin antibody (goat), final concentration 3 μg/ml (Vector Laboratories, Burlingame, CA). This was followed by a staining step with a streptavidin-phycoerythrin conjugate, final concentration 2 μg/μl (Molecular Probes, Eugene, OR) in 6× SSPE-T for 30 min at 25° and 10 washes in 6× SSPE-T at 25°. The probe arrays were scanned at 560 nm using a confocal microscope (Hewlett Packard GeneArray Scanner G2500A).
Statistical analysis:
The data were analyzed using programs developed in R, a programming language and developer environment for statistical computing and graphics (http://www.r-project.org/). Preprocessing of expression values was performed using the robust multi-array analysis (GCRMA) algorithm (Irizarry et al. 2003; Wu et al. 2004). In this algorithm, raw intensity values are background corrected on the basis of a model using sequence information followed by a quantile normalization and a robust multichip fit with median polish (Wu et al. 2004). This algorithm combines the strengths of stochastic-model-based algorithms and physical models and has been shown to be superior in accuracy and precision to other normalization methods such as microarray analysis suite, RMA, and PerfectMatch (Wu and Irizarry 2004). To exclude genes that could not be confidently detected in the data analysis probe, sets with less than three present calls within at least one of the three treatments were excluded (a transcript must be represented on at least three chips within the control, fast inbreeding, or slower inbreeding treatments). The filtered gene set contained 8884 transcripts.
Differential expression was assessed using significance analysis of microarrays (SAM) proposed by Tusher et al. (2001). An overall test of significance for a gene was performed using moderated F-statistics in a multiclass analysis. For each gene the three contrasts (control-slow, control-fast, and slow-fast) were tested for differentially expressed gene transcripts. The moderated F-statistic tests whether any of the contrasts are nonzero for that gene, i.e., whether that gene is differentially expressed on any contrast. Specific treatment contrasts (control vs. slow, control vs. fast, and slow vs. fast) were also tested on the basis of a modified t-statistic using the two-class unpaired analysis. Multiple testing was accounted for by controlling the false discovery rate at 20% for both the multiclass and two-class analyses. The SAM analysis was performed as implemented in the R package called siggenes (Schwender 2004).
Groups of genes being differentially expressed were annotated on the basis of the biological process ontology directed by the Gene Ontology (GO) database (Gene Ontology Consortium 2001). The expression analysis systematic explorer (EASE) application on the DAVID homepage (http://david.niaid.nih.gov/david/ease.htm) (Hosack et al. 2003) was used to test for overrepresentation of genes in given annotation categories. EASE scores were calculated for the likelihood of overrepresentation in the annotation categories.
The probability that the overlap of genes being differentially expressed with both types of inbreeding is different from the number expected by chance was calculated by using Monte Carlo simulations. In each simulation the gene list within each treatment was permuted and the overlap of induced genes was determined. A Kolmogorov-Smirnov test (Conover 1971) was used to determine if the distribution of the within-gene variances in gene expression levels differs significantly among the three breeding treatments.
RESULTS
The Affymetrix array contained 13,966 probe sets representing ∼13,000 unique genes and 8884 genes were left after the filtering process.
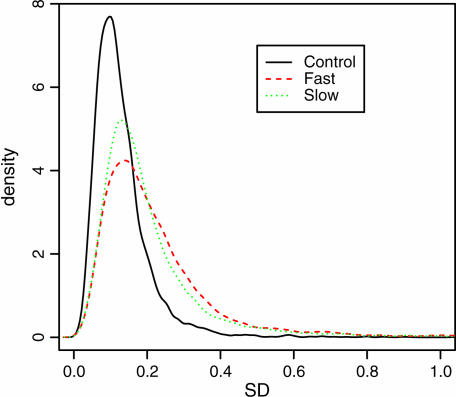
The within-gene variance in gene expression levels within both inbred treatments was higher than that within the control treatment (control vs. fast, D = 0.354, P < 2.2e-16; control vs. slow, D = 0.296, P < 2.2e-16; see Figure 1). The within-gene variance in gene expression in the fast inbred treatment was higher than that in the slower inbred treatment (fast vs. slow, D = 0.081, P < 2.2e-16; see Figure 1).
Figure 1.
Distribution plots of standard deviations for within-gene variances in gene expression levels for the three treatments. Kolmogorov-Smirnov tests showed that the distribution of variances in gene expression levels was significantly higher across lines with inbreeding, and more so with fast inbreeding, compared to control lines.
