Abstract
Dorsoventral (DV) patterning is essential for growth of the Drosophila eye. Recent studies suggest that ventral is the default state of the early eye, which depends on Lobe (L) function, and that the dorsal fate is established later by the expression of the dorsal selector gene pannier (pnr). However, the mechanisms of regulatory interactions between L and dorsal genes are not well understood. For studying the mechanisms of DV patterning in the early eye disc, we performed a dominant modifier screen to identify additional genes that interact with L. The criterion of the dominant interaction was either enhancement or suppression of the L ventral eye loss phenotype. We identified 48 modifiers that correspond to 16 genes, which include fringe (fng), a gene involved in ventral eye patterning, and members of both Hedgehog (Hh) and Decapentaplegic (Dpp) signaling pathways, which promote L function in the ventral eye. Interestingly, 29% of the modifiers (6 enhancers and 9 suppressors) identified either are known to interact genetically with pnr or are members of the Wingless (Wg) pathway, which acts downstream from pnr. The detailed analysis of genetic interactions revealed that pnr and L mutually antagonize each other during second instar of larval development to restrict their functional domains in the eye. This time window coincides with the emergence of pnr expression in the eye. Our results suggest that L function is regulated by multiple signaling pathways and that the mutual antagonism between L and dorsal genes is crucial for balanced eye growth.
IMAGINAL discs of Drosophila are sac-like epithelial structures that represent developing fields. The patterning of a developing field involves generation of compartments that are specified by the domain-specific expression of “selector” genes (Curtiss et al. 2002; Mann and Carroll 2002; Blair 2003b; Milan and Cohen 2003). The boundary between the two compartments is the site of complex signaling that results in the growth and differentiation of an undifferentiated imaginal disc to its adult counterpart. Anteroposterior compartments are formed prior to dorsoventral (DV) patterning in most imaginal discs (Cohen 1993). However, in the eye disc, the formation of the DV axis is the first lineage restriction event (Baker 1978; Dominguez and de Celis 1998; Singh and Choi 2003). Therefore, it is important to understand how DV patterning and growth is regulated in the early eye disc.
The eye imaginal disc differentiates into the adult compound eye comprising precise arrays of ∼800 ommatidia. Each ommatidium contains eight photoreceptors that are arranged in a hexagonal lattice. At the end of the second larval instar, retinal differentiation initiates from the posterior margin of the eye disc and progresses anteriorly during the third instar stage as a wave of differentiation referred to as the morphogenetic furrow (MF) (Wolff and Ready 1993). The photoreceptor clusters of ommatidia in the dorsal and ventral halves of the eye exhibit opposite chiralities and generate a mirror-image symmetry at the DV midline of the eye called the equator (Wolff and Ready 1993).
In the dorsal eye, a GATA-type zinc (Zn)-finger domain transcription factor, Pnr, which initiates Wg signaling, is expressed (Maurel-Zaffran and Treisman 2000). Wg signaling in turn promotes the expression of members of the Iroquois complex (Iro-C), namely araucan (ara), caupolican (caup), and mirror (mirr) (Heberlein et al. 1998). pnr and members of Iro-C act as selectors for the dorsal eye fate (Gomez-Skarmeta et al. 1996; McNeill et al. 1997; Cavodeassi et al. 1999). The boundary between the dorsal and ventral compartments is the site of Notch (N) signaling, which is required for cell proliferation and differentiation of the eye (Cho and Choi 1998; Dominguez and de Celis 1998; Papayannopoulos et al. 1998; Dominguez and Casares 2005). The activation of N signaling at the DV boundary is mediated by the function of Fringe (Fng), a glycosyl transferase preferentially expressed in the ventral domain of the early eye disc (Cho and Choi 1998; Dominguez and de Celis 1998; Papayannopoulos et al. 1998; Moloney et al. 2000; Haines and Irvine 2003). Iro-C genes restrict the expression of fng to the ventral eye, thereby establishing the DV boundary (Cho and Choi 1998; Dominguez and de Celis 1998). Thus, DV patterning is a crucial event for the growth of the early eye disc.
Apart from the genes expressed in a domain-specific pattern, there are genes that are expressed ubiquitously in the eye but exhibit a domain-specific function (Chern and Choi 2002; Singh and Choi 2003; Singh et al. 2004). For example, Lobe (L), which encodes a novel protein, is expressed ubiquitously in the eye disc but is required only for ventral eye development. Genetic epistasis analysis suggests that L acts downstream of N signaling and is required for expression of Serrate (Ser), which is expressed preferentially in the ventral eye (Cho and Choi 1998) and is essential for ventral eye disc growth and development (Chern and Choi 2002; Singh and Choi 2003). Earlier studies showed that prior to the onset of pnr gene expression, the entire eye disc is ventral in fate and requires L and Ser gene function. Subsequently, pnr expression appears in the dorsal margin of the early second instar eye disc to establish the DV pattern (Singh and Choi 2003). Thus, the functional domain of L and Ser is either the entire eye field or restricted to the ventral domain, depending on the time of initiation of pnr expression in the dorsal eye margin (Singh and Choi 2003).
This raises two questions: (1) How is the ventral eye domain maintained without being entirely dorsalized by the function of the dorsal selector gene? and (2) How does L regulate the survival and growth of tissue in the ventral eye disc? To address these questions, it is necessary to identify additional genetic components that either antagonize or promote L functions. One of the major hurdles to employing conventional classical genetic screens that rely on the loss-of-function (LOF) phenotypes is that nearly 66% of the Drosophila genes are functionally redundant and phenotypically silent (Rubin et al. 2000). Thus, it is possible that heterozygous conditions for LOF mutations in potential modifier genes may be insufficient to alter the L mutant phenotype. However, these functionally redundant genes can be identified by an alternative and powerful gain-of-function approach (Blair 2003a). This approach uses a collection of EP element lines (Rorth 1996), which allows misexpression of genes in specific tissues in a sensitized genetic background to reveal phenotypic enhancement or suppression.
EP lines harbor a P-transposable element containing the upstream activation sequence (UAS) that binds GAL4 and a basal promoter to direct the expression of genes downstream from the EP insertion site (Rorth 1996). Using specific GAL4 driver strains, EP-mediated targeted misexpression can generate more recognizable phenotypes than the ones elicited by LOF mutations in the same gene (Rorth 1996; Rorth et al. 1998; Huang and Rubin 2000; Abdelilah-Seyfried et al. 2001; Kraut et al. 2001; Pena-Rangel et al. 2002; Tseng and Hariharan 2002). Another advantage of this gain-of-function (GOF) strategy is that the genes associated with EP insertion lines can be rapidly identified (Blair 2003a). Therefore, once the modifier genes are identified, it is possible to test for reciprocal genetic interactions using available LOF mutations in these identified genes. On the basis of this rationale, we devised a GOF strategy to identify genes affecting L function in ventral eye growth and development. Since L is necessary for DV patterning and growth of the ventral eye tissue, genes interacting with L may be grouped into two main categories: (i) the genes that show asymmetric expression or function in the dorsal or ventral domain and (ii) the genes that either regulate or are regulated by L to promote eye growth. The genes in the second category may not necessarily function asymmetrically in the dorsal and ventral domains. Both groups of genes would provide important clues to understanding the mechanism of L function in patterning and growth control.
In this study, we performed an EP-based genetic screen to look for modifiers of L gene function in DV patterning and/or growth. Since the majority of the modifiers identified in our screen are known to interact genetically with pnr or are members of the downstream Wg signaling pathway, we further analyzed the genetic interaction between L and pnr. Here, we show that during DV patterning of the eye, L and pnr act antagonistically to each other in the time window spanning through the second instar of larval development to restrict the functional domains of each other. Furthermore, this relation coincides with the initiation of pnr expression in the dorsal eye margin. Our results suggest that the antagonistic interactions between genes involved in patterning and growth of DV domains are essential for balanced eye growth and that the L function in eye growth is regulated by interaction with Hh and Dpp signaling pathways, which have not been documented previously.
MATERIALS AND METHODS
Fly stocks used:
Stocks used were ey-GAL4 (Hazelett et al. 1998), UAS-GFP, y w; L2/CyO, Lsi/CyO, y w; FRT42D, and Lrev6-3/CyO. L2 is a dominant mutation, which is lethal when homozygous or when heterozygous over Df (2R) trix deficiency. Mutant alleles used in this study are emc12, an amorph allele of emc (Brown et al. 1995); Bar1(B1)/FM7, a dominant gain-of-function mutation generated by duplication of the 16A region containing BarH1 and BarH2 genes, referred to as Bar (B); Df(1)Bar263-20, a deficiency in which both B genes are deleted (Higashijima et al. 1992); sgg1/FM7a Dp(1;2;4) w+, a lethal recessive X-ray allele of sgg (Bourouis et al. 1990); Df (2L) BSC7/CyO, UAS-pnrD4 (Haenlin et al. 1997); y w; FRT82, pnrvx6/TM6B (Heitzler et al. 1996); UAS-pnrENR, a dominant-negative form of pnr, which contains the repressor domain from Engrailed (amino acids 2–298) (Klinedinst and Bodmer 2003); UAS-ush (Fossett et al. 2001); Df (3L) Iro-CDFM3 FRT80/TM6B, a deficiency for ara, caup, and promoter for mirr, UAS-ara (Gomez-Skarmeta et al. 1996); UAS-mirr (McNeill et al. 1997); y w ey-FLP (Newsome et al. 2000); and pnr-GAL4 (Calleja et al., 1996). All stocks are described in FlyBase (http://flybase.bio.indiana.edu).
Temperature-shift regimen:
We followed a commonly used strategy of exploiting the temperature sensitivity of GAL4 expression (Cho and Choi 1998; Kumar and Moses 2001; Singh and Choi 2003) to block pnr gene function by overexpressing ush (ey-GAL4, UAS-ush) during different stages of eye development. Eggs were collected for 2 hr from the cross of L2/CyO; ey-GAL4 flies with y w, UAS-ush flies. Each egg collection was divided into several batches. These independent batches were reared at 18° (blue line, Figure 9A) except for a single shift to 29° (red line, Figure 9A) in a 24-hr time window during different periods of development spanning from embryogenesis to the late third instar. From each egg collection we generated a set of cultures. For controls, we maintained (a) ey-GAL4/UAS-ush (ey>ush) and (b) L2/+; ey-GAL4/UAS-ush (L2/+; ey>ush) cultures at 18°, 25°, and 29° during the entire course of eye development.
