Abstract
The poly(A)-binding protein (PABP) is a unique translation initiation factor in that it binds to the mRNA 3′ poly(A) tail and stimulates recruitment of the ribosome to the mRNA at the 5′ end. PABP activity is tightly controlled by the PABP-interacting protein 2 (Paip2), which inhibits translation by displacing PABP from the mRNA. Here, we describe a close interplay between PABP and Paip2 protein levels in the cell. We demonstrate a mechanism for this co-regulation that involves an E3 ubiquitin ligase, EDD, which targets Paip2 for degradation. PABP depletion by RNA interference (RNAi) causes co-depletion of Paip2 protein without affecting Paip2 mRNA levels. Upon PABP knockdown, Paip2 interacts with EDD, which leads to Paip2 ubiquitination. Supporting a critical role for EDD in Paip2 degradation, knockdown of EDD expression by siRNA leads to an increase in Paip2 protein stability. Thus, we demonstrate that the turnover of Paip2 in the cell is mediated by EDD and is regulated by PABP. This mechanism serves as a homeostatic feedback to control the activity of PABP in cells.
Keywords: EDD, translation initiation, ubiquitination
Introduction
mRNA translation plays an important role in the regulation of gene expression and is implicated in the control of cell growth, proliferation and differentiation (reviewed in Gingras et al, 1999; Mathews et al, 2000). The mRNA 5′ cap structure (m7GpppN, where N is any nucleotide) interacts with eukaryotic initiation factor 4F (eIF4F) (Hershey and Merrick, 2000). eIF4F is a protein complex composed of three subunits: eIF4E, the cap binding protein; eIF4A, a bidirectional ATP-dependent RNA helicase, and eIF4G, a modular scaffolding protein that binds eIF4E, eIF4A, eIF3 and poly(A)-binding protein (PABP). Simultaneous interactions between eIF4G, eIF4E and PABP bring about mRNA circularization and promote translation initiation (Tarun and Sachs, 1996; Le et al, 1997; Imataka et al, 1998; Gebauer and Hentze, 2004; Kahvejian et al, 2005).
PABP is a phylogenetically conserved protein that contains four RNA recognition motifs (RRMs) in its N-terminal domain, of which RRMs 1 and 2 bind to poly(A) with high specificity (for reviews, see Sachs, 2000; Kahvejian et al, 2001; Mangus et al, 2003). The carboxy terminal third of PABP contains an unstructured region and PABC, an α-helical peptide-binding domain (Kozlov et al, 2001). The activity of PABP is modulated by two PABP-binding proteins, Paip1 and Paip2 (Craig et al, 1998; Khaleghpour et al, 2001b), which share two conserved PABP-interacting motifs (PAMs): PAM1 and PAM2 (Khaleghpour et al, 2001a; Roy et al, 2002). PAM1 consists of a region rich in acidic amino acids that binds strongly to the RRM region of PABP. PAM2 consists of a 15-amino-acid sequence that binds to helices 2, 3 and 5 of PABC as a series of two β-turns (Kozlov et al, 2004).
In the ubiquitin–proteasome degradation pathway, substrates are marked for degradation by covalent linkage to ubiquitin (Hochstrasser, 1996; Hershko and Ciechanover, 1998; Glickman and Ciechanover, 2002). The ubiquitinated substrate proteins are then recognized and degraded by the 26S proteasome. Ubiquitination generally requires three proteins: the ubiquitin activating enzyme (E1), the ubiquitin-carrier or conjugating enzyme (E2), and the ubiquitin ligase (E3), which transfers (or assists the transfer of) activated ubiquitin to the protein substrate. Specificity of the ubiquitination reaction is determined by the E3 ubiquitin ligase (Hershko and Ciechanover, 1998; Pickart, 2001). EDD (also known as rat100) is the mammalian ortholog of the Drosophila melanogaster hyperplastic disc (HYD) protein and is a member of the HECT (homology to E6-AP carboxy terminus) domain family of E3 ubiquitin ligases (Huibregtse et al, 1995; Callaghan et al, 1998; Oughtred et al, 2002). Surprisingly, EDD contains a PABC domain near its carboxy terminus, suggesting a role in mRNA metabolism (Callaghan et al, 1998; Deo et al, 2001; Kozlov et al, 2001; Oughtred et al, 2002). Significantly, the PABC domain of EDD is functional as it binds to Paip1 (Deo et al, 2001).
In this study, we show that Paip2 also binds to the PABC domain of EDD, and that the binding results in Paip2 ubiquitination and its subsequent degradation. This process is controlled by PABP levels. Thus, EDD modulates translation initiation through the control of the amount of the translational regulatory molecule Paip2.
Results
siRNA-mediated PABP depletion causes a decrease in the level of Paip2
To study the biological role of PABP, we depleted it from cells using small interfering RNA (siRNA) (Elbashir et al, 2001