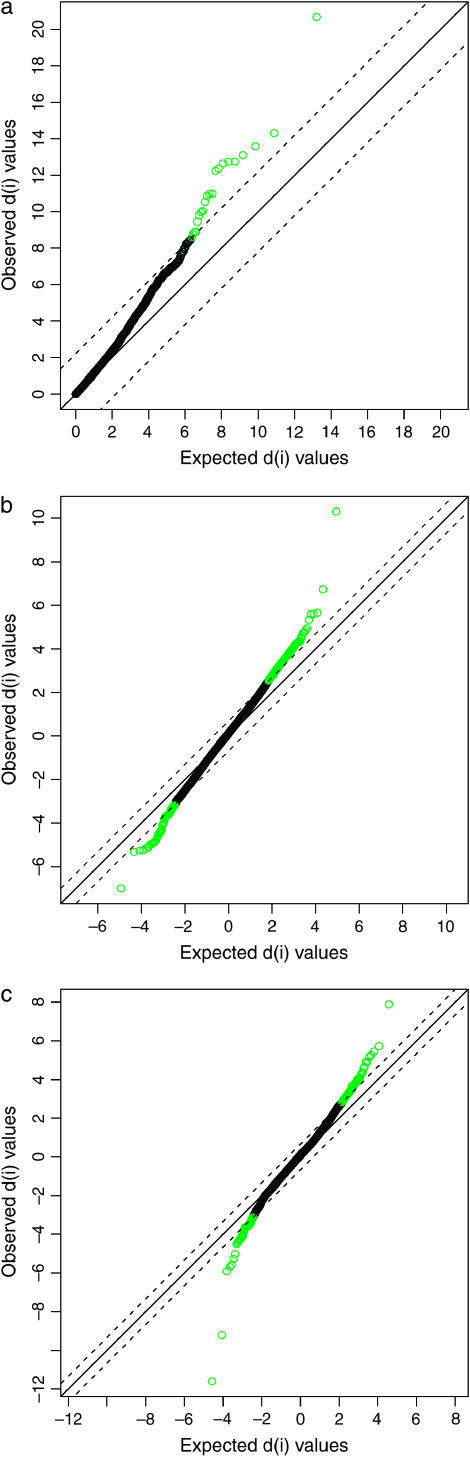
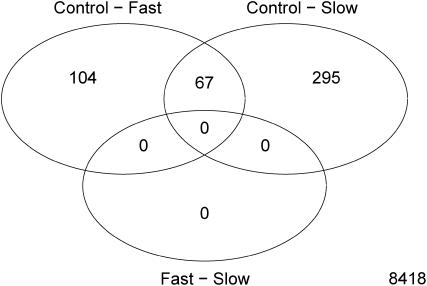
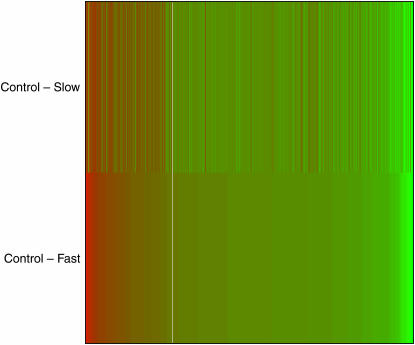
The moderated F-statistics in the multiclass analysis revealed 21 genes as being differentially expressed (Table 2, Figure 2). Of these 12 were significantly downregulated within both the fast and the slower inbred treatments and the remaining 9 genes were significantly upregulated within both inbred treatments (Table 2, Figure 2). Several genes were differentially expressed for the contrasts control vs. fast and control vs. slower inbred lines whereas no transcripts were differentially expressed between fast and slower inbred lines (Tables 3–5, Figure 2). The results are summarized in a Venn diagram (Figure 3). Sixty-seven genes are differentially expressed with both control vs. fast and control vs. slower inbreeding. The probability of the observed overlap arising by chance is small (P < 0.00001) under the assumption that all genes are liable to change with treatment. To assess the potential contribution of a smaller gene pool, one can diminish the number of genes that are capable of changing with the two types of inbreeding while simulating the overlap. If the set of genes is reduced to 5000 it is still highly unlikely to observe 67 genes overlapping (P < 0.00001). The 67 genes differentially expressed in both control vs. fast and control vs. slow inbreeding were all either up- or downregulated in both comparisons. Fifty genes were upregulated and 17 were downregulated. Given that all genes are either up- or downregulated within both inbred treatments inbreeding, per se, not the intensity of inbreeding, appears to determine the up or down change in transcript level. Genes that are significantly differentially expressed with fast or slower inbreeding compared to the controls are in the great majority of cases either up- or downregulated with both types of inbreeding (Figure 4). The log twofold change of the 67 genes being differentially expressed was not affected by the type of inbreeding; 34 genes had a fold change that was higher in the control vs. fast inbreeding comparison whereas 33 genes had a fold change that was higher in the control vs. slower inbreeding comparison.
TABLE 2.
Genes (identified by their AFFYID) significantly differentially expressed on the basis of a multiclass SAM analysis
| AFFYID | Control vs. fast | Control vs. slow |
|---|---|---|
| 141233_at | + | + |
| 146991_at | + | + |
| 147059_s_at | + | + |
| 147114_at | + | + |
| 149631_at | + | + |
| 151967_at | + | + |
| 152851_at | + | + |
| 153761_at | + | + |
| 154711_at | + | + |
| 141242_at | − | − |
| 141315_at | − | − |
| 141511_at | − | − |
| 142893_at | − | − |
| 143005_at | − | − |
| 144191_at | − | − |
| 144701_at | − | − |
| 144845_at | − | − |
| 146793_at | − | − |
| 149039_at | − | − |
| 150482_at | − | − |
| 154821_at | − | − |
Plus (+) and minus (−) indicate whether genes are respectively up- or downregulated in inbred lines compared to control lines (control vs. fast inbreeding and control vs. slow inbreeding).
Figure 2.
Scatter plots for identification of genes with significant changes in expression based on results from the F-statistics in (a) the multiclass analysis and from (b) a modified t-test for the contrasts control vs. slow inbreeding and (c) control vs. fast inbreeding. The scatter plots are of the observed and expected relative differences d(i) in gene expression levels (Tusher et al. 2001). Each gene is represented by a circle. The solid lines indicate where the observed and the expected relative differences are identical. Genes represented by a green color (above the top and below the bottom dotted lines) are differentially expressed assuming a false discovery rate at 20%.
TABLE 3.