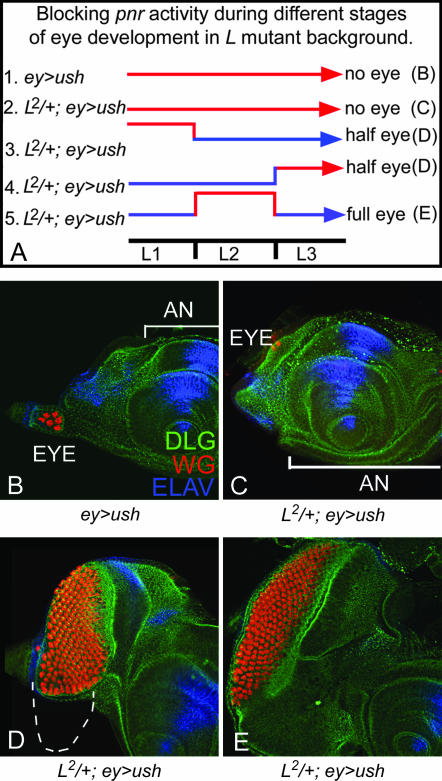
Figure 9.
Antagonistic interaction of pnr and L is crucial for DV patterning during second instar of larval eye development: (A) Schematic for the conditional blocking of pnr activity by overexpression of ush in the eye by ey-GAL4 (ey>ush). Temperature sensitivity of GAL4 drivers was used to control the time window of expression of Ush. Cultures were shifted to 29° to induce ush expression in a specific time window. Cultures were otherwise maintained at 18°. (A) Five different experimental conditions listed as 1–5 were used to block pnr activity. (B) Control showing overexpression of ush in the eye. ey>ush (A, experiment 1) throughout eye development showed no eye in 80% of the eye discs observed. ELAV was used as a marker for eye fate. (C) Overexpression of ush in the L2/+ eye (L2/+; ey>ush) throughout development completely eliminates eye field in nearly 100% of flies observed (A, experiment 2). (D) Blocking pnr activity by ush overexpression in L2/+ background during first or third instar of larval eye development results in loss of the ventral eye (A, experiments 3 and 4), a phenotype similar to that of the L2/+ mutant eye. (E) Blocking pnr activity during second instar of larval eye development by overexpression of ush results in rescue of the L mutant's ventral eye phenotype (A, experiment 5).
Strategy of EP screen:
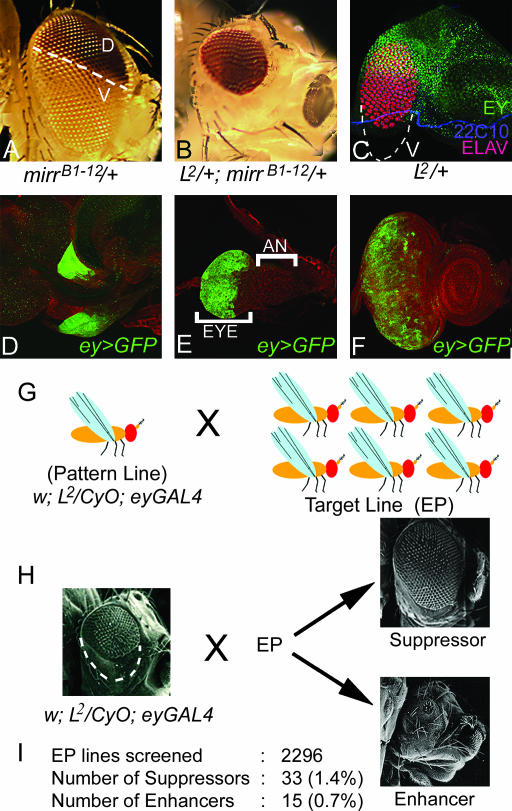
This GOF screen employed the GAL4/UAS system for overexpression of EP lines (Brand and Perrimon 1993; Rorth 1996). The EP transgenic line collection comprising 2296 EP lines (Rorth 1996) was obtained from the European Drosophila Stock Center and the Bloomington Stock Center. In our screen, y w; L2/CyO; ey-GAL4 flies were mated individually to EP lines (Figure 1, G and H). We scored F1 progeny from each cross for any modification in the loss of the ventral eye phenotype of the L2/+ mutant. EP lines showing the complete elimination or reduction of the L2/+ eye phenotype were classified as enhancers, and the lines that significantly rescued the loss of the ventral eye phenotype were classified as suppressors of the L mutant phenotype (Figure 1H). Since the L2 mutation was balanced over CyO, we could compare experimental (L2/+; ey>EP) vs. control (+/CyO; ey>EP) phenotypes within the same progeny population. We found that ey-GAL4-driven UAS-GFP (ey>GFP) is expressed in the entire eye during all stages of eye disc development (Figure 1, D–F). We noted that GAL4 expression by ey-GAL4 in the third instar eye disc differs from EY protein localization, which is expressed anterior to the morphogenetic furrow in undifferentiated cells (Halder et al. 1998; Singh et al. 2002). We checked if this ey>GFP expression is due to perdurance of GFP by using overexpression of homothorax (hth) as a test. Hth, a transcription factor, which is expressed anterior to the furrow, blocks eye development when misexpressed in the differentiating eye (Pai et al. 1998). We found that ey-GAL4-driven UAS-hth (ey>hth) results in complete elimination of the eye, a phenotype similar to the one exhibited when hth is ectopically induced in the differentiated eye. These results suggest that, in ey-GAL4 flies, GAL4 proteins are present in the region posterior to the furrow where photoreceptor differentiation takes place during third instar larval eye development.
Figure 1.
Schematic of the gain-of-function screen. (A) Adult eye of mirrB1-12, an enhancer trap line showing dorsal-eye-specific expression of the mini-white reporter gene. (B) In L2/+; mirrB1-12 /+ flies, almost all w− ventral ommatidia are lost showing only w+ dorsal ommatidia. (C) Eye imaginal disc of L2/+ mutant shows selective loss of ventral eye pattern. (D–F) (ey>GFP), ey-GAL4 driven UAS-GFP reporter gene expression in (D) first-, (E) second-, and (F) third instar larval eye imaginal disc. D shows a pair of early eye imaginal discs. GFP is expressed in the entire eye primordium during all stages. Unlike Ey protein that expresses anteriorly to the furrow in the third instar disc, ey>GFP is expressed in the entire eye disc. (G) Schematic of F1 screen where flies of pattern line (L2/CyO; ey-GAL4) were crossed to various EP target lines. (H) The criteria used for classification of the L modifiers into enhancer and suppressor categories. The modifiers that upon overexpression enhance the L mutant phenotype to no or significantly reduced eyes were referred to as enhancers whereas the ones that suppress the L mutant phenotype from partial rescue of the ventral eye to near wild-type eye were classified as suppressors. (I) Summary of the data generated from the screen. All eye discs are oriented with anterior on the right and dorsal at the top. AN, antenna.
Scanning electron microscopy:
We prepared the flies for scanning electron microscopy by dehydration through a series of increasing concentrations of ethanol. Dehydrated flies were dried in a Samdri-780 critical point dryer and mounted for electron microscopy. Fly samples were coated with silver using a Denton vacuum sputter coater and analyzed using a Jeol-6100 scanning electron microscope.
LOF clones:
LOF clones were generated using the FLP/FRT system of mitotic recombination (Xu and Rubin 1993). To generate LOF clones of L in the eye (Newsome et al. 2000), ey-FLP, FRT42 ubi-GFP females were crossed to FRT42D, Lrev6-3 males. To generate the clones of pnrvx6 males of y w; FRT82, pnrvx6/TM6B were crossed to ey-FLP; FRT82 ubi-GFP females.
Immunohistochemistry:
Eye-antenna discs were dissected from wandering third instar larvae and stained as described earlier (Singh et al. 2002). The antibodies used were rabbit anti-DLG (1:200) (a gift from Kyung-Ok Cho), rabbit anti-HTH (1:300) (a gift from Y. Henry Sun), rabbit anti-EY (1:500) (a gift from Uwe Walldorf), rat anti-ELAV (1: 20), mouse anti-WG (1: 20), and mouse anti-22C10 (Developmental Studies Hybridoma Bank). Secondary antibodies (Jackson Laboratories) were donkey anti-rat IgG conjugated with Cy5 (1: 200), donkey anti-rabbit IgG conjugated with Cy3 (1:400), or goat anti-mouse IgG conjugated with Alexa Fluor 488 (1:200). pnr expression was detected using pnr-GAL4>UAS-GFP (Pichaud and Casares 2000; Singh and Choi 2003).
RESULTS
Gain-of-function strategy to identify EP lines that suppress the L mutant phenotype:
We used the L2 mutant, which exhibits a strong, consistent phenotype of preferential loss of the ventral eye pattern in heterozygotes (Figure 1, B and C; Singh and Choi 2003). To check the dorsal vs. ventral fate of ommatidia in the L2/+ mutant eye, we used mirrB1-12, an enhancer trap line with a P-element insertion in the mirr gene (Choi et al. 1996), which serves as an excellent marker for dorsal eye fate (Figure 1A). We found that except for a few cells along the ventral eye margin, nearly all ommatidia in L2/+ mutant eyes are dorsal in fate (Figure 1B). This loss of ventral eye pattern in the L2/+ mutant can also be observed in the eye imaginal disc (Figure 1C) and adult eye sections (data not shown).
The genes represented in the EP lines collection (Rorth 1996) were overexpressed in L2/+ background using the ey-GAL4 driver (Hazelett et al. 1998), which drives GFP reporter gene expression in the entire eye (Figure 1, D–F) in both differentiating cells posterior to the morphogenetic furrow and undifferentiated proliferating cells anterior to the morphogenetic furrow (Figure 1F; see also materials and methods). Of the 2296 EP lines tested, 33 lines (∼1.4%) with EP insertions in 10 genes were identified as modifiers that can suppress the L2/+ mutant eye phenotype upon overexpression (Figure 1, H and I; Table 1). Nearly 27% (9/33) of these modifiers either were found to interact with pnr (Pena-Rangel et al. 2002) (Table 1) or were members of the Wg signaling pathway. To eliminate the possibility of nonspecific interactions, we retested these genetic interactions with LOF mutations in the identified modifier genes. Since the initial screen was based on the interaction of L2/+ and EP-mediated overexpression of modifier genes, we would expect to see opposite genetic interactions with LOF mutations. Here we present the phenotypes of a few selected suppressors.
TABLE 1.
