Abstract
We analyzed the effects of mating system and recombination rate on single nucleotide polymorphisms using 14 single-copy nuclear loci from single populations of five species of wild tomatoes (Solanum section Lycopersicon). The taxa investigated comprise two self-compatible (SC) and three self-incompatible (SI) species. The observed reduction in nucleotide diversity in the SC populations compared to the SI populations is much stronger than expected under the neutral effects of the mating system on effective population size. Importantly, outgroup sequences available for 11 of the 14 loci yield strong positive correlations between silent nucleotide diversity and silent divergence, indicative of marked among-locus differences in mutation rates and/or selective constraints. Furthermore, using a physical estimate of local recombination rates, we find that silent nucleotide diversity (but not divergence) is positively correlated with recombination rate in two of the SI species. However, this correlation is not nearly as strong as in other well-characterized species (in particular, Drosophila). We propose that nucleotide diversity in Lycopersicon is dominated mainly by differences in neutral mutation rates and/or selective constraints among loci, demographic processes (such as population subdivision), and background selection. In addition, we hypothesize that the soil seed bank plays an important role in the maintenance of the large genetic diversity in the SI species (in particular L. peruvianum).
WHETHER diversity levels in the genomes of animals and plants are shaped mainly by neutral processes or selection is a recurrent theme in population genetics. In many organisms, genes in regions of low recombination exhibit reduced levels of DNA polymorphism, while divergence among species remains relatively unaffected. These observations may be explained by genetic hitchhiking, involving advantageous mutations sweeping through a population (Maynard Smith and Haigh 1974) and/or by background selection eliminating unconditionally deleterious mutations (Charlesworth et al. 1993, 1995). Depending on the effective recombination rate in the regions subjected to either of the above processes and on the strength of natural selection, linked neutral variation is reduced under both selection scenarios, while interspecific divergence is not influenced.
A positive correlation between DNA diversity and recombination rate has been found in many organisms, both animals (Aguadé et al. 1989; Stephan and Langley 1989; Begun and Aquadro 1992; Nachman 1997; Nachman et al. 1998) and plants (Dvorak et al. 1998; Kraft et al. 1998; Stephan and Langley 1998). Such patterns were usually attributed to the action of natural selection. However, neutral explanations for an observed positive correlation between recombination and diversity have also been proposed, when, in addition to intraspecific variation, divergence also scaled with recombination rate (e.g., Wang et al. 1997; Hellmann et al. 2003).
Among plants, maize (Zea mays ssp. mays L.) and its wild ancestor were the focus of recent studies examining the interplay between nucleotide variability, recombination, and selection (Tenaillon et al. 2001, 2002, 2004). In maize, single nucleotide polymorphism (SNP) and recombination rate, as measured by a quantitative recombination nodule map, were found to be uncorrelated. However, a positive correlation was found for one of the sequence-based estimators of the population recombination rate, which estimates recombination rate on the basis of linkage disequilibrium (Hudson 1987). Clearly, a thorough understanding of the evolutionary factors underlying the relationship between recombination rates and levels of nucleotide diversity in plants—and even ascertainment of such an association—requires much more sequence data obtained from genomic regions spanning a range of known recombination rates.
Here we report our findings from a multilocus survey of another well-studied plant model system, the clade Lycopersicon, encompassing the cultivated tomato and its progenitor, as well as several other wild species. Native to western South America, the wild relatives of the cultivated tomato represent suitable model organisms to study genealogical footprints of speciation and demographic processes (Städler et al. 2005). The species complex appears to be relatively young yet ecologically well differentiated (Rick and Lamm 1955; Rick 1979; Taylor 1986), and the genetics of the cultivated tomato have long been under intense investigation (Rick and Yoder 1988; Tanksley et al. 1992; Ganal et al. 1998). Collectively, the wild taxa represent a wide spectrum of mating system variation, including both self-compatible, partially self-fertilizing and self-incompatible, obligately outcrossing species (Rick et al. 1976, 1977, 1979; Rick 1983; Igic et al. 2004).
In an exploratory study based on five nuclear genes, Baudry et al. (2001) found very strong effects of the mating system on levels of silent nucleotide diversity among five wild tomato species, while levels of recombination did not have significant effects. A shortcoming of this study, however (shared by many other such studies), was the lack of sufficiently diverged outgroup sequences and hence the inability to distinguish between intra- and interspecific effects on sequence evolution. We have obtained sequence data for nine additional nuclear genes and outgroup sequences for 11 of the 14 loci. Using this much-expanded data set, we are now able to explore the interrelationships among nucleotide diversity, interspecific divergence, genomic recombination rates, and the possible role of natural selection in more depth and with much more confidence. With the addition of 9 more loci, we are also able to investigate the effect of the mating system on levels of DNA diversity in more detail.
Our study addresses the following questions: (i) What is the effect of the mating system on levels of silent nucleotide diversity?, (ii) Are nucleotide diversity and interspecific divergence correlated with each other, as expected under neutral evolution?, (iii) Are recombination rate and nucleotide diversity and/or interspecific divergence correlated with each other?, and (iv) What are the relative strengths of natural selection vs. demographic (and possibly other, neutral) processes in shaping genetic variation in wild tomatoes, and how do these compare to other model organisms for which the data are currently much more complete (in particular Drosophila)?
MATERIALS AND METHODS
Plant material:
The clade Lycopersicon encompasses the cultivated tomato Lycopersicon esculentum and its wild relatives. Although the former genus Lycopersicon has been taxonomically reassigned to Solanum section Lycopersicon (Spooner et al. 1993; Peralta and Spooner 2001), we will retain the old nomenclature for consistency with our and other authors' previous work. Native to western South America, wild tomatoes are herbaceous perennials, with individual plants often bearing receptive, insect-pollinated flowers and fruits simultaneously. Annual recruitment of seedlings may occur in more mesic environments, but seedling establishment might be restricted to favorable years in more xeric habitats (e.g., southern Peru and northern Chile; Rick 1979; T. Städler, personal observations).
Our sampling scheme encompasses a wide spectrum of mating systems in the clade Lycopersicon, from self-compatible (SC) to self-incompatible (SI) species (Igic et al. 2004). The species in our study include L. peruvianum (= Solanum peruvianum; SI; accession no. LA2744) from Tarapaca and L. chilense (= S. chilense; SI; LA2884) from Antofagasta, both located in northern Chile. Our sample of L. hirsutum (= S. habrochaites; SI; LA1775) is from Ancash in central Peru. For geographical details on the distribution and locations of the SI samples, see Städler et al. (2005). We also sampled the following SC species: L. chmielewskii (= S. chmielewskii; SC; LA3653) from Apurimac, south-central Peru, and L. pimpinellifolium (= S. pimpinellifolium; SC; LA1583) from Lambayeque, northern Peru. L. pimpinellifolium is distributed along the coast of Peru and Ecuador and restricted to Andean river valleys (Taylor 1986; Caicedo and Schaal 2004), whereas L. chmielewskii prefers more mesic habitats in interior Peru with a fairly narrow geographic distribution (Rick et al. 1976). Although no estimates of outcrossing rates are available for our two SC source populations, morphological data on flower size and the degree of stigma exsertion indicate facultative outcrossing (http://tgrc.ucdavis.edu). Some populations of L. pimpinellifolium are thought to have outcrossing rates up to 37% (Rick et al. 1977, 1978; Georgiady et al. 2002).
