Abstract
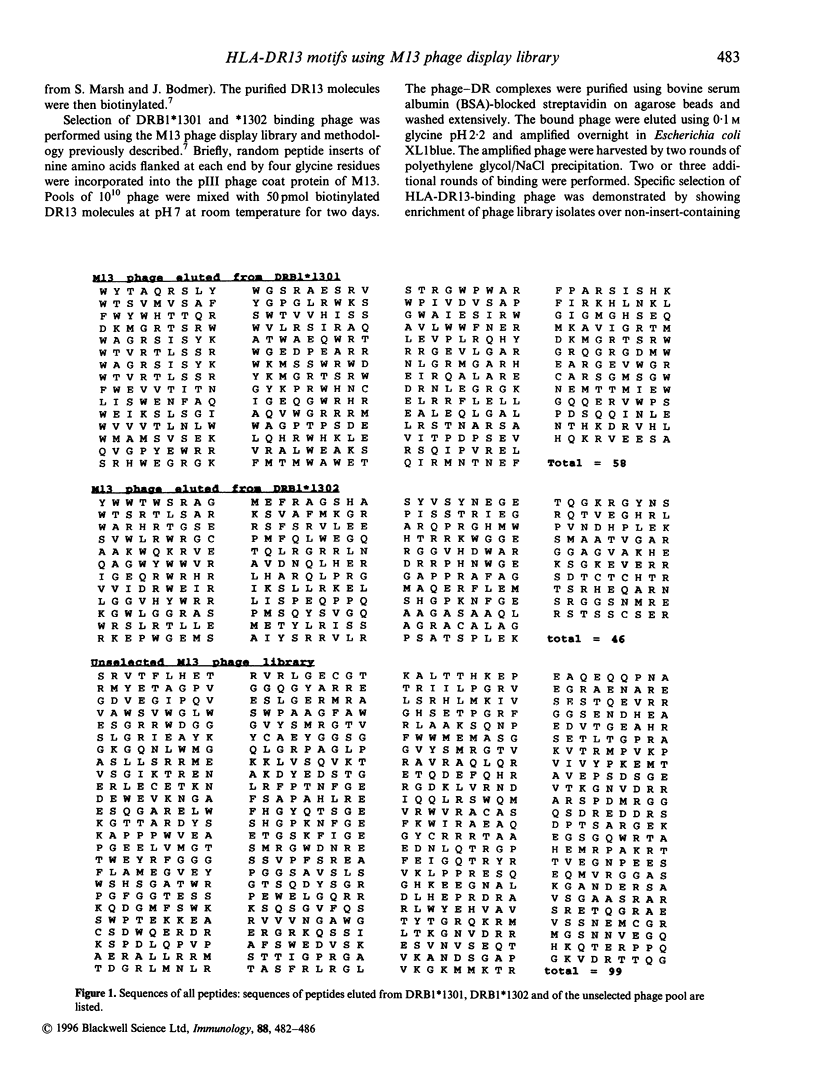
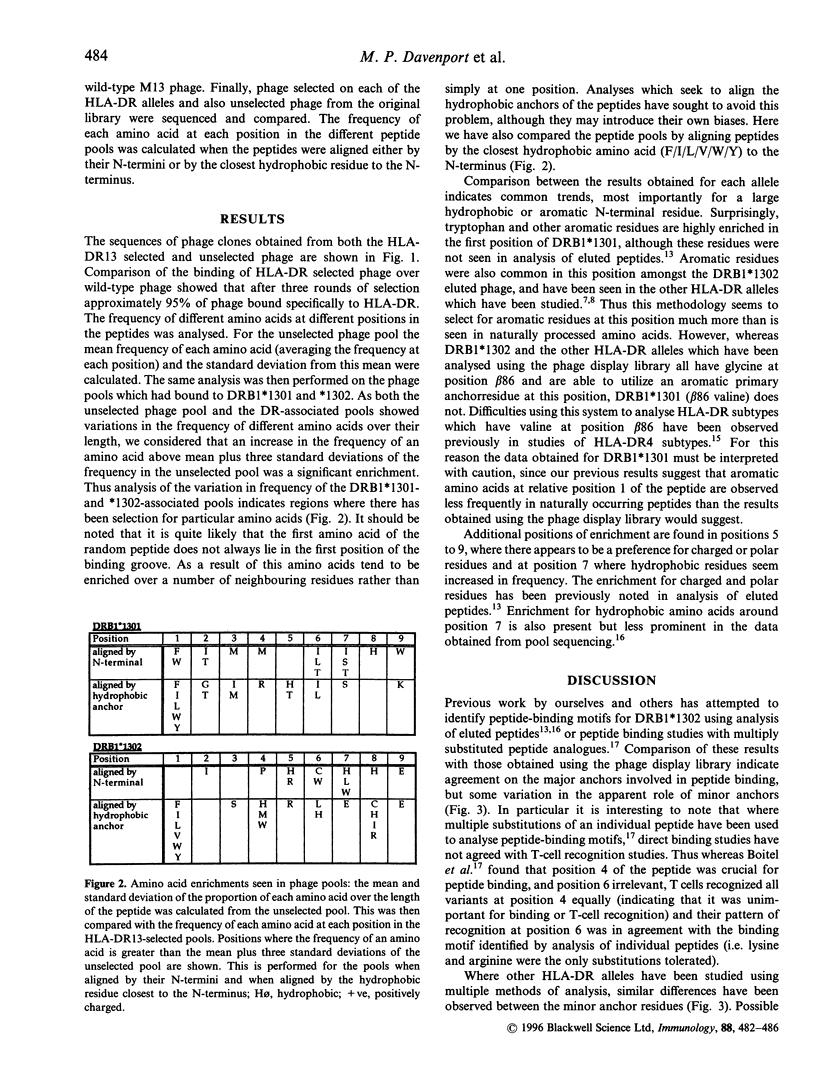
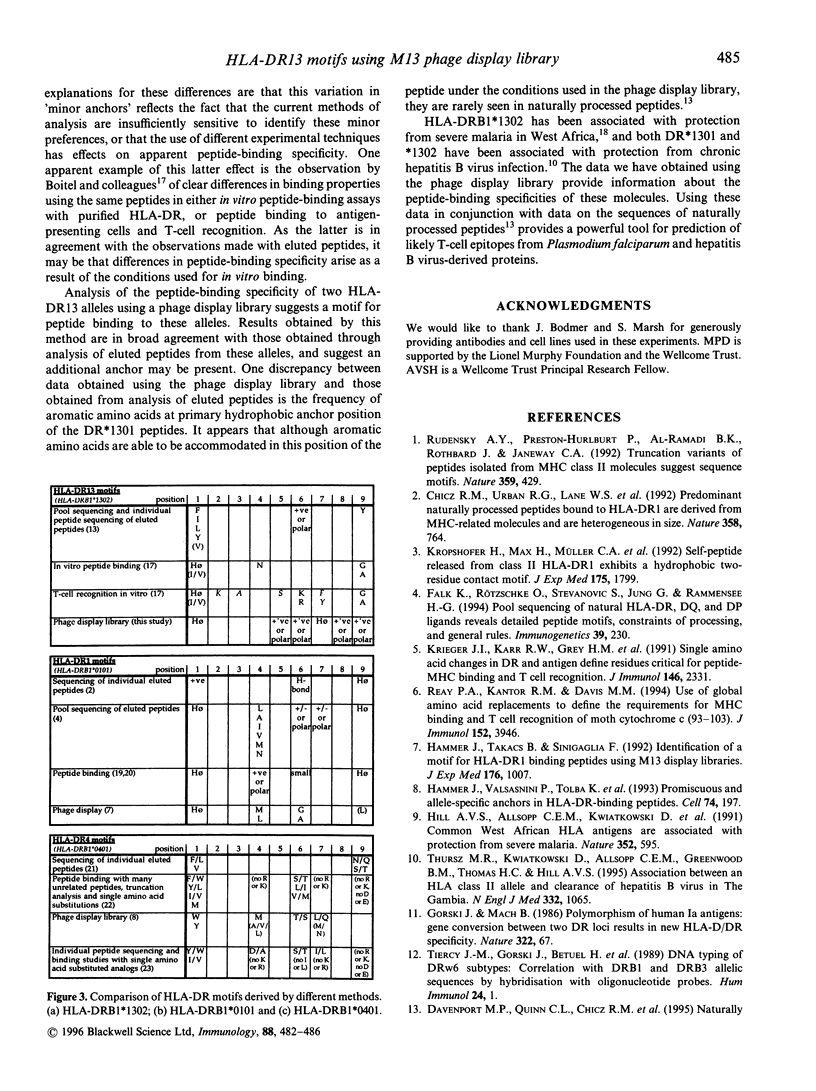
Major histocompatibility complex (MHC) molecules bind peptides bearing an appropriate 'sequence motif' for MHC binding. The use of phage display libraries exploits the ability of MHC class II molecules to exchange peptides in solution and thus select out peptide sequences with high-affinity binding from a large array of random peptides. We have analysed the peptide binding motifs of HLA-DRB1*1301 and *1302 using affinity purified HLA-DR13 molecules to purify sequentially HLA-DR13-binding peptides from a large random library of M13 phage containing nonamer inserts in the pIII coat protein. These DR13 alleles differ only at position 86 of the HLA-DR beta chain, where they contain valine and glycine residues respectively. These alleles were chosen because of their association with protection from severe malaria and chronic hepatitis B virus infection in West Africa. Analysis of the phage bound to these DR molecules suggests binding motifs. We compare the results derived from the use of the phage display library with results obtained from analysis of eluted peptides and peptide-binding studies. This analysis shows that although there is a common theme to motifs derived using different methods, there are also subtle variations between them.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altmann D. M., Heyes J. M., Ikeda H., Sadler A. M., Wilkinson D., Madrigal J. A., Bodmer J. G., Trowsdale J. Fine mapping of HLA class II monoclonal antibody specificities using transfected L cells. Immunogenetics. 1990;32(1):51–55. doi: 10.1007/BF01787329. [DOI] [PubMed] [Google Scholar]
- Boitel B., Blank U., Mège D., Corradin G., Sidney J., Sette A., Acuto O. Strong similarities in antigen fine specificity among DRB1* 1302-restricted tetanus toxin tt830-843-specific TCRs in spite of highly heterogeneous CDR3. J Immunol. 1995 Apr 1;154(7):3245–3255. [PubMed] [Google Scholar]
