Abstract
Chromatin insulators have been implicated in the establishment of independent gene expression domains and in the nuclear organization of chromatin. Post-translational modification of proteins by Small Ubiquitin-like Modifier (SUMO) has been reported to regulate their activity and subnuclear localization. We present evidence suggesting that two protein components of the gypsy chromatin insulator of Dorsophila melanogaster, Mod(mdg4)2.2 and CP190, are sumoylated, and that SUMO is associated with a subset of genomic insulator sites. Disruption of the SUMO conjugation pathway improves the enhancer-blocking function of a partially active insulator, indicating that SUMO modification acts to regulate negatively the activity of the gypsy insulator. Sumoylation does not affect the ability of CP190 and Mod(mdg4)2.2 to bind chromatin, but instead appears to regulate the nuclear organization of gypsy insulator complexes. The results suggest that long-range interactions of insulator proteins are inhibited by sumoylation and that the establishment of chromatin domains can be regulated by SUMO conjugation.
Keywords: chromatin, insulator, nuclear organization, SUMO, transcription
Introduction
Eukaryotic genomes are thought to be organized into independent domains of gene expression. The maintenance of this autonomy is considered to be important for the proper execution of complex developmental programs and cellular responses to stimuli. Chromatin insulators are characterized by two key properties suggestive of an involvement in the organization of independent expression domains. First, insulators are able to block enhancer–promoter communication when positioned between these elements (Geyer and Corces, 1992; Kellum and Schedl, 1992). Second, insulators can protect transgenes from the effects of surrounding chromatin, allowing for their position-independent expression (Kellum and Schedl, 1991; Chung et al, 1993).
The gypsy insulator of Dorsophila melanogaster consists of a ∼350 bp DNA sequence that binds a protein complex of at least three components, Su(Hw), Mod(mdg4)2.2 and CP190. Su(Hw) and CP190 can bind DNA directly via their zinc-finger domains (Spana et al, 1988; Pai et al, 2004), whereas Mod(mdg4)2.2 does not bind DNA directly but is recruited to the gypsy insulator sequence through physical interactions with Su(Hw) and CP190 (Gerasimova et al, 1995; Pai et al, 2004). The gypsy insulator was identified originally as the enhancer-blocking element within the gypsy retrotransposon (Geyer and Corces, 1992), but hundreds of endogenous binding sites for the gypsy protein complex exist throughout the Drosophila genome (Gerasimova and Corces, 1998). Analysis of highly replicated polytene chromosomes reveals that gypsy insulator proteins are found preferentially at the borders between condensed and decondensed chromatin, suggestive of their role in partitioning independent chromatin domains (Labrador and Corces, 2002; Pai et al, 2004). However, in diploid nuclei, gypsy insulator proteins coalesce into large complexes, termed insulator bodies (Gerasimova and Corces, 1998; Byrd and Corces, 2003). These bodies are thought to represent the meeting places of distant insulator complexes, which loop out the chromatin fiber and thus delineate chromatin domains. Both Mod(mdg4)2.2 and CP190 contain a conserved BTB/POZ domain capable of self-interactions (Dhordain et al, 1995; Ghosh et al, 2001), which has been proposed to mediate clustering of insulator complexes. The integrity of insulator bodies has been correlated functionally with gypsy insulator activity. For instance, mutations in insulator components that disrupt the enhancer-blocking activity of gypsy also interfere with insulator body formation (Gerasimova and Corces, 1998).
Chromatin insulators may thus play an important role in structurally demarcating domains of independently occurring transcriptional activity. Expectedly, such domains are often subject to developmental or environmental regulation, which implies that insulators may themselves be regulated to allow for a variety of gene expression programs of an organism. Regulatory mechanisms that can influence insulator activity have been described for the vertebrate insulator protein CTCF. The parent-specific enhancer-blocking activity of CTCF at the H19/Igf2 locus is controlled by differential methylation of its binding sites within this imprinted locus (Bell and Felsenfeld, 2000; Hark et al, 2000). Specifically, methylation of binding sites on the paternal chromosome only prevents the binding of CTCF to allow activation of Igf2 expression. In this case, the insulator activity of CTCF is regulated at the level of its DNA-binding ability. A second regulatory mechanism of CTCF activity employs post-translational modification by poly-ADP-ribose (PAR), which appears to be involved in positive regulation of insulator activity (Yu et al, 2004). Conjugation of PAR to CTCF does not alter its DNA-binding properties, but may be necessary for protein–protein interactions involved in setting up chromatin domains (Klenova and Ohlsson, 2005).
Conjugation to Small Ubiquitin-like Modifier (SUMO) serves as another post-translational modification that regulates the activity of multiple nuclear factors. Similar to ubiquitin, SUMO is covalently attached to target proteins by a cascade of enzymes, including the activating enzyme E1, which is a heterodimer of Aos1/SAE1 and Uba2/SAE2, the conjugating enzyme E2, also known as Ubc9, and a variety of the specificity-enhancing E3 ligases (Dohmen, 2004; Hay, 2005). SUMO conjugation has been detected most often within a sumoylation consensus motif ψKxE (where ψ is a large hydrophobic amino-acid residue and x is any residue). SUMO attachment has been linked to a variety of functional outputs, including regulation of transcriptional activity, subnuclear targeting and formation of nuclear compartments. Frequently, sumoylation of proteins has been shown to alter their ability to bind DNA or other protein factors.