From each species, five individuals (10 alleles) were sampled from one population, except for L. hirsutum where only three or four plants were included, depending on the locus. We studied the same individual plants from one population per species as in previous work (Baudry et al. 2001; Städler et al. 2005), representing one accession per species from the Tomato Genetics Resource Center at the University of California at Davis (http://tgrc.ucdavis.edu).
DNA sequencing and haplotyping:
We sequenced the same 8 loci (CT093, CT114, CT099, CT066, CT166, CT179, CT148, and CT198) in the two SC species previously sequenced in the three SI species by Städler et al. (2005). One additional locus (CT189) was added for all five taxa. Furthermore, we used previously published sequence data for five loci (CT143, CT208, CT251, CT268, and Sucr) (Baudry et al. 2001). For some of these loci, we slightly modified the alignment and/or the exon-intron boundary annotation (see Städler et al. 2005). Moreover, we obtained homologous sequences from one of two outgroup species (S. lycopersicoides or S. ochranthum) for 11 of the 14 loci in this study (see below). Repeated attempts to amplify the loci CT114, CT148, and CT208 from the outgroup taxa failed. Summed across all genes, this amounts to ∼20.5 kb of sequence per “allele” or 41 kb per individual plant. The loci are single-copy nuclear genes and are distributed across 9 of the 12 tomato chromosomes (Table 1). They were chosen from regions representing low, intermediate, and high recombination rates (RN) according to a recombination nodule map (Stephan and Langley 1998). PCR primers, designed on the basis of cDNA sequences for the cultivated tomato L. esculentum (= S. lycopersicum), are available from the Tomato Gene Index at The Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/lgi/). PCR primers and reaction conditions can be accessed at http://www.zi.biologie.uni-muenchen.de/evol/index.html.
TABLE 1.
Chromosome location, putative function, and recombination rate (RN) of sequenced loci
| Locus | Chromosome | Length (bp) | Putative encoded protein | RNb (×10−8) |
|---|---|---|---|---|
| Sucra | 3 | 1575 | Vacuolar invertase | 0.00 |
| CT208a | 9 | 1767 | Alcohol dehydrogenase, class III | 0.00 |
| CT093 | 5 | 1415 | S-adenosylmethionine decarboxylase proenzyme | 0.00 |
| CT114 | 7 | 1169 | Phospho-glycerate kinase | 0.00 |
| CT189 | 12 | 1463 | 40S ribosomal protein S19 | 0.35 |
| CT251a | 2 | 1779 | At5g37260 gene | 0.46 |
| CT099 | 12 | 1354 | Unknown | 0.88 |
| CT066 | 10 | 1346 | Arginine decarboxylase | 0.93 |
| CT166 | 2 | 2673 | Ferredoxin-NADP reductase | 1.61 |
| CT179 | 3 | 995 | Tonoplast intrinsic protein Δ-type | 1.97 |
| CT148 | 8 | 1497 | Copper/zinc superoxide dismutase | 2.00 |
| CT198 | 9 | 779 | Submergence induced protein 2-like | 2.10 |
| CT268a | 1 | 1887 | Receptor-like protein kinase | 2.33 |
| CT143a | 9 | 1821 | Sterol C-14 reductase | 2.73 |
Except for Sucr (sucrose accumulator gene), locus designations refer to particular EST sequences that have been integrated into longer “tentative contigs” in the TIGR Tomato Gene Index (http://www.tigr.org/tdb/lgi/). The length per locus is given across the total alignment of all five tomato species (without outgroup), including indels.
From Baudry et al. (2001).
Derived from Stephan and Langley (1998); recombination rate per site per generation.
Haplotype phase was fully resolved for the newly sequenced nine loci, but not for L. peruvianum in the previously published five loci (Baudry et al. 2001). PCR products were sequenced directly on both strands with a MegaBACE 1000 automated sequencer (Amersham Pharmacia, Freiburg, Germany). Distinct haplotypes within heterozygous individuals were resolved by applying a suite of haplotype-specific sequencing primers. In most cases, we exploited putative or confirmed SNPs to anchor the 3′-end of 18-bp sequencing primers that were intended to resolve the heterogeneous PCR products. This approach enabled us to verify SNPs (and indel variation) and establish haplotype phase on the basis of overlapping information (usually) supported by multiple, differential primer pairs. In a few technically challenging cases (caused by extensive indel polymorphism), we cloned the PCR product (Invitrogen, Karlsruhe, Germany) and then sequenced at least 10 clones per plant. Sequences were edited and initially aligned in Sequence Navigator (Applied Biosystems, Darmstadt, Germany). Interspecific alignments were performed with ClustalW (Thompson et al. 1994) and adjusted manually in MacClade, version 3.07 (Maddison and Maddison 1992).
Estimation of recombination rates and levels of diversity and divergence:
The genomic recombination rate RN was estimated from a genetic linkage map and a frequency map of recombination nodules based on a cross between L. esculentum and L. pennellii (= S. pennellii) (Stephan and Langley 1998); this sequence-independent estimator of recombination is given as the recombination fraction per site per generation, c. From our sequence data, we also estimated the population recombination parameter (C = 4Nec, where Ne denotes the effective population size) by the following methods: (i) The estimator ρ was calculated with the composite-likelihood coalescent method of Hudson (2001), as implemented in the program LDhat (McVean et al. 2002), and (ii) γ, a maximum-likelihood estimator that uses subsets of four sequences, was calculated with the program Sites (Hey and Wakeley 1997). All reported estimates are per-site values.
Due to the observed haplotype structure in L. chilense and L. hirsutum (see Städler et al. 2005) and too few nucleotide polymorphisms in L. chmielewskii and L. pimpinellifolium, we estimated ρ and γ only from L. peruvianum sequences. This approach is justified because L. peruvianum is the most polymorphic species and the studied population appears to be close to demographic equilibrium (Städler et al. 2005). Sites with observed multiple hits were excluded from the estimation of γ and ρ.
As an estimator of nucleotide diversity (θ = 4Neμ, where μ is the mutation rate per site per generation), we calculated Watterson's (1975) θW for silent sites (denoted as θsil). Divergence at silent sites (Ksil) was measured as the average number of nucleotide substitutions per silent site between species, using Jukes-Cantor correction (Nei 1987) as implemented in DnaSP version 4.0 (Rozas et al. 2003). This analysis was performed for the 11 loci for which we obtained outgroup sequences from either S. ochranthum (Sucr, CT251, CT189, CT099, CT066, CT166, CT179, CT198, and CT143) or S. lycopersicoides (CT093 and CT268). All 11 Ksil estimates were used for correlation analyses, despite the fact that we used different outgroups.
