Abstract
Recent studies have highlighted the dangers of using haplotypes reconstructed directly from population data for a fine-scale mapping analysis. Family data may help resolve ambiguity, yet can be costly to obtain. This study is concerned with the following question: How much family data (if any) should be used to facilitate haplotype reconstruction in a population study? We conduct a simulation study to evaluate how changes in family information can affect the accuracy of haplotype frequency estimates and phase reconstruction. To reconstruct haplotypes, we introduce an EM-based algorithm that can efficiently accommodate unrelated individuals, parent-child trios, and arbitrarily large half-sib pedigrees. Simulations are conducted for a diverse set of haplotype frequency distributions, all of which have been previously published in empirical studies. A wide variety of important results regarding the effectiveness of using pedigree data in a population study are presented in a coherent, unified framework. Insight into the different properties of the haplotype frequency distribution that can influence experimental design is provided. We show that a preliminary estimate of the haplotype frequency distribution can be valuable in large population studies with fixed resources.
THERE is currently a strong interest in how best to use linkage disequilibrium (LD) information for fine-scale mapping and association analysis of complex traits. A growing number of studies demonstrate that haplotype-based approaches may provide more power and accuracy in locating quantitative trait loci (QTL) and causative disease variants than single-locus methods (see, e.g., Fallin et al. 2001; Morris et al. 2002; Zhao et al. 2003). Since haplotypes are typically not observed in vitro, haplotype-based studies are likely to follow a two-step procedure: first, haplotypes are inferred from a sample of phase-unknown genotypes using a computational algorithm, and second, inferred haplotypes are fed into a multilocus LD model, where they are treated as having been directly observed.
There are two principal approaches to inferring haplotypes from population data, both with potential drawbacks. One approach is to use family data, which may be able to deterministically resolve phase for genotypes featuring multiple heterozygous loci. However, ascertaining this information can be costly. When resources are fixed, it may actually be more efficient to use a genotype-based mapping model rather than reallocate resources to ascertain family data for haplotype reconstruction (Grapes et al. 2004).
A second approach is to infer haplotypes directly from population data. A variety of statistical algorithms exist for random-mating populations, and good comparative surveys are available (see, e.g., Stephens and Donnelly 2003; Zhao et al. 2003). A problem with reconstructing haplotypes using these models is that there may be considerable uncertainty associated with the inferred haplotypes. It was recently demonstrated that failing to account for this uncertainty can result in unreliable location estimates in a subsequent mapping analysis (Morris et al. 2004). These results indicate that, when possible, haplotype-based analyses should be modified to efficiently accommodate this uncertainty. When the two-stage procedure must be used, it is important to understand the factors that will limit its effectiveness. One important factor is likely to be the haplotype frequency distribution. This is because, under the standard assumption of random mating, the data can be regarded as independent samples from this distribution.
In this article, we illustrate the different ways in which the haplotype frequency distribution can affect the accuracy of both the phase assignments and the haplotype frequency estimates. We also examine the effectiveness of using family data to improve accuracy for different frequency profiles.
To facilitate our analysis, we begin by introducing an EM-based haplotype reconstruction model that can accommodate outbred half-sib pedigrees, unrelated individuals, and family-child trios. The method is efficient for arbitrarily large sibships with missing marker data and will be of interest to studies of hierarchical population structures, including those in populations of many natural and domestic animal species.
The remainder of the article is devoted to simulating and analyzing results, using a diverse set of published haplotype frequencies. We describe summary statistics that can be calculated directly from inferred frequency data and that can be used to predict the accuracy of phase assignment and the usefulness of family data. A wide variety of results that either extend or complement existing analysis are presented in a coherent, unified framework.
METHODS
Notation and modeling assumptions:
We consider a candidate region in the genome characterized by L tightly linked biallelic loci. Let h = h1, … , hM denote the M = 2L possible haplotypes, and let Θ = (θ1, … , θM) denote corresponding haplotype frequencies in the target population.
For a large, panmictic population, we can specify the probability of observing a given phase configuration, z = (hi, hj) as
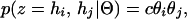 |
(1) |
where c = 1 if the individual is a homozygote (i.e., i = j) or 2 if the individual is a heterozygote (i.e., i ≠ j).
Similarly, the probability of observing a given phase-unknown genotype, y, in a panmictic population is
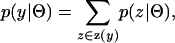 |
(2) |
where z(y) is the set of all possible phase configurations that can resolve y. Note that the size of z(y) increases exponentially with the number of heterozygous loci.
Haplotype reconstruction via the EM algorithm:
Let y = y1, … , yN denote a sample of N phase-unknown genotypes. The objective of the EM-based approach to haplotype reconstruction is to calculate the maximum-likelihood estimate of Θ given y. Haplotype frequency estimates can then be used to reconstruct phase.
An important initial assumption concerns any underlying pedigree structure of the data. It is useful to make these assumptions explicit by characterizing the observed data using two parameters: the marker data, y, and a representation of the underlying pedigree structure, ℱ. When individuals are unrelated, or no additional family information is included, we set 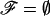 .
.
A central challenge of any likelihood-based pedigree model is achieving reasonable computational complexity for a given ℱ, and one of the principal contributions of our method is that we provide an efficient algorithm for sparse half-sib pedigrees. It is useful to first review the EM-based approach for unrelated individuals.