Multiple SUMO E3 ligases, which do not seem to be required for but enhance sumoylation of specific substrates both in vivo and in vitro, have been characterized. Members of the Siz/PIAS family of SUMO E3 ligases contain a domain homologous to the RING domain of ubiquitin E3 ligases (Johnson and Gupta, 2001; Schmidt and Muller, 2002). Recently, dTopors, which harbors a RING domain, has been characterized as a E3 ubiquitin ligase involved in gypsy insulator activity (Capelson and Corces, 2005), but several reports also implicate homologs of dTopors, human Topors and viral ICP0, in the SUMO pathway (Muller and Dejean, 1999; Weger et al, 2003, 2005; Lee et al, 2004). We thus investigated the possibility that gypsy insulator proteins are regulated by SUMO modification as well as the potential involvement of dTopors in this process, perhaps as an E3 SUMO ligase. Here, we present evidence suggesting that two components of the gypsy insulator complex, Mod(mdg4)2.2 and CP190, are sumoylated in vivo and in vitro, and that sumoylation negatively attenuates gypsy insulator activity. Specifically, SUMO conjugation interferes with nuclear coalescence of insulator bodies, suggesting that establishment of higher-order chromatin domains can be regulated by post-translational modification of insulator proteins.
Results
Insulator proteins are sumoylated in vitro
Topors, the mammalian homolog of dTopors, has been recently shown to interact with the SUMO E2-conjugating enzyme Ubc9 (Weger et al, 2003). This association appears to be evolutionarily conserved, as we have also detected an interaction between Drosophila Ubc9 and dTopors in the yeast two-hybrid assay (data not shown). To determine whether dTopors functions as an E3 SUMO ligase for insulator proteins, Su(Hw), Mod(mdg4)2.2 and CP190 were tested as substrates in an in vitro sumoylation reaction, in the presence or absence of dTopors. All three proteins contain lysines found within a SUMO modification consensus motif ψKxE, and can thus be potentially modified by SUMO. For each sumoylation reaction, in vitro-transcribed and -translated 35S-labeled substrate protein was incubated with the E1, E2 enzymes, SUMO and in vitro-generated or -purified recombinant dTopors. Both CP190 and Mod(mdg4)2.2 can be modified by SUMO, as higher molecular weight bands appear when the SUMO conjugation machinery is added (Figures 1A and B). The addition of dTopors not only does not promote, but appears to disrupt the sumoylation of CP190 and Mod(mdg4)2.2, as assessed by decreased levels of the higher migrating modified forms of either protein in the presence of dTopors (Figures 1A and B). No disruption of sumoylation is observed upon introduction of equivalent amounts of another substrate protein in the reaction, such as the addition of Mod(mdg4)2.2 to a reaction with CP190 as a substrate (data not shown). The same pattern can be observed when detection of SUMO conjugation is carried out by using anti-SUMO antisera (Figure 1C). Again, the appearance of Mod(mdg4)2.2-specific SUMO conjugates is counteracted by the addition of recombinant purified dTopors protein.
Figure 1.
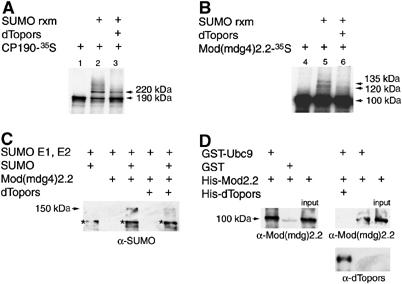
Mod(mdg4)2.2 and CP190 are modified by SUMO in vitro. The size of various protein bands is indicated in kDa. The unmodified form of Mod(mdg4)2.2 runs at 100 kDa, and the unmodified form of CP190 is 190 kDa. The sumoylated form of CP190 is 220 kDa and Mod(mdg4)2.2 shows two different sumoylated bands of 120 and 135 kDa. (A, B) In vitro sumoylation reactions with 35S-labeled CP190 (A) or Mod(mdg4)2.2 (B) used as substrate, in the presence or absence of SUMO reaction components (SUMO rxm), including E1, E2 enzymes, SUMO and ATP, or of dTopors. Lane 1, CP190 alone; lane 2, CP190 with SUMO rxm; lane 3, CP190 with SUMO rxm and in vitro-generated dTopors; lane 4, Mod(mdg4)2.2 alone; lane 5, Mod(mdg4)2.2 with SUMO rxm; lane 6, Mod(mdg4)2.2 with SUMO rxm and in vitro-generated dTopors. Arrows point to sumoylated forms of CP190 and Mod(mdg4)2.2. (C) In vitro sumoylation reactions in the presence or absence of SUMO E1 and E2, SUMO, Mod(mdg4)2.2 or dTopors monitored with α-SUMO antibodies. The arrow points to the Mod(mdg4)2.2-specific SUMO-GST conjugate. The lower molecular weight band marked with an asterisk corresponds to Ubc9-SUMO-GST. (D) GST-Ubc9 or GST, bound to glutathione beads, were mixed with His6-Mod(mdg4)2.2 in the presence or absence of His6-dTopors. The precipitated fractions and input proteins were resolved by SDS–PAGE and Western blotted with α-Mod(mdg4)2.2 or α-dTopors antibodies.
Like many identified substrates for SUMO conjugation, CP190 and Mod(mdg4)2.2 do not seem to require the presence of an E3 ligase in vitro in order to be sumoylated, suggesting that they are able to bind Ubc9 directly. We confirmed this association for Mod(mdg4)2.2 and Ubc9 using a GST pull-down assay (Figure 1D, left panel). SUMO E3 ligases are thought to function as adaptor surfaces, binding the catalytic E2 and the target protein simultaneously to promote conjugation. If dTopors acts as a SUMO E3 ligase, it should be able to form a complex with both Ubc9 and Mod(mdg4)2.2 and facilitate the interaction between them. However, in agreement with the results of in vitro sumoylation assays, addition of dTopors to a mixture of purified GST-Ubc9 and Mod(mdg4)2.2 interferes with their interaction and results in decreased levels of Mod(mdg4)2.2 recovered in the pulled down fraction (Figure 1D, right panel). These results suggest that dTopors may antagonize sumoylation by disrupting the association between Ubc9 and Mod(mdg4)2.2 or CP190. The ability of dTopors to reduce sumoylation of CP190 and Mod(mdg4)2.2 in vitro is reminiscent of the in vivo properties of its viral homolog ICP0, which inhibits the modification of PML and Sp100 proteins by SUMO (Muller and Dejean, 1999). Together, these findings suggest that CP190 and Mod(mdg4)2.2 are sumoylated and argue against dTopors functioning as an E3 SUMO ligase for insulator proteins. Instead, dTopors may regulate the modification of these proteins by inhibiting their sumoylation.
