Abstract
The dominance of deleterious mutations has important consequences for phenomena such as inbreeding depression, the evolution of diploidy, and levels of natural genetic variation. Kacser and Burns' metabolic theory provides a paradigmatic explanation for why most large-effect mutations are recessive. According to the metabolic theory, the recessivity of large-effect mutations is a consequence of a diminishing-returns relationship between flux through a metabolic pathway and enzymatic activity at any step in the pathway, which in turn is an inevitable consequence of long metabolic pathways. A major line of support for this theory was the demonstration of a negative correlation between homozygous effects and dominance of mutations in Drosophila, consistent with a central prediction of the metabolic theory. Using data on gene deletions in yeast, we show that a negative correlation between homozygous effects and dominance of mutations exists for all major categories of genes analyzed, not just those encoding enzymes. The relationship between dominance and homozygous effects is similar for duplicated and single-copy genes and for genes whose products are members of protein complexes and those that are not. A complete explanation of dominance therefore requires either a generalization of Kacser and Burns' theory to nonenzyme genes or a new theory.
MOST major-effect mutations are recessive; i.e., the wild-type allele is almost always the dominant allele (Fisher 1928; Wright 1934; Simmons and Crow 1977; Orr 1991; Nanjundiah 1993; Wilkie 1994). This is central to several important evolutionary phenomena. Recessive deleterious mutations are a major cause for the phenomenon of inbreeding depression (Charlesworth and Charlesworth 1999), and diploidy may have evolved to mask the effects of recessive deleterious mutations (Kondrashov and Crow 1991). A necessary condition for some genetic load theories of the evolution of sex is that most deleterious mutations should be recessive (Kondrashov 1982; Chasnov 2000). The dominance of deleterious mutations thus represents an important parameter in evolutionary biology.
The rediscovery of Mendel's laws and with it dominance attracted a great deal of attention, including that of the major architects of the modern evolutionary theory. Fisher (1928) made an early attempt to explain the ubiquity of genetic dominance. According to him, new mutations have additive effects when they first occur but gradually become recessive through the accumulation of dominance modifiers, which reflect natural selection against the deleterious heterozygous effects of recurrent mutations. Wright and Haldane criticized Fisher's theory, arguing that unrealistically high selection pressures acting over long periods of time were required for the evolution of dominance modifiers (Wright 1929, 1934; Haldane 1930). Haldane pointed out that the frequency of heterozygotes in a population would be high during the course of fixation of a beneficial allele and that dominance modification was more likely under such circumstances (Haldane 1956). Wright (1934), on the other hand, proposed a physiological theory of dominance, according to which the relationship between a phenotype and gene activity could be described as a hyperbolic curve of diminishing returns. Haldane and Muller agreed with Wright's model and suggested that wild-type alleles with high levels of gene activity are selected to provide a factor of safety against genetic and environmental fluctuations (Haldane 1930, 1939; Muller 1932). Kacser and Burns (1981), using metabolic control analysis (MCA), developed a theoretical model for dominance along the lines of Wright's physiological theory. Kacser and Burns hypothesized—on mathematical grounds and citing abundant empirical data—that the relationship between flux through a long metabolic pathway and enzyme activity at any single step in the pathway is a curve of diminishing returns. They also showed that the wild-type levels of enzyme activity are usually at or near the plateau of the curve. Consequently, reducing enzyme activity by much as 50% has a negligible effect on flux through the pathway. If flux represents a phenotype, the recessivity of mutations is, therefore, an inevitable consequence of long metabolic pathways.
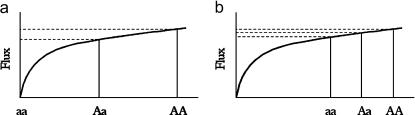
Kacser and Burns's metabolic theory (henceforth referred to as “metabolic theory”) makes a clear prediction about the relationship between dominance (h) and selection coefficients (s) of mutations. (A mutation's selection coefficient is the amount of decrease in fitness it causes in the mutant homozygote, relative to the wild-type homozygote. A mutation's dominance coefficient is the ratio of its heterozygous to homozygous effects. A dominance coefficient of h = 0 means that a mutation is completely recessive, h = 1 means that it is completely dominant, and h = 0.5 means that it has an additive effect.) An outcome of Kacser and Burns's curve relating flux to enzyme activity is that mutations having large fitness effects (large s) should have relatively small effects in heterozygotes (small h) and mutations having small fitness effects (small s) should have approximately additive effects in heterozygotes (large h). To understand the above prediction, first consider a mutation with a large homozygous effect on enzyme activity, which causes a large reduction in the flux through the pathway (Figure 1a). According to Kacser and Burns's curve relating flux to enzyme activity, the flux through the pathway for an intermediate enzyme activity of the heterozygote should be similar to that of the wild type because the level of flux in the heterozygote lies near the plateau of the curve. A mutation of large homozygous effect (large s) should hence be recessive when heterozygous (small h). Now consider a mutation with a small homozygous effect on enzyme activity, which causes a small reduction in the flux through the pathway (Figure 1b). According to the Kacser and Burns's curve relating flux to enzyme activity, the flux through the pathway for an intermediate enzyme activity of the heterozygote should now be intermediate to that of the homozygous mutant and that of the wild type because the level of flux in the heterozygote lies on the linear portion of the curve. A mutation of small homozygous effect (small s) should hence be approximately additive (large h) in the heterozygote. Kacser and Burns's theory thus predicts a negative correlation between the dominance (h) and the homozygous effect (s) of mutations. Wright's physiological theory also implicitly makes the same prediction since it is based on a similar hyperbolic curve of diminishing returns.