EM algorithm for unrelated individuals:
The EM algorithm for unrelated individuals has been developed and evaluated in many different contexts (see, e.g., Hill 1974; Terwilliger and Ott 1994; Excoffier and Slatkin 1995; Hawley and Kidd 1995; Fallin and Shorck 2000; Kirk and Cardon 2002; Qin et al. 2002). The general approach entails augmenting the observed phase-unknown genotypes for each member in the sample by the corresponding phase configurations. We denote these latent phase configurations by z = z1, … , zN. Each iteration of the EM algorithm requires calculating the expected log-likelihood of the augmented data,
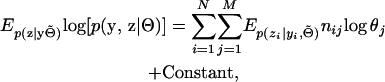 |
(3) |
where 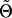 denotes the current estimate of Θ and nij refers to the number of times haplotype j appears in the phase configuration of individual i. Once (3) has been calculated, the result is maximized with respect to Θ and the process is repeated until
denotes the current estimate of Θ and nij refers to the number of times haplotype j appears in the phase configuration of individual i. Once (3) has been calculated, the result is maximized with respect to Θ and the process is repeated until 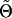 converges at a maximum,
converges at a maximum, 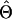 .
.
Note that without the expectation, (3) is a straightforward description of “gene counting.” The added complexity incurred from using the EM algorithm is therefore attributed to calculating 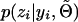 . This quantity is given by dividing Equation 1 by Equation 2.
. This quantity is given by dividing Equation 1 by Equation 2.
The complexity is dominated by calculating the terms in (2), which increase linearly with the number of possible phase configurations. However, the number of possible phase configurations increases exponentially with the number of heterozygous genotypes, which limits the number of SNPs that can reasonably be evaluated. This constraint will apply to any EM-based approach that requires estimating the distribution of phase given marker data, including the one we are about to describe for outbred half-sib pedigrees. It is worth mentioning that one way to accommodate a large number of markers is to use the partition-ligation algorithm introduced by Niu et al. (2002). While this algorithm was developed as part of a Bayesian model, it has since been incorporated into the EM framework (Qin et al. 2002).
EM algorithm for outbred half-sib pedigrees:
We now consider the case where members of y may be related through one of P ≤ N sires. If P < N, then the likelihood in (3) is no longer valid since the summation over N follows only from assuming that each animal is unrelated.
One EM-based approach that accommodates nuclear family information is described by Rohde and Fuerst (2001). The model treats the parents as independent and augments the marker data by the latent parental phase configurations. The E-step entails enumerating phase configurations that are consistent with parent and progeny marker data and then evaluating each configuration. This enumerative approach is sufficient for small sibships, but it is computationally infeasible for a sib structure of moderate size. For sparse family structures, such as half-sib pedigrees with untyped parents, it is not a realistic strategy.
The starting point for our model follows a generic approach described by O'Connell (2000), and entails augmenting the data by the latent phase configurations of the founders. The E-step would then require calculating the marginal distribution of each founder given the marker data specific to that founder's pedigree.
Let the latent phase configurations of the sires and dams be denoted s and d, respectively. The expectation of the augmented data (omitting the constant term) is
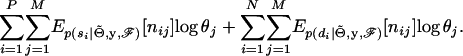 |
(4) |
The challenge is therefore how to calculate 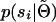 and
and 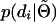 . This can be done in any number of ways, but many potentially relevant techniques have been developed in the context of human linkage analysis. In the appendix we provide a formal derivation of the approach we use for calculating the marginal distributions of the sire and dams. Briefly, inferring
. This can be done in any number of ways, but many potentially relevant techniques have been developed in the context of human linkage analysis. In the appendix we provide a formal derivation of the approach we use for calculating the marginal distributions of the sire and dams. Briefly, inferring 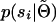 follows a straightforward application of the peeling algorithm (Elston and Stewart 1971). Inferring
follows a straightforward application of the peeling algorithm (Elston and Stewart 1971). Inferring 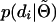 can be achieved by adopting general principles used to simulate pedigree data (Ploughman and Boehnke 1989). Using these techniques results in a computational complexity that is cubic in the number of phase configurations. This is still two orders of magnitude worse than the complexity for unrelated individuals. In the appendix, we also demonstrate how to achieve quadratic complexity for both sire and dam. In the context of haplotype reconstruction, the corresponding reduction in computational resources can be substantial.
can be achieved by adopting general principles used to simulate pedigree data (Ploughman and Boehnke 1989). Using these techniques results in a computational complexity that is cubic in the number of phase configurations. This is still two orders of magnitude worse than the complexity for unrelated individuals. In the appendix, we also demonstrate how to achieve quadratic complexity for both sire and dam. In the context of haplotype reconstruction, the corresponding reduction in computational resources can be substantial.
The methods of both O'Connell (2000) and Rohde and Fuerst (2001) assume nonrecombinant haplotypes, which is realistic for small genomic regions or over regions lacking recombination hotspots. We make the same assumption and note that O'Connell (2000) outlines a strategy for dealing with recombinant regions that is similar to the partition-ligation algorithm (Niu et al. 2002) for accommodating large numbers of loci.
Our algorithm also provides a unified framework to evaluate arbitrarily large half-sib pedigrees, parent-child trios (a sibship of size one), and unrelated individuals. When no additional family data are provided, results are the same as using the standard EM algorithm for unrelated individuals.