CP190 and Mod(mdg4)2.2 are sumoylated in vivo
To determine whether CP190 and Mod(mdg4)2.2 are sumoylated in vivo, we analyzed endogenous proteins from larval protein extracts prepared with and without two inhibitors of SUMO isopeptidases, N-ethylmalemide (NEM) and iodoacetamide (IAA) (Eloranta and Hurst, 2002). Lysis of cells in the presence of NEM and IAA results in higher levels of recovered SUMO conjugates compared to extracts prepared without the inhibitors, as demonstrated by Western analysis with SUMO antisera (Figure 2A, left panel). Similarly, this treatment results in the appearance of higher molecular weight conjugates of CP190 and Mod(mdg4)2.2 (Figure 2A, right panels). We normally detect one or two additional bands of Mod(mdg4)2.2 in protein extracts or in in vitro assays, which is consistent with the presence of two strong consensus sumoylation sites in the amino-acid sequence of Mod(mdg4)2.2. CP190 appears to have one predominant sumoylated form in vivo or in vitro, although its sequence contains five predicted consensus sites. No higher molecular weight forms of Su(Hw) were observed to correlate with the addition of the inhibitors, regardless of the length of the Western blot exposure.
Figure 2.
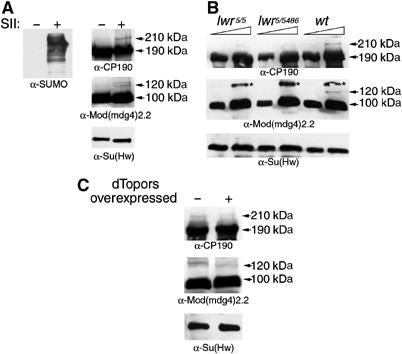
Mod(mdg4)2.2 and CP190 are sumoylated in vivo. The size of various protein bands is indicated in kDa. The unmodified form of Mod(mdg4)2.2 runs at 100 kDa, and the unmodified form of CP190 is 190 kDa. The sumoylated form of CP190 is 220 kDa and Mod(mdg4)2.2 shows two different sumoylated bands of 120 and 135 kDa. (A) Protein extracts from Drosophila larvae were prepared in the presence or absence of SUMO Isopeptidase Inhibitors (SII) NEM and IAA, resolved by SDS–PAGE and Western blotted with antibodies to SUMO and indicated gypsy insulator proteins. (B) Larval protein extracts from wild-type organisms or indicated lwr mutants were prepared in the presence of SII, resolved by SDS–PAGE and Western blotted with antibodies to indicated proteins. Triangles indicate increasing amounts of extracts loaded on the gels. Asterisk indicates a non-Mod(mdg4)2.2 band recognized by the antibody (Mongelard et al, 2002). (C) Protein extracts from wild-type (−) or UAS-dTopors/ActGAL4 (+) larvae were prepared in the presence of SII, resolved by SDS–PAGE and Western blotted with antibodies to indicated proteins.
The identity of these slower migrating forms of CP190 and Mod(mdg4)2.2 was verified further by examining mutants for the SUMO conjugating E2 enzyme Ubc9, which in Drosophila is encoded by the gene lesswright (lwr). Protein extracts from larvae of two hypomorphic allelic combinations of lwr, the homozygous lwr5/lwr5 and the transheterozyous lwr5/lwr5486 (Apionishev et al, 2001), display a significant reduction in the higher molecular weight forms of CP190 and Mod(mdg4)2.2 as compared to wild type (Figure 2B). As expected, no variation in Su(Hw) was detected in these mutants.
Since dTopors was found to interfere with sumoylation in vitro, its ability to affect the sumoylated forms of CP190 and Mod(mdg4)2.2 was also examined in vivo. To this end, overexpression of dTopors was induced in larvae carrying a UAS-dTopors transgenic construct by an actin-GAL4 (ActGAL4) driver. Elevated levels of dTopors result in a reduction of the sumoylated forms of CP190 and Mod(mdg4)2.2, as judged by Western analysis of protein extracts obtained from larvae with induced versus uninduced dTopors (Figure 2C). Together, the observed decrease of these slower migrating protein forms in larvae mutant for the SUMO E2 ligase or larvae overexpressing dTopors reinforces the idea that they represent sumoylated forms of CP190 and Mod(mdg4)2.2.
SUMO is associated with chromatin binding sites of insulator proteins
To examine the functional relevance of the SUMO modification of insulator proteins, we analyzed whether SUMO is present at chromatin binding sites of the gypsy insulator complex. For this purpose, the distribution of SUMO on polytene chromosomes of Drosophila third instar larvae was compared to that of Mod(mdg4)2.2 and CP190. Consistent with its involvement in multiple nuclear processes and as described previously (Lehembre et al, 2000), SUMO localizes to hundreds of discrete binding sites throughout the genome. Furthermore, it appears to associate with different chromatin states of polytene chromosomes, including the heterochromatic chromocenter, the euchromatic decondensed interbands and the border regions between interbands and the condensed bands. A fraction of Mod(mdg4)2.2 sites, which are found exclusively at the border regions, colocalize with SUMO, as revealed by costaining of polytene chromosomes with anti-SUMO and anti-Mod(mdg4)2.2 antisera (Figure 3A). Similarly, SUMO is detected at a subset of CP190 chromatin binding sites (Figure 3B). As described previously, all binding sites of Mod(mdg4)2.2 overlap with CP190, but CP190 binds to additional sites that do not overlap with Mod(mdg4)2.2 and Su(Hw) (Pai et al, 2004). Although the total number of bands that overlap with SUMO is higher for CP190 than for Mod(mdg4)2.2, we estimate the percentage of sites that colocalize with SUMO is approximately 10–15% for both of the analyzed insulator proteins. These sites presumably correspond to regions where bound CP190 or Mod(mdg4)2.2 may be modified by SUMO. The presence of SUMO modification at a fraction of insulator sites suggests that sumoylation may be associated with a specialized insulator state.