Taxonomic studies based on a variety of molecular markers have identified either S. lycopersicoides or S. ochranthum as the sister group to the tomato clade (Spooner et al. 1993, 2005; Peralta and Spooner 2001). The estimated divergence time between S. ochranthum and Lycopersicon is between 5.8 and 18.6 million years (Rose 2002), while the divergence time between S. lycopersicoides and Lycopersicon has not been estimated. However, S. lycopersicoides is a close relative of the tomato clade and extensive synteny between the chromosomes is observed (Rick 1979; Chetelat et al. 2000). We explored the possible error introduced by using two outgroup species and found that in correlation analyses both always indicated the same trends.
Tests of neutrality:
To test for deviations from the standard neutral model of evolution (Kimura 1983), we performed the following tests using the programs DnaSP and HKA (http://lifesci.rutgers.edu/∼heylab/): (i) the McDonald-Kreitman test (McDonald and Kreitman 1991), which compares the pattern of within-species polymorphism and between-species divergence at synonymous and nonsynonymous sites in coding regions of a gene; (ii) the Hudson-Kreitman-Aguadé (HKA) test (Hudson et al. 1987), which tests for heterogeneity in the ratio of polymorphism to divergence among loci; (iii) Tajima's D (Tajima 1989) for all sites (Dall) and for silent sites (Dsil), which tests the neutral prediction that the estimators π and θW should measure the same quantity, θ; and (iv) Fu and Li's D test (Fu and Li 1993), which is based on the differences between the total number of mutations in external branches of the genealogy and the total number of mutations. Fu and Li's D is constructed in a similar way to Tajima's D, but takes into account whether mutations are ancestral or derived. For both tests, negative values indicate an excess of polymorphisms with low frequency, whereas positive values indicate an excess of polymorphisms with an intermediate frequency.
To explore the ratio of nonsynonymous to synonymous substitutions, we calculated intraspecific nucleotide diversity at nonsynonymous (πa) and synonymous sites (πs) (Nei 1987) and nucleotide divergence to the outgroup at nonsynonymous (Ka) and synonymous sites (Ks); the latter estimates were obtained with Jukes-Cantor correction (Nei 1987).
Estimation of effective population size:
Effective population size (Ne) for the five Lycopersicon species was estimated using the following formulas: θsil = 4Neμ and μ = Ksil/2t, with t being the divergence time in generations. We assume that the split between Lycopersicon and S. ochranthum occurred between 5.8 and 18.6 million years ago (Rose 2002) and calculate θsil as the weighted average silent diversity per species and Ksil as the weighted average silent divergence. To account for uncertainties regarding generation time (see above), we assume two rather extreme cases: one generation per year and one generation every seven years. The latter estimate is based on the assumption that the natural habitats of Lycopersicon and the outgroup species have been affected by the El Niño southern oscillation (ENSO), which determines moisture availability and consequently seed germination and plant establishment (see discussion).
RESULTS
Levels of diversity and mating system:
Table 2 summarizes levels of silent diversity (θsil) for all 14 loci across the five study species. We observed only very little variation in the two SC species L. chmielewskii and L. pimpinellifolium (mean θsil = 0.027% and 0.160%, respectively), whereas we found the three SI species to be highly polymorphic (L. peruvianum mean θsil = 2.233%, L. chilense mean θsil = 0.642%, and L. hirsutum mean θsil = 0.577%). Thus, average levels of variation in the SI species are 3.6- to 14-fold higher than those in the partially selfing L. pimpinellifolium, and 21- to 83-fold higher than those in L. chmielewskii.
TABLE 2.
Levels of silent diversity at 14 loci in five Lycopersicon species
| Locus | L. peruvianum | L. chilense | L. hirsutum | L. pimpinellifolium | L. chmielewskii |
|---|---|---|---|---|---|
| Sucra | 3.189 | 0.944 | 0.475 | 0.103 | 0.000 |
| CT208a | 1.259 | 0.118 | 0.000 | 0.060 | 0.000 |
| CT093 | 1.224 | 0.406 | 0.063 | 0.058 | 0.000 |
| CT114 | 1.407 | 0.488 | 0.354 | 0.054 | 0.055 |
| CT189 | 1.866 | 0.186 | 0.000 | 0.094 | 0.031 |
| CT251a | 2.631 | 0.197 | 0.440 | 0.000 | 0.000 |
| CT099 | 3.452 | 1.632 | 2.722 | 0.429 | 0.000 |
| CT066 | 3.513 | 1.812 | 0.465 | 0.106 | 0.000 |
| CT166 | 1.982 | 0.861 | 0.572 | 0.169 | 0.000 |
| CT179 | 2.001 | 1.663 | 1.692 | 0.413 | 0.149 |
| CT148 | 2.299 | 0.778 | 0.898 | 0.000 | 0.000 |
| CT198 | 4.292 | 0.481 | 1.145 | 0.997 | 0.207 |
| CT268a | 2.840 | 1.787 | 1.150 | 0.163 | 0.000 |
| CT143a | 2.244 | 0.000 | 0.153 | 0.123 | 0.049 |
| Means | 2.233 | 0.642 | 0.577 | 0.160 | 0.027 |
All estimates are per-site θsil values, expressed in percentages. Silent diversity in the three SI species (the first three species) was reported by Städler et al. (2005), except for locus CT189. The bottom row reports the weighted mean θsil across all 14 loci.
Estimates deviating from those given in Baudry et al. (2001) are due to minor alignment and/or annotation changes.
Estimates of effective population size also indicate a 3- to 83-fold reduction between SI and SC species (Table 3). The observed differences in neutral diversity between SC and SI populations cannot be attributed to the difference in mating system alone. Additional forces such as natural selection and/or forces affecting the entire genome, such as demographic factors or life-history traits influencing Ne (e.g., metapopulation dynamics or a soil seed bank) must be invoked to explain these results (see discussion).
TABLE 3.
Estimates of effective population size in five Lycopersicon species
| Assumed divergence time
|
||
|---|---|---|
| Species | 5.8 MYA | 18.6 MYA |
| L. peruvianum | 1.165 | 3.736 |
| L. chilense | 0.335 | 1.073 |
| L. hirsutum | 0.308 | 0.987 |
| L. pimpinellifolium | 0.097 | 0.310 |
| L. chmielewskii | 0.014 | 0.046 |
The range of estimated divergence times between the clade Lycopersicon and the outgroup S. ochranthum is based on Rose (2002). Estimates of effective population size (×106) assume one generation per year and would be only one-seventh of these estimates under a generation time of 7 years (but see text).