Figure 1.
Relationship between flux and the enzyme activity at a single step in a pathway according to the metabolic theory. The wild-type allele is denoted by “A” and the mutant allele by “a.” (a) Case for a mutant allele that causes a large reduction in the enzyme activity. (b) Case for a mutant allele that causes a small reduction in the enzyme activity. See Introduction for a detailed explanation.
In contrast, for Fisher's theory of the evolution of dominance, the intensity of selection on a dominance modifier depends only on the mutation rate and the magnitude of the effect a modifier has on dominance and is independent of the homozygous effects of mutations (Wright 1934). Thus, no correlation between dominance (h) and homozygous (s) effects of mutation is expected under Fisher's theory (Charlesworth 1979). The same argument applies to the evolutionary theories proposed by Haldane (1930; Charlesworth 1979).
Empirical studies of mutations affecting viability in Drosophila (reviewed in Simmons and Crow 1977) have yielded estimates of dominance for lethal mutations of h = 0.02 and for mildly deleterious mutations of h = 0.35. This has been taken as evidence for a negative correlation between dominance and selection coefficients, consistent with Kacser and Burns's prediction. In addition, a survey by Orr (1991) found that most mutations that have visible effects are recessive in artificial diploids of the naturally haploid alga Chlamydomonas. Since selection for dominance modifiers is impossible in a haploid organism, the dominance of the wild-type alleles is unlikely to have resulted from the evolution of modifiers in Chlamydomonas. The metabolic theory has, therefore, gained widespread acceptance (Charlesworth 1979; Orr 1991; Keightley 1996; Porteous 1996; Hartl and Clark 1997; Lynch and Walsh 1998; Bourguet 1999; Turelli and Orr 2000).
Kacser and Burns viewed the organism as an enzyme system consisting of “a large array of specific and saturable catalysts organized into diverging and converging pathways, cycles and spirals all transforming molecular species and resulting in a flow of metabolites” (Kacser and Burns 1981, p. 641). The elegance and generality of their theory appealed widely (Charlesworth 1979; Orr 1991; Keightley 1996; Porteous 1996; Hartl and Clark 1997; Lynch and Walsh 1998; Bourguet 1999; Turelli and Orr 2000; however, see also Cornish-Bowden 1987; Savageau and Sorribas 1989; Savageau 1992; Grossniklaus et al. 1996; Omholt et al. 2000; Bagheri and Wagner 2004). Nonetheless, the theory has a major limitation: because it was derived from consideration of the kinetic properties of enzymes, it does not make predictions about dominance of mutations in nonenzyme genes (see also Wilkie 1994; Omholt et al. 2000). Kacser and Burns argued that the general predictions of their theory would hold for mutations in genes whose products have “quasicatalytic” activity, for which the rate of some process is proportional to gene product concentration. Genes involved in transport or signaling might fall into this class. For “noncatalytic” genes, on the other hand, Kascer and Burns speculated that mutations might act additively. Another reason for predicting that the negative correlation between h and s will extend to categories of genes other than enzymes is that many nonenzymatic genes (e.g., transcription factors, chaperones) influence enzyme concentrations or activity. To the extent that mutations in nonenzyme genes exert their effects on fitness in this manner, they might be expected to have dominance properties in their phenotypic effects similar to those of mutations in enzyme genes.
While there is evidence that mutations in genes encoding structural proteins in both humans and yeast show relatively high dominance (Kondrashov and Koonin 2004), the relationship between h and s has not been systematically quantified for mutations in specific categories of nonenzymatic genes. Here, we use data from precise gene deletions (Steinmetz et al. 2002) in Saccharomyces cerevisiae to study how the relationship between h and s varies across several functional categories of genes. Our key finding is that the relationship between h and s is remarkably similar for precise gene deletions in every gene category, with h declining sharply as s increases. Moreover, the relationship between h and s differs little between duplicate and single-copy genes and between genes whose products physically interact in complexes and those not known to interact in complexes (cf. Papp et al. 2003).