SIMULATIONS
We conduct a simulation study based on independent half-sib pedigrees, using empirically derived haplotype data. Our simulation strategy is divided into the following three steps: (1) specification of a haplotype frequency distribution for the parental population, (2) simulation of genotypes for independent half-sib pedigrees, and (3) estimation of haplotype frequencies and phase configurations using alternate categories of family data.
We fix the number of sampled individuals at 100 and consider sib sizes of 1, 2, 5, 10, and 25. The categories of family information that are used with each sample when reconstructing haplotypes are given in Table 1. Family sizes are exact, and therefore results for samples featuring a family of size 1 correspond to unrelated individuals or, if parental genotypes are provided, to parent-child trios. One consequence of using this simulation strategy is that increasing the size of a sibship will reduce the number of independent haplotypes in a given sample. This allows us to evaluate whether the resolving power from additional pedigree data compensates for the loss in independent haplotypes (i.e., whether the improved quality of the data compensates for the reduced quantity).
TABLE 1.
Categories of family data that can be included with a sample of phase-unknown genotypes in a simulation study
| Category | Description |
|---|---|
| UN | No information—all animals assumed unrelated |
| P | Pedigree structure only (parents untyped) |
| PS | Pedigree structure + sire genotype |
| PSD | Pedigree structure + sire and dam genotypes |
The most important parameter in the simulation study is the parental haplotype frequency distribution. Our analysis is based on three empirically derived haplotype frequency distributions. The first two frequency profiles, APOE1 and APOE2, were provided by Fallin et al. (2001) and correspond to two sets of marker data for a control group used in an association study for Alzheimer's disease. The third data set, IL8E, was presented by Hull et al. (2001) and corresponds to haplotype frequency estimates of a European sample for six biallelic loci spanning a 7.6-kb region within the IL8 locus.
One of the central results from the simulation study is that the expected accuracy for any EM-based quantity will be different for each of the population frequency distributions. It is useful to identify relevant summary statistics that capture the relative performance that can be expected for random samples from each of the populations.
Qin et al. (2002) demonstrate that the expected information in a random sample of phase-unknown genotypes can be expressed as the sum of two components: the first component reflects the variance of 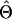 if phase configurations are observed, while the second component reflects the loss of information because of unknown phase configurations.
if phase configurations are observed, while the second component reflects the loss of information because of unknown phase configurations.
The first component can be approximated by the gene diversity, 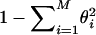 , which is a measure of the uniformity of the frequency distribution.
, which is a measure of the uniformity of the frequency distribution.
To describe the additional uncertainty from the unknown phase configurations, we use the expected error rate using the most likely phase configuration. Consider a given phase-unknown genotype, y. The probability that the most likely phase configuration is the correct one is given by max p(z|y, Θ). A measure of the uncertainty from not knowing phase for this genotype is 1 − max p(z|y, Θ). The expectation of incorrectly assigning phase for a random sample is therefore
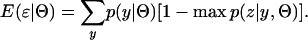 |
(5) |
Appreciating the relevance of Equation 5 in the context of accurate EM-based phase reconstruction cannot be overstated. The expression describes the number of incorrect phase assignments that is expected in a population sample when the most likely phase configuration is used and haplotype frequencies are known. It can therefore be considered a lower bound on the number of errors that are calculated from haplotype frequencies inferred by the EM algorithm.
These two statistics are presented in Table 2. If the population haplotype frequency for APOE1 were known with certainty, we would expect to get no greater than 82% of the sample correct if the most likely phase criterion is used. By contrast, phase assignment using the most likely phase criterion would be virtually error free for population samples generated from the IL8E distribution, even though the expected number of ambiguous genotypes (i.e., genotypes with two or more heterozygous loci) is similar to that in APOE1. Although there are multiple phase configurations that can, in theory, resolve an ambiguous genotype sampled from IL8E, the vast majority of these will feature at least one haplotype that does not actually segregate in the population. This demonstrates that family data may be unnecessary for accurate phase reconstruction, even when a sample features many ambiguous genotypes.
TABLE 2.
Summary statistics for haplotype frequency estimates used in data analysis
| APOE1 | APOE2 | IL8E | |
|---|---|---|---|
| No. of loci | 4 | 4 | 6 |
| No. of haplotypes | 13 | 10 | 9 |
| Ambiguous genotypesa | 0.52 | 0.41 | 0.53 |
| Gene diversity | 0.86 | 0.8 | 0.56 |
| Frequency-known error rateb | 0.18 | 0.12 | 0.0003 |
The expected proportion of a population sample that is heterozygous for at least two loci.
The expected proportion of phase configurations that will be incorrectly resolved in a population sample when the most likely phase criterion is used and haplotype frequencies are known.
RESULTS
We describe the results from our simulation study using two standard summary statistics based on haplotype frequency estimates and phase accuracy. For assessing haplotype frequency estimates, we use the discrepancy metric (Excoffier and Slatkin 1995; Kirk and Cardon 2002), which is defined as
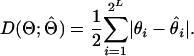 |
(6) |
For the phase configurations, we use the estimated frequencies to assign each individual its most likely phase configuration and then calculate the percentage of individuals that are incorrectly assigned. This metric is appropriate since it is the typical criterion on which haplotypes are assigned for use in a fine-scale mapping analysis.