Silent diversity and silent divergence:
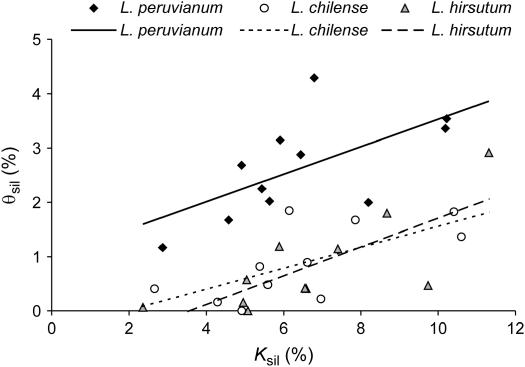
Figure 1 shows that for all three SI species, θsil and Ksil are positively correlated (L. peruvianum: Spearman's R = 0.718, P = 0.013; L. chilense: R = 0.655, P = 0.029; L. hirsutum: R = 0.682, P = 0.021). This positive correlation between θsil and Ksil conforms to the pattern expected under neutrality and clearly suggests that neutral mutation rates vary among loci. Nevertheless, the multilocus HKA test, which assesses the neutral prediction that the ratio of polymorphism to divergence across loci should be constant, is significant for L. chilense, L. hirsutum, and L. pimpinellifolium (Table 4). Loci with the greatest contribution to the overall test statistic differ among species. In L. chilense, CT143 contributes greatly to the significant multilocus HKA statistic; diversity at this locus is strongly reduced in both L. chilense and L. hirsutum, and CT189 has a similar effect in L. hirsutum. For L. pimpinellifolium, there is an excess of polymorphism at locus CT198. Removal of these single loci from the multilocus HKA test causes the test statistic to drop below the critical value in each of these species.
Figure 1.
Scatter plots of levels of silent divergence and silent diversity in the three SI species (n = 11 loci). Regression lines are also shown for each species (Figures 1–4). L. peruvianum: Spearman's R = 0.718, P = 0.013; L. chilense: R = 0.655, P = 0.029; L. hirsutum: R = 0.682, P = 0.021.
TABLE 4.
Summary of multilocus neutrality tests
| Species | Tajima's Dall | Tajima's Dsil | Fu and Li's D | HKAall | HKAsil |
|---|---|---|---|---|---|
| L. peruvianum | −0.37 (−0.35) | −0.39 (−0.34) | −0.53 | 0.99 | 0.98 |
| L. chilense | 0.58* (0.84)*** | 0.60* (0.88)*** | 0.83* | 0.02* | 0.01* |
| L. hirsutum | 0.24 (0.35) | 0.32 (0.42)* | 0.56* | 0.04* | 0.02* |
| L. pimpinellifolium | 0.18 (0.08) | −0.01 (−0.07) | 0.25 | 0.004** | 0.004** |
| L. chmielewskii | −0.15 (−0.13) | −0.003 (0.002) | −0.044 | — | — |
Tajima's Dall (based on all sites) and Dsil (based on silent sites) values are averages across the 9 new loci, followed by the mean values using all 14 loci in parentheses (i.e., including Sucr, CT208, CT251, CT268, and CT143 from Baudry et al. 2001). Average Tajima's Dsil values based on 13 loci (without CT189) in the three SI species were presented by Städler et al. (2005). Analyses in the other three columns are based on the 11 loci for which an outgroup sequence is available (Sucr, CT093, CT189, CT251, CT099, CT066, CT166, CT179, CT198, CT268, and CT143). Numbers in the HKA columns are probability values from the χ2 distribution with 10 d.f.; L. chmielewskii harbors too few polymorphisms for these analyses. *P < 0.05; **P < 0.01; ***P < 0.001 (one-tailed tests).
Other neutrality tests:
We applied several other neutrality tests to probe for signatures of selection and/or demographic processes in our data. In L. peruvianum, CT198 displays the highest silent nucleotide diversity among all loci (θsil = 4.29%) and shows a significant McDonald-Kreitman test (Fisher's exact test, P = 0.033). However, this result rests on only four fixed differences among species, three of which are replacement substitutions. Tajima's D and Fu and Li's D are significantly positive for this locus in L. pimpinellifolium (CT 198; Tajima's Dall = 2.42, P < 0.01; Fu and Li's D = 1.76, P < 0.02). CT099 displays the greatest apparent deviation in L. hirsutum, with a significantly positive Tajima's D (Dall = 1.48, P = 0.039) and Fu and Li's D (D = 1.78, P = 0.023) and unusually high polymorphism for this species (θsil = 2.72%; Table 2). The average values of Tajima's D and Fu and Li's D in the three SI species are consistent with parameter estimates obtained under the simple “isolation” model of speciation; both L. chilense and L. hirsutum are inferred to have undergone population-size contractions compared to the common, ancestral species (Städler et al. 2005).
Although several loci yield significant results for various tests in one or more species, the above results were obtained without correcting for multiple testing; hence we see no conclusive evidence that one of the loci has recently been subject to natural selection. The lack of nucleotide diversity at some loci may simply be due to our limited sample size or due to demographic effects. A further aspect to consider is that mean values for Tajima's D for all sites and silent sites across 14 loci are not significantly different (Mann-Whitney U-test: L. peruvianum, P = 0.98; L. chilense, P = 1.00; L. hirsutum, P = 0.95; L. chmielewskii, P = 0.93; L. pimpinellifolium, P = 0.85). Thus we find no indication that the frequency spectrum of nonsynonymous polymorphisms is significantly different from that of silent polymorphisms.
Recombination rate and silent genetic diversity:
We analyzed the effect of recombination rate on levels of DNA diversity for 14 genes in three self-incompatible wild tomato species using a physical estimate of the genomic recombination rate (RN), which is based on detailed recombination nodule maps of all 12 tomato chromosomes (Stephan and Langley 1998). In addition, we compared our results for RN estimates with sequence-based estimators of the population recombination parameter, ρ (Hudson 2001) and γ (Hey and Wakeley 1997).
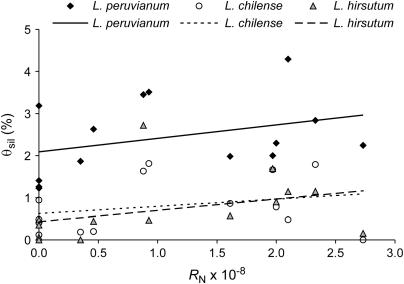
On the basis of 14 loci, Figure 2 presents correlations between θsil and RN for the three outcrossing species. In both L. peruvianum and L. hirsutum, θsil tends to be positively correlated with RN (Spearman's R = 0.471, P = 0.089 and R = 0.528, P = 0.052, respectively). In L. chilense, however, we find no positive correlation (R = 0.198, P = 0.498; Table 5).
Figure 2.
Scatter plots of recombination rate per site and silent nucleotide diversity in the three SI species (n = 14 loci). L. peruvianum: Spearman's R = 0.471, P = 0.089; L. chilense: R = 0.198, P = 0.498; L. hirsutum: R = 0.528, P = 0.052.
TABLE 5.
Correlations between diversity levels (θsil) and estimates of recombination rate
| Species | θsilvs. RN | θsilvs. γ | θsilvs. ρ |
|---|---|---|---|
| L. peruvianum | 0.471 (0.089) | 0.617 (0.077) | 0.600 (0.088) |
| L. chilense | 0.198 (0.498) | 0.050 (0.898) | 0.267 (0.488) |
| L. hirsutum | 0.528 (0.052) | 0.567 (0.112) | 0.617 (0.077) |
All values represent Spearman's coefficient of rank correlation; P-values (two-tailed) follow in parentheses. All 14 loci were used for the analyses involving the sequence-independent estimate of recombination, RN. Our ρ and γ estimates were obtained from L. peruvianum sequences, and the correlation analyses shown are based on the 9 loci with experimentally established haplotype phase (see text).