Biological process ontology of genes (identified by their AFFYID) with (a) significant fast inbreeding and (b) slower inbreeding-dependent transcript representation
| Functional ontology | Up (%) | AFFYID | Down (%) | AFFYID |
|---|---|---|---|---|
| a. | ||||
| Metabolism | 26 (23) | 142157_at, 142162_at, 142335_at, 142767_at, 142932_at, 143062_at, 143303_at, 143341_at, 144037_at, 146745_at, 147189_at, 148274_at, 151348_f_at, 151767_at, 151967_at, 152078_at, 152355_at, 152801_at, 153129_at, 153194_at, 153290_at, 153303_at, 153323_at, 153636_at, 154644_at, 154978_at | 4 (6.9) | 142251_at, 142911_at, 143198_at, 148640_at |
| Cellular physiological process | 10 (8.8) | 142932_at, 143283_at, 146745_at, 148274_at, 149631_at, 150547_at, 151989_at, 153194_at, 153432_at, 154454_at | 3 (5.2) | 143198_at, 153385_at, 153844_at |
| Response to stimulus | 7 (6.2) | 142657_at, 143283_at, 143303_at, 143443_at, 147473_at, 153432_at, 153636_at | 3 (5.2) | 143191_at, 143198_at, 148358_at |
| Morphogenesis | 6 (5.3) | 142932_at, 143283_at, 152549_at, 153290_at, 153432_at, 154454_at | — | |
| Organismal physiological process | 5 (4.4) | 142657_at, 143341_at, 143443_at, 147473_at, 153432_at | 3 (5.2) | 142251_at, 143198_at, 144191_at |
| Cell communication | 3 (2.7) | 142932_at, 143283_at, 154978_at | — | |
| Regulation of cellular process | 3 (2.7) | 153432_at, 154454_at, 154978_at | — | |
| Unclassified | 77 (68.1) | 49 (84.5) | ||
| b. | ||||
| Metabolism | 58 (20.1) | 141231_at, 142162_at, 142196_at, 142335_at, 142758_at, 142904_at, 142926_at, 143062_at, 143250_at, 143299_at, 143314_at, 143450_at, 143729_at, 143736_at, 143775_at, 144037_at, 144358_at, 144561_at, 145027_at, 145098_at, 145934_at, 146084_at, 149085_at, 150001_at, 150466_at, 150697_at, 151767_at, 151832_at, 151967_at, 152078_at, 152088_at, 152117_at, 152559_at, 152801_at, 153122_at, 153194_at, 153215_at, 153298_at, 153303_at, 153314_at, 153332_at, 153369_at, 153515_at, 153867_at, 154176_at, 154229_at, 154264_at, 154275_at, 154355_at, 154521_at, 154538_at, 154644_at, 154659_at, 154826_at, 154910_at, 154911_at, 154926_at, 155116_at | 14 (19.2) | 141810_at, 142251_at, 142336_at, 143198_at, 146747_at, 149735_at, 150131_at, 151662_s_at, 151666_s_at, 152031_at, 152392_at, 152658_at, 152685_at, 152964_at |
| Cellular physiological process | 18 (6.2) | 142904_at, 142926_at, 143060_f_at, 143450_at, 143958_at, 144336_at, 146590_s_at, 149631_at, 151989_at, 152088_at, 153181_at, 153194_at, 153432_at, 154659_at, 154910_at, 154926_at, 155116_at, AFFX-Dros-ACTIN_M_r_at | 6 (8.2) | 143198_at, 143391_i_at, 146494_at, 152031_at, 152685_at, 153412_at |
| Response to stimulus | 12 (4.2) | 142657_at, 143443_at, 143607_at, 143609_at, 143958_at, 145970_at, 145971_at, 146590_s_at, 148460_at, 153432_at, 154910_at, 154926_at | 4 (5.5) | 143127_at, 143198_at, 143391_i_at, 152031_at |
| Morphogenesis | 6 (2.1) | 142926_at, 146590_s_at, 153432_at, 153515_at, 153867_at, 155116_at | 5 (6.8) | 141511_at, 143391_i_at, 150268_at, 150269_at, 151662_s_at |
| Organismal physiological process | 9 (3.1) | 142657_at, 143443_at, 143607_at, 143609_at, 145970_at, 145971_at, 146590_s_at, 153432_at, 155116_at | 3 (4.1) | 142251_at, 143198_at, 144191_at |
| Cell differentiation | 4 (1.4) | 143450_at, 153515_at, 153867_at, 155116_at | 3 (4.1) | 141511_at, 151666_s_at, 152031_at |
| Embryonic development | — | 3 (4.1) | 141511_at, 143198_at, 151666_s_at | |
| Pattern specification | 3 (1) | 153515_at, 153867_at, 155116_at | 3 (4.1) | 141511_at, 143198_at, 151666_s_at |
| Reproduction | 3 (1) | 143450_at, 152072_at, 155116_at | 3 (4.1) | 143198_at, 151666_s_at, 152031_at |
| Unclassified | 214 (74) | 51 (69.9) |
Only categories with three or more classified genes are represented in the table to reduce the risk of including false positives. Percentages of total are in parentheses.
TABLE 4.