Modifiers affecting the L mutant's phenotype of loss of the ventral half of eye
| EP line | Cytological location | Gene | Description | Effect of modifier |
|---|---|---|---|---|
| 1576 | 3B1-3B1 | sgg | Glycogen synthase kinase 3, protein serine/threonine kinase activity | + |
| 1643 | 7B2 | sarcoplasmic calcium-binding protein | Calcium ion-binding activity | + |
| 950,1149 1179, 1410 | 8F9 | nej CBP | Acetyl transferase, CREB-binding protein | − |
| 1235 1427 | 9E2 | raspberry (ras) | Ionosine-5′ monophosphate dehydrogenase activity, interacts genetically with pnr | + |
| 1353 1508 | 12A9-B2 | Misexpression Suppressor of Ras 1 (NFAT) | Transcription factor involved in cellular defense response | ++ |
| 1324 | 15E2-3 | CG16700 | Gamma amino butyric acid transporter activity | ++ |
| 1350 | 16A1 | B | Transcription factor | − − |
| 1220 | 21B7 | smo | G-protein-coupled receptor activity, Hedgehog receptor activity | + |
| 2500, 598 | 22D1 | anterior open (aop) or yan | ETS-binding domain transcription factor, negative regulator of EGFR pathway | − − |
| 2454 | 27B1 | nrv1 | Sodium/potassium-exchanging ATPase activity | +++ |
| 655 | 34A1 | kekkon-1 (kek-1) | Transmembrane receptor protein, serine/ threonine kinase activity | + |
| 2173 | 35B2 | no ocelli (noc) | Zn finger, C2H2 type, transcription factor | + |
| 633, 683 2159, 2370 2408 | 35D2 | esg | Zn finger, C2H2 type, transcription factor involved in neuroblast cell division, interacts with pnr | +++ |
| 2518 | 44B1 | cul-4 | Ubiquitin protein ligase activity | ++ |
| 2208 | 45D5 | wunen (wun); wunen 2 (wun2) | Phosphatidate phosphatase activity, G-protein-coupled receptor protein | ++ |
| 2316 | 52B2 | EP1229 | Unknown, interacts genetically with N and pnr | ++ |
| 2012 2031 | 53F8-9 | Glutathione S transferase S1 (Gst S1) | Glutathione transferase and glutathione peroxidase, interact genetically with pnr | + |
| 2452 | 60E1 | CG16936 | Glutathione transferase activity | +++ |
| 415, 3166 3614, 3620 3087 | 61C9 | emc | Helix-loop-helix dimerization domain for protein-binding activity, interacts genetically with pnr | − |
| 3168 | 66E1-3 | dally | Cell adhesion molecule, interacts genetically with wg | − |
| 3152 | 68F5-6 | RhoGAP68F | Rho GTPase activator | + |
| 3622 | 69E2-3 | CG17667 | Cell adhesion molecule, serine threonine kinase activity | + |
| 3123 | 73E4 | CG6664 | Encodes a protein with chromatin binding and involved in cell cycle | + |
| 3082 | 78A1 | fng | UDP-glycosyl transferase, dorso-ventral polarity of eye | ++ |
| 3387 | 89A6 | CG4225 | ATP-binding cassette, transporter activity | + |
| 3196 | 89E11 | Dad | Domain A of Dwarfin protein, SMAD/FHA domain, antagonist of Dpp signaling | − − |
| 1230 | 93D2 | CG3308 | Deoxyribonuclease activity | ++ |
| 3521 | 94E1 | hh | Progression of MF | ++ |
| 3285 | 95C8 | Transcription factor- IIA-S (TfIIA-S) | Transcription factor IIA | ++ |
| 3096 | 99D1 | CG7920 | 4-Hydroxybutyrate CoA-transferase | ++ |
| 3280 | 100B2 | dco | Casein kinase activity, serine/threonine kinase activity | + |
| 3408 | 100C6 | pygo | PHD finger domain, transcriptional regulator, Wg signaling pathway | − − |
| 3309 | 100B8 | CG12054 | Unknown | + |
Suppressors (+): +++, ∼40% of flies show complete rescue of eye; ++, ∼30% of flies show 75% rescue of ventral eye; +, ∼30–40% flies show weak rescue. Enhancers (−)—Note that loss-of-function mutations of these enhancer genes act as suppressors of the L phenotype: − −, no eye, high penetrance (∼40%), very strong pupal lethality; −, no eye (∼20%) and 30% show a very small eye.
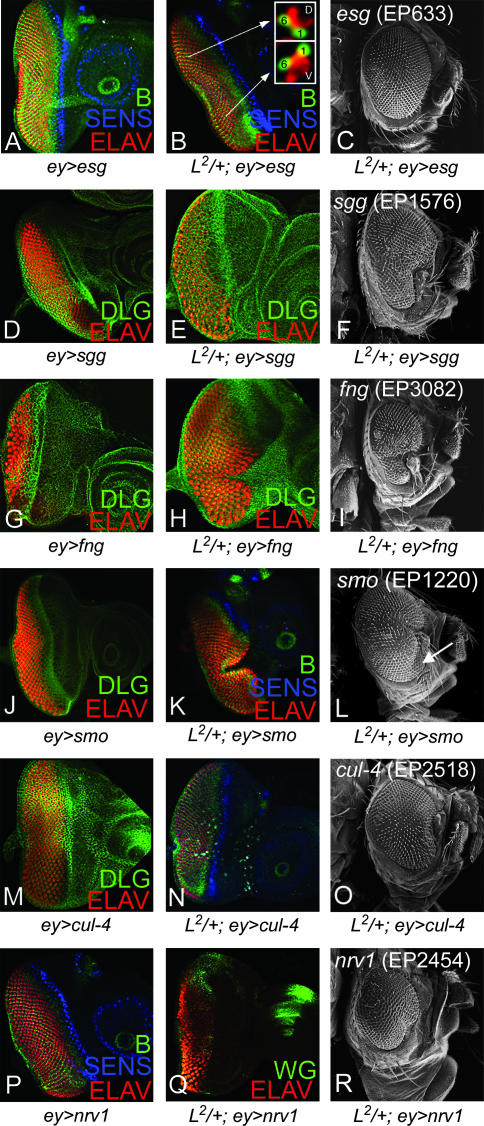
The overexpression of escargot (esg), using EP insertion in an esg gene, rescued the L2/+ ventral eye loss phenotype to a near wild-type eye (Figure 2, B and C) whereas the overexpression of esg alone does not affect the eye size (Figure 2A). esg encodes a Zn-finger transcription factor involved in imaginal disc development (Hayashi et al. 1993; Fuse et al. 1994). We stained the eye discs (L2/+; ey>esg) with anti-Bar antibody, a marker for R1 and R6 fate, and confirmed that the planar polarity of the ommatidia in the rescued portion of the eye disc is ventral (Figure 2B inset). We also verified the rescue of the ventral eye phenotype on the basis of the ommatidial polarity in the adult eye sections (data not shown). All five independent EP insertions in the esg gene (EP633, EP683, EP2159, EP2370, and EP2408) showed suppression of the L2/+ mutant phenotype, supporting the fact that the interaction of L and esg is specific (Table 1). Nearly 20% of the adult progeny showed complete eyes whereas the remaining 80% partially suppressed the L2/+ mutant phenotype to varying degrees. These results suggest that esg may act downstream or function in parallel to L. Furthermore, reducing levels of esg in a heterozygous background of esg mutant allele (esgP3/+) enhanced the L2/+ mutant phenotype of ventral eye loss to no eye in ∼21% (10/48) of flies observed.
Figure 2.
Modifiers that suppress the L mutant phenotype in the ventral eye. (A–C) Overexpression of esg (EP633) in eye (A) (ey>esg), (B and C) in L2/+; ey>esg can fully rescue the ventral eye loss phenotype of the L2/+ mutant in (B) the eye disc and (C) the adult eye. Inset in B shows that ventral eye pattern is restored in the rescued eyes upon esg overexpression on the basis of Bar (B) staining, which marks R1 and R6 photoreceptor cells in the eye. R1 and R6 cells demonstrate mirror-image symmetry in the dorsal and ventral eye domains and B expression marking these photoreceptor cells is commonly used to distinguish DV fate in the eye disc. Overexpression of modifiers that suppress the L2/+ mutant phenotype are (D–F) EP1576 (sgg), (G–I) EP3082 (fng), (J–L) EP1220 (smo), (M–O) EP2518 (cul-4), and (P–R) EP2454 (nrv 1). For controls, (D) ey>cul-4, (J) ey>sgg, (M) ey>nrv 1, and (P) ey>smo have no visible effect on the eye whereas (G) ey>fng results in a small eye. Neuronal marker ELAV that marks photoreceptor nuclei was used for marking the eye fate. Other markers used were DLG (membrane), B (R1 and R6), Sens (R8), and Wg.
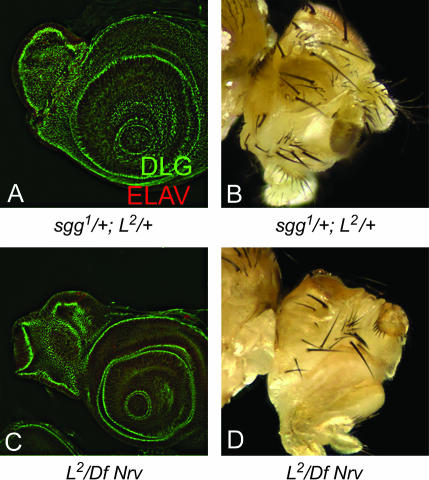
The overexpression of EP1576, an insertion at the shaggy (sgg)/Drosophila GSK-3β gene, could suppress the L2/+ mutant eye phenotype; however, the overexpression of sgg (ey>sgg) in the eye results in a normal looking eye (Figure 2D). sgg acts as an antagonist of the Wg signaling pathway (Peifer et al. 1994; Siegfried et al. 1994). The suppression did not extend completely to the anterior ventral margin of the eye disc (Figure 2E) and was also observed in ∼16% (11/69) of adult flies (Figure 2F). The genetic interaction of L and sgg was also confirmed by using a LOF mutation of sgg. The genetic interaction of sgg1 and L2 in heterozygous combination (sgg1/+; L2/+) resulted in the complete elimination of the eye field in ∼15% (7/48) of observed flies (Figure 3, A and B), whereas sgg1/+; Lrev6-3/+ mutant female flies showed a loss of the ventral eye phenotype (data not shown). Notably, Lrev6-3 in a heterozygous combination (Lrev6-3/+) shows a normal wild-type eye. These results suggest that Wg signaling acts antagonistically to L function.
Figure 3.