To assess consistency between the different estimators of recombination, we performed correlation analyses and compared the results for the entire data set (14 loci) with results for the 9 newly sequenced loci. This comparison was done to assess the possible error introduced by the lack of reliable haplotype-phase information in L. peruvianum for the 5 previously studied loci. RN is positively correlated with γ (9 loci: Spearman's R = 0.669, P = 0.049; 14 loci: R = 0.476, P = 0.086), but not with ρ (9 loci: R = 0.326, P = 0.391; 14 loci: R = 0.302, P = 0.294). The values of γ and ρ exhibit strong positive correlations among loci (9 loci: R = 0.733, P = 0.025; 14 loci: R = 0.648, P = 0.012). Results for both analyses (using 14 and 9 loci, respectively) are thus consistent with each other, suggesting that the assignment of “quasi-haplotypes” at 5 L. peruvianum loci does not introduce marked biases in our analyses.
Additionally, results for correlations between RN and θsil (14 loci) and between θsil and the sequence-based estimators (9 loci) are consistent; γ and ρ show a strong positive correlation with θsil for both L. peruvianum and L. hirsutum, but not for L. chilense (Table 5). Since sequence-based estimators of recombination rate are believed to be biased upward in regions of high genetic diversity (Wall 2000), in the following sections we concentrate on results based on the physical estimate of recombination, RN.
Recombination rate and interspecific divergence:
If neutral processes explain the positive correlation between nucleotide diversity and recombination rate (e.g., mutagenic effects of the recombination process), as was suggested by the study of Hellmann et al. (2003), divergence between species is also expected to be positively correlated with recombination. We analyzed the correlation between recombination rate (RN) and levels of silent divergence (Ksil), calculated for 11 loci with outgroup sequences from either S. ochranthum (n = 9 loci) or S. lycopersicoides (n = 2 loci); no significant correlation between RN and Ksil was detected (L. peruvianum: Spearman's R = 0.159, P = 0.640; L. chilense: R = 0.064, P = 0.852; L. hirsutum: R = 0.036, P = 0.915) (Figure 3).
Figure 3.
Scatter plots of recombination rate per site and silent divergence for the three SI species (n = 11 loci). L. peruvianum: Spearman's R = 0.159, P = 0.640; L. chilense: R = 0.064, P = 0.852; L. hirsutum: R = 0.036, P = 0.915.
θsil/Ksil and recombination rate:
If the weakly positive correlation between recombination rate and neutral genetic diversity is in fact a signature of diversity-reducing selection in regions of low recombination, we would also expect a positive trend between recombination rate and diversity corrected for interspecific divergence (i.e., θsil/Ksil). Although a positive trend remains, θsil/Ksil is not significantly correlated with RN in any of the three SI species (L. peruvianum: Spearman's R = 0.123, P = 0.719; L. chilense: R = 0.064, P = 0.852; L. hirsutum: R = 0.405, P = 0.216) (Figure 4). This is consistent with our findings of significant positive correlations between θsil and Ksil (Figure 1). Thus, selection does not seem to play a significant role in shaping levels of genetic diversity in the genomes of wild tomato species. In the case of positive selection, we would expect that regions with very low recombination harbor very little silent genetic diversity (Innan and Stephan 2003). Yet in both L. peruvianum and L. chilense we detected notable levels of θsil in regions of low recombination (Figure 2). L. hirsutum also retains appreciable amounts of genetic diversity at some loci with zero recombination; only locus CT208 (RN = 0) displays zero silent genetic diversity. Since this observation for CT208 is based on only six alleles in L. hirsutum, sampling effects could account for this result. The steepest slope for the regression between θsil/Ksil and RN was seen in L. hirsutum (Figure 4); the same trend was previously observed by Stephan and Langley (1998).
Figure 4.
Scatter plots of recombination rate per site and silent diversity corrected for divergence (n = 11 loci). L. peruvianum: Spearman's R = 0.123, P = 0.719; L. chilense: R = 0.064, P = 0.852; L. hirsutum: R = 0.405, P = 0.216.
DISCUSSION
We examined intraspecific nucleotide diversity at 14 loci in five species of wild tomatoes with different mating systems and interspecific divergence to an outgroup at 11 of these 14 loci. In comparison to the previous study by Baudry et al. (2001), this study considerably increases the amount of available sequence data in Lycopersicon and permits a more detailed examination of the effect of recombination rate differences on levels of silent nucleotide diversity. Importantly, the outgroup sequences for 11 of our loci (not previously available) enable us to distinguish between intraspecific and interspecific effects influencing sequence evolution in wild tomatoes.
Mating system differences:
The mating systems of our study species vary from partial self-fertilization in the SC taxa (i.e., L. chmielewskii and L. pimpinellifolium) to obligate outcrossing in the SI populations (i.e., L. hirsutum, L. peruvianum, and L. chilense). Evolutionary change in mating system from SI to SC is common and irreversible in the Solanaceae and self-incompatibility is thought to be ancestral (Igic et al. 2004). Previous studies in wild tomatoes have shown that genetic variation within species decreases with an increasing degree of selfing (Rick et al. 1976, 1977, 1979; Caicedo and Schaal 2004). Given that our study encompasses both SI and SC populations, we can show directly how partial selfing and/or demographic processes manifest themselves at the population level. All other things being equal, population-genetic theory predicts a twofold reduction in levels of neutral variation between outcrossing and completely selfing populations (Pollak 1987; Nordborg and Donnelly 1997; Charlesworth 2003; and references therein) and slighter differences for partially selfing populations. Clearly, average levels of variation in the SC (but not necessarily highly selfing) wild tomato populations are lower than expected under effects of the mating system on effective population size per se.
Plant mating systems are well known to have significant impacts on the levels and distribution of genetic diversity, either directly via differences in effective population size due to (partial) selfing or indirectly via life-history features that are associated with mating-system differences, such as colonization potential (Ingvarsson 2002; Barrett 2003). The relationship between mating systems and levels of genetic variation has been studied empirically in animals and plants (e.g., Liu et al. 1999; Graustein et al. 2002; Wright et al. 2002), and until recently was dominated by studies using allozymes to quantify levels of diversity within and among local populations (Hamrick and Godt 1990; Schoen and Brown 1991). In recent studies on closely related angiosperm species with different mating systems, the discrepancies in levels of diversity between inbred and outbred taxa were attributed to asymmetric introgression from selfing to outcrossing species (Sweigart and Willis 2003), demographic effects (Savolainen et al. 2000), and/or natural selection (Baudry et al. 2001). Since selfing reduces the effective recombination rate, the reduction of nucleotide diversity due to selective sweeps or background selection should be more pronounced in partially self-fertilizing plants than in outcrossers. The SC populations in our study show a moderate-to-high reduction in nucleotide diversity compared to the SI species, which exceeds the theoretical twofold prediction despite the fact that our SC populations are not completely selfing (Rick et al. 1976, 1977, 1978).