Results for these two measures of accuracy are presented in Table 3. Table 3 is structured to highlight a wide variety of trends, some of which are indexed by letters that are referenced in the text. When referring to an entry in Table 3 indexed by X, we use the notation (X). We include only indexes for the APOE1 results since annotating each table would have obscured trends. It is contextually clear which distribution(s) is relevant to supporting a given statement. Similarly, we do not include standard errors. Comparative statements were verified at the 95% significance level using a paired t-test.
TABLE 3.
Impact of family size, family information, and parental haplotype distribution on estimated haplotype frequencies, as measured by the discrepancy statistic (top) and on phase reconstruction error (bottom)
| Family size | UN | P | PS | PSD | ||
|---|---|---|---|---|---|---|
| Haplotype frequency discrepancies | ||||||
| APOE1 | 1 | 0.116 A | 0.116 A, B, F | 0.078 B | 0.063 B | |
| 2 | 0.123 | 0.117 | 0.095 | 0.070 | ||
| 5 | G | 0.143 | 0.119 | 0.109 F | 0.077 | |
| 10 | 0.162 | 0.119 E | 0.116 E | 0.081 | ||
| 25 | 0.212 | 0.122 C | 0.122 C, D | 0.082 D | ||
| APOE2 | 1 | 0.082 | 0.082 | 0.061 | 0.049 | |
| 2 | 0.091 | 0.088 | 0.077 | 0.060 | ||
| 5 | 0.110 | 0.097 | 0.090 | 0.065 | ||
| 10 | 0.132 | 0.103 | 0.099 | 0.066 | ||
| 25 | 0.180 | 0.104 | 0.104 | 0.070 | ||
| IL8E | 1 | 0.047 | 0.047 | 0.037 | 0.032 | |
| 2 | 0.054 | 0.054 | 0.051 | 0.040 | ||
| 5 | 0.065 | 0.060 | 0.058 | 0.043 | ||
| 10 | 0.085 | 0.065 | 0.063 | 0.046 | ||
| 25 | 0.110 | 0.068 | 0.068 | 0.047 | ||
| Phase reconstruction error rate | ||||||
| APOE1 | 1 | 0.21 A′ | 0.21 B′, F′ | 0.12 B′, H | 0.06 B′, I | |
| 2 | 0.20 | 0.17 | 0.10 | 0.03 | ||
| 5 | G′ | 0.19 | 0.12 H | 0.06 F′, I | 0.02 | |
| 10 | 0.18 J | 0.07 J | 0.05 | 0.02 | ||
| 25 | 0.15 J | 0.06 J | 0.06 | 0.02 | ||
| APOE2 | 1 | 0.13 | 0.13 | 0.08 | 0.04 | |
| 2 | 0.13 | 0.11 | 0.07 | 0.03 | ||
| 5 | 0.13 | 0.09 | 0.05 | 0.02 | ||
| 10 | 0.11 | 0.06 | 0.05 | 0.02 | ||
| 25 | 0.11 | 0.05 | 0.04 | 0.02 | ||
| IL8E | 1 | 0.00 | 0.00 | 0.00 | 0.00 | |
| 2 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| 5 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| 10 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| 25 | 0.00 | 0.00 | 0.00 | 0.00 | ||
Each entry for a given metric corresponds to the average value for 100 replicates. Italic entries denote estimates that were obtained using the standard EM algorithm for unrelated individuals, and underlined italic entries denote estimates obtained by treating related individuals as unrelated. Letters are referenced in the text. See Table 1 for category definitions.
Table 3 also features results from a standard analysis using the EM algorithm for unrelated individuals (column in italics). These will be useful when discussing the results from treating related individuals as unrelated (underlined italics). Note that using our algorithm gives the same results as those of the EM algorithm for unrelated individuals when no family data are provided (Table 3, A). Hence either entry can be used to describe accuracy for 100 unrelated individuals, which is often useful as a base comparison to other scenarios that use family data.
Broadly, we are interested in how changes in family data, sample size, and frequency distribution affect each of the two measures of accuracy. We are also interested in whether trends observed for one measure of accuracy apply to the other one. Since the number of progeny is fixed at 100, we measure sample size by the number of independent haplotypes segregating in the sample.
We begin by focusing exclusively on the accuracy of haplotype frequency estimation. We then provide a similar analysis for phase reconstruction accuracy, highlighting how the two metrics differ in sensitivity to family data. These two sections collectively illustrate the importance of the true haplotype frequency distribution in determining the magnitude of reconstruction error as well as the effectiveness of reallocating resources for family data. In the last section, we discuss the sensitivity of a popular case-control association test to biased frequency estimates that are inferred by treating related individuals as unrelated.
Impact of family data on haplotype frequency estimation:
We start by observing that for a given family size, increasing family information typically results in an improvement in accuracy (i.e., discrepancy decreases along a given row). However, adding family information does not always contribute to accuracy, as can been seen in the case of adding the genotype from a single parent (Table 3, C). This is because the number of progeny is sufficient to explain the parental phase and therefore the sire genotype provides redundant information. By contrast, there is always an improvement in discrepancy if both parental genotypes are included (Table 3, D). This is because the second parental genotype will always provide information regarding an additional independent haplotype (which follows from our assumption of one progeny per dam).