Genetic interaction of loss-of-funtion mutations in L and suppressors. Reducing gene function levels of suppressors, namely sgg (sgg1/+) (A and B) and Nrv (Df (2L) BSC7/+) (C and D) enhances the L2/+ mutant phenotype of ventral eye loss to no eye as seen in the eye disc marked by DLG and ELAV in A and C and in the adult eye in B and D. Note that sgg1 is a hemizygous lethal mutation and that these phenotypes have been observed only in the eyes of female flies.
We also identified fringe (fng), a member of the N signaling pathway involved in DV patterning of the eye, as a suppressor of the L2/+ mutant phenotype (Table 1). The overexpression of fng using the EP3082 line partially rescued the loss of the ventral eye phenotype of the L2/+ mutant as seen in the eye disc and the adult eye (Figure 2, H and I), whereas the overexpression of fng (ey>fng) results in a small eye (Figure 2G; Cho and Choi 1998). These results suggest that fng might act downstream from or parallel to L during early eye growth.
Our data also suggest that L genetically interacts with the Hh signaling pathway. First, EP1220, an insertion at smoothened (smo) that encodes a receptor of the Hedgehog (Hh) signaling pathway, can suppress the L2/+ eye phenotype (Figure 2, K and L). The overexpression of smo (ey>smo) in a wild-type background did not affect the eye (Figure 2J). Second, the overexpression of hh with EP3521, an insertion at hh, also suppressed the L2/+ mutant phenotype (Table 1). Furthermore, the double mutant of L2/+ and hh, (L2 /CyO; hh1/hh1), showed complete elimination of the eye. Similar phenotypes were seen with Lrev6-3/CyO; hh1/hh1 (data not shown), which further supports the fact that Hh signaling promotes L function.
Cullin-4 (Cul-4) is a key component of the E3 ubiquitin ligase complex that is required for DNA damage control (Zhong et al. 2003). The overexpression of cul-4 by EP2518, an insertion at the cul-4 gene, rescued the L2/+ mutant phenotype (Figure 2, N and O), whereas ey>cul-4 did not affect eye size (Figure 2M). Trans-heterozygous combinations of cul-4 and L mutants (cul-4/L2) resulted in variable reduction of the adult eye when a different L allele was used (data not shown). When cul-4 mutant clones were generated by the FLP/FRT method, the mutant cells could not grow in either dorsal or ventral domain and were out-competed by the surrounding wild-type cells, suggesting that Cul-4 is essential for cell survival and/or growth (data not shown).
EP2454, an insertion in the nervana 1 (nrv 1) gene that encodes a subunit of sodium/potassium ATPase (Sun and Salvaterra 1995), rescued the ventral eye loss of the L2/+ mutant in the eye disc (Figure 2Q) and the adult eye (Figure 2R). ey>nrv 1 in a wild-type background did not affect eye size (Figure 2P). We found a dominant genetic interaction between various mutant alleles of L and Df (2L) BSC7, a deficiency that uncovers nrv 1. In the heterozygous combination of L2/Df (2L) BSC7, the eye field was completely eliminated (Figure 3, C and D). Although the mechanisms of specific interactions of cul-4 and nrv 1 with L remain to be studied, their strong genetic interactions with L by LOF and GOF approaches suggest that they may be involved in L-mediated eye growth function directly or through a common target gene.
EP lines that enhance the L mutant phenotype:
We screened 2296 EP lines and identified 15 (∼0.7%) modifiers in 6 genes, which, when overexpressed in the eye, enhanced the severity of the L2/+ mutant eye phenotype (Figure 1E; Table 1). These modifiers are referred to as enhancer modifiers throughout the text. As seen in the case of modifiers that suppress the L2/+ mutant phenotype, 54% (7/13) of enhancer modifiers interact genetically with pnr (Pena-Rangel et al. 2002). We carried out additional tests to see whether the L2/+ phenotype can be suppressed by LOF mutations in the enhancer genes (Figure 5) and verified the specific interaction of the modifiers using their LOF mutations. We present here only the modifiers whose LOF mutations dominantly enhanced the phenotypes of the various L mutant alleles. These enhancer modifiers include extramacrochaetae (emc), B, nejire (nej), pygopus (pygo), and division abnormally delayed (dally) (Table 1). Of these, pygo and dally are members of the Wg signaling pathway.
Figure 5.
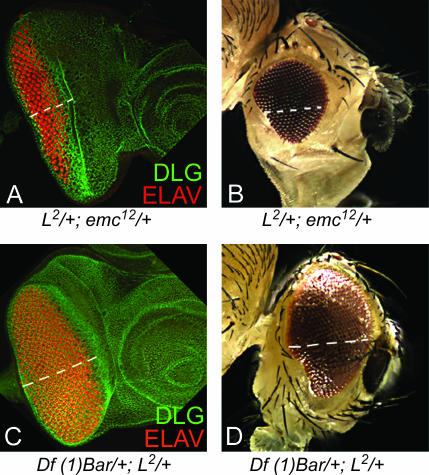
Genetic interaction of loss-of-function mutations in L and enhancer modifiers. (A–D) Trans-heterozygous combinations of the L2/+ mutant with (A and B) the emc mutant (L2/+; emc12/+) and (C and D) the deficiency of B (Df (1) Bar/+; L2/+) result in partial rescue of the ventral eye loss phenotype as seen in the eye disc (A and C) and in the adult eye (B and D).
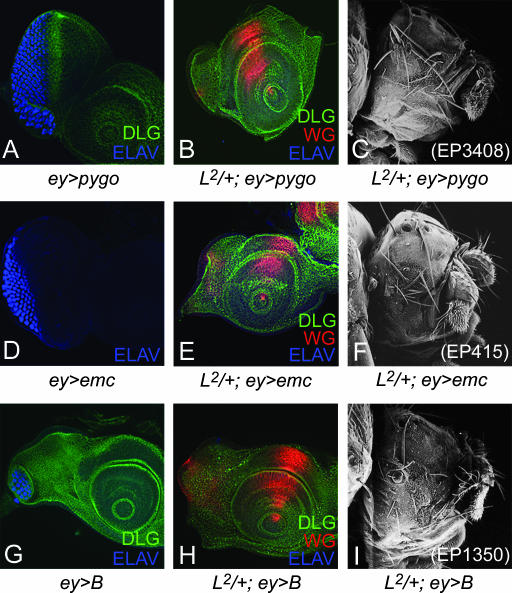
The overexpression of EP 3408, an insertion at the pygo gene, strongly enhanced the L2/+ mutant phenotype to no eye (Figure 4, B and C). However, ey>pygo in wild-type background reduced the eye size but did not generate no-eye phenotypes (Figure 4A). Pygo contains a plant homeodomain (PHD) finger at its C terminus, a motif often found in chromatin-remodeling factors, and is required for Wg signaling throughout Drosophila development (Thompson et al. 2002). We also checked the interaction using the LOF mutant allele of pygo and L and observed the rescue of the ventral eye loss phenotype seen in the L mutant (data not shown).
Figure 4.
Modifiers that enhance the L mutant phenotype in eye. (A) Overexpression of EP3408, an insertion at pygo in wild-type disc (ey>pygo) causes reduction in eye size. (B) Overexpression of pygo in L2/+ mutant eye disc (L2/+; ey>pygo) enhances the ventral eye loss phenotype to no eye in eye disc (B) and adult eye (C). Overexpression of (D–F) emc using EP415 and (G–I) B using EP1350 in the eye. (D) ey>emc results in a small eye. (E and F) L2/+; ey>emc enhances the L2/+ mutant phenotype to no eye. (G) Overexpression of B in eye (ey>B) strongly reduces the eye size. (H and I) L2/+; ey>emc strongly enhances the L mutant phenotype to no eye.
Five independent insertions in the emc gene (EP415, EP3166, EP3614, EP3620, and EP3087) enhanced the L mutant phenotype. emc encodes a helix-loop-helix protein and acts as a negative regulator of the morphogenetic furrow (Brown et al. 1995). The overexpression of emc using EP lines or UAS-emc enhanced the L2/+ mutant phenotype of ventral eye loss to no eye with normal antennae (Figure 4, E and F). ey>emc resulted only in small eyes (Figure 4D). We also verified these interactions using bi-GAL4 (data not shown). bi-GAL4 is an insertion at the optomotor blind (omb) locus, which expresses on both the dorsal and the ventral polar margins of the eye (Calleja et al.1996; Singh et al. 2002). Furthermore, reducing the levels of emc gene function in trans-heterozygous combinations of L and emc mutants (L2/+; emcE12/+) resulted in partial rescue of ventral eye loss generated by L2/+ mutations (Figure 5, A and B). These results suggest that the observed genetic interactions are not due to an additive effect of overexpression of emc and the L2/+ mutant background and that emc acts as an antagonist of L gene function.
We found that the overexpression of the homeodomain gene B using the EP1350 insertion strongly enhanced the L2/+ mutant phenotype and caused a very strong pupal lethality. These pharate adults had a no-eye phenotype, whereas the antennal field was not affected (Figure 4, H and I). We used B mutant alleles to check the specificity of the genetic interaction. The B1 is a GOF mutant (Higashijima et al. 1992). B1/+; L2/+ females exhibited a no-eye phenotype (data not shown) similar to the one observed by overexpression of EP1350 in the L2/+ mutant background. In contrast, the reduction in B gene dosage by removing one wild-type copy of B in a deficiency of the B locus, Df (1) Bar263-20 w+/+, significantly rescued the loss of the ventral eye pattern in the L2/+ mutant (Figure 5, C and D). The B mutants also demonstrated similar interactions with the Lsi and Lrev mutations (data not shown).
We identified B, emc, and members of the Wg signaling pathway as enhancers, whereas esg, fng, cul-4, and members of Dpp and Hh signaling pathway were suppressors of the L mutant phenotype. An important outcome from our screen was that nearly 29% of the modifiers of both the enhancer and the suppressor category were either genes that interact with pnr or members of the downstream Wg signaling pathway. Thus, it would be interesting to study the genetic interaction between L and pnr.
L and pnr are mutually antagonistic:
It has been shown that L is essential for growth of the ventral eye tissue but is dispensable in the dorsal region specified by pnr function (Singh and Choi 2003). However, it has not been tested whether overexpression of pnr is sufficient to antagonize L function in the ventral eye and whether L can act antagonistically to the dorsal eye selectors to restrict their functional domain to the dorsal eye. Therefore, we tested the genetic interaction of pnr and L using combinations of GOF and LOF approaches.