Biological process ontology of genes [identified by their AFFYID and names (when known)] with significant transcript representation being common for both fast and slower inbreeding
| Functional ontology | Up (%) | AFFYID | Down (%) | AFFYID |
|---|---|---|---|---|
| Metabolism | 11 (22) | 142162_at (α-mannosidase II), 142335_at, 143062_at (adenosine 3), 144037_at (iron regulatory protein 1B), 151767_at (NAD-dependent methyl enetetrahydrofolate dehydrogenase), 151967_at, 152078_at, 152801_at, 153194_at, 153303_at (proteasome 25-kD subunit), 154644_at (proteasome α-subunit) | — | |
| Cellular physiological process | 4 (8) | 149631_at, 151989_at, 153194_at, 153432_at (Thor) | — | |
| Organismal physiological process | 3 (6) | 142657_at, 143443_at (Diptericin), 153432_at (Thor) | 3 (17.6) | 142251_at (foraging), 143198_at (Hsp83), 144191_at (vacuolar H+-ATPase SFD subunit) |
| Response to stimulus | 3 (6) | 142657_at, 143443_at (Diptericin), 153432_at (Thor) | — | |
| Unannotated | 34 (68) | 14 (82.4) |
Only categories with at least three classified genes are included. Percentages of total are in parentheses.
TABLE 5.
EASE scores for groups of genes being differentially expressed within the control vs. fast inbreeding comparison and the control vs. slower inbreeding comparison and for genes being differentially expressed with both fast and slower inbreeding
| Contrasts | System | Category | LH | LT | PH | PT | EASE scores |
|---|---|---|---|---|---|---|---|
| Control vs. fast inbreeding | Molecular function | Catalytic activity | 60 | 93 | 2998 | 6508 | 0.0003 |
| Biological process | Regulation of biological process | 6 | 48 | 110 | 4221 | 0.0070 | |
| Cellular component | Lysosyme | 3 | 45 | 17 | 3883 | 0.0153 | |
| Cellular component | Lytic vacuole | 3 | 45 | 17 | 3883 | 0.0153 | |
| Cellular component | Vacuole | 4 | 45 | 51 | 3883 | 0.0194 | |
| Molecular function | Anion transporter activity | 4 | 93 | 41 | 6508 | 0.0198 | |
| Molecular function | Inorganic transporter activity | 3 | 93 | 16 | 6508 | 0.0209 | |
| Molecular function | Mannosidase activity | 3 | 93 | 16 | 6508 | 0.0209 | |
| Biological process | Antibacterial humoral response | 3 | 48 | 22 | 4221 | 0.0243 | |
| Biological process | Response to stress | 7 | 48 | 211 | 4221 | 0.0281 | |
| Molecular function | Transferase activity | 19 | 93 | 804 | 6508 | 0.0307 | |
| Cellular component | Proteasome core complex | 3 | 45 | 25 | 3883 | 0.0319 | |
| Biological process | Carboxylic acid metabolism | 5 | 48 | 121 | 4221 | 0.0444 | |
| Biological process | Organic acid metabolism | 5 | 48 | 121 | 4221 | 0.0444 | |
| Molecular function | Methyltransferase activity | 4 | 93 | 57 | 6508 | 0.0463 | |
| Biological process | Physiological process | 44 | 48 | 3438 | 4221 | 0.0470 | |
| Molecular function | Transferase activity, transferring one-carbon groups | 4 | 93 | 58 | 6508 | 0.0483 | |
| Control vs. slow inbreeding | Biological process | Organic acid metabolism | 13 | 96 | 121 | 4221 | 0.0000 |
| Biological process | Carboxylic acid metabolism | 13 | 96 | 121 | 4221 | 0.0000 | |
| Biological process | Physiological process | 92 | 96 | 3438 | 4221 | 0.0000 | |
| Biological process | Metabolism | 72 | 96 | 2302 | 4221 | 0.0000 | |
| Molecular function | Catalytic activity | 114 | 190 | 2998 | 6508 | 0.0001 | |
| Molecular function | Oxidoreductase activity | 26 | 190 | 421 | 6508 | 0.0002 | |
| Biological process | Pyruvate metabolism | 4 | 96 | 10 | 4221 | 0.0012 | |
| Molecular function | Transaminase activity | 5 | 190 | 17 | 6508 | 0.0012 | |
| Molecular function | Transferase activity, transferring nitrogenous groups | 5 | 190 | 17 | 6508 | 0.0012 | |
| Biological process | Ubiquitin-dependent protein catabolism | 5 | 96 | 22 | 4221 | 0.0013 | |
| Biological process | Antibacterial humoral response | 5 | 96 | 22 | 4221 | 0.0013 | |
| Biological process | Modification-dependent protein catabolism | 5 | 96 | 24 | 4221 | 0.0018 | |
| Biological process | Amino acid metabolism | 8 | 96 | 79 | 4221 | 0.0018 | |
| Cellular component | Cytoplasm | 59 | 104 | 1642 | 3883 | 0.0026 | |
| Cellular component | Mitochondrial matrix | 11 | 104 | 145 | 3883 | 0.0046 | |
| Molecular function | Cyclohydrolase activity | 3 | 190 | 4 | 6508 | 0.0048 | |
| Molecular function | Glycine hydroxymethyltransferase activity | 3 | 190 | 4 | 6508 | 0.0048 | |
| Biological process | ATP-dependent proteolysis | 4 | 96 | 17 | 4221 | 0.0060 | |
| Biological process | Defense response to bacteria | 5 | 96 | 36 | 4221 | 0.0082 | |
| Molecular function | Ligase activity, forming carbon nitrogen bonds | 7 | 190 | 63 | 6508 | 0.0095 | |
| Biological process | Amino acid and derivative metabolism | 8 | 96 | 107 | 4221 | 0.0098 | |
| Molecular function | Inositol-1(or 4)-monophosphatase activity | 3 | 190 | 6 | 6508 | 0.0117 | |
| Cellular component | Intracellular | 84 | 104 | 2732 | 3883 | 0.0119 | |
| Biological process | Lipid metabolism | 6 | 96 | 63 | 4221 | 0.0130 | |
| Biological process | Amine metabolism | 8 | 96 | 113 | 4221 | 0.0130 | |
| Molecular function | Oxidoreductase activity, acting on CH-OH group of donors | 9 | 190 | 110 | 6508 | 0.0143 | |
| Biological process | Response to bacteria | 5 | 96 | 43 | 4221 | 0.0152 | |
| Biological process | Serine family amino acid metabolism | 3 | 96 | 9 | 4221 | 0.0163 | |
| Biological process | Nucleotide metabolism | 4 | 96 | 25 | 4221 | 0.0177 | |