- Chicz R. M., Urban R. G., Gorga J. C., Vignali D. A., Lane W. S., Strominger J. L. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993 Jul 1;178(1):27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicz R. M., Urban R. G., Lane W. S., Gorga J. C., Stern L. J., Vignali D. A., Strominger J. L. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992 Aug 27;358(6389):764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- Davenport M. P., Ho Shon I. A., Hill A. V. An empirical method for the prediction of T-cell epitopes. Immunogenetics. 1995;42(5):392–397. doi: 10.1007/BF00179401. [DOI] [PubMed] [Google Scholar]
- Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H. G. Pool sequencing of natural HLA-DR, DQ, and DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics. 1994;39(4):230–242. doi: 10.1007/BF00188785. [DOI] [PubMed] [Google Scholar]
- Gorski J., Mach B. Polymorphism of human Ia antigens: gene conversion between two DR beta loci results in a new HLA-D/DR specificity. Nature. 1986 Jul 3;322(6074):67–70. doi: 10.1038/322067a0. [DOI] [PubMed] [Google Scholar]
- Hammer J., Takacs B., Sinigaglia F. Identification of a motif for HLA-DR1 binding peptides using M13 display libraries. J Exp Med. 1992 Oct 1;176(4):1007–1013. doi: 10.1084/jem.176.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer J., Valsasnini P., Tolba K., Bolin D., Higelin J., Takacs B., Sinigaglia F. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993 Jul 16;74(1):197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- Hill A. V., Allsopp C. E., Kwiatkowski D., Anstey N. M., Twumasi P., Rowe P. A., Bennett S., Brewster D., McMichael A. J., Greenwood B. M. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991 Aug 15;352(6336):595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- Hill A. V., Elvin J., Willis A. C., Aidoo M., Allsopp C. E., Gotch F. M., Gao X. M., Takiguchi M., Greenwood B. M., Townsend A. R. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature. 1992 Dec 3;360(6403):434–439. doi: 10.1038/360434a0. [DOI] [PubMed] [Google Scholar]
- Krieger J. I., Karr R. W., Grey H. M., Yu W. Y., O'Sullivan D., Batovsky L., Zheng Z. L., Colón S. M., Gaeta F. C., Sidney J. Single amino acid changes in DR and antigen define residues critical for peptide-MHC binding and T cell recognition. J Immunol. 1991 Apr 1;146(7):2331–2340. [PubMed] [Google Scholar]