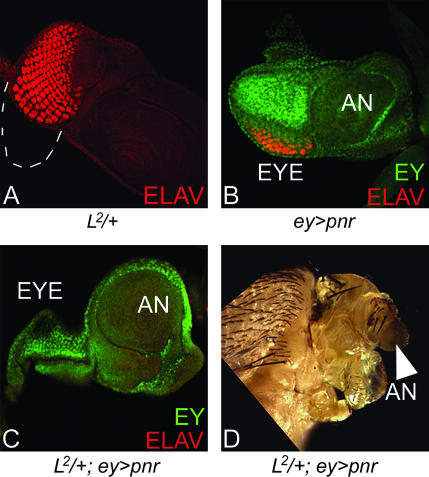
We used the L2/+ mutant, which exhibits preferential loss of the ventral eye pattern (Figure 6A and Figure 1E). Misexpression of pnr in the entire eye using ey-GAL4 driver (ey>pnr) results in a small or no eye with 30% penetrance (Figure 6B) (Maurel-Zaffran and Treisman 2000). It has been suggested that loss-of-eye phenotype by overexpression of pnr is due to elimination of the DV boundary of the eye as the entire eye field is dorsalized (Maurel-Zaffran and Treisman 2000), but these flies do not show pupal lethality.
Figure 6.
Overexpression of pnr can enhance the L mutant phenotype. (A) L2/+ eye disc showing selective loss of ventral eye indicated by dotted line. (B) Overexpression of pnr in the wild-type eye, using ey-GAL4 driver (ey>pnr), resulted in reduction of the eye field. (C and D) Overexpression of pnr in the L2/+ mutant eye background (L2/+; ey>pnr) resulted in loss of complete eye field as seen in the eye imaginal disc (C) and the in adult eye (D). Note that antennal structures (arrowhead) and the mouthparts of adult flies are not affected.
The no-eye phenotype of pnr overexpression (ey>pnr) was further aggravated from 30 to 100% when L gene function (L2/+; ey>pnr) was reduced. The overexpression of pnr in the L2/+ mutant background resulted in the loss of the eye field (Figure 6, C and D). In these cultures very strong pupal lethality was observed as nearly 59% of the progeny failed to eclose. Only ∼41% (36/88) of the flies of the L2/+; ey>pnr genotype eclosed, and these flies had highly reduced heads with no-eyes but normal antennae (Figure 6D, arrowhead). This suggests that L and pnr may have a mutually antagonistic relation. Thus, in the presence of L+ wild-type function, the overexpression of pnr cannot effectively dorsalize the ventral region of the eye disc. We verified these results using bi-GAL4. The overexpression of pnr on the DV margins of the eye by bi-GAL4 (bi >pnr) resulted in a very small eye but in the L2/+ mutant background it enhanced the phenotype to no eye (data not shown).
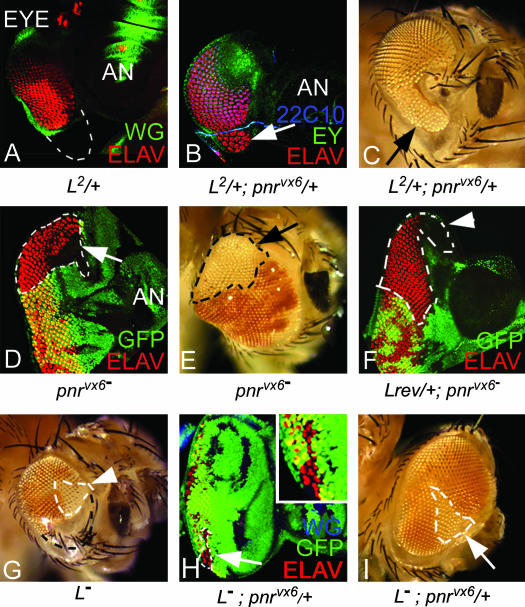
To further test the specificity of the antagonistic relation of Pnr and L, we checked whether the loss of the ventral eye phenotype of the L mutants can be rescued by reducing the level of pnr gene function. We used pnrvx6, a null allele of pnr for these studies (Heitzler et al. 1996). In pnrvx6 heterozygotes (pnrvx6/+) where pnr gene function is reduced to half, eye development occurs normally. However, the loss of ventral eye phenotype of L2/+ (Figure 7A) was partially rescued by pnrvx6/+ as seen in eye discs (Figure 7B, arrow) as well as in ∼22% (5/23) of observed adult eyes (Figure 7C, arrow). The sections of the adult eye of the L2/+; pnrvx6/+ flies showed rescue of the ventral eye on the basis of the planar polarity of ommatidia in the ventral eye (data not shown). We also verified this relationship using the weak allele Lsi (data not shown).
Figure 7.
pnr acts as an antagonist of ventral eye growth. (A) L2/+ eye imaginal disc. (B and C) In trans-heterozygous combination of L2/+; pnrvx6/+, the L2/+ mutant phenotype of ventral eye loss showed partial rescue in (B) eye disc and (C) adult eye (arrow). LOF clones generated by the FLP-FRT approach (Xu and Rubin 1993) were marked by loss of GFP reporter in eye imaginal disc and by loss of mini-white reporter gene expression in adult eye. (D and E) LOF clones of pnr in the dorsal (D) eye disc and (E) adult eye showed dorsal eye enlargements (arrows). Clone boundaries are marked by dotted lines. However, LOF clones of pnr in the ventral clones have no effect. (F) In the Lrev/+ background, the LOF half of eye of pnr no longer showed dorsal eye enlargements. (G) LOF clones of L in adult eye cause preferential elimination of the ventral eye pattern. Note that the LOF clone of Lrev (shown as L−) in the dorsal eye has no effect (clone boundary marked by white dotted line). (H and I) In 30% of eye discs containing LOF clones of L in the pnrvx6/+ background show ventral eye tissue and retinal differentiation within the clones as seen in the eye disc (H) and the adult eye (I). (H) The inset shows ELAV-positive cells within the clone in the ventral eye. Note that nearly 100% of Lrev clones show loss of the ventral eye.
LOF clones of pnr change the fate of dorsal eye cells to ventral and result in dorsal eye enlargements due to the generation of an ectopic equator (Maurel-Zaffran and Treisman 2000; Singh and Choi 2003) (Figure 7, D and E, arrows). However, there is no effect of pnr LOF clones in the ventral eye as pnr is not expressed in the ventral eye. To test whether the pnr function in DV patterning depends on the L+ function, we generated LOF clones of pnr in the heterozygous background of a null allele of L (Lrev6-3/+). When LOF clones of pnr were generated in the Lrev6-3/+ background, they failed to induce such enlargements in the dorsal eye (Figure 7F). These results suggest that the dorsal eye cells, which acquire ventral eye fate upon loss of pnr gene function, become sensitive to the reduced L gene dosage in the Lrev6-3/+ background. Thus the L function may be inhibited by pnr in the dorsal eye.
LOF clones of L result in elimination of the ventral eye (Figure 7G; clone boundary is marked by a black dotted line), whereas LOF clones in the dorsal eye have no effect (Figure 7G, arrowhead) (Chern and Choi 2002; Singh and Choi 2003). However, the heterozygosity of pnrvx6 (pnrvx6/+) prevented the loss of the ventral eye phenotype in nearly 22% of the L LOF clones (Figure 7H; clone boundary marked by dotted line; inset shows ELAV-positive photoreceptors in the magnified ventral clone). Similarly, the phenotype of the LOF clones of L in the adult ventral eye was suppressed by the pnrvx6/+ background (Figure 7I, arrow). These results suggest that the levels of pnr are instrumental in the manifestation of the L mutant phenotype and that L and pnr act antagonistically to each other in the eye.
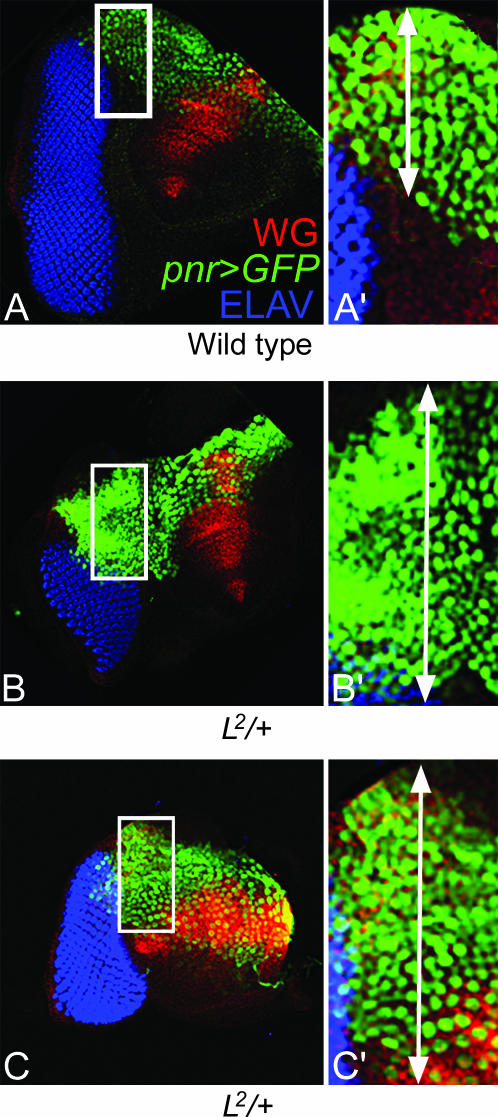
Pnr expression broadens in the L mutant background:
Pnr is expressed in the dorsal margin of the eye in the peripodial membrane (Figure 8A) (Maurel-Zaffran and Treisman 2000; Singh and Choi 2003). One possible mechanism for L-mediated inhibition of pnr function may be restricting the pnr expression domain to the dorsal margin. We tested this hypothesis by checking pnr expression in the L mutant background. We used the pnr-GFP reporter gene construct to examine pnr expression in the eye disc (Pichaud and Casares 2000; Singh and Choi 2003). The GFP-positive nuclei of the pnr-expressing cells were counted in wild-type and L mutant eye discs for approximate quantitation of the pnr expression domain. We found that in the L2/+ mutant background pnr expression in the dorsal margin broadens significantly (Figure 8, B and C). On the basis of the average taken from 17 wild-type eye discs, pnr expression was confined to 16 ± 3 cell widths from the dorsal margin. In contrast, the pnr expression domain extends to 29 ± 4 cells in the L mutant discs (on the basis of the average from 16 eye discs). This corresponds to a nearly 1.8-fold increase in the width of the pnr expression domain. We also used the “image J” program to count the number of nuclei in the control vs. experimental eye discs and found similar results. Other L mutant alleles also showed similar expansion of the pnr expression domain (data not shown). In the wild-type eye disc, the pnr expression in a small group of dorsal margin cells is sufficient to dorsalize the entire dorsal domain by activating Wg signaling (Maurel-Zaffran and Treisman 2000). Thus, the near 1.8-fold expansion of the Pnr expression domain in L mutant eye discs may be sufficient to affect the ventral domain, resulting in the loss of ventral eye as seen in L mutants.