Various factors could contribute to our observations. Ecological adaptations (e.g., the presence of soil seed banks in adverse environments) might differ among the study species. Since SI species appear to be less prone to extinction due to high diversity and gene flow that reduce genetic isolation despite high local extinction and recolonization rates (Igic et al. 2004), characteristics and longevity of their soil seed banks could differ from those of SC species. Second, stronger population substructure in partially selfing taxa may cause DNA variation to be largely distributed among populations, whereas in outcrossing species to a large extent it is found within local demes (Awadalla and Ritland 1997; Charlesworth 2003; Sweigart and Willis 2003). Since our sequence data are derived from single populations per species, we cannot offer a conclusive assessment of this factor. Moreover, selfing in species with high rates of population turnover may cause reductions in levels of DNA diversity that greatly exceed the magnitude expected without such metapopulation dynamics (Ingvarsson 2002).
Diversity-reducing forces and variation in mutation rates and selective constraints:
We found positive correlations between a physical estimate of the recombination rate (RN) and silent nucleotide diversity (θsil) in two of three SI taxa, L. peruvianum and L. hirsutum. Studies in Drosophila found much stronger positive correlations between levels of nucleotide polymorphism and recombination rate (Aguadé et al. 1989; Stephan and Langley 1989; Begun and Aquadro 1992) and attributed these findings to an interplay between recombination and selection. In Drosophila melanogaster, positive directional selection (i.e., selective sweeps) seems to be the predominant cause for the strong positive correlation between silent genetic diversity and recombination rate. In regions of low recombination, selection removes more neutral genetic variation around the selected site than in regions of high recombination. In particular, levels of variation are very low in regions of very low recombination, as expected under directional selection (Innan and Stephan 2003). In Lycopersicon, the situation appears to be different. In the three SI species, we observed only a weak correlation between RN and Ksil, which is pronounced enough, however, that the ratio θsil/Ksil exhibits no positive correlation with RN.
Tenaillon et al. (2001, 2002) studied 21 loci in maize (Z. mays ssp. mays L.) and found no correlation between SNP diversity and their physical estimate of recombination rates. Sequence-based estimates of recombination rate, however, suggested a different picture. Hudson's (1987) and Wall's (2000) estimators were correlated with nucleotide diversity, whereas ρ (Hudson 2001) was not correlated with diversity. This appears to be in contrast to our findings in Lycopersicon, where both physical estimates of recombination rate (RN) and sequence-based estimators (ρ and γ) are positively correlated with silent diversity (in two of three SI species) and are consistent with each other.
An important insight gained through the availability of sufficiently diverged outgroup sequences is that silent nucleotide diversity and silent divergence are strongly correlated, as expected if neutral mutation rates and/or selective constraints vary among loci or genomic regions (Figure 1). In a recent study, Tenaillon et al. (2004) found no correlation between diversity and any measure of recombination in the wild ancestor of maize, teosinte (Z. mays ssp. parviglumis). However, comparable to our findings in wild tomatoes, silent diversity and silent divergence were positively correlated. This suggests that in both of these plant model systems, mutation rate (and/or selective constraint) differences among loci drive this correlation (at least partly). Since silent diversity was not corrected for divergence in the earlier maize studies (Tenaillon et al. 2001, 2002), we assume that the positive correlation between recombination rate and nucleotide diversity would be considerably weaker if corrected for divergence.
From our data, we can provide several observations that may be interpreted as a signature of background selection removing deleterious mutations (Charlesworth et al. 1993, 1995). This interpretation rests on the assumption that the positive correlation between RN and θsil is real and not spurious, which is consistent with previous RFLP data based on a larger number of nuclear loci (Stephan and Langley 1998). First, the regression lines are relatively flat for all SI species (RN vs. θsil; Figure 2). Second, nucleotide variation in regions of very low recombination does not drop down to zero; this is best seen in L. peruvianum. Third, there is a slight positive trend between θsil/Ksil and RN for all three SI species, in particular for L. hirsutum. Furthermore, the relative paucity of segregating nonsynonymous variation at all sequenced loci is a clear signal of purifying selection. In particular, the ratios πa/πs and Ka/Ks are much lower than one for all loci (Figure 5), indicating that selection has removed deleterious nonsynonymous mutations within lineages. Likewise, none of the sequenced alleles in our study appear to be nonfunctional. Although we did not sequence all exons for any of the genes, we would nevertheless expect mutations resulting in frameshifts and premature stop codons to occur under neutral evolution. The background selection model was also favored by Innan and Stephan (2003) who found that in RFLP data (Miller and Tanksley 1990), levels of polymorphism in L. pimpinellifolium and L. chmielewskii are best explained by a concave function of the recombination rate in regions of low recombination. According to their analysis, under hitchhiking associated with positive selection, the shape of this function would be expected to be convex in regions of low recombination, as was seen in data from D. melanogaster.
Figure 5.
Ratios of average pairwise differences per nonsynonymous site to synonymous site in L. peruvianum (πa/πs; 14 loci) and of average pairwise divergence to the outgroup (Ka/Ks; 11 loci). The dotted line represents Ka/Ks = 1 and πa/πs = 1.
Life-history traits and demographic processes affecting Ne:
Life-history traits such as the presence of a seed bank can have important consequences for effective population size in plants, especially in the presence of population turnover (Nunney 2002). Fluctuations in population size are likely to be common in the natural habitats of our study populations. The west coast of South America is affected by the ENSO, and this recurrent meteorological phenomenon affects not only populations of adult plants but also the replenishment of the seed bank (Gutiérrez and Meserve 2003). The onset, frequency, and strength of the ENSO before historical times are difficult to establish (DeVries 1987; Tudhope and Collins 2003), but there is reason to believe that the current arid climate has shaped the vegetation of coastal western South America for considerable lengths of time (Gregory-Wodzicki 2000; Hartley 2003).
We attempted to estimate Ne for the wild tomato species using levels of silent diversity and silent divergence (see materials and methods). Assuming one generation per year, estimates for Ne in L. peruvianum vary between 1.165 × 106 and 3.736 × 106, depending on the timing of the split between S. ochranthum and the tomato clade. If we assume only one generation every 7 years, our estimates are reduced to one-seventh of the above values (but see below). Estimates for the other species are much lower, in accordance with the lower levels of nucleotide variation in these species (Table 3). Our estimates of effective population size imply that substitution rates between 1.6 × 10−9 and 5.2 × 10−9 silent substitutions/year (weighted over all loci), depending on the timing of the split between the tomato clade and the outgroup. These estimates are not substantially different from the previously estimated synonymous substitution rates in several angiosperm genes (Moniz de Sá and Drouin 1996; Dvornyk et al. 2002).
Our Ne estimates seem extraordinarily high for plant species characterized by patchy distribution in temporally fluctuating environments (T. Städler, personal observations). Estimates for African D. melanogaster yielded a similar Ne of ∼106 (Li et al. 1999), yet the census size for Drosophila and wild tomatoes differs dramatically. Likewise, in loblolly pine, an outcrossing gymnosperm that is abundant in the southeastern United States, Brown et al. (2004) calculated an effective population size of only 5.6 × 105 and explained the apparent discrepancy between census population size and Ne by glacial population fluctuations.