MATERIALS AND METHODS
Steinmetz et al. (2002) measured growth rates of strains with precise deletions of nearly each gene in the yeast genome using a parallel molecular bar-coding strategy. We used their data (available at http://www-deletion.stanford.edu/YDPM/YDPM_index.html) for nonlethal gene deletion strains grown in YPD, YPG, YPDGE, YPE, and YPL media. For these media, two replicate competition experiments were performed for homozygous strains and at least one for heterozygous strains. Although one cannot estimate absolute dominance (h) of deletion mutations from these data sets, one can estimate the relative dominance h* = kh, where k > 1, as described below. Information on relative dominance is sufficient to detect a negative correlation between h and s. We consider only nonlethal mutations for which homozygous and heterozygous growth rate data are available on all media.
The selection coefficient for a mutation is estimated as (Wwt − Whom), and dominance is estimated as (Wwt − Whet)/(Wwt − Whom), where Wwt, Whet, and Whom are wild-type, heterozygous mutant, and homozygous mutant fitness, respectively (Simmons and Crow 1977). A limitation of the yeast deletion data set is that the wild-type progenitor of the deletion strains was not included in the competition experiments. To estimate what the wild-type growth rate would have been in a given replicate, we averaged the growth rates of strains that were in the fastest growing 5% in the independent homozygous replicate on the same medium. For example, to estimate wild-type growth rates in homozygous and heterozygous replicate 1 on YPD medium, the mean growth rates in those replicates of strains with the fastest growth on YPD homozygous replicate 2 were used. Using data from the other replicate to identify control strains is preferable to using data from the same replicate, which would tend to overestimate wild-type growth rates.
With only one or two replicate measurements per strain on a given medium, it is not possible to calculate meaningful estimates of dominance for individual genes. Instead, we estimated the average dominance of deciles of genes by dividing the average heterozygous effect (wild-type growth rate minus heterozygous growth rate) of strains in a decile by the average homozygous effect (wild-type growth rate minus homozygous growth rate) of the decile. Deciles were defined on the basis of the growth rate rankings of strains in the independent homozygous replicate on the same medium.
Our dominance estimates have three potential sources of bias that need to be considered. First, the ratio of average heterozygous effects to average homozygous effects (s) strictly speaking estimates average dominance weighted by s. This weighting is a relatively trivial source of bias, because the deciles necessarily have relatively little variation in s. Second, because the 5% of strains used to estimate wild-type growth rates contained mutations, albeit small-effect ones, we probably slightly underestimated wild-type fitness and thus slightly underestimated heterozygous and homozygous effects of mutations. If we instead use the maximum growth rate observed in a replicate as the estimate of wild-type growth rate in that replicate, a procedure that almost certainly overestimates wild-type growth rate, our conclusions are unchanged. Finally, growth rates of homozygous and heterozygous strains were estimated in separate competition experiments, so that the two types of growth rates are in effect on different scales. Because deletion heterozygotes are more fit on average than deletion homozygotes, it is likely that the heterozygous experiments provided a more competitive environment than the homozygous experiments and therefore amplified small fitness differences relative to the homozygous experiments. For this reason, we consider our estimates of dominance to be estimates of relative dominance h* = kh, where k is probably >1. This is supported by the finding that for genes with small s, average h* is usually >0.5 and sometimes >1 (see results).
To test statistically whether dominance is negatively correlated with s, we calculated Spearman rank correlations between h* for a decile and decile rank for homozygous viability (1–9, with 1 being the lowest; decile 10 was not used, as it included the strains used for estimating wild-type fitness). Because decile ranks were based on one homozygous replicate, while h was calculated from the other, this method avoids possible spurious correlations that could come about from using the same data to estimate both h and s.
The above methods were applied to heterozygous replicates 1 and 2 in YPD medium, to heterozygous replicate 1 in YPG medium, and to the single heterozygous replicates in YPDGE, YPE, and YPL media. Heterozygous replicate 2 in YPG medium was disregarded because growth rates from this replicate showed an anomalous negative correlation with growth rates of the corresponding strains in YPG homozygous replicate 1 (Spearman rank correlation rs = −0.11, P < 0.0001). In contrast, the six other correlations of homozygous replicate 1 (2) with heterozygous replicate 2 (1) in a given medium were all significantly positive (rs = 0.21–0.33, P < 0.0001), as expected. This suggests that some error or artifact may have affected the results of YPG heterozygous replicate 2.