| Molecular function | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 7 | 190 | 77 | 6508 | 0.0239 | |
| Molecular function | Methyltransferase activity | 6 | 190 | 57 | 6508 | 0.0242 | |
| Molecular function | Lyase activity | 9 | 190 | 122 | 6508 | 0.0252 | |
| Molecular function | Transferase activity, transferring one-carbon groups | 6 | 190 | 58 | 6508 | 0.0259 | |
| Cellular component | Mitochondrion | 21 | 104 | 484 | 3883 | 0.0270 | |
| Cellular component | Proteasome core complex | 4 | 104 | 25 | 3883 | 0.0273 | |
| Molecular function | Oxidoreductase activity, acting on the aldehyde or oxo group of donors | 4 | 190 | 23 | 6508 | 0.0278 | |
| Molecular function | Hydrolyase activity | 5 | 190 | 40 | 6508 | 0.0279 | |
| Molecular function | Structural constituent of cytoskeleton | 5 | 190 | 42 | 6508 | 0.0327 | |
| Molecular function | Chaperone activity | 7 | 190 | 87 | 6508 | 0.0402 | |
| Cellular component | Proteasome complex | 5 | 104 | 50 | 3883 | 0.0423 | |
| Biological process | Antimicrobial humoral response | 5 | 96 | 60 | 4221 | 0.0450 | |
| Molecular function | Oxidoreductase activity, acting on the CH-NH2 group of donors | 3 | 190 | 12 | 6508 | 0.0457 | |
| Biological process | Response to stress | 10 | 96 | 211 | 4221 | 0.0464 | |
| Molecular function | Carbon-oxygen lyase activity | 5 | 190 | 48 | 6508 | 0.0497 | |
| Control vs. inbreeding | Molecular function | Methyltransferase activity | 4 | 35 | 57 | 6508 | 0.0031 |
| Molecular function | Catalytic activity | 25 | 35 | 2998 | 6508 | 0.0033 | |
| Molecular function | Transferase activity, transferring one-carbon groups | 4 | 35 | 58 | 6508 | 0.0033 | |
| Biological process | Regulation of biological process | 4 | 21 | 110 | 4221 | 0.0142 | |
| Biological process | Carboxylic acid metabolism | 4 | 21 | 121 | 4221 | 0.0183 | |
| Biological process | Organic acid metabolism | 4 | 21 | 121 | 4221 | 0.0183 | |
| Biological process | Organismal physiological process | 6 | 21 | 411 | 4221 | 0.0387 |
The genes were grouped by “system” (molecular function, biological process, or cellular component) and “category” within the systems by the EASE application on the DAVID homepage (http://david.niaid.nih.gov/david/ease.htm). Categories with significant EASE scores (<0.05) are presented here. LH represents number of genes in gene list assigned to category; LT represents number of genes in gene list assigned to system; PH represents the number of all known genes assigned to category; and PT represents the number of all known genes in that system. The representation of each category of genes (only categories represented by at least three genes are included) was evaluated by the EASE-score criteria. The test calculates the probability of detecting the actual detected number of genes in a category, by evaluating the proportion of genes in each gene list belonging to a category vs. the proportion of genes belonging to this category out of all known genes. Thus, the ratio LH/LT is compared to the ratio PH/PT.
Figure 3.
Venn diagram of differential gene expression in control, slow inbred, and fast inbred flies. In total 171 genes were differentially expressed with fast inbreeding and 362 genes were differentially expressed with slower inbreeding in the microarray analysis. In total 67 genes were differentially expressed with both fast and slower inbreeding. The observed overlap is significantly higher than that expected by chance alone (see text for details).
Figure 4.
Genes that are differentially expressed with inbreeding (either fast or slower) are mostly responding in the same direction. The heat diagram shows color-coded gene expression profiles in which rows correspond to treatment comparison (control vs. slow, control vs. fast) and columns correspond to genes. Genes are sorted by differential expression in the control vs. fast comparison. The heat diagram was drawn using the R package limma (Smyth 2004).
EASE scores for annotation categories with more genes than expected by chance among the genes being significantly differentially expressed with fast and slower inbreeding and with both are given in Table 5. Classes of genes involved in metabolism, immune, and stress responses are overrepresented.
DISCUSSION
The large number of genes differentially expressed in this study means that a detailed description of the changes on a gene-by-gene basis would be too extensive to list (for a complete list of differentially expressed genes see supplementary material at http://www.genetics.org/supplemental/). However, we here present gene groups defined by function.
Genes involved in stress resistance and metabolism are disproportionately affected by inbreeding (Tables 2–5). Some groups of genes associated with these biological processes are upregulated, whereas a few are downregulated with inbreeding. Most genes being differentially expressed with either fast or slower inbreeding in this study responded in the same direction with both types of inbreeding (Figure 4). Furthermore, the overlap between genes being differentially expressed with both types of inbreeding is much larger than an expected overlap arising by chance, and all 67 genes responded in the same direction. Given the high level of replication (10 inbred lines and 5 control lines) these results indicate that there is a general effect of inbreeding on gene expression patterns.