This example demonstrates how increasing the number of independent haplotypes or the amount of family information improves accuracy. The question of whether resources intended for population data should be reallocated for family data is concerned with whether one should be increased at the expense of the other. This question was addressed for the case of nuclear families vs. unrelated individuals in several studies, which showed that the optimal allocation decision will be frequency dependent (Becker and Knapp 2002; Schaid 2002). Our results illustrate that these frequency-dependent trade-offs between the quality and quantity of population data can be found for many pedigree configurations. Specifically, we compare the accuracy of 200 independent haplotypes from a sample of unrelated individuals to 140 independent haplotypes segregating in 20 half-sib pedigrees of size 5 with a typed sire (Table 3, F). For the APOE1 distribution, better accuracy is achieved from using more family data and fewer independent haplotypes, while for the APOE2 and IL8E distributions more independent data are preferable to family data. As discussed in the previous section, a random sample generated from the APOE1 distribution will have the most uncertainty associated with phase assignments and therefore will benefit most from family data.
We also observe that when family data are ignored (i.e., related individuals are treated as unrelated) discrepancy increases with family size (Table 3, G). This follows since increasing family size (i.e., increasing the number of conditional dependencies in the data) implies further deviation from the assumption of unrelated (independent) individuals. We evaluate these results squarely in the context of association analysis at the end of this section.
Impact of family data on phase reconstruction accuracy:
The most striking result is the uniformly perfect phase reconstruction given by IL8E, which provides an example of a distribution where family data are redundant despite >50% of a sample containing ambiguous genotypes. These results are consistent with the frequency-known error rate given in Table 2. We note that for all three distributions, the observed error rate (Table 3, A′) is fairly close to the frequency-known error rate, which is the best-case average error rate that can be achieved when using the EM algorithm. In this context, it is reasonable to claim that EM-based phase assignments are accurate.
For the APOE1 and APOE2 distributions, increasing sib size and adding parental marker data always improve phase reconstruction accuracy. Specifically, as we move down a given column or across a row for either distribution, we observe a gradual decrease in phase reconstruction error rate. It should be noted that when resources are fixed, increasing family size decreases the number of independent haplotypes used in the subsequent study, and therefore this gain in phase reconstruction accuracy may not be justified.
While phase reconstruction error decreases with family information, it is not eliminated. Even for very large sib sizes, there is a small, but significant error when both parental genotypes are provided. Note also that for both distributions, there is also a discernable increase in the error rate when only the genotype for the common parent is provided. However, results for the APOE1 distribution are consistently worse than those for the APOE2 distribution. These observations highlight the importance that both the frequency distribution and the pedigree structure have in determining whether resources should be allocated to ascertain family data.
Although increasing both sib size and parental marker information will improve phase reconstruction accuracy, obtaining parental marker data is more efficient than adding more half-sibs. For both distributions, introducing genotype data for an untyped parent is more efficient than introducing as many as five additional half-sibs (Table 3, H and I). This complements the results that show that two full-sibs with untyped parents can be very inefficient in the context of optimal frequency estimation (Schaid 2002).
It is important to recognize that reconstruction accuracy for progeny does not extend to parents. This means that parental phase may still be incorrectly reconstructed even when reconstruction is accurate for progeny. Adding half-sibs will help reconstruct phase for an untyped common parent, yet our results show that the total number of half-sibs needed to make this parental genotype redundant can be quite large. For each of the three distributions, we see that a typed sire still provides a small, but significant, improvement in discrepancy when as many as 10 progeny are available (Table 3, E). Introducing sibs without genotyping parents can actually be worse than reconstruction from population data if this information is to be used in a subsequent LD model that relies on haplotype accuracy of both parents and progeny (Meuwissen et al. 2002; Lee and van der Werf 2004).
Note that the frequency-dependent trade-off between independent haplotypes and family size that was observed for optimal haplotype frequency estimation (Table 3, F) is not applicable to optimal phase reconstruction accuracy (Table 3, F′). Although increasing family information tends to improve both haplotype reconstruction accuracy and phase reconstruction accuracy the impact of family data differs. For example, the gain achieved when introducing the genotype for one parent rather than for both is larger when the first parent's genotype is provided in the context of haplotype frequency estimation (Table 3, B), yet each parent makes roughly equal contribution to accuracy in the context of phase reconstruction accuracy (Table 3, B′).
There is an actual contradiction in trends between frequency estimation and phase assignment when related individuals are treated as unrelated. Specifically, we see that phase accuracy improves as the number of related individuals that are treated as unrelated increases (Table 3, G′). This paradox can be explained by noting that for more closely related individuals, there is a higher probability that two haplotypes are identical by descent (i.e., more homozygosity) in the genotype data. Fallin and Shorck (2000) made a similar observation when investigating the robustness of the EM algorithm to departures from Hardy-Weinberg Equilibrium by imposing homozygosity on the haplotype data. When individuals are related, however, the appropriate comparison is not the “benefit” in phase reconstruction accuracy relative to unrelated individuals, but the loss by not properly accounting for the conditional dependencies in the data. This loss can be quite large as seen in the case of APOE1, where >10 haplotypes are incorrectly assigned by not accounting for underlying pedigree structure (Table 3, J).