We next categorized genes according to their molecular function as described in the gene ontology database (Gene Ontology Consortium 2000) (http://www.geneontology.org) and analyzed the effects of the gene deletions separately for the different classes of genes. We used the five largest functional categories of genes described for yeast (Kondrashov and Koonin 2004)—enzymes, structural proteins, transcription regulators, binding proteins, and transport proteins. For these analyses, wild-type fitness estimates from the entire data set were used, but deciles were defined on the basis of the subset only. We also examined whether single-copy genes showed a different relationship between h* and s than duplicate genes; for this, we used the list of single-copy and duplicate genes compiled by Gu et al. (2003). Finally, we examined whether genes whose products physically interact with other gene products as parts of complexes showed a different relationship between h* and s than the entire data set showed. Following Papp et al. (2003), we compiled a list of genes whose products physically interact as parts of complexes from the MIPS comprehensive yeast genome database (CYGD) (Mewes et al. 1997) catalog of known protein complexes and from protein complexes detected by high-throughput mass spectrometry using either the TAP procedure (Gavin et al. 2002; Krogan et al. 2004) or yeast two-hybrid screen (Ho et al. 2002). For our analysis, a gene detected as a part of a complex in any of the four data sets was classified as a protein involved in complexes; excluding genes from any individual data set yielded results similar to that of the larger data set. All analyses were done using SAS version 8.
RESULTS
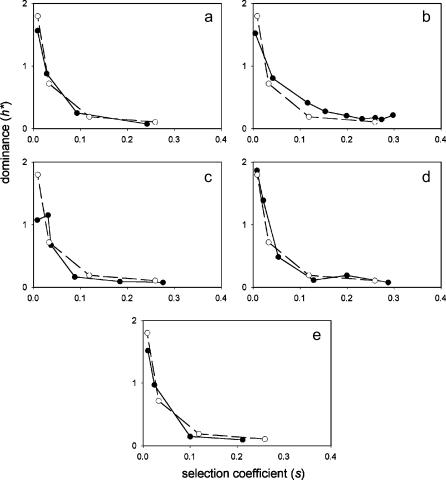
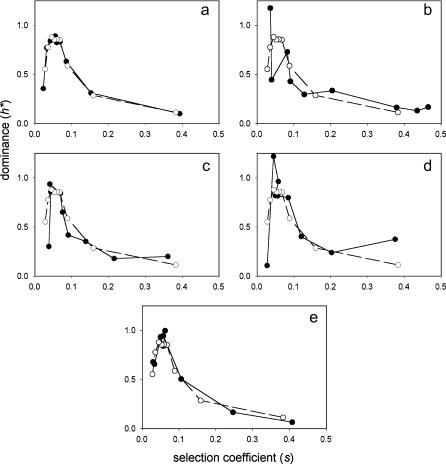
The relationship between relative dominance (h*) and selection coefficients (s) for deletions in five functional categories of genes is shown in Figure 2 for the first replicate on YPD medium and in Figure 3 for YPL medium. Results for the other three media, and the second YPD replicate, are similar and are given in the supplementary figures at http://www.genetics.org/supplemental/. Surprisingly, there is a negative correlation between h* and s for all five categories. All plots closely overlap that for the entire data set, except for structural genes, which tend to show higher h*-values for a given s, particularly for large s (Figures 2 and 3, Figures S1–S4 at http://www.genetics.org/supplemental/). This is consistent with the observation that mutations in structural genes are more often “haplo-insufficient” than mutations in other types of genes (Maroni 2001; Kondrashov and Koonin 2004). (We restrict ourselves to qualitative conclusions about differences between gene categories in average h* for a given s; the nonlinear relationship between the two variables hampers formal statistical comparison of the height of the curves.)
Figure 2.
Dominance of growth rate effects of single-gene deletions in yeast (YPD medium, replicate 1). Solid lines and solid symbols give the results from individual gene ontology (GO) functional categories; dashed lines and open symbols show results for all genes for comparison. (a) Enzyme activity (GO: 0003824). (b) Structural molecule activity (GO: 0005198). (c) Transcription regulator activity (GO: 0030528). (d) Binding activity (GO: 0005488). (e) Transporter activity (GO: 0005215). Points represent the means of deciles of genes, with the deciles defined on the basis of growth rate rankings in an independent replicate. Deciles with mean selection coefficients <0.02 are pooled for clarity. See Table 1 for sample sizes for each gene category.
Figure 3.
Dominance of growth rate effects of single-gene deletions in yeast (YPL medium). See Figure 2 legend for explanation.
Table 1 gives correlations between h* and decile ranking for homozygous growth rate; a positive correlation implies a negative correlation between h and s. For all five functional categories, correlations from each medium are positive, with the most significant at P < 0.05. At least one correlation in each category is significant after controlling for the entire set of tests.
TABLE 1.