Figure 3.
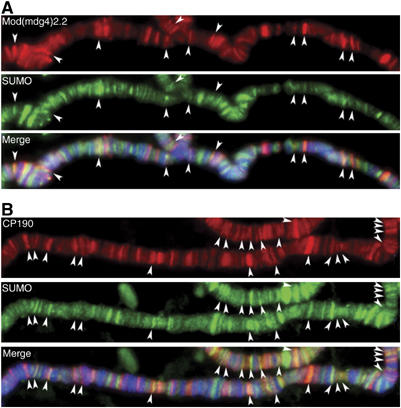
SUMO is associated with a fraction of gypsy protein complexes on chromatin. White arrows point to places of colocalization between SUMO and insulator proteins. DNA is stained with DAPI (blue). (A) Immunostaining of polytene chromosomes of third instar larvae with antibodies to Mod(mdg4)2.2 (red) and SUMO (green). (B) Immunostaining of polytene chromosomes with antibodies to CP190 (red) and SUMO (green).
The SUMO modification pathway antagonizes gypsy insulator activity
In order to understand the functional consequences of sumoylation of gypsy insulator proteins, we analyzed mutations in components of the SUMO conjugation pathway for effects on the enhancer-blocking activity of gypsy. To this end, three gypsy retrotransposon-induced mutations were used as phenotypic indicators of gypsy insulator function. The y2 allele contains an insertion of gypsy between the body enhancer and the promoter of the yellow (y) gene, which results in low expression of the yellow gene product in the cuticle of adult flies (Figure 4, top panel). Similarly, the cut6 (ct6) mutant allele is caused by the insertion of gypsy between the promoter and the wing margin enhancer of the cut gene. Phenotypically, this is manifested by a jagged or cut appearance of the fly wing edge (Figure 4, top panel). Finally, the ombP1-D11 mutation results from the insertion of gypsy between the regulatory region of the omb gene and the promoter of a white transgene inserted at this locus (Tsai et al, 1997). Control of white gene expression by the omb regulatory region produces a characteristic expression of the red pigment in dorsal and ventral patches of the fly eye. Insertion of the gypsy insulator prevents a silencing element of the omb regulatory region from acting on the white gene promoter, resulting in a broader distribution of the red pigment throughout the eye (Figure 4, top panel).
Figure 4.
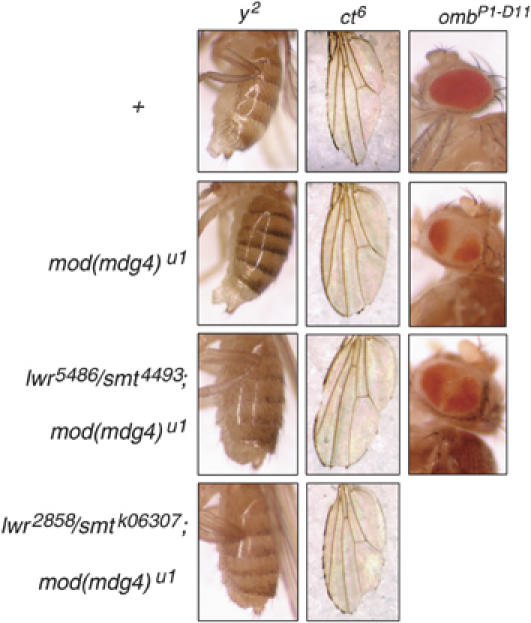
Mutations in components of the SUMO conjugation pathway promote activity of the gypsy insulator. Shown are abdomens, wings and eyes of y2ombP1-D11ct6; +; +, y2ombP1-D11ct6; +; mod(mdg4)u1, y2ombP1-D11ct6; lwr5486/smt34493; mod(mdg4)u1 and y2ombP1-D11ct6; lwr2858/smt3k06307; mod(mdg4)u1 female flies.
In order to test for positive or negative effects of sumoylation on insulator activity, mutations in SUMO pathway components were examined in the sensitized background of a compromised insulator created by a null mutation in Mod(mdg4)2.2, mod(mdg4)u1 (Mongelard et al, 2002). Lack of Mod(mdg4)2.2 partially disrupts insulator activity, thus increasing enhancer–promoter communication. At y2 and ct6, this is manifested by an increase in gene product expression, which results in a more pigmented abdomen or a smoother wing edge, respectively (Figure 4, second from top panel). At the ombP1-D11 locus, the mod(mdg4)u1 mutation yields an opposite effect on transcription, since in this case, the insulator interferes with a silencer–promoter communication. Thus, the result is less white gene product or smaller areas of red pigmentation in the eye (Figure 4, second from top panel).