For wild tomato species, the existence of soil seed banks could explain the apparent incongruence of small, isolated local populations and our high Ne estimates. Nunney (2002) showed that the effective size of a local population is proportional to the generation time, T, for species with seed banks. Average generation times may very well vary between the more mesic habitats of L. hirsutum and the hyperarid conditions experienced by both L. peruvianum and L. chilense in southern Peru and northern Chile, perhaps approaching the period between successive El Niño events in the latter region. One should also keep in mind that species such as L. peruvianum consist of many subpopulations. Thus, assuming the presence of a soil seed bank buffering against genetic bottlenecks and assuming a large number of subpopulations may help to explain our large θsil and Ne estimates.
The above considerations of demographic processes and life-history traits may help to explain why positive directional selection is not a significant force causing reductions of silent diversity in regions of low recombination in wild tomatoes. Even if selective sweeps occur locally in restricted geographic areas, their effects on diversity in the total population are expected to be weak, because the selected alleles may be unable to spread fast through the entire species range.
Conclusions:
Sequence evolution in Lycopersicon seems to be dominated to a large extent by demographic processes, variation in neutral mutation rates, and/or selective constraints among genes, and most likely, background selection. This is in contrast to findings in Drosophila where selective sweeps are likely to be the underlying cause for a strong positive correlation between recombination rate and nucleotide diversity. In wild tomato species, a comparably strong positive correlation is not detected. Despite small census size of local isolated populations, wild outcrossing tomatoes are able to maintain high effective total population sizes. Both observations may be explained by the presence of soil seed banks and extensive population substructure. Demographic processes and life-history traits presumably contribute mostly to the marked differences in diversity levels between partially selfing and obligately outcrossing tomato species. Further studies of additional populations are needed to understand how population substructure, extinction-recolonization processes, and soil seed banks interact to shape levels of diversity in wild tomatoes.
Acknowledgments
We are grateful to G. Feldmaier-Fuchs for invaluable expert laboratory assistance. We thank R. Chetelat and L. Rose for suggestions and discussion and O. Savolainen and two anonymous reviewers whose comments improved the final version of this article. This work was funded by the Deutsche Forschungsgemeinschaft through its Priority Program “Radiations—Origins of Biological Diversity” (SPP-1127) grant Ste 325/5 to W.S.
References
- Aguadé, M., N. Miyashita and C. H. Langley, 1989. Reduced variation in the yellow-achaete-scute region in natural populations of Drosophila melanogaster. Genetics 122: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla, P., and K. Ritland, 1997. Microsatellite variation and evolution in the Mimulus guttatus species complex with contrasting mating systems. Mol. Biol. Evol. 14: 1023–1034. [DOI] [PubMed] [Google Scholar]
- Barrett, S. C. H., 2003. Mating strategies in flowering plants: the outcrossing-selfing paradigm and beyond. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry, E., C. Kerdelhué, H. Innan and W. Stephan, 2001. Species and recombination effects on DNA variability in the tomato genus. Genetics 158: 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and C. F. Aquadro, 1992. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356: 519–520. [DOI] [PubMed] [Google Scholar]
- Brown, G. R., G. P. Gill, R. J. Kuntz, C. H. Langley and D. B. Neale, 2004. Nucleotide diversity and linkage disequilibrium in loblolly pine. Proc. Natl. Acad. Sci. USA 101: 15255–15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo, A. L., and B. A. Schaal, 2004. Population structure and phylogeography of Solanum pimpinellifolium inferred from a nuclear gene. Mol. Ecol. 13: 1871–1882. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., 2003. Effects of inbreeding on the genetic diversity of populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1051–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., B. Charlesworth and M. T. Morgan, 1995. The pattern of neutral molecular variation under the background selection model. Genetics 141: 1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat, R. T., V. Meglic and P. Cisneros, 2000. A genetic map of tomato based on BC1 Lycopersicon esculentum × Solanum lycopersicoides reveals overall synteny but suppressed recombination between these homeologous genomes. Genetics 154: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries, T. J., 1987. A review of geological evidence for ancient El Niño activity in Peru. J. Geophys. Res. 92: 14471–14479. [Google Scholar]
- Dvorak, J., M.-C. Luo and Z.-L. Yang, 1998. Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics 148: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvornyk, V., A. Sirviö, M. Mikkonen and O. Savolainen, 2002. Low nucleotide diversity at the pal1 locus in the widely distributed Pinus sylvestris. Mol. Biol. Evol. 19: 179–188. [DOI] [PubMed] [Google Scholar]
- Fu, Y.-X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal, M. W., R. Czihal, U. Hannappel, D.-U. Kloos, A. Polley et al., 1998. Sequencing of cDNA clones from the genetic map of tomato (Lycopersicon esculentum). Genome Res. 8: 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiady, M. S., R. W. Whitkus and E. M. Lord, 2002. Genetic analysis of traits distinguishing outcrossing and self-pollinating forms of currant tomato, Lycopersicon pimpinellifolium (Jusl.) Mill. Genetics 161: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graustein, A., J. M. Gaspar, J. R. Walters and M. F. Palopoli, 2002. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 161: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory-Wodzicki, K. M., 2000. Uplift history of the central and northern Andes: a review. GSA Bull. 112: 1091–1105. [Google Scholar]
- Gutiérrez, J. R., and P. L. Meserve, 2003. El Niño effects on soil seed bank dynamics in north-central Chile. Oecologia 134: 511–517. [DOI] [PubMed] [Google Scholar]
- Hamrick, J. L., and M. J. W. Godt, 1990. Allozyme diversity in plant species, pp. 43–63 in Plant Population Genetics, Breeding, and Genetic Resources, edited by A. H. D. Brown, M. T. Clegg, A. L. Kahler and B. S. Weir. Sinauer Associates, Sunderland, MA.
- Hartley, A. J., 2003. Andean uplift and climate change. J. Geol. Soc. Lond. 160: 7–10. [Google Scholar]
- Hellmann, I., I. Ebersberger, S. E. Ptak, S. Pääbo and M. Przeworski, 2003. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72: 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., and J. Wakeley, 1997. A coalescent estimator of the population recombination rate. Genetics 145: 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 50: 245–250. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., 2001. Two-locus sampling distributions and their application. Genetics 159: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igic, B., L. Bohs and J. R. Kohn, 2004. Historical inferences from the self-incompatibility locus. New Phytol. 161: 97–105. [Google Scholar]
- Ingvarsson, P. K., 2002. A metapopulation perspective on genetic diversity and differentiation in partially self-fertilizing plants. Evolution 56: 2368–2373. [DOI] [PubMed] [Google Scholar]
- Innan, H., and W. Stephan, 2003. Distinguishing the hitchhiking and background selection models. Genetics 165: 2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- Kraft, T., T. Säll, I. Magnusson-Rading, N.-O. Nilsson and C. Halldén, 1998. Positive correlation between recombination rates and levels of genetic variation in natural populations of sea beet (Beta vulgaris subsp. maritima). Genetics 150: 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. J., Y. Satta and N. Takahata, 1999. Paleo-demography of the Drosophila melanogaster subgroup: application of the maximum likelihood method. Genes Genet. Syst. 74: 117–127. [DOI] [PubMed] [Google Scholar]
- Liu, F., D. Charlesworth and M. Kreitman, 1999. The effect of mating system differences on nucleotide diversity at the phosphoglucose isomerase locus in the plant genus Leavenworthia. Genetics 151: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, W. P., and D. R. Maddison, 1992. MacClade3: Analysis of Phylogeny and Character Evolution. Sinauer Associates, Sunderland, MA.