Figure 8.
L restricts the expression domain of pnr. (A) pnr>GFP expression (green) marks the dorsal lateral margin of the third instar eye imaginal disc. (A′) In the dorsal eye margin, the pnr expression stripe is composed of approximately 16 ± 3 cells, based on an average from 17 eye discs. (B and C) In the L2/+ mutant background, the pnr expression domain is broadened. (B′ and C′) Thickness of the pnr expression stripe in the dorsal eye margin, which comprises nearly 29 ± 4 cells, based on an average from 11 eye discs, shows an ∼1.8 fold increase from that in the wild-type pnr expression domain.
Time window of Pnr and L interaction:
We looked for the time window when the antagonistic interaction between Pnr and L is crucial for DV patterning of the eye. We blocked the activity of pnr during different developmental time windows by misexpressing U-shaped (Ush), an inhibitor of Pnr activity. Ush, a zinc-finger protein that is normally not expressed in the eye (Maurel-Zaffran and Treisman 2000; Fossett et al. 2001), dimerizes with Pnr and acts as a negative regulator of Pnr transcriptional activity (Haenlin et al. 1997). Misexpression of ush in the eye has been used to inhibit pnr activity. It was shown earlier that ey-GAL4 used to drive UAS-ush (ey>ush) generates no-eye phenotypes in 80% of the flies whereas 20% of the flies show extremely small eyes (Fosset et al. 2001; Singh and Choi 2003). We exploited the temperature sensitivity of the GAL4 driver (Brand and Perrimon 1993; Cho and Choi 1998; Kumar and Moses 2001; Singh and Choi 2003) to misexpress ush conditionally at desired times during eye development (see materials and methods).
To conditionally regulate pnr activity, we used 29° (red lines in Figure 9A) and 18° (blue lines in Figure 9A) for high and low levels of pnr expression, respectively. For controls, we maintained the cultures of ey>ush at 29° throughout development (Figure 9A, experiment 1), which resulted in a very small eye field, whereas the antennal field was relatively normal (Figure 9B). When pnr activity was blocked throughout eye development in the L2/+ mutant background (Figure 9A, experiment 2), the frequency of the no-eye phenotype as seen in the ey>ush eye imaginal discs increased from 80 to nearly 98% (Figure 9C) and the adult flies failed to eclose.
In experimental samples, we blocked pnr activity by exposing each batch of cultures to a single shift at 29° in a 24-hr time window. This allowed us to test the effects of temperature shifts in different time windows extending from embryo to late third instar larval stages. We found that the L2/+ mutant phenotype of ventral eye loss was not affected when pnr activity was blocked from embryogenesis until the first instar of larval development (Figure 9A, experiment 3) or during the third instar of larval development (Figure 9A, experiment 5) and resulted in a phenotype similar to that of the L2/+ mutant phenotype of ventral eye loss (Figure 9D). However, when pnr function was blocked during the second instar stage (Figure 9A, experiment 4), the L2/+ mutant phenotype was significantly rescued in the eye (Figure 9E). We also found similar results by overexpressing PnrEnR, a dominant-negative form of Pnr (Klinedinst and Bodmer 2003), which blocks pnr activity in the eye discs (data not shown). These results suggest that the second instar of larval development is the time window for mutually antagonistic interaction between L and Pnr during DV patterning of eye.
Iro-C genes act antagonistically to L:
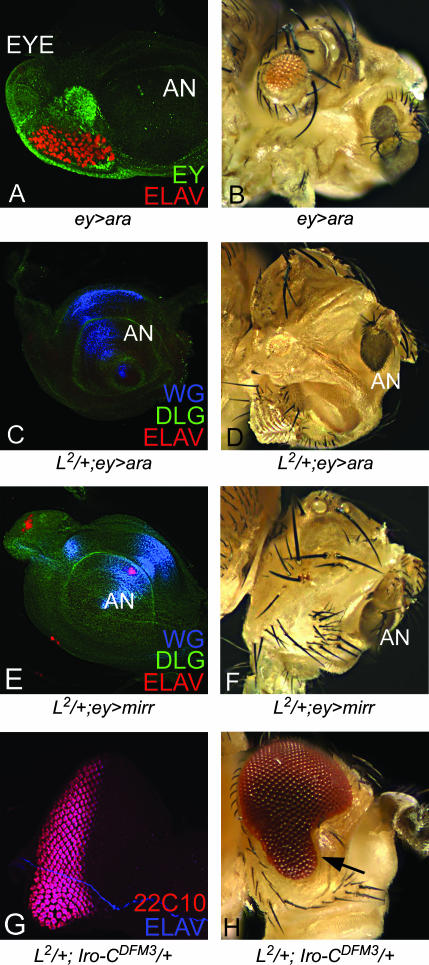
We tested whether the antagonistic relation of L and pnr is exclusive or if it extends to other downstream dorsal eye selectors such as members of Iro-C. Since all three members of Iro-C showed similar effects upon overexpression, we present the overexpression phenotype of ey>ara as a representative control, which shows small eye phenotypes in nearly 33% of eye discs and adult eye (Figure 10, A and B). Overexpression of ara (Figure 10, C and D) or mirr (Figure 10, E and F) using ey-GAL4 in the L2/+ mutant background resulted in no-eye phenotype in the disc and in the adult with high penetrance. In the case of ara overexpression (L2/+; ey>ara), ∼96% (67/70) of the flies showed similar no-eye phenotypes.
Figure 10.
Members of Iro-C act as antagonists of L function in the ventral eye. (A and B) Overexpression of ara in eye (ey>ara) results in small eye as seen in the eye imaginal disc (A) and in the adult eye (B). In the L2/+ mutant background, ey-GAL4-driven overexpression of ara (L2/+; ey>ara) (C and D) and mirr (L2/+; ey>mirr) (E and F) results in elimination of the eye field in eye disc as well as in adult eye. (G and H) Trans-heterozygous combination of L2/+; Iro-CDFM3/+, a deficiency uncovering ara, caup, and the promoter of mirr rescues the ventral eye loss phenotype of L mutant in eye disc (G) and in adult eye (H). 22C10 is a neuronal marker marking photoreceptor clusters.
We also tested whether the eye phenotype of the L2/+ mutant can be suppressed by reducing the Iro-C function. Iro-CDFM3 deficiency, which uncovers ara, caup, and mirr, was used to reduce the dosage of all three Iro-C genes (Gomez-Skarmeta et al. 1996). We found that the loss of the ventral eye phenotype in the L2/+ mutant eye disc (Figure 7A) as well as in the adult eye (Figure 1E) could be partially rescued in the L2/+; Iro-C DFM3/+ background (Figure 10, G and H). Nearly 18% (7/39) of the flies showed rescue of the L2/+ eye phenotype. The homozygous phenotype of a weaker allele, Lsi, was suppressed at a higher frequency (64%, 32/50 flies observed). These results suggest that L function is antagonistic to Pnr as well as to Iro-C. Our studies demonstrate that the mutually antagonistic relationship between the dorsal eye selectors and the ventral eye growth control genes is crucial for regulating the DV patterning in the eye. The time window for their antagonistic relationship is during the second instar of larval eye development and at that stage optimum levels of these genes are required for DV patterning of the eye.
DISCUSSION
Axial patterning plays a crucial role in organizing growth and in differentiating developing fields. To understand how the DV pattern is established in the Drosophila eye, we analyzed the genetic relationships between dorsal and ventral eye genes. We also identified a group of new genes that modify the L mutant eye phenotype not only by misexpression but also by reduced gene function.
fng act as a modifier of L function:
In the early eye disc, fng is preferentially expressed in the ventral eye (Cho and Choi 1998). The DV domain specification by Fng is also important for growth of the eye disc as its ubiquitous overexpression in the eye disc blocks eye development (Cho and Choi 1998). Even though L and fng play important roles during ventral eye growth and patterning, the developmental interaction between the two has been unknown. Here, we showed that overexpression of fng can partially compensate for the loss of L gene function in the eye (Figure 2, H and I). This suggests that fng works either downstream or parallel to L in the growth of the ventral eye. It is possible that L and fng interact through the induction of a common target, Ser, in the eye (Figure 11).
Figure 11.
A model for early DV patterning in Drosophila eye. Pnr expression in the dorsal eye disc margin induces Wg, which is required for expression of Iro-C genes in the dorsal half of the eye disc (Heberlein et al. 1998; Maurel-Zaffran and Treisman 2000). Iro-C genes restrict Fng expression to the ventral eye and establish dorsal and ventral domains (Cho and Choi 1998; Dominguez and de Celis 1998). Our results show that pnr and L act antagonistically to each other and restrict their functional domains to the dorsal and ventral eye, respectively. The antagonistic behavior of Pnr can be direct or mediated through Wg signaling. Partial rescue of the L mutant phenotype by overexpression of fng suggests that L may act upstream of fng during early DV patterning. In the ventral eye, fng and L can regulate Ser expression (Chern and Choi 2002). (Earlier known relations are shown in black whereas new additions from our studies are shown in red.) Arrows do not necessarily indicate direct actions.
Wg signaling pathway negatively affects the ventral eye growth function of L:
Like several other pathways, Wg signaling has multiple functions during eye development. We identified Sgg, a serine/threonine kinase (also know as Zeste-white 3), as a modifier that suppresses the L mutant phenotype upon overexpression (Figure 2, K and L). Sgg is known to inhibit the Wg signaling pathway by downregulating Armadillo (Arm) via ubiquitin-mediated proteosomal degradation (Peifer et al. 1994; Siegfried et al. 1994; Aberle et al. 1997). We also identified other components of the Wg signaling pathway such as pygo (Figure 4, B and C) and dally as modifiers, which, upon overexpression in the eye, enhanced the L mutant phenotype (Table 1). Our results suggest that Wg signaling acts antagonistically to L function in the ventral eye. The genetic interaction of these EP lines with the L mutations represents specific enhancement rather than additive effects, since antagonists of Wg signaling were identified as suppressors, whereas members required for Wg signaling were identified as enhancers of the L mutant phenotype in our EP screen.
Gain of Hh and Dpp signaling suppresses the L mutant phenotype:
In this screen, we found that the overexpression of Daughters against Dpp (Dad), an antagonist of Dpp signaling (Tsuneizumi et al. 1997), enhances the L mutant phenotype (data not shown), whereas EP insertions at hh and its receptor gene smo were identified as suppressors of the L mutant phenotype. The members of these two signaling pathways are known to be involved in eye growth and differentiation (Greenwood and Struhl 1999). These results raise another interesting possibility of the possible role of Hh and Dpp signaling pathways in early eye growth and patterning.