- Kropshofer H., Max H., Müller C. A., Hesse F., Stevanovic S., Jung G., Kalbacher H. Self-peptide released from class II HLA-DR1 exhibits a hydrophobic two-residue contact motif. J Exp Med. 1992 Jun 1;175(6):1799–1803. doi: 10.1084/jem.175.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan D., Arrhenius T., Sidney J., Del Guercio M. F., Albertson M., Wall M., Oseroff C., Southwood S., Colón S. M., Gaeta F. C. On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J Immunol. 1991 Oct 15;147(8):2663–2669. [PubMed] [Google Scholar]
- O'Sullivan D., Sidney J., Del Guercio M. F., Colón S. M., Sette A. Truncation analysis of several DR binding epitopes. J Immunol. 1991 Feb 15;146(4):1240–1246. [PubMed] [Google Scholar]
- Reay P. A., Kantor R. M., Davis M. M. Use of global amino acid replacements to define the requirements for MHC binding and T cell recognition of moth cytochrome c (93-103). J Immunol. 1994 Apr 15;152(8):3946–3957. [PubMed] [Google Scholar]
- Rudensky AYu, Preston-Hurlburt P., al-Ramadi B. K., Rothbard J., Janeway C. A., Jr Truncation variants of peptides isolated from MHC class II molecules suggest sequence motifs. Nature. 1992 Oct 1;359(6394):429–431. doi: 10.1038/359429a0. [DOI] [PubMed] [Google Scholar]
- Sette A., Sidney J., Oseroff C., del Guercio M. F., Southwood S., Arrhenius T., Powell M. F., Colón S. M., Gaeta F. C., Grey H. M. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. J Immunol. 1993 Sep 15;151(6):3163–3170. [PubMed] [Google Scholar]
- Thursz M. R., Kwiatkowski D., Allsopp C. E., Greenwood B. M., Thomas H. C., Hill A. V. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995 Apr 20;332(16):1065–1069. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- Tiercy J. M., Gorski J., Bétuel H., Freidel A. C., Gebuhrer L., Jeannet M., Mach B. DNA typing of DRw6 subtypes: correlation with DRB1 and DRB3 allelic sequences by hybridization with oligonucleotide probes. Hum Immunol. 1989 Jan;24(1):1–14. doi: 10.1016/0198-8859(89)90042-6. [DOI] [PubMed] [Google Scholar]