Spearman rank correlations between mean dominance (h*) and homozygous fitness rank of deciles
| Gene category | No. of genes | YPD-1 | YPD-2 | YPG-1 | YPL-1 | YPE-1 | YPDGE-1 |
|---|---|---|---|---|---|---|---|
| Enzyme | 1074 | 0.58 | 0.58 | 0.53 | 0.47 | 0.70 | 1.00a |
| Structural | 203 | 0.82 | 0.60 | 0.93a | 0.95a | 0.93a | 0.98a |
| Transcription regulation | 185 | 0.23 | 1.00a | 0.92a | 0.72 | 0.73 | 0.95a |
| Binding | 356 | 0.87a | 0.98a | 0.87a | 0.53 | 0.63 | 0.93a |
| Transport | 298 | 0.43 | 0.50 | 0.45 | 0.50 | 0.53 | 0.88a |
| Singleton genes | 858 | 0.93a | 0.97a | 0.95a | 0.78 | 0.95a | 0.98a |
| Duplicate genes | 974 | 0.40 | 0.00 | 0.37 | 0.63 | 0.75 | 0.83 |
| Involved in complexes | 1715 | 0.85 | 0.92a | 0.85 | 0.70 | 0.82 | 0.98a |
| Not involved in complexes | 2814 | 0.23 | 0.60 | 0.38 | 0.35 | 0.53 | 0.98a |
| All | 4529 | 0.53 | 0.82 | 0.60 | 0.47 | 0.67 | 1.00a |
Critical values (one-tailed, n = 9): rs > 0.467, P < 0.10; rs > 0.583, P < 0.05; rs > 0.767, P < 0.01; rs > 0.900, P < 0.001.
Significant after sequential Bonferroni correction (Rice 1989) for entire set of tests.
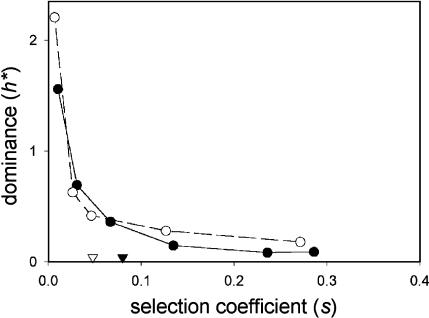
Two other factors that have been hypothesized to influence the dominance of mutations are the presence of gene duplicates (Wagner 2000; Gu et al. 2003) and the extent to which a gene product interacts with other proteins (Veitia 2002; Papp et al. 2003). Using the yeast deletion data set, Gu et al. (2003) found that mutations in single-copy genes have greater homozygous effects than those in duplicate genes, but did not compare the dominance of mutations in the two classes. Using their list of single-copy genes, we find that single-copy genes show a similar relationship between h and s as do duplicate genes (Figure 4, Figure S5 at http://www.genetics.org/supplemental/). Although mutations in single-copy genes have significantly higher s on average (Figure 4, Figure S5; two sample t-tests, P < 0.001 for all media), as Gu et al. (2003) found, the dominance of these genes, for a given s, is similar to that of duplicate genes.
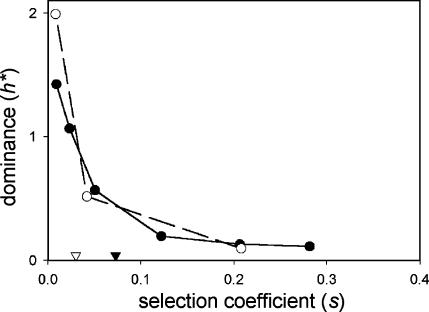
Figure 4.
Dominance of growth rate effects of deletions in single-copy genes (solid line and solid symbols) and all other genes (dashed line and open symbols). Mean s-values for the two categories are indicated by triangles. See Figure 2 for details.
According to the gene dosage balance hypothesis (GDBH) (Veitia 2002), deleterious mutations in genes whose products physically interact with other proteins should be more dominant than those without protein interactors (Papp et al. 2003), as the former are thought to be particularly sensitive to gene dosage. Papp et al. (2003) confirmed this prediction for homozygous lethal mutations in the yeast knockout data set, but did not consider nonlethal mutations. We find that knockout mutations in nonessential genes whose products physically interact with other proteins show similar dominance for a given s as do mutations in genes that do not have known interactors (Figure 5, Figure S6 at http://www.genetics.org/supplemental/). Nonetheless, mutations in genes whose products physically interact with other proteins have significantly greater homozygous effects (s) on average than do mutations in genes with no known physical interactions (Figure 5, Figure S6; P < 0.001 for all media).
Figure 5.
Dominance of growth rate effects of deletions of genes whose products participate in protein complexes (solid line and solid symbols) and all other genes (dashed line and open symbols). Mean s-values for the two categories are indicated by triangles. See Figure 2 for details.