During Drosophila eye development, Hh controls progression of the furrow by inducing the expression of dpp and atonal (ato), a proneural gene responsible for R8 photoreceptor formation (Jarman et al. 1994; Dominguez 1999). In the eye, hh and dpp are involved in a positive feedback loop for the initiation and movement of the MF whereas Wg signaling acts antagonistically to Dpp signaling to block MF movement and progression (Ma and Moses 1995; Treisman and Rubin 1995; Royet and Finkelstein 1997). This antagonistic relation may be present even during early eye development (Royet and Finkelstein 1997; Kenyon et al. 2003) since wg and dpp are localized to opposing regions of the undifferentiated younger eye primordia: dpp along the posterior margin and wg across the dorsal anterior region (Cho et al. 2000). These results suggest that the early function of Hh and Dpp signaling is to promote L-mediated ventral eye growth whereas Wg signaling acts as an antagonist.
Other modifiers of the L mutant phenotype:
We identified BarH1 and BarH2 as enhancers of the L mutant phenotype. BarH1 and BarH2 are a pair of homeobox proteins that express in a subset of photoreceptors (Higashijima et al. 1992) and in the basal undifferentiated cells of the eye disc (Lim and Choi 2003). B is required for the negative regulation of eye development by repressing the expression of the proneural gene ato (Lim and Choi 2003). However, it is not known whether B plays a role in early eye growth, prior to retinal differentiation. Clonal analysis has not yet revealed evidence for B function in DV asymmetric eye patterning. However, our data showed genetic interactions of L mutants with GOF and LOF mutants of B (Figure 4, H and I; Figure 5, C and D). The suppression of the L2/+ eye phenotype by a LOF mutation of B (B−/+; L2/+), which by itself (B−/+) has no defects in the eye, raises the possibility that B itself may not have DV asymmetric function but needs to be downregulated by L for normal growth of the early eye disc. This is also consistent with the dramatic eye reduction observed when ey-GAL4 drive B (ey>B) is overexpressed during early eye development (Figure 4, G–I). It has been shown that Wg and B have both positive and negative regulatory relationships in prepatterning of the notum. B expression is activated by Wg in the scutum whereas B represses Wg expression in the most anterior part of the notum (Sato et al. 1999). L mutants respond to GOF and LOF of both Wg signaling and B in a similar fashion, suggesting that Wg and B may be regulating each other positively during early eye development.
L is known to act downstream of N (Chern and Choi 2002). In the eye, emc and h, the repressors of ato, are downregulated by N (Baonza and Freeman 2001). During eye development, emc acts in collaboration with hairy (h) as the negative regulator of the morphogenetic furrow by repressing ato (Brown et al. 1995). Therefore, identification of emc as an antagonist of L-mediated early ventral eye growth seems possible. Interestingly, both emc and B have also been identified as modifiers of pnr (Pena-Rangel et al. 2002).
Some of the genes that we have identified as L modifiers, such as B, emc, and smo, have been well characterized, but their roles in early eye disc growth and/or DV asymmetric function have not been studied. We also identified genes involved in cell survival and growth such as disc over grown (dco), a member of the serine/threonine protein kinases family (Kloss et al. 1998), and genes involved in vesicular trafficking, including RhoGAP68F, an ion transport such as nrv 1 (Sun and Salvaterra 1995), and the acetyl transferase nej (Kumar et al. 2004). It is possible that potential DV asymmetric function of these genes might have been missed by LOF analysis because of functional redundancy or these genes may be modifying the early growth function of L in the eye. More in-depth studies will be necessary to explore these possibilities. However, it is important to note that both GOF and LOF of these genes exhibit specific genetic interactions with L mutant backgrounds. In addition to the well-characterized genes, we have identified a few novel genes like EP1229 and EP1595 whose functions are not known. These genes were not listed here as we have not tested the specificity of their genetic interaction with L by using LOF mutations.
Antagonistic relation of dorsal eye selectors and ventral eye growth genes:
Our results demonstrate that the level of pnr gene function is a crucial factor for DV patterning of the eye as increased levels of pnr gene function enhance the L mutant phenotype of ventral eye loss to no eye (Figure 6, C and D), whereas reduction of pnr gene function rescued the loss of the ventral eye phenotype of the L mutant (Figure 7, B and C). Further, the phenotypes of LOF clones of L where only the ventral cells are lost (Figure 7G; Singh and Choi 2003) can be rescued by reducing the levels of pnr gene function (Figure 7, H and I). These results suggest that pnr acts antagonistically to the ventral eye growth function of L. However, we also found that the antagonism of pnr and L is mutual (Figure 11). This conclusion is based on the fact that the gain-of-function phenotype of pnr in the eye is significantly enhanced when L function is reduced (Figure 6). We also validated our conclusions by showing that the dorsal eye enlargements associated with LOF clones of pnr can be prevented by reducing the levels of L gene function (Figure 7F). These results suggest that optimal levels of pnr and L are necessary for DV patterning and growth of the eye. We also found that the downstream dorsal eye selectors, Iro-C members (ara, caup, mirr) are involved in a mutually antagonistic relationship with L (Figure 10). Our studies demonstrate that the antagonism of L holds true for key components involved in dorsal fate selection during early eye development.
Developmental time window of the pnr and L antagonistic relationship:
We also identified the time window of the second instar of larval development during which mutual antagonistic interaction of L and pnr is required for DV patterning and growth in the eye (Figure 9). Previously, we have shown that the pnr function in eye development is critically required during the second instar larval stage (Singh and Choi 2003). This time window is coincident with the one that is required for the antagonism of pnr and L as shown in this study, suggesting that a major function of L in early eye development is to establish the DV domains by negatively regulating the dorsal selectors. Our studies also support the physiological relevance of this mutually antagonistic interaction in DV patterning.
It is not known how L antagonizes Pnr function. One possibility is that L may be required for restricting the pnr expression domain to the dorsal margin of the eye disc. It was difficult to check whether L is cell-autonomously required for pnr repression because LOF clones of L result in the elimination of the entire or ventral eye, depending on the time when the clones are generated (Singh and Choi 2003). Alternatively, we studied the effect of a L mutation on pnr expression (Figure 8, B and C). Interestingly, pnr expression, which is restricted to the dorsal eye margin in wild type eye discs (Figure 8A), shows a nearly twofold expansion in L2/+ mutant discs (Figure 8, B and C). It remains to be studied whether L is required for the repression of pnr expression or for the inhibition of growth of pnr-expressing cells. On the basis of our data we suggest that during early DV patterning, the onset of pnr expression might restrict the functional domain of L and Ser to the ventral eye. It is possible that pnr may also suppress L gene function via the Wg signaling pathway (Figure 11).
Our results support the view that various developmental pathways cross-talk with each other to define the final form of a developing eye field. Such genes are likely to interact with both pnr and L. It is interesting to note that we identified several pnr-interacting genes (Table 1) as L modifiers in our screen. This illustrates the importance of the interaction of L and pnr pathways and also the efficacy of our screen. Further study of new modifiers of L may provide important clues to the mechanism of pnr-L interactions in the control of growth and/or DV patterning of the eye. Since the compound eye of Drosophila shares some similarities with the vertebrate eye (Hartenstein and Reh 2002; Peters and Cepko 2002) and genetic machinery is highly conserved, it would be interesting to see if these antagonistic interactions between the dorsal eye selectors and the ventral eye genes play roles in the DV patterning and growth of vertebrate eyes.
Acknowledgments
We thank the European Drosophila Research Center and the Bloomington Stock Center for the Drosophila strains; G. Struhl, G. Morata, J. Treisman, Y. Henry Sun, U. Walldorf, and K. Cho for fly stocks; and the Developmental Studies Hybridoma Bank for the antibodies. We thank K. Cho, G. Roman, M. Kango-Singh, members of our lab, and two unknown referees for their thoughtful comments to improve the quality of the manuscript. We thank J. Barish for technical support in scanning electron microscopy. Confocal microscopy was supported by a Core Grant for Vision Research from the National Institutes of Health (NIH) to D. B. Jones. This work was supported by an NIH grant to K.-W. Choi.