Combined mutations of the SUMO E2-conjugating enzyme lwr and of the SUMO gene smt3 suppress the effect of mog(mdg4)u1 on gypsy-induced phenotypes, suggesting that SUMO conjugation reduces gypsy insulator activity. Two different heterozygous combinations of lwr and smt3, lwr5486/smt34493 and lwr2858/smt3k06307, produced similar effects on y2, ct6 and ombP1-D11. The phenotype of homozygotes of these alleles could not be assessed as none of them survive to adulthood. A decreased expression of yellow or a lighter abdomen in y2; lwr5486/smt34493; mod(mdg4)u1 or y2; lwr2858/smt3k06307; mod(mdg4)u1 flies was observed when compared to y2; mod(mdg4)u1 controls (Figure 4, bottom two panels), indicative of improved enhancer-blocking activity of the gypsy insulator. Similarly, increased insulator activity is detected for ct6, judging from the more jagged and discontinuous wing edge in flies that bear SUMO pathway mutations in the mod(mdg4)u1 background relative to mod(mdg4)u1 flies (Figure 4, bottom two panels). The eyes of ombP1-D11; lwr5486/smt34493; mod(mdg4)u1 mutants display broader distribution of red pigment than those of the ombP1-D11; +; mod(mdg4)u1 controls (Figure 4, second from bottom panel), indicative of increased insulator activity, implying that the SUMO conjugation pathway is antagonistic to normal gypsy insulator function. The observed phenotypic changes are subtle due to the fact that only heterozygous combinations of these alleles could be tested. The effects observed in ct6 and ombP1-D11 are consistent within the population of mutant flies and the photographs shown in Figure 4 are a good representation of the observed effects. Phenotypic changes at y2 in the lwr/smt3 mutants occur in approximately 30% of flies of either genotype (the remaining 70% display a continuum of less pronounced effects). Interestingly, all of these genetic interactions are particularly pronounced in females, although they are detectable in both sexes. The effects of the smt3k06307 mutation on ombP1-D11 could not be assessed because this mutation is caused by insertion of a P-element carrying the white gene. These results suggest that lwr and the SUMO conjugation pathway are involved in negative regulation of gypsy insulator function.
Sumoylation of insulator proteins does not regulate their binding to DNA
It has been reported that sumoylation can interfere with the DNA-binding ability of some transcription factors or with their recruitment to certain chromatin locations (Goodson et al, 2001; Chalkiadaki and Talianidis, 2005). Similarly, sumoylation of CP190 or Mod(mdg4)2.2 may disrupt their binding to chromatin. To investigate this possibility, we analyzed the effects of varying levels of Ubc9 on the binding of insulator proteins to polytene chromosomes. The localization of Mod(mdg4)2.2 and CP190 appears unchanged in polytene chromosomes of lwr mutant larvae or larvae overexpressing UAS-lwr induced by the ActGAL4 driver, as compared to wild type (Figure 5A). No significant changes in the levels of chromatin-associated insulator proteins or in their global binding patterns were observed.
Figure 5.
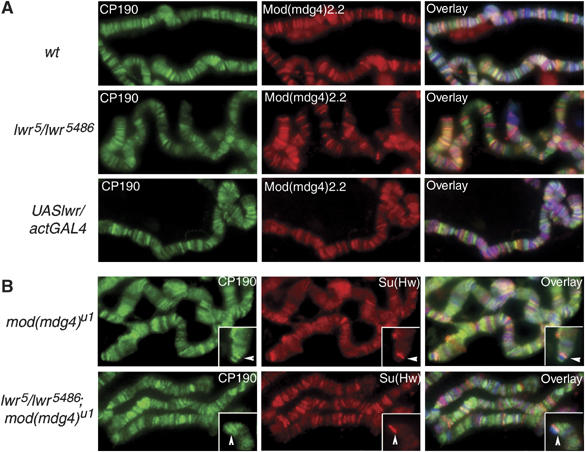
Sumoylation does not affect the binding of CP190 and Mod(mdg4)2.2 to chromatin. DNA is stained with DAPI (blue). (A) Immunostaining of polytene chromosomes from wild-type, UASlwr/ActGAL4 and lwr5/5486 larvae with antibodies to Mod(mdg4)2.2 (red) and CP190 (green). (B) Immunostaining of polytene chromosomes from y2; +; mod(mdg4)u1 and y2; lwr5/5486; mod(mdg)u1 larvae with antibodies to Mod(mdg4)2.2 (red) and CP190 (green). Arrows point to the y2 locus (insets).
Since phenotypic consequences of mutations in the SUMO pathway were analyzed in the mod(mdg4)u1 background, we also examined the binding of CP190 to chromosomes in lwr5/lwr5486; mod(mdg4)u1 and mod(mdg4)u1 mutants. No variation in the overall binding of CP190 to polytene chromosomes was detected in larvae mutant for lwr and mod(mdg4) as compared to larvae mutant for mod(mdg4) alone (Figure 5B). Since we had not observed sumoylation of Su(Hw), its binding was not expected to be altered in the absence of the SUMO E2 ligase. Consequently, similar levels of Su(Hw) are found at the y2 locus in the lwr5/lwr5486; mod(mdg4)u1 genetic background relative to mod(mdg4)u1 (Figure 5B, inset). Likewise, CP190 is detected in comparable amounts at the y2 locus in these mutants, suggesting that sumoylation does not affect the ability of insulator proteins to associate with chromatin.
Sumoylation disrupts nuclear clustering of insulator proteins
SUMO conjugation has also been described to regulate protein–protein interactions, where sumoylation of proteins causes them to lose certain associations or to gain new ones (Seeler et al, 2001; Girdwood et al, 2003). As a consequence of this, sumoylation has been shown to induce subnuclear reorganization, such that sumoylated proteins are associated with nuclear compartments different from those of their unmodified counterparts (Kim et al, 1999; Ross et al, 2002). Since nuclear organization of gypsy insulator proteins is functionally linked to insulator activity (Gerasimova and Corces, 1998; Pai et al, 2004; Capelson and Corces, 2005), we wanted to investigate whether SUMO modification of Mod(mdg4)2.2 and CP190 can affect their ability to form insulator bodies.
Overexpression of Ubc9 by driving the expression of UAS-lwr with ActGAL4 causes a dramatic dispersal of insulator bodies, as assessed by immunostaining of diploid cells of third instar larvae with anti-Mod(mdg4) antiserum (Figure 6A). Staining of DNA with DAPI is shown as a control for nuclear integrity. These findings are consistent with the observed genetic interference of sumoylation with the enhancer-blocking function of gypsy. Furthermore, they suggest that SUMO conjugation negatively regulates insulator activity by interfering with nuclear coalescence of insulator proteins.