DISCUSSION
Kacser and Burns' metabolic theory is a widely accepted explanation for why most large-effect mutations are recessive (Charlesworth 1979; Orr 1991; Keightley 1996; Porteous 1996; Hartl and Clark 1997; Lynch and Walsh 1998; Bourguet 1999; Turelli and Orr 2000). However, the metabolic theory has not been without criticisms. It has repeatedly been suggested that some of the assumptions of the metabolic theory may not always hold, and the generality of the theory has often been doubted. For example, Cornish-Bowden (1987) showed that pathways in which all enzymes are more than half-saturated are at least theoretically possible, and in such pathways, changes in enzyme concentrations cause substantial changes in metabolic fluxes. According to Cornish-Bowden, such pathways are not common due to natural selection for factors of safety. Savageau (Savageau and Sorribas 1989; Savageau 1992) outlined several cases in which flux through biochemical pathways is sensitive to changes in enzyme concentrations. They rejected the metabolic theory and instead advocated natural selection for robustness against altered enzyme levels as the cause of dominance. Other criticisms of the metabolic theory have also been made (e.g., Grossniklaus et al. 1996; Bourguet 1999; Omholt et al. 2000; Bagheri and Wagner 2004).
Kacser and Burns's theory relates the effects of mutations of differing magnitudes in a single-enzyme gene to the total amount of flux through a metabolic pathway. Their theory thus implicitly concerns a negative correlation between dominance and selection coefficients for different mutations within a gene. Unlike Kacser and Burns, we consider the correlation between h and s among null mutations in different genes. This extension of Kacser and Burns' prediction follows if either or both of two assumptions are made. First, if a function (e.g., catalytic step) can be performed by products of two or more genes (an assumption that probably holds true for a significant fraction of the genes we consider, all of which are nonessential), then knocking out a gene that makes a relatively small (large) contribution to the function should have an effect similar to a small-effect (large-effect) mutation in a single gene essential for a function. Second, even if a single gene (e.g., an enzyme-encoding gene) is essential for a function, the level of functional gene product is likely to be influenced by the products of other genes (e.g., transcription factors); a gene knockout that causes a relatively small (large) reduction in level of the first gene's product should therefore have an effect similar to a small-effect (large-effect) mutation in the gene itself.
Our first finding is that there is a strong negative correlation between dominance and selection coefficients for precise gene deletions in yeast. Most surprisingly, this correlation is not restricted to enzyme genes, as a strict interpretation of Kacser and Burns' metabolic theory would suggest (Hodgkin 1993; Wilkie 1994; Mayo and Burger 1997; Gilchrist and Nijhout 2001). There are at least four possible explanations of this result:
The dominance properties of mutations in nonenzyme genes may mirror those of mutations in enzyme genes because many of the former ultimately exert their phenotypic effects by influencing enzymes. Transport proteins, chaperones, and transcription factors all play roles in determining the concentration of a catalytically active enzyme in a particular cellular location at a particular time; thus mutations in any one of these gene categories could potentially have effects on activity of an enzyme similar to those of mutations in the enzyme gene itself.
Most gene products may function in a quasicatalytic manner sensu Kacser and Burns, such that the rates of nonenzymatic processes involving multiple steps (e.g., transport) can be described by equations similar to those used by Kascer and Burns to model flux through metabolic pathways. This hypothesis predicts that these processes should show a similar diminishing-returns relationship between rate of the process and concentration of individual gene products as does flux through metabolic pathways.
The growth rate of yeast and other organisms may be related to the rate of individual molecular processes (e.g., cell wall synthesis, chromosome replication) by a curve of diminishing returns similar to that describing the relationship between flux through a metabolic pathway and enzyme activity at a particular step. If the role of most nonessential genes is to increase the rate of essential molecular processes (Thatcher et al. 1998), a negative correlation between h and s would be expected even if mutations in the genes have additive effects on the rates of the processes they influence. While we are unaware of data on this subject, it seems likely that a diminishing-returns relationship would exist between the rate of individual molecular processes and growth rate in culture, because as the efficiency of any one process is increased, growth rate would become limited by the rate of other processes.
If large-effect genes, but not small-effect genes, are regulated such that the effects of halving the gene dose are compensated by the production of extra functional gene product (e.g., through feedback regulation), then a negative correlation between h and s would be expected. While it is widely assumed that halving the gene dose results in the production of half the amount of functional gene product, there are scattered examples of nearly complete compensation in heterozygotes (Takahashi et al. 2002, 2005; Hurst and Pal 2005). Nonetheless, the expected halving of gene product is observed in enough examples (Harris 1980) to make us doubt that compensation could provide a complete explanation for our results, although more investigation of this issue is warranted.