The data presented here have not been validated by quantitative (Q)RT-PCR or Northern blot. However, Park et al. (2004) showed that the ratios of gene expression obtained from Affymetrix platforms and QRT-PCR analyses are highly correlated (r = 0.93). Park et al. (2004) and Yuen et al. (2002) also showed that results obtained from Affymetrix platforms underestimated the real expression change as detected by QRT-PCR. This latter result shows that our results can be interpreted as being of conservative nature. Moreover, the consistency in direction and magnitude of altered transcript levels (inbreeding vs. control) for the numerous inbreeding replicates suggests that the observed differential transcript levels are not an artifact. A thorough comparison of other published data with ours would require a full treatment of the raw data for common normalization, similar statistical analysis, etc. Nevertheless, we have performed a simple analysis using our own data set and the list of differentially expressed genes in the study by Landis et al. (2004) investigating differentially expressed genes in response to aging and oxidative stress. The present microarray study revealed that a total of 466 genes were differentially expressed with inbreeding whereas Landis et al. (2004) observed that 913 genes were differentially expressed with aging and that 593 genes were differentially expressed with oxidative stress. Monte Carlo simulations were used to test whether the observed overlap between treatments (inbreeding, aging, or oxidative stress) was higher than expected by chance. The overlap was in all comparisons significantly higher than expected by chance even when the number of genes liable to change with treatment was reduced to 5000 (P < 0.001 in all cases). Thirty-four genes are differentially expressed under all of the conditions investigated in both studies (inbreeding, aging, and oxidative stress), including stress response genes [e.g., Hsp83 (Affymetrix probe identifier, AFFYID: 143198_at), Diptericin (AFFYID: 147473_at), and Defensin (AFFYID: 143607_at)] and metabolism genes [e.g., adenosine (AFFYID: 143062_at) and NAD-dependent methylenetetrahydrofolate dehydrogenase (AFFYID: 151767_at)]. Seventy-two and 78 genes overlap in the comparison of aged and inbred individuals or between oxidative-stressed and inbred individuals, respectively. Heat-shock protein- and immune-response genes appear to be differentially expressed in these comparisons [e.g., Hsp60 (AFFYID: 152031_at), Hsc70 (AFFYID: 143191_at), Thor (AFFYID: 153432_at), and Drosocin (AFFYID: 143609_at)]. This indicates that effects of different kinds of stresses (such as inbreeding, aging, and oxidative stress) may bear similarities and those genes being differentially expressed under such conditions may act to maintain homeostasis in organisms exposed to diverse stresses. Clearly these genes are candidate genes that should be investigated in more detail by investigating their protein products and by performing knockout studies to obtain knowledge about their function.
Transcription of a number of genes coding for antibacterial peptides is upregulated with inbreeding (Tables 3–5). These include Defensin (AFFYID: 143607_at), Drosocin (AFFYID: 143609_at), Diptericins (AFFYID, 143443_at; AFFYID, 147473_at) and Thor (AFFYID: 153432_at), which all have well-described antibacterial functions (Bulet et al. 1999; Beutler 2003; Ganz 2003). In this study inbred and noninbred lines were kept under the same laboratory conditions and infection pressure is not expected to differ between control and inbred lines. However, there is a possibility that the inbred lines are more susceptible to infection and that upregulation of antibacterial peptide gene transcripts is a defense mechanism induced due to bacterial infection. Alternatively, the protein products of this group of genes have more general stress resistance functions and are part of a general stress response. In accordance with this idea, upregulated transcription of antibacterial genes has also been observed in response to aging in D. melanogaster and Caenorhabditis elegans (Pletcher et al. 2002; Murphy et al. 2003; Landis et al. 2004) and in response to environmental stress in D. melanogaster (Kayo et al. 2001; Pletcher et al. 2002; Landis et al. 2004).
A number of studies have investigated the association between heterozygosity and disease resistance and susceptibility (Hedrick et al. 2001; Giese and Hedrick 2003; Reid et al. 2003), but no consistent pattern emerges from those studies. Giese and Hedrick (2003) recently showed that noninbred populations of the Gila topminnow (Poeciliopsis occcidentalis) have significantly higher disease susceptibility compared to inbred populations. This result may be understood partly in light of our results showing an upregulation of genes coding for antibacterial peptides in inbred populations, whereby immunity toward pathogens may be increased in inbred populations.
Several gene transcripts coding for molecular chaperones such as Hsp60 (AFFYID: 152031_at), Hsp83 (AFFYID: 143198_at), and the heat-shock protein cognate 1 (AFFYID: 143191_at) are differentially expressed within one or both of the inbred treatments in this study. Heat-shock proteins are well known for their importance as part of the cellular stress response apparatus (Feder and Hofmann 1999; Sørensen et al. 2003), and Hsp70 protein level in inbred D. melanogaster and D. buzzatii lines has previously been shown to be higher than that in noninbred lines (Kristensen et al. 2002; Pedersen et al. 2005).