An application to association analysis:
We now evaluate the discrepancy that arises from treating related individuals as unrelated (Table 3, G), in the context of an association analysis. This is relevant for many natural and domestic populations, where, in the absence of pedigree information, a sample may be treated as unrelated even through the sampled members can be closely related. A popular test that utilizes haplotype frequencies directly is a chi-square test, which is used to detect association in a case-control study (Zhao et al. 2000). While variants of the test have been proposed (Fallin et al. 2001), the underlying principle remains that if a causative variant is linked to a marker haplotype, then case haplotype frequencies should differ from those of the control frequencies. A type I error occurs when the reconstructed frequencies for case and control data are considered significantly different, yet the markers are actually unlinked with a causative variant. Treating related individuals as unrelated might be expected to increase the probability of committing a type I error, since variability that naturally arises between two groups sampled from the same neutral region will be accentuated when the data are dependent. To investigate the impact of using these reconstructed frequencies on a type I error, we simulate neutral marker data for hypothetical cases and controls from two repetitions of the simulation process described above. We estimate frequencies under the assumption that individuals are unrelated and then calculate the chi-square statistic on the basis of the reconstructed frequencies. The results are presented in Table 4, columns 4 and 5. They reveal that an inflated type I error is likely to be realized for a family size of 5 for each of the distributions. Even though the actual discrepancy statistic is uniformly greatest for the APOE1 and least for IL8E, the rate at which significance increases with family size is fairly consistent for each of the three distributions: the principal difference lies in the standard deviation. We also ran a standard single-locus Hardy-Weinberg test for each of the loci and present the results in Table 4, columns 2 and 3. This reveals that data sets that are sufficiently related to exhibit a type I error in case-control association analysis will not be detected using the standard single-locus Hardy-Weinberg test.
TABLE 4.
Impact of treating half-sibs as unrelated on two nonparametric tests: single-locus tests for Hardy-Weinberg Disequilibrium and nonhomogeneity of haplotype frequency profiles for case control data in a neutral genomic region
| Single-locus analysis for Hardy-Weinberg disequilibriuma
|
Association test using haplotype frequency estimates
|
||||
|---|---|---|---|---|---|
| Family size | χ2 (1) | SD | χ2 (8) | SD | |
| IL8E | 1 | 1.69 | 2.23 | 9.32 | 4.05 |
| 5 | 1.71 | 1.87 | 14.53* | 6.57 | |
| 10 | 1.67 | 1.74 | 18.78** | 9.22 | |
| 25 | 2.17 | 2.42 | 26.17*** | 16.37 | |
| 50 | 3.55* | 4.27 | — | — | |
| APOE2 | 1 | 2.47 | 2.05 | 9.37 | 4.12 |
| 5 | 2.29 | 1.99 | 14.76* | 7.09 | |
| 10 | 2.24 | 1.87 | 21.35** | 9.69 | |
| 25 | 2.79 | 2.61 | 35.73*** | 17.91 | |
| 50 | 4.50** | 4.42 | — | — | |
| APOE1 | 1 | 2.43 | 2.11 | 12.51 | 4.66 |
| 5 | 2.43 | 1.99 | 21.41* | 7.47 | |
| 10 | 2.36 | 2.00 | 27.66*** | 12.24 | |
| 25 | 2.99 | 2.77 | 47.68*** | 19.77 | |
| 50 | 5.58** | 4.95 | — | — | |
*Significance at 90%; **significance at 95%; ***significance at 99%. Numbers of loci are in parentheses.
For each replicate, the test statistic for the locus exhibiting the highest disequilbirum was used.
DISCUSSION
The results of this study have significant implications for an experimental design using two-stage haplotype analysis. The effectiveness of a two-stage haplotype analysis will be contingent on two factors: (1) the magnitude of the estimation error, which we have shown depends on the haplotype frequency distribution, and (2) the sensitivity of the subsequent haplotype-based analysis to this estimation error, which can be determined from simulation studies.
EM-based haplotype frequency estimates are often considered accurate for sample sizes consisting of ∼100 individuals (Fallin and Shorck 2000; Qin et al. 2002). For smaller samples sizes, or for samples featuring large amounts of missing genotype data, a Bayesian approach may be more appropriate (Stephens et al. 2001; Niu et al. 2002). The EM algorithm is also considered to provide accurate results under departures from the random-mating assumption (Fallin and Shorck 2000). However, our results demonstrate that failing to account for related individuals in a sample (by treating each member in the sample as independent) can lead to an appreciable increase in type I error in a study where two populations are contrasted using EM-based haplotype frequencies. In natural populations featuring sibships, it is important to ascertain pedigree information, e.g., by sibship reconstruction methods described by Thomas and Hill (2002), before using this test.
The results of Morris et al. (2004) clearly demonstrate that phase assignments based on the most likely phase criterion should not be blindly used in a subsequent haplotype-based analysis. We show that while reconstruction errors may be unavoidable (i.e., independent of sample size), this error rate can be calculated directly from the frequency distribution. We also provide an example where the most likely phase criterion will yield perfectly accurate results, even for a sample containing a large proportion of ambiguous genotypes.
Practically, we can calculate the error rate from an estimated frequency distribution that is based on a preliminary sampling of the population. This estimated error rate can then be compared against a predetermined threshold denoting the minimum level of phase accuracy. If the predicted phase reconstruction error exceeds this threshold, then either pedigree data are required or an alternative to the two-stage approach must be used.