More theoretical and empirical work is needed to evaluate the above hypotheses, and doubtless other hypotheses could be advanced. Nonetheless, it is clear that our results cannot be fully explained by the simplest and traditional version of Kacser and Burns's theory.
We also found that mutations in single-copy genes show a similar negative correlation between h and s as do mutations in duplicate genes. Thus, our results are not an artifact of the widespread degree of gene duplication in the yeast genome (Wolfe and Shields 1997; Gu et al. 2002). Furthermore, deletions in genes whose products physically interact in complexes show a similar relationship between h and s as do those that are not known to physically interact in complexes. The GDBH (Papp et al. 2003) predicts that mutations in genes whose products physically interact with other proteins should be more deleterious and show higher dominance than those without protein interactors. Our results lend support to the GDBH in that mutations in genes whose products interact in complexes have larger homozygous effects, but we find little evidence for more dominant effects of mutations in such genes, in contrast to the situation for essential genes (Papp et al. 2003). This suggests that the dominance properties of mutations in essential genes may not be a good guide to those of nonessential genes. These results must, however, be interpreted with some caution since methods for detecting genes whose products interact with other proteins might be biased and may not detect all interactions (von Mering et al. 2002).
In conclusion, our results give further evidence against Fisher's theory for the evolution of dominance, which predicts no correlation between h and s among deleterious mutations (Charlesworth 1979). Our finding of such a correlation for enzyme genes is in agreement with Kacser and Burns's metabolic theory. That a similar correlation appears for other functional categories of genes might be explained by a generalization of Kacser and Burns' theory or might require a different explanation. Nonetheless, other current theories of dominance, such as the gene dosage balance hypothesis, make no predictions about the relationship between h and s. Determining whether Kacser and Burns' theory can be formally generalized to explain the negative correlations between h and s for nonenzyme genes, or whether a new theory of dominance is required, will require additional theoretical and empirical work.
Acknowledgments
We thank H. Allen Orr and Charles Aquadro for valuable suggestions and discussion, Zhenglong Gu for providing the list of single-copy and duplicate genes, Michael Whitlock for his suggestion on the differential regulation of small- and large-effect genes, and Homayoun Bagheri for insightful comments on the manuscript. This work was supported by National Science Foundation grant DEB-0108730 to J.D.F. and National Institutes of Health grant GM51932 to H. A. Orr.
References
- Bagheri, H. C., and G. P. Wagner, 2004. Evolution of dominance in metabolic pathways. Genetics 168: 1713–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguet, D., 1999. The evolution of dominance. Heredity 83: 1–4. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1979. Evidence against Fisher's theory of dominance. Nature 278: 848–849. [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 1999. The genetic basis of inbreeding depression. Genet. Res. 74: 329–340. [DOI] [PubMed] [Google Scholar]
- Chasnov, J. R., 2000. Mutation-selection balance, dominance and the maintenance of sex. Genetics 156: 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden, A., 1987. Dominance is not inevitable. J. Theor. Biol. 125: 335–338. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A., 1928. The possible modification of the response of the wild type to recurrent mutations. Am. Nat. 62: 115–126. [Google Scholar]
- Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch et al., 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium, 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist, M. A., and H. F. Nijhout, 2001. Nonlinear developmental processes as sources of dominance. Genetics 159: 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., M. S. Madhusudhan and V. Nanjundiah, 1996. Nonlinear enzyme kinetics can lead to high metabolic flux control coefficient: implications for the evolution of dominance. J. Theor. Biol. 182: 299–302. [DOI] [PubMed] [Google Scholar]
- Gu, Z., A. Cavalcanti, F.-C. Chen, P. Bouman and W.-H. Li, 2002. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 19: 256–262. [DOI] [PubMed] [Google Scholar]
- Gu, Z., L. M. Steinmetz, X. Gu, C. Scharfe, R. W. Davis et al., 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1930. A note of Fisher's theory of the origin of dominance and linkage. Am. Nat. 64: 87–90. [Google Scholar]
- Haldane, J. B. S., 1939. The theory of the evolution of dominance. J. Genet. 37: 365–374. [Google Scholar]
- Haldane, J. B. S., 1956. The theory of selection for melanism in Lepidoptera. Proc. R. Soc. Lond. Ser. B 145: 303–306. [DOI] [PubMed] [Google Scholar]
- Harris, H., 1980. The Principles of Human Biochemical Genetics, Ed. 3. Elvesier/North-Holland Biomedical Press, Amsterdam.
- Hartl, D. L., and A. G. Clark, 1997. Principles of Population Genetics. Sinauer Associates, Sunderland, MA.
- Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore et al., 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., 1993. Fluxes, doses and poisons—molecular perspectives on dominance. Trends Genet. 9: 1–2. [DOI] [PubMed] [Google Scholar]
- Hurst, L. D., and C. Pal, 2005. Dissecting dispensability. Nat. Genet. 37: 214–215. [DOI] [PubMed] [Google Scholar]
- Kacser, H., and J. A. Burns, 1981. The molecular basis of dominance. Genetics 97: 639–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley, P. D., 1996. A metabolic basis for dominance and recessivity. Genetics 143: 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1982. Selection against harmful mutations in large sexual and asexual populations. Genet. Res. 40: 325–332. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., and J. F. Crow, 1991. Haploidy or diploidy: Which is better? Nature 351: 314–315. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F. A., and E. V. Koonin, 2004. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 7: 287–290. [DOI] [PubMed] [Google Scholar]
- Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw et al., 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13: 225–239. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Maroni, G., 2001. Molecular and Genetic Analysis of Human Traits. Blackwell, London.
- Mayo, O., and R. Burger, 1997. The evolution of dominance: A theory whose time has passed? Biol. Rev. 72: 97–110. [Google Scholar]
- Mewes, H. W., K. Albermann, K. Heumann, S. Liebl and F. Pfeiffer, 1997. MIPS: a database for protein sequences, homology data and yeast genome information. Nucleic Acids Res. 25: 28–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, H. J., 1932. Further studies on the nature and causes of gene mutations. Proc. Sixth Int. Congr. Genet. 1: 213–255. [Google Scholar]
- Nanjundiah, V., 1993. Why are most mutations recessive? J. Genet. 72: 85–97. [Google Scholar]
- Omholt, S. W., E. Plahte, L. Oyehaug and K. Xiang, 2000. Gene regulatory networks generating the phenomena of additivity, dominance and epistasis. Genetics 155: 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1991. A test of Fisher's theory of dominance. Proc. Natl. Acad. Sci. USA 88: 11413–11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp, B., C. Pal and L. D. Hurst, 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197. [DOI] [PubMed] [Google Scholar]
- Porteous, J. W., 1996. Dominance—one hundred and fifteen years after Mendel's paper. J. Theor. Biol. 182: 223–232. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 1989. Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Savageau, M. A., 1992. Dominance according to metabolic control analysis: Major achievement or house of cards? J. Theor. Biol. 154: 131–136. [DOI] [PubMed] [Google Scholar]
- Savageau, M. A., and A. Sorribas, 1989. Constraints among molecular and systematics properties: implications for physiological genetics. J. Theor. Biol. 141: 93–115. [DOI] [PubMed] [Google Scholar]
- Simmons, M. J., and J. F. Crow, 1977. Mutations affecting fitness in Drosophila populations. Annu. Rev. Genet. 11: 49–78. [DOI] [PubMed] [Google Scholar]
- Steinmetz, L. M., C. Scharfe, A. M. Deutschbauer, D. Mokranjac, Z. S. Herman et al., 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31: 400–404. [DOI] [PubMed] [Google Scholar]
- Takahashi, N., H. L. Brooks, J. B. Wade, W. Liu, Y. Kondo et al., 2002. Posttranscriptional compensation for heterozygous disruption of the kidney-specific NaK2Cl cotransporter gene. J. Am. Soc. Nephrol. 13: 604–610. [DOI] [PubMed] [Google Scholar]
- Takahashi, N., M. L. Lopez, J. E. Cowhig, Jr., M. A. Taylor, T. Hatada et al., 2005. Ren1c homozygous null mice are hypotensive and polyuric, but heterozygotes are indistinguishable from wild-type. J. Am. Soc. Nephrol. 16: 125–132. [DOI] [PubMed] [Google Scholar]
- Thatcher, J. W., J. M. Shaw and W. J. Dickinson, 1998. Marginal fitness contributions of nonessential genes in yeast. Proc. Natl. Acad. Sci. USA 95: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 2000. Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154: 1663–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia, R. A., 2002. Exploring the etiology of haploinsufficiency. BioEssays 24: 175–184. [DOI] [PubMed] [Google Scholar]
- von Mering, C., R. Krause, B. Snel, M. Cornell, S. G. Oliver et al., 2002. Comparative assessment of large scale datasets of protein-protein interactions. Nature 417: 399–403. [DOI] [PubMed] [Google Scholar]
- Wagner, A., 2000. Robustness against mutations in genetic networks of yeast. Nat. Genet. 24: 355–361. [DOI] [PubMed] [Google Scholar]
- Wilkie, A. O., 1994. The molecular basis of genetic dominance. J. Med. Genet. 31: 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H., and D. C. Shields, 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1929. Fisher's theory of dominance. Am. Nat. 63: 274–279. [Google Scholar]
- Wright, S., 1934. Physiological and evolutionary theories of dominance. Am. Nat. 68: 24–53. [Google Scholar]