References
- Abdelilah-Seyfried, S., Y. M. Chan, C. Zeng, N. J. Justice, S. Younger-Shepherd et al., 2001. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 157: 455–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberle, H., A. Bauer, J. Stappert, A. Kispert and R. Kemler, 1997. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16: 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, W. K., 1978. A clonal analysis reveals early developmental restrictions in the Drosophila head. Dev. Biol. 62: 447–463. [DOI] [PubMed] [Google Scholar]
- Baonza, A., and M. Freeman, 2001. Notch signalling and the initiation of neural development in the Drosophila eye. Development 128: 3889–3898. [DOI] [PubMed] [Google Scholar]
- Blair, S. S., 2003. a Genetic mosaic techniques for studying Drosophila development. Development 130: 5065–5072. [DOI] [PubMed] [Google Scholar]
- Blair, S. S., 2003. b Lineage compartments in Drosophila. Curr. Biol. 13: R548–R551. [DOI] [PubMed] [Google Scholar]
- Bourouis, M., P. Moore, L. Ruel, Y. Grau, P. Heitzler et al., 1990. An early embryonic product of the gene shaggy encodes a serine/threonine protein kinase related to the CDC28/cdc2+ subfamily. EMBO J. 9: 2877–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brown, N. L., C. A. Sattler, S. W. Paddock and S. B. Carroll, 1995. Hairy and emc negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell 80: 879–887. [DOI] [PubMed] [Google Scholar]
- Calleja, M., E. Moreno, S. Pelaz and G. Morata, 1996. Visualization of gene expression in living adult Drosophila. Science 274: 252–255. [DOI] [PubMed] [Google Scholar]
- Cavodeassi, F., R. Diez Del Corral, S. Campuzano and M. Dominguez, 1999. Compartments and organising boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development 126: 4933–4942. [DOI] [PubMed] [Google Scholar]
- Chern, J. J., and K.-W. Choi, 2002. Lobe mediates Notch signaling to control domain-specific growth in the Drosophila eye disc. Development 129: 4005–4013. [DOI] [PubMed] [Google Scholar]
- Cho, K. O., and K.-W. Choi, 1998. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature 396: 272–276. [DOI] [PubMed] [Google Scholar]
- Cho, K. O., J. Chern, S. Izzadost and K.-W. Choi, 2000. Novel signaling from the peripodial membrane is essential for eye disc patterning in Drosophila. Cell 103: 331–342. [DOI] [PubMed] [Google Scholar]
- Choi, K.-W., B. Mozer and S. Benzer, 1996. Independent determination of symmetry and polarity in the Drosophila eye. Proc. Natl. Acad. Sci. USA 93: 5737–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. M., 1993. Imaginal disc development, pp. 747–841 in The Development of Drosophila melanogaster, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Curtiss, J., G. Halder and M. Mlodzik, 2002. Selector and signalling molecules cooperate in organ patterning. Nat. Cell Biol. 4: E48–E51. [DOI] [PubMed] [Google Scholar]
- Dominguez, M., 1999. Dual role for hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development 126: 1175–1187. [DOI] [PubMed] [Google Scholar]
- Dominguez, M., and F. Casares, 2005. Organ specification-growth control connection: new in-sights from the Drosophila eye-antennal disc. Dev. Dyn. 232: 673–684. [DOI] [PubMed] [Google Scholar]
- Dominguez, M., and J. F. de Celis, 1998. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature 396: 276–278. [DOI] [PubMed] [Google Scholar]
- Fossett, N., S. G. Tevosian, K. Gajewski, Q. Zhang, S. H. Orkin et al., 2001. The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse, N., S. Hirose and S. Hayashi, 1994. Diploidy of Drosophila imaginal cells is maintained by a transcriptional repressor encoded by escargot. Genes Dev. 8: 2270–2281. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta, J. L., R. Diez Del Corral, E. De La Calle-Mustienes, D. Ferre-Marco and J. Modolell, 1996. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 85: 95–105. [DOI] [PubMed] [Google Scholar]
- Greenwood, S., and G. Struhl, 1999. Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development 126: 343–354. [DOI] [PubMed] [Google Scholar]
- Haenlin, M., Y. Cubadda, F. Blondeau, P. Heitzler, Y. Lutz et al., 1997. Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 11: 3096–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines, N., and K. D. Irvine, 2003. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 10: 786–797. [DOI] [PubMed] [Google Scholar]
- Halder, G., P. Callaerts, S. Flister, U. Walldorf, U. Kloter et al., 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125: 2181–2191. [DOI] [PubMed] [Google Scholar]
- Hartenstein, V., and T. A. Reh, 2002. Homologies between vertebrate and invertebrate eyes, pp. 219–251 in Drosophila Eye Development, edited by K. Moses. Springer-Verlag, Heidelberg, Germany. [DOI] [PubMed]
- Hayashi, S., S. Hirose, T. Metcalfe and A. D. Shirras, 1993. Control of imaginal cell development by the escargot gene of Drosophila. Development 118: 105–115. [DOI] [PubMed] [Google Scholar]
- Hazelett, D. J., M. Bourouis, U. Walldorf and J. E. Treisman, 1998. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 125: 3741–3751. [DOI] [PubMed] [Google Scholar]
- Heberlein, U., E. R. Borod and F. A. Chanut, 1998. Dorsoventral patterning in the Drosophila retina by wingless. Development 125: 567–577. [DOI] [PubMed] [Google Scholar]
- Heitzler, P., M. Haenlin, P. Ramain, M. Calleja and P. Simpson, 1996. A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Genetics 143: 1271–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima, S., T. Kojima, T. Michiue, S. Ishimaru, Y. Emori et al., 1992. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 6: 50–60. [DOI] [PubMed] [Google Scholar]
- Huang, A. M., and G. M. Rubin, 2000. A misexpression screen identifies genes that can modulate RAS1 pathway signaling in Drosophila melanogaster. Genetics 156: 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman, A. P., E. H. Grell, L. Ackerman, L. Y. Jan and Y. N. Jan, 1994. atonal is the proneural gene for Drosophila photoreceptors. Nature 369: 398–400. [DOI] [PubMed] [Google Scholar]
- Kenyon, K. L., S. S. Ranade, J. Curtiss, M. Mlodzik and F. Pignoni, 2003. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell 5: 403–414. [DOI] [PubMed] [Google Scholar]
- Klinedinst, S. L., and R. Bodmer, 2003. Gata factor Pannier is required to establish competence for heart progenitor formation. Development 130 (13): 3027–3038. [DOI] [PubMed] [Google Scholar]
- Kloss, B., J. L. Price, L. Saez, J. Blau, A. Rothenfluh et al., 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase I epsilon. Cell 94: 97–107. [DOI] [PubMed] [Google Scholar]
- Kraut, R., K. Menon and K. Zinn, 2001. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11: 417–430. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., and K. Moses, 2001. The EGF receptor and notch signaling pathways control the initiation of the morphogenetic furrow during Drosophila eye development. Development 128: 2689–2697. [DOI] [PubMed] [Google Scholar]
- Kumar, J. P., T. Jamal, A. Doetsch, F. R. Turner and J. B. Duffy, 2004. CREB binding protein functions during successive stages of eye development in Drosophila. Genetics 162: 877–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J., and K.-W. Choi, 2003. Bar homeodomain proteins are anti-proneural in the Drosophila eye: transcriptional repression of atonal by Bar prevents ectopic retinal neurogenesis. Development 130: 5965–5974. [DOI] [PubMed] [Google Scholar]
- Ma, C., and K. Moses, 1995. Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development 121: 2279–2289. [DOI] [PubMed] [Google Scholar]
- Mann, R. S., and S. B. Carroll, 2002. Molecular mechanisms of selector gene function and evolution. Curr. Opin. Genet. Dev. 12: 592–600. [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran, C., and J. E. Treisman, 2000. pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development 127: 1007–1016. [DOI] [PubMed] [Google Scholar]
- McNeill, H., C. H. Yang, M. Brodsky, J. Ungos and M. A. Simon, 1997. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes Dev. 11: 1073–1082. [DOI] [PubMed] [Google Scholar]
- Milan, M., and S. M. Cohen, 2003. A re-evaluation of the contributions of Apterous and Notch to the dorsoventral lineage restriction boundary in the Drosophila wing. Development 130: 553–562. [DOI] [PubMed] [Google Scholar]
- Moloney, D. J., V. M. Panin, S. H. Johnston, J. Chen, L. Shao et al., 2000. Fringe is a glycosyltransferase that modifies Notch. Nature 406: 369–375. [DOI] [PubMed] [Google Scholar]
- Newsome, T. P., B. Asling and B. J. Dickson, 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127: 851–860. [DOI] [PubMed] [Google Scholar]
- Pai, C. Y., T. S. Kuo, T. J. Jaw, E. Kurant, C. T. Chen et al., 1998. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 12: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos, V., A. Tomlinson, V. M. Panin, C. Rauskolb and K. D. Irvine, 1998. Dorsal-ventral signaling in the Drosophila eye. Science 281: 2031–2034. [DOI] [PubMed] [Google Scholar]
- Peifer, M., D. Sweeton, M. Casey and E. Wieschaus, 1994. wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development 120: 369–380. [DOI] [PubMed] [Google Scholar]
- Pena-Rangel, M. T., I. Rodriguez and J. R. Riesgo-Escovar, 2002. A misexpression study examining dorsal thorax formation in Drosophila melanogaster. Genetics 160: 1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, M. A., and C. L. Cepko, 2002. The dorsal-ventral axis of the neural retina is divided into multiple domains of restricted gene expression which exhibit features of lineage compartments. Dev. Biol. 251: 59–73. [DOI] [PubMed] [Google Scholar]
- Pichaud, F., and F. Casares, 2000. homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech. Dev. 96: 15–25. [DOI] [PubMed] [Google Scholar]
- Rorth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth, P., K. Szabo, A. Bailey, T. Laverty, J. Rehm et al., 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Royet, J., and R. Finkelstein, 1997. Establishing primordia in the Drosophila eye-antennal imaginal disc: the roles of decapentaplegic, wingless and hedgehog. Development 124: 4793–4800. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., M. D. Yandell, J. R. Wortman, G. L. Gabor Miklos, C. R. Nelson et al., 2000. Comparative genomics of the eukaryotes. Science 287: 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., T. Kojima, T. Michiue and K. Saigo, 1999. Bar homeobox genes are latitudinal prepattern genes in the developing Drosophila notum whose expression is regulated by the concerted functions of decapentaplegic and wingless. Development 126: 1457–1466. [DOI] [PubMed] [Google Scholar]
- Siegfried, E., E. L. Wilder and N. Perrimon, 1994. Components of wingless signalling in Drosophila. Nature 367: 76–80. [DOI] [PubMed] [Google Scholar]
- Singh, A., and K.-W. Choi, 2003. Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development 130: 6351–6360. [DOI] [PubMed] [Google Scholar]
- Singh, A., M. Kango-Singh and Y. H. Sun, 2002. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development 129: 4271–4280. [DOI] [PubMed] [Google Scholar]
- Singh, A., M. Kango-Singh, K.-W. Choi and Y. H. Sun, 2004. Dorsal-ventral asymmetric functions of teashirt in Drosophila eye development depend on spatial cues provided by early DV patterning genes. Mech. Dev. 121: 365–370. [DOI] [PubMed] [Google Scholar]
- Sun, B., and P. M. Salvaterra, 1995. Two Drosophila nervous system antigens, Nervana 1 and 2, are homologous to the beta subunit of Na+,K(+)-ATPase. Proc. Natl. Acad. Sci. USA 92: 5396–5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, B., F. Townsley, R. Rosin-Arbesfeld, H. Musisi and M. Bienz, 2002. A new nuclear component of the Wnt signalling pathway. Nat. Cell Biol. 4: 367–373. [DOI] [PubMed] [Google Scholar]
- Treisman, J. E., and G. M. Rubin, 1995. wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development 121: 3519–3527. [DOI] [PubMed] [Google Scholar]
- Tseng, A. S., and I. K. Hariharan, 2002. An overexpression screen in Drosophila for genes that restrict growth or cell-cycle progression in the developing eye. Genetics 162: 229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneizumi, K., T. Nakayama, Y. Kamoshida, T. B. Kornberg, J. L. Christian et al., 1997. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature 389: 539–551. [DOI] [PubMed] [Google Scholar]
- Wolff, T., and D. F. Ready, 1993. Pattern formation in the Drosophila retina, pp. 127–132 in The Development of Drosophila melanogaster, edited by M. Bate and A. Martinez-Arias. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Xu, T., and G. M. Rubin, 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zhong, W., H. Feng, F. E. Santiago and E. T. Kipreos, 2003. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423: 885–889. [DOI] [PubMed] [Google Scholar]