The effectiveness of pedigree data will also depend on the haplotype frequency distribution, as well as on the type of pedigree information provided. We considered two kinds of information: increasing the family size and introducing parental marker data. For half-sibs, introducing parental genotypes is more efficient than increasing family size and leaving parents untyped. In general, assessing the effectiveness of pedigree structure and frequency distribution on haplotype inference can be done through a simulation study based on the inferred frequencies.
In conclusion, we have confirmed that the biased haplotype frequency estimates that result from treating related individuals as unrelated can affect association analysis, and we have provided the appropriate statistical methods to efficiently accommodate some important population structures. We have also demonstrated how to forecast the accuracy of phase reconstruction on the basis of the most likely phase criterion, which will determine whether pedigree data are warranted. We find that the impact of pedigree data depends upon the actual haplotype structure in the population. Since this structure will vary throughout the genome, a single optimal strategy for accurate haplotype reconstruction from markers used in genomewide scans is unlikely. Rather, the impact of various pedigree data on particular subsets of markers can be assessed through a simulation study based upon initial frequency estimates.
Acknowledgments
The authors thank Sarah Blott and two anonymous reviewers for their valuable suggestions. Michael Schouten also thanks Michalis Titsias for his technical insight and Alan Stubbs and Vivian Fu for their comments on the manuscript. This research was supported by Sygen International and by the Biotechnology and Biological Sciences Research Council.
APPENDIX: AN EFFICIENT ALGORITHM FOR EVALUATING HALF-SIB PEDIGREES
Our objective is efficient computation of the posterior distributions for sire and dam phase configurations, which are required to calculate the complete-data log-likelihood given in (4). We begin by describing a straightforward peeling approach to obtain the posterior for the sire. We then demonstrate how the results obtained during the peeling process can be used for efficient calculation of the dam distribution. Both of these approaches achieve cubic complexity in the number of possible phase configurations. We conclude by demonstrating how complexity can be reduced to quadratic complexity, which, as described in the text, can result in substantial savings in computational resources.
To minimize notational clutter we drop explicit references to 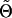 and ℱ, and it should be understood that this information is given. Furthermore, since we are talking about the distribution for a specific sire or dam, we can drop any index that refers to an order in the sample. Hence,
and ℱ, and it should be understood that this information is given. Furthermore, since we are talking about the distribution for a specific sire or dam, we can drop any index that refers to an order in the sample. Hence,
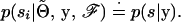 |
(A1) |
Finally, we need to reference other members in a given sibship. For a sibship of size K, we assign an arbitrary ordering to all progeny and dam pairs and use (yi, di), i = 1, … , K, to refer to the ith pair.
Calculating the distribution of the sire phase:
We begin by writing the objective as
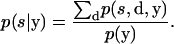 |
(A2) |
Using the key insight of Elston and Stewart (1971), the joint distribution is expressed as a telescopic sum,
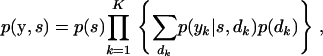 |
(A3) |
where
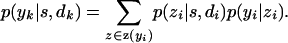 |
(A4) |
Peeling the jth family first entails calculating
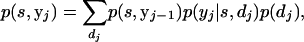 |
(A5) |
where yj−1 = y0, y1, … , yj−1 and y0 = ∅. The likelihood is then updated to
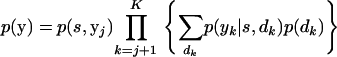 |
Each family is iteratively peeled and, after the final family has been peeled, the resulting expression, p(y, s), can be used to calculate p(s|y). p(y) can then be obtained by summing out s.
Calculating the posterior for the dam:
First, we can rewrite our objective as
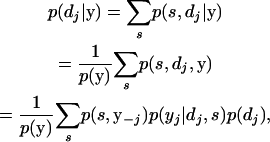 |
(A6) |
where efficient calculation of p(y) is given above and the definition of p(s, y−j) is given by
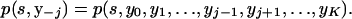 |
To calculate p(s, y−j), we must store each of the K expressions given by (A5), i.e.,
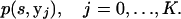 |
(A7) |
These are sufficient to calculate p(s, y−j) since
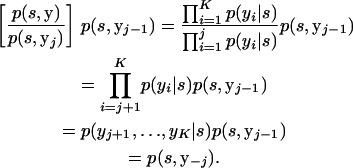 |
(A8) |
Further reduction in complexity:
From Equations A4 and A8, we see that the complexity of the phase distribution for both sire and dam is dominated by the expression p(y|s, d), which is cubic in the number of phase configurations. To achieve quadratic complexity, we show how to calculate this expression by evaluating p(y|s) and p(y|d) directly.
Consider first evaluating the relevant expression for the sire where the genotype of the dam is unknown. Rather than summing over all the dam configurations, as suggested by (A3), we attempt to directly evaluate
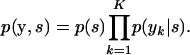 |
(A9) |
A given set of phase configurations, z ∈ z(yk), can be expressed as
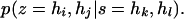 |
(A10) |
Let t1(s) denote the event that the sire transmitted hk and let t2(s) denote the event that the sire transmitted hl. Then (A10) can be written as
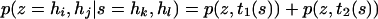 |
(A11) |
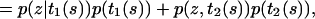 |
(A12) |
where p(t1(s)) = p(t2(s)) = 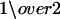 . Importantly, p(z|t1(s)) and p(z|t2(s)) can be calculated directly from
. Importantly, p(z|t1(s)) and p(z|t2(s)) can be calculated directly from 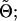 for example,
for example,
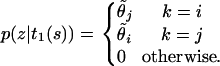 |
(A13) |
When the genotype of the dam is known, the probabilities in (A13) can be calculated from the distribution 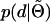 . Evaluating
. Evaluating 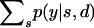 follows an analogous procedure.
follows an analogous procedure.