Figure 6.
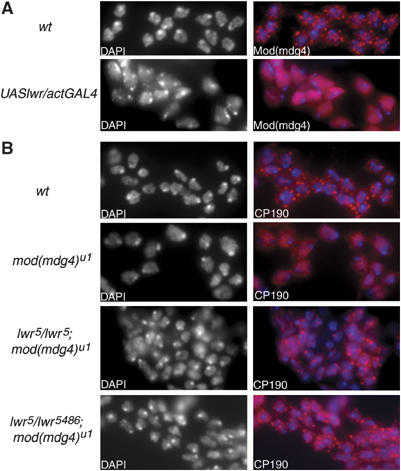
SUMO conjugation antagonizes nuclear coalescence of insulator proteins. (A) Immunostaining of diploid cells from brains and imaginal discs of wild-type and UASlwr/ActGAL4 larvae with antibodies to Mod(mdg4) (red). (B) Immunostaining of diploid cells of wild-type, mod(mdg4)u1, lwr5/5; mod(mdg)u1 and lwr5/5486; mod(mdg)u1 larvae with antibodies to CP190 (red). DAPI alone is shown at left and in blue in overlay.
This conclusion is further substantiated by the effect of lower levels of Ubc9 on the distribution of gypsy insulator bodies. In the absence of Mod(mdg4)2.2, the formation of insulator bodies is disrupted due to the loss of one of the bridging components of the insulator complex (Gerasimova and Corces, 1998). For both of the analyzed combinations of mutant alleles of lwr in the mod(mdg4)u1 background, lwr5/lwr5; mod(mdg4)u1 and lwr5/lwr5486; mod(mdg4)u1, insulator bodies, marked with anti-CP190 antibodies, are seen to reform in a pattern similar to that observed in wild-type cells (Figure 6B). The correlation of improved nuclear clustering of insulator proteins with lower levels of their modification by SUMO suggests that sumoylation may interfere with self-interactions of distant insulator complexes (Figure 7). This is consistent with the presence of consensus sumoylation sites within the BTB domain of Mod(mdg4)2.2 and immediately preceding the BTB domain of CP190, since these domains are involved in mediating interactions between these two proteins (Pai et al, 2004).
Figure 7.
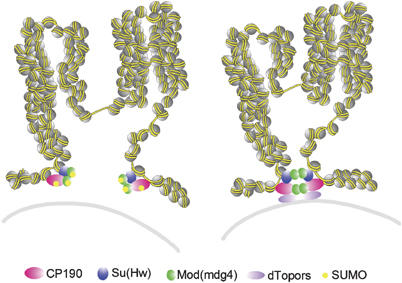
Model for the effect of SUMO modification on chromatin domain formation. Chromatin loop domains may be established through interactions via distant gypsy insulator complexes and further stabilized through the tethering function of dTopors (right). Sumoylation of CP190 and Mod(mdg4)2.2 interferes with their self-interactions, resulting in a breakdown of chromatin domains (left). Gray line represents the nuclear lamina, and yellow spheres represent nucleosomes.
Discussion
Two protein components of the gypsy chromatin insulator, Mod(mdg4)2.2 and CP190, were found to be modified by SUMO in vitro and in vivo. dTopors was observed to interfere with their sumoylation by possibly disrupting the contacts between the SUMO E2 enzyme Ubc9 and substrate insulator proteins. The inhibitory effect of dTopors, although relatively subtle, is consistent across the various assays utilized such that any time dTopors was introduced at higher levels, either by direct addition in vitro or by increasing expression in vivo, it was found to result in reduced sumoylation of Mod(mdg4)2.2 and CP190. Disruption of SUMO conjugation by mutations in genes coding for Ubc9 and SUMO exerts a positive effect on gypsy insulator activity, suggesting that the normal role of SUMO modification is to antagonize insulator function. A fraction of chromatin-bound insulator proteins appears to be associated with SUMO, yet mutations in the SUMO pathway are not seen to affect the chromatin-binding properties of CP190 or Mod(mdg4)2.2. Instead, sumoylation interferes with the formation of nuclear insulator bodies, such that overexpression of Ubc9 leads to breakdown of nuclear insulator structures, whereas lower levels of Ubc9 and sumoylation result in a partial recovery of coalescence lost in the absence of Mod(mdg4)2.2.
These findings suggest that modification of CP190 and Mod(mdg4)2.2 by SUMO may prevent self-association and thus interfere with long-range interactions between distant insulator complexes required to form insulator bodies (Figure 7). Thereby, sumoylation may preclude formation of closed chromatin loops and the consequent establishment of autonomous gene expression domains.
Multiple lines of evidence point to a role for SUMO modification in transcriptional repression. Sumoylation of histones has been characterized as a mark of repressed chromatin (Shiio and Eisenman, 2003), whereas SUMO conjugation to certain transcriptional regulators leads to their association with histone deacetylases, which remove the active acetylation marks from histones (Girdwood et al, 2003; Yang and Sharrocks, 2004). SUMO modification of the Polycomb group (PcG) protein SOP-2 is required for its function in stable repression of Hox genes (Zhang et al, 2004), and another PcG repressor, Pc2, acts as a SUMO E3 ligase (Kagey et al, 2003). Modification of gypsy insulator proteins by SUMO does not seem to associate them exclusively with transcriptional repression, as reduction of sumoylation in lwr/smt3 mutants results in the upregulation of expression from the ombP1-D1 locus, but in the downregulation of transcription at y2 and ct6. In these cases, transcriptional output appears to correlate only with the enhancer-blocking activity of the insulator. Nevertheless, it is possible that one of the roles of sumoylation involves association of selected insulator sites in the genome with transcriptional repression. Sumoylated insulator complexes may not participate in the formation of expression domains, but instead, could target silencing factors to the surrounding chromatin.