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- McVean, G., P. Awadalla and P. Fearnhead, 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. C., and S. D. Tanksley, 1990. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 80: 437–448. [DOI] [PubMed] [Google Scholar]
- Moniz de Sá, M., and G. Drouin, 1996. Phylogeny and substitution rates of angiosperm actin genes. Mol. Biol. Evol. 13: 1198–1212. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W., 1997. Patterns of DNA variability at X-linked loci in Mus domesticus. Genetics 147: 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M. W., V. L. Bauer, S. L. Crowell and C. F. Aquadro, 1998. DNA variability and recombination rates at X-linked loci in humans. Genetics 150: 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nordborg, M., and P. Donnelly, 1997. The coalescent process with selfing. Genetics 146: 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney, L., 2002. The effective size of annual plant populations: the interaction of a seed bank with fluctuating population size in maintaining genetic variation. Am. Nat. 160: 195–204. [DOI] [PubMed] [Google Scholar]
- Peralta, I. E., and D. M. Spooner, 2001. Granule-bound starch synthase (GBSSI) gene phylogeny of wild tomatoes (Solanum L. section Lycopersicon [Mill.] Wettst. subsection Lycopersicon). Am. J. Bot. 88: 1888–1902. [PubMed] [Google Scholar]
- Pollak, E., 1987. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics 117: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, C. M., 1979. Biosystematic studies in Lycopersicon and closely related species of Solanum, pp. 667–678 in The Biology and Taxonomy of the Solanaceae, edited by J. G. Hawkes, R. N. Lester and A. D. Skelding. Academic Press, New York.
- Rick, C. M., 1983. Evolution of mating systems: evidence from allozyme variation, pp. 215–221 in Genetics: New Frontiers (Proc. XV International Congress of Genetics). Oxford and IBH Publishing, New Delhi.
- Rick, C. M., and R. Lamm, 1955. Biosystematic studies on the status of Lycopersicon chilense. Am. J. Bot. 42: 663–675. [Google Scholar]
- Rick, C. M., and J. I. Yoder, 1988. Classical and molecular genetics of tomato: highlights and perspectives. Annu. Rev. Genet. 22: 281–300. [DOI] [PubMed] [Google Scholar]
- Rick, C. M., E. Kesicki, J. F. Fobes and M. Holle, 1976. Genetic and biosystematic studies on two new sibling species of Lycopersicon from Interandean Peru. Theor. Appl. Genet. 47: 55–68. [DOI] [PubMed] [Google Scholar]
- Rick, C. M., J. F. Fobes and M. Holle, 1977. Genetic variation in Lycopersicon pimpinellifolium: evidence of evolutionary change in mating system. Plant Syst. Evol. 127: 139–170. [Google Scholar]
- Rick, C. M., M. Holle and R. W. Thorp, 1978. Rates of cross-pollination in Lycopersicon pimpinellifolium: impact of genetic variation in floral characters. Plant Syst. Evol. 129: 31–44. [Google Scholar]
- Rick, C. M., J. F. Fobes and S. D. Tanksley, 1979. Evolution of mating systems in Lycopersicon hirsutum as deduced from genetic variation in electrophoretic and morphological characters. Plant Syst. Evol. 132: 279–298. [Google Scholar]
- Rose, L., 2002. The population genetics and functional analysis of the Pto disease resistance gene in Lycopersicon spp. and the RPP13 gene in Arabidopsis thaliana. Ph.D. Thesis, University of California, Davis, CA.
- Rozas, J., J. C. Sanchéz-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Savolainen, O., C. H. Langley, B. P. Lazzaro and H. Fréville, 2000. Contrasting patterns of nucleotide polymorphism at the alcohol dehydrogenase locus in the outcrossing Arabidopsis lyrata and the selfing Arabidopsis thaliana. Mol. Biol. Evol. 17: 645–655. [DOI] [PubMed] [Google Scholar]
- Schoen, D. J., and A. H. D. Brown, 1991. Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc. Natl. Acad. Sci. USA 88: 4494–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner, D. M., G. J. Anderson and R. K. Jansen, 1993. Chloroplast DNA evidence for the interrelationships of tomatoes, potatoes, and pepinos (Solanaceae). Am. J. Bot. 80: 676–688. [Google Scholar]
- Spooner, D. M., I. E. Peralta and S. Knapp, 2005. Comparison of AFLPs with other markers for phylogenetic inference in wild tomatoes. [Solanum L. section Lycopersicon (Mill.) Wettst.]. Taxon 54: 43–61. [Google Scholar]
- Städler, T., K. Roselius and W. Stephan, 2005. Genealogical footprints of speciation processes in wild tomatoes: demography and evidence for historical gene flow. Evolution 59: 1268–1279. [PubMed] [Google Scholar]
- Stephan, W., and C. H. Langley, 1989. Molecular genetic variation in the centromeric region of the X chromosome in three Drosophila ananassae populations. I. Contrasts between the vermilion and forked loci. Genetics 121: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, W., and C. H. Langley, 1998. DNA polymorphism in Lycopersicon and crossing-over per physical length. Genetics 150: 1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart, A. L., and J. H. Willis, 2003. Patterns of nucleotide diversity in two species of Mimulus are affected by mating system and asymmetric introgression. Evolution 57: 2490–2506. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S. D., M. W. Ganal, J. P. Prince, M. C. de Vicente, M. W. Bonierbale et al., 1992. High density molecular linkage map of the tomato and potato genomes. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, I. B., 1986. Biosystematics of the tomato, pp. 1–34 in The Tomato Crop: A Scientific Basis for Improvement, edited by J. G. Atherton and J. Rudich. Chapman & Hall, London.
- Tenaillon, M. I., M. C. Sawkins, A. D. Long, R. L. Gaut, J. F. Doebley et al., 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, L. K. Anderson, S. M. Stack, J. Doebley et al., 2002. Patterns of diversity and recombination along chromosome 1 of maize (Zea mays ssp. mays L.). Genetics 162: 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., J. U'Ren, O. Tenaillon and B. S. Gaut, 2004. Selection versus demography: a multilocus investigation of the domestication process in maize. Mol. Biol. Evol. 21: 1214–1225. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudhope, S., and M. Collins, 2003. The past and future of El Niño. Nature 424: 261–262. [DOI] [PubMed] [Google Scholar]
- Wall, J. D., 2000. A comparison of estimators of the population recombination rate. Mol. Biol. Evol. 17: 156–163. [DOI] [PubMed] [Google Scholar]
- Wang, R. L., J. Wakeley and J. Hey, 1997. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics 147: 1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., B. Lauga and D. Charlesworth, 2002. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19: 1407–1420. [DOI] [PubMed] [Google Scholar]