References
- Becker, T., and M. Knapp, 2002. Efficiency of haplotype frequency estimation when nuclear family information is included. Hum. Hered. 54: 45–53. [DOI] [PubMed] [Google Scholar]
- Elston, R., and J. Stewart, 1971. A general model for the genetic analysis of pedigree data. Hum. Hered. 21: 523–542. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., and M. Slatkin, 1995. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 12: 921–927. [DOI] [PubMed] [Google Scholar]
- Fallin, D., and N. Shorck, 2000. Accuracy of haplotype frequency estimation for biallelic loci, via the expectation-maximization algorithm for unphased diploid genotype data. Am. J. Hum. Genet. 67: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallin, D., A. Cohen, L. Essioux, I. Chumakov, M. Blumenfeld et al., 2001. Genetic analysis of case/control data using haplotype frequencies: application to APOE locus variation and Alzheimer's disease. Genet. Res. 11: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapes, L., J. Dekkers, M. Rothschild and R. Fernando, 2004. Comparing linkage disequilibrium-based methods for fine mapping of quantitative trait loci. Genetics 166: 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, M., and K. Kidd, 1995. HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J. Hered. 86: 409–411. [DOI] [PubMed] [Google Scholar]
- Hill, W. G., 1974. Estimation of linkage disequilibrium in randomly mating populations. Heredity 33: 229–239. [DOI] [PubMed] [Google Scholar]
- Hull, J., H. Ackerman, K. Isles, S. Usen, M. Pinder et al., 2001. Unusual haplotypic structure of IL8, a susceptibility locus for a common respiratory virus. Am. J. Hum. Genet. 69: 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk, K. M., and L. R. Cardon, 2002. The impact of genotyping error on haplotype reconstruction and frequency estimation. Eur. J. Hum. Genet. 10: 616–622. [DOI] [PubMed] [Google Scholar]
- Lee, S. H., and J. H. van der Werf, 2004. The efficiency of designs for fine-mapping of quantitative trait loci using combined linkage disequilibrium and linkage. Genet. Sel. Evol. 36: 145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen, T. H. E., A. Karlsen, S. Lien, I. Olsaker and M. E. Goddard, 2002. Fine mapping of a quantitative trait locus for twinning rate using combined linkage and linkage disequilibrium mapping. Genetics 161: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, A., J. Whittaker and D. Balding, 2002. Fine-scale mapping of disease loci via shattered coalescent modeling of genealogies. Am. J. Hum. Genet. 70: 686–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, A., J. Whittaker and D. Balding, 2004. Little loss of information due to unknown phase for fine-scale linkage-disequilibirium with single-nucleotide-polymorphism genotype data. Am. J. Hum. Genet. 74: 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, T., Z. S. Qin, X. Xu and J. S. Liu, 2002. Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am. J. Hum. Genet. 70: 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, J. R., 2000. Zero-recombinant haplotyping: applications to fine mapping using SNPs. Genet. Epidemiol. 19: S582–S587. [DOI] [PubMed] [Google Scholar]
- Ploughman, L. M., and M. Boehnke, 1989. Estimating the power of a proposed linkage study for a complex genetic trait. Am. J. Hum. Genet. 44: 543–551. [PMC free article] [PubMed] [Google Scholar]
- Qin, Z. S., T. Niu and J. S. Liu, 2002. Partial-ligation-expectation-maximization for haplotype inference with single nucleotide polymorphisms. Am. J. Hum. Genet. 71: 1242–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, K., and R. Fuerst, 2001. Haplotyping and estimation of haplotype frequencies for closely linked biallelic multilocus genetic phenotypes including nuclear family information. Hum. Mutat. 17: 289–295. [DOI] [PubMed] [Google Scholar]
- Schaid, D. J., 2002. Relative efficiency of ambiguous vs. directly measured haplotype frequencies. Genet. Epidemiol. 23: 426–443. [DOI] [PubMed] [Google Scholar]
- Stephens, M., and P. Donnelly, 2003. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73: 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, M., N. J. Smith and P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, J., and J. Ott, 1994. Handbook of Human Linkage Analysis. Johns Hopkins University Press, Baltimore.
- Thomas, S. C., and W. G. Hill, 2002. Sibship reconstruction in hierarchical population structures using Markov chain Monte Carlo techniques. Genet. Res. 79: 227–234. [DOI] [PubMed] [Google Scholar]
- Zhao, H., R. Pfeiffer and M. H. Gail, 2003. Haplotype analysis in population genetics and association studies. Pharmacogenomics 4: 171–178. [DOI] [PubMed] [Google Scholar]
- Zhao, J. H., D. Curtis and P. C. Sham, 2000. Model-free analysis and permutation tests for allelic associations. Hum. Hered. 50: 133–139. [DOI] [PubMed] [Google Scholar]