The reason why different rates of inbreeding were investigated in this study was that slower inbreeding is expected to be less deleterious than faster inbreeding for the same level of inbreeding (Robertson 1952; Ehiobu et al. 1989; Day et al. 2003; Reed et al. 2003; Pedersen et al. 2005). This is because with slower inbreeding there are more generations and greater opportunity for selection to act before a given inbreeding level is reached. The fast inbred lines investigated here have been tested for fertility and heat resistance in another study (Pedersen et al. 2005). Pedersen et al. (2005) showed that fertility, but not heat resistance, was significantly affected by the rate of inbreeding with the slower inbred lines having higher fertility. We hypothesized that purging of deleterious alleles within the slower inbred lines would cause changes in gene expression patterns between the two inbred treatments. More gene transcripts were differentially expressed with slower inbreeding, but there were no genes being significantly differentially expressed between inbreeding treatments (Figure 3). One reason for the higher number of differentially expressed genes in the slower compared to the fast inbred treatment probably is that the variance in gene expression is higher within the fast inbred treatment. This means that for a gene to be significantly differentially expressed a higher-fold change between the control and fast inbred treatments is needed compared to the situation for the control and slower inbred comparison (Figure 1). Another reason for the apparent lack of difference in expression may be that the slower inbreeding treatment investigated here is actually still extreme compared to most situations in nature or in domestic livestock. It would be informative to perform the same experiment on lines being inbred fast and slower than the “slow inbreeding” regime investigated here.
Between-line variance in phenotype is expected to increase with inbreeding (Falconer and Mackay 1996; Lynch and Walsh 1998; Kristensen et al. 2005). However, this is the first experiment showing this on the level of gene expression (Figure 1). The reason why the variance in gene expression is higher with fast compared to slower inbreeding may be that selection is more efficient the slower the rate of inbreeding, while drift will become more important the faster the rate of inbreeding. Given that selection regimes were similar across treatments and lines, this would cause slower inbred lines to have more similar gene expression patterns than fast inbred lines, and therefore the observation made in this study confirms theoretical predictions.
Genetic drift is expected to cause fixation of different genes within the different inbred lines. Given the number of replicate lines and the fact that the set of genes found to be differentially expressed is not a random sample of the gene pool but primarily related to metabolism and stress resistance, we find it unlikely that drift alone can explain our results. This emphasizes that there is a general effect of inbreeding that is an indirect result of the change in genotype frequencies. Perhaps the cumulative fixation of deleterious alleles results in net physiological duress to which the organism responds in a standard manner. Our results also show that transcriptional responses to inbreeding overlap with microarray studies of aging and oxidative stress (Pletcher et al. 2002; Landis et al. 2004). We argue that those genes found to be differentially expressed with inbreeding may be candidate genes for stress resistance in general, and we expect the results to have important implications for other disciplines such as medicine, animal breeding, and conservation biology.
Acknowledgments
We are grateful to Jesper Dahlgaard, Just Justesen, Morten Mulig Nielsen, and Jesper Givskov Sørensen for fruitful discussions on various parts of this experiment; to Doth Andersen and Bente Devantié for excellent technical assistance; to Stuart Barker, Richard Frankham, Lawrence Harshman, Bob Krebs, and two anonymous reviewers for critical comments on earlier versions of the article; and to the Danish National Research Council for financial support via a centre grant.
References
- Beutler, B., 2003. Innate immune responses to microbial poisons: Discovery and function of the toll-like receptors. Annu. Rev. Pharmacol. 43: 609–628. [DOI] [PubMed] [Google Scholar]
- Bijlsma, R., J. Bundgaard and W. F. van Putten, 1999. Environmental dependence of inbreeding depression and purging in Drosophila melanogaster. J. Evol. Biol. 12: 1125–1137. [Google Scholar]
- Bulet, P., C. Hetru, J. L. Dimarcq and D. Hoffmann, 1999. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 23: 329–344. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., and B. Charlesworth, 1987. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 18: 237–268. [Google Scholar]
- Conover, W. J., 1971. Practical Nonparametric Statistics. John Wiley & Sons, New York.
- Crnokrak, P., and D. A. Roff, 1999. Inbreeding depression in the wild. Heredity 83: 260–270. [DOI] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970. An Introduction to Population Genetics Theory. Harper & Row, New York.
- Day, S. B., E. H. Bryant and L. M. Meffert, 2003. The influence of variable rates of inbreeding on fitness, environmental responsiveness, and evolutionary potential. Evolution 57: 1314–1324. [DOI] [PubMed] [Google Scholar]
- Dyrskjot, L., T. Thykjaer, M. Kruhøffer, J. L. Jensen, N. Marcussen et al., 2003. Identifying distinct classes of bladder carcinoma using microarrays. Nat. Genet. 33: 90–96. [DOI] [PubMed] [Google Scholar]
- Ehiobu, N. G., M. E. Goddard and J. F. Taylor, 1989. Effect of the rate of inbreeding on inbreeding depression in Drosophila melanogaster. Theor. Appl. Genet. 77: 123–127. [DOI] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Longman, Harlow, UK.