In mammalian nuclei, the homolog of dTopors localizes to PML bodies, which are enriched in the SUMO conjugation machinery (Rasheed et al, 2002). If inhibition of sumoylation is also a property of mammalian Topors, it may play a role in preventing further sumoylation of factors that are targeted to these nuclear compartments. In this manner, ICP0 also localizes to the PML bodies, where it causes desumoylation of two primary components, PML and SP100 (Muller and Dejean, 1999). It was recently reported that Topors may function as a SUMO E3 ligase for the tumor suppressor p53 protein (Weger et al, 2005). This apparent contradiction with our results may be due to several reasons. Topors and dTopors may have diverged their functions regarding the SUMO pathway, such that Topors functions as a SUMO E3 while dTopors interferes with SUMO addition due to its conserved interaction with Ubc9. Alternatively, the involvement of dTopors in the SUMO pathway may be substrate-specific, since it may bind to Ubc9 in ways that allow for interaction with a given target protein or prevent it. In the context of the gypsy insulator, the interference of dTopors with sumoylation is consistent with previous observations that dTopors promotes insulator activity (Capelson and Corces, 2005), whereas sumoylation appears to disrupt it.
It has been suggested that SUMO conjugation may affect the function of the modified protein even after the SUMO tag itself has been removed, creating a cellular memory for protein regulation (Hay, 2005). This idea has arisen partly to explain the commonly observed contradiction between the small percentage of a given protein that is modified by SUMO and the dramatic consequences of the modification on the protein's cellular function. Sumoylation may be needed for proteins to enter stable complexes or functional states, but the persistence of the SUMO modification may not be required after the initial establishment. Thus, the actual effect of sumoylation may far exceed that of the detectable sumoylated population since the function of a much larger proportion of molecules has been altered by SUMO conjugation and subsequent deconjugation. Similarly to other reported cases, the sumoylated forms of Mod(mdg4)2.2 and of CP190 represent a small fraction of the total pool of the insulator proteins, yet the phenotypic effects of the loss of these forms are quite striking. It is possible that SUMO attachment regulates the initial organization of chromatin domains, perhaps in earlier development or following mitosis, yet once established, the domains may be stably maintained without SUMO. Additionally, the rapid conjugation and deconjugation cycle of the SUMO tag implies that sumoylation may be used by processes that require reassembly upon signal. In that sense, SUMO modification seems particularly suitable for the regulation of gene expression domains as it can result in ‘remembered' yet flexible states.
Materials and methods
In vitro sumoylation
For reactions with radioactively labeled substrates, CP190, Mod(mdg4)2.2 or Su(Hw) were in vitro-transcribed and -translated with 35S-methionine (using TNT Coupled Rabbit Reticulocyte Lysate System; Promega). A measure of 3 μl of each TNT reaction were mixed with 150 ng of SAE1/SAE2, 1000 ng of Ubc9, 1000 ng of SUMO (all components of the sumoylation kit from LAE Biotech # K007), with or without 1000 ng of recombinant purified His6-dTopors or 3 μl of in vitro-transcribed/translated dTopors. Reactions were carried out in a buffer containing 20 mM HEPES, pH 7.5, 5 mM MgCl2, 5 mM ATP for 60 min at 37°C, resolved on 7.5% SDS–PAGE gels and visualized by autoradiography. To study the effect of dTopors on sumoylation of Mod(mdg4)2.2 and Cp190 in vitro, we used increasing concentrations of in vitro-transcribed/translated protein ranging from 1 to 5 μl. No effect was observed below 3 μl, and this was the concentration used in the experiments shown in Figure 1. For reactions to be detected by Western blotting, 400 ng of recombinant purified Drosophila Ubc9-GST (the pGEX-Ubc9 construct was a gift from Dr L Griffith) and 150 ng of SAE1/SAE2 (LAE # P006) were combined in a buffer containing 50 mM Tris, pH 7.5, 2.5 mM MgCl2, 0.1 mM DTT, 5 mM ATP, with or without 3 μg of recombinant purified Drosophila SUMO-GST (the pGEX-SMT3gg construct was a gift from Dr A Courey), 400 ng of recombinant purified His6-Mod(mdg4)2.2 and 1000 ng of recombinant purified His6-dTopors, and the reactions were carried out for 120 min at 30°C. Western blotting was carried out with α-SUMO antibodies (a gift from Dr A Dejean) at 1/10 000 dilution.
Recombinant protein purification and GST pull-down assays
Cultures of GST-Ubc9, GST-SUMO, GST, His6-Mod(mdg4)2.2 and His6-dTopors, transformed into the Rosetta bacterial strain (Novagen), were induced with 0.1 mM IPTG, grown for 3 h, lysed by sonication in PBS with 1% Triton X and purified by either glutathione or Ni chromatography. For GST pull-down assays, 0.5 ml of lysate of cultures expressing GST-Ubc9 or GST were incubated with glutathione-conjugated beads for 3 h at 4°C, washed once with PBS and combined with purified His6-Mod(mdg4)2.2, with or without equimolar amounts of His6-dTopors. After overnight incubation, bound proteins were washed with PBS, eluted by boiling and analyzed by Western blotting.
Preparation of protein extracts and Western blot analysis
Protein extracts from third instar larvae were prepared as described (Capelson and Corces, 2005), with or without SUMO isopeptidase inhibitors NEM at 80 mM and IAA at 0.2 mM. Proteins were resolved on 7.5% SDS–PAGE and transferred to PVDF membranes in glycine buffer with 7% methanol. Blots were probed as described (Capelson and Corces, 2005).
Fly strains and crosses
Fly stocks were maintained in standard medium at 25°C. Stocks of smt34493 and smt3k06307 mutations were obtained from the Bloomington Stock Center. The lwr5486, lwr5, lwr2858 mutant strains and the UAS-lwr transgenic strain were a gift from Dr S Tanda.
Immunohistochemistry
Immunostaining of polytene chromosomes and diploid cells of larval imaginal discs and brains was carried out as described previously (Gerasimova and Corces, 1998; Gerasimova et al, 2000). Rabbit and rat α-CP190 antibodies were used at 1:400 and at 1:100 dilutions, respectively, and rabbit α-SUMO at 1:50 dilution.
Acknowledgments
We thank Dr S Tanda for providing us with fly strains of lwr mutants and transgenics, Dr A Dejean and Dr J Seeler for α-SUMO antibodies, Dr A Courey for the SUMO-GST and other constructs, Dr L Griffith for the Ubc9-GST construct, Dr A Spradling for helpful suggestions and Drs EP Lei and M Labrador for long nights of illuminating discussions. This work was supported by US Public Health Service Award GM35463 from the National Institutes of Health.
References
- Apionishev S, Malhotra D, Raghavachari S, Tanda S, Rasooly RS (2001) The Drosophila UBC9 homologue lesswright mediates the disjunction of homologues in meiosis I. Genes Cells 6: 215–224 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Byrd K, Corces VG (2003) Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol 162: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG (2005) The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell 20: 105–116 [DOI] [PubMed] [Google Scholar]
- Chalkiadaki A, Talianidis I (2005) SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol Cell Biol 25: 5095–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Ansieau S, Koken MH, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert JP, Leprince D (1995) The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene 11: 2689–2697 [PubMed] [Google Scholar]
- Dohmen RJ (2004) SUMO protein modification. Biochim Biophys Acta 1695: 113–131 [DOI] [PubMed] [Google Scholar]
- Eloranta JJ, Hurst HC (2002) Transcription factor AP-2 interacts with the SUMO-conjugating enzyme UBC9 and is sumoylated in vivo. J Biol Chem 277: 30798–30804 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG (2000) A chromatin insulator determines the nuclear localization of DNA. Mol Cell 6: 1025–1035 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG (1995) A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position–effect variegation. Cell 82: 587–597 [DOI] [PubMed] [Google Scholar]
- Geyer PK, Corces VG (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873 [DOI] [PubMed] [Google Scholar]
- Ghosh D, Gerasimova TI, Corces VG (2001) Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J 20: 2518–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, Garcia-Wilson E, Perkins ND, Hay RT (2003) P300 transcriptional repression is mediated by SUMO modification. Mol Cell 11: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD (2001) Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem 276: 18513–18518 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Hay RT (2005) SUMO: a history of modification. Mol Cell 18: 1–12 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113: 127–137 [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P (1991) A position–effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950 [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12: 2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Choi CY, Kim Y (1999) Covalent modification of the homeodomain-interacting protein kinase 2 (HIPK2) by the ubiquitin-like protein SUMO-1. Proc Natl Acad Sci USA 96: 12350–12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova E, Ohlsson R (2005) Poly(ADP-ribosyl)ation and epigenetics. Is CTCF PARt of the plot? Cell Cycle 4: 96–101 [DOI] [PubMed] [Google Scholar]
- Labrador M, Corces VG (2002) Setting the boundaries of chromatin domains and nuclear organization. Cell 111: 151–154 [DOI] [PubMed] [Google Scholar]
- Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH (2004) Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol 78: 6527–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehembre F, Badenhorst P, Muller S, Travers A, Schweisguth F, Dejean A (2000) Covalent modification of the transcriptional repressor tramtrack by the ubiquitin-related protein Smt3 in Drosophila flies. Mol Cell Biol 20: 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelard F, Labrador M, Baxter EM, Gerasimova TI, Corces VG (2002) Trans-splicing as a novel mechanism to explain interallelic complementation in Drosophila. Genetics 160: 1481–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Dejean A (1999) Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol 73: 5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG (2004) The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748 [DOI] [PubMed] [Google Scholar]
- Rasheed ZA, Saleem A, Ravee Y, Pandolfi PP, Rubin EH (2002) The topoisomerase I-binding RING protein, topors, is associated with promyelocytic leukemia nuclear bodies. Exp Cell Res 277: 152–160 [DOI] [PubMed] [Google Scholar]
- Ross S, Best JL, Zon LI, Gill G (2002) SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell 10: 831–842 [DOI] [PubMed] [Google Scholar]
- Schmidt D, Muller S (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 99: 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler JS, Marchio A, Losson R, Desterro JM, Hay RT, Chambon P, Dejean A (2001) Common properties of nuclear body protein SP100 and TIF1alpha chromatin factor: role of SUMO modification. Mol Cell Biol 21: 3314–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN (2003) Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100: 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C, Harrison DA, Corces VG (1988) The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev 2: 1414–1423 [DOI] [PubMed] [Google Scholar]
- Tsai SF, Jang CC, Prikhod'ko GG, Bessarab DA, Tang CY, Pflugfelder GO, Sun YH (1997) Gypsy retrotransposon as a tool for the in vivo analysis of the regulatory region of the optomotor-blind gene in Drosophila. Proc Natl Acad Sci USA 94: 3837–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger S, Hammer E, Engstler M (2003) The DNA topoisomerase I binding protein topors as a novel cellular target for SUMO-1 modification: characterization of domains necessary for subcellular localization and sumolation. Exp Cell Res 290: 13–27 [DOI] [PubMed] [Google Scholar]
- Weger S, Hammer E, Heilbronn R (2005) Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett 579: 5007–5012 [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD (2004) SUMO promotes HDAC-mediated transcriptional repression. Mol Cell 13: 611–617 [DOI] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, Oshimura M, Feinberg AP, Lobanenkov V, Klenova E, Ohlsson R (2004) Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet 36: 1105–1110 [DOI] [PubMed] [Google Scholar]
- Zhang H, Smolen GA, Palmer R, Christoforou A, van den Heuvel S, Haber DA (2004) SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat Genet 36: 507–511 [DOI] [PubMed] [Google Scholar]