- Feder, M. E., and G. E. Hofmann, 1999. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61: 243–282. [DOI] [PubMed] [Google Scholar]
- Fowler, K., and M. C. Whitlock, 1999. The variance in inbreeding depression and the recovery of fitness in bottlenecked populations. Proc. R. Soc. Lond. Ser. B Biol. Sci. 266: 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. B., G. Namkoong and J. E. Carlson, 1998. Comparison of breeding strategies for purging inbreeding depression via simulation. Conserv. Biol. 12: 856–864. [Google Scholar]
- Ganz, T., 2003. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3: 710–720. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium, 2001. Creating the gene ontology resource: design and implementation. Genome Res. 11: 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese, A. R., and P. W. Hedrick, 2003. Genetic variation and resistance to a bacterial infection in the endangered Gila topminnow. Anim. Conserv. 6: 369–377. [Google Scholar]
- Girardot, F., V. Monnier and H. Tricoire, 2004. Genome wide analysis of common and specific stress responses in adult Drosophila melanogaster. BMC genomics 5: 74 (http://www.biomedcentral.com/1471-2164/5/74). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W., 1994. Purging inbreeding depression and the probability of extinction – full sib mating. Heredity 73: 363–372. [DOI] [PubMed] [Google Scholar]
- Hedrick, P. W., and S. T. Kalinowski, 2000. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 31: 139–152. [Google Scholar]
- Hedrick, P. W., T. J. Kim and K. M. Parker, 2001. Parasite resistance and genetic variation in the endangered Gila topminnow. Anim. Conserv. 4: 103–109. [Google Scholar]
- Hosack, D. A., G. Dennis, B. T. Sherman, H. C. Lane and R. A. Lempicki, 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis et al., 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Kayo, T., D. B. Allison, R. Weindruch and T. A. Prolla, 2001. Influences of aging and caloric restriction on the transcriptional profile of skeletal muscle from rhesus monkeys. Proc. Natl. Acad. Sci. USA 98: 5093–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, L. F., and D. M. Waller, 2002. Inbreeding effects in wild populations. Trends Ecol. Evol. 17: 230–241. [Google Scholar]
- Kristensen, T. N., and A. C. Sørensen, 2005. Inbreeding – lessons from animal breeding, evolutionary biology and conservation genetics. Anim. Sci. 80: 121–133. [Google Scholar]
- Kristensen, T. N., J. Dahlgaard and V. Loeschcke, 2002. Inbreeding affects Hsp70 expression in two species of Drosophila even at benign temperatures. Evol. Ecol. Res. 4: 1209–1216. [Google Scholar]
- Kristensen, T. N., J. Dahlgaard and V. Loeschcke, 2003. Effects of inbreeding and environmental stress on fitness – using Drosophila buzzatii as a model organism. Conserv. Genet. 4: 453–465. [Google Scholar]
- Kristensen, T. N., A. C. Sørensen, D. Sorensen, K. S. Pedersen, J. G. Sørensen et al., 2005. A test of quantitative genetic theory using Drosophila—effects of inbreeding and rate of inbreeding on heritabilities and variance components. J. Evol. Biol. 18: 763–770. [DOI] [PubMed]
- Lacy, R. C., and J. D. Ballou, 1998. Effectiveness of selection in reducing the genetic load in populations of Peromyscus polionotus during generations of inbreeding. Evolution 52: 900–909. [DOI] [PubMed] [Google Scholar]
- Landis, G. N., D. Abdueva, D. Skvortsov, J. Yang, B. E. Rabin et al., 2004. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 101: 7663–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Murphy, C. T., S. A. Mccarroll, C. I. Bargmann, A. Fraser, R. S. Kamath et al., 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–284. [DOI] [PubMed] [Google Scholar]
- Park, P. J., Y. A. Cao, S. Y. Lee, J. W. Kim, M. S. Chang et al., 2004. Current issues for DNA microarrays: platform comparison, double linear amplification, and universal RNA reference. J. Biotechnol. 112: 225–245. [DOI] [PubMed] [Google Scholar]
- Pedersen, K. S., T. N. Kristensen and V. Loeschcke, 2005. Effects of inbreeding and rate of inbreeding in Drosophila melanogaster—Hsp70 expression and fitness. J. Evol. Biol. 18: 756–762. [DOI] [PubMed] [Google Scholar]
- Pletcher, S. D., S. J. Macdonald, R. Margurerie, U. Certa, S. C. Stearns et al., 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12: 712–723. [DOI] [PubMed] [Google Scholar]
- Reed, D. H., D. A. Briscoe and R. Frankham, 2002. Inbreeding and extinction: the effect of environmental stress and lineage. Conserv. Genet. 3: 301–307. [Google Scholar]
- Reed, D. H., E. H. Lowe, D. A. Briscoe and R. Frankham, 2003. Inbreeding and extinction: effects of rate of inbreeding. Conserv. Genet. 4: 405–410. [Google Scholar]
- Reid, J. M., P. Arcese and L. F. Keller, 2003. Inbreeding depresses immune response in song sparrows (Melospiza melodia): direct and inter-generational effects. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270: 2151–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, A., 1952. The effect of inbreeding on the variation due to recessive genes. Genetics 37: 189–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender, H., 2004. Identifying differentially expressed genes with siggenes. (http://bioconductor.org/).
- Smyth, G. K., 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 (1): Article 3. (http://www.bepress.com/sagmb/vol3/iss1/art3). [DOI] [PubMed]
- Sørensen, J. G., T. N. Kristensen and V. Loeschcke, 2003. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6: 1025–1037. [Google Scholar]
- Tusher, V. G., R. Tibshirani and G. Chu, 2001. Significance analysis of microarrays. Applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen, C. J., and R. Bijlsma, 2004. Characterization of conditionally expressed mutants affecting age-specific survival in inbred lines of Drosophila melanogaster: lethal conditions and temperature-sensitive periods. Genetics 167: 1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., W. G. Hill, D. Charlesworth and B. Charlesworth, 1999. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genet. Res. 74: 165–178. [DOI] [PubMed] [Google Scholar]
- Wu, Z. J., R. A. Irizarry, R. Gentleman, F. M. Murillo and F. Spencer, 2004. A model based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99: 909–917. [Google Scholar]
- Wu, Z. J., and R. A. Irizarry, 2004. Preprocessing of oligonucleotide array data. Nat. Biotechnol. 22: 656–658. [DOI] [PubMed] [Google Scholar]
- Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole and S. C. Sealfon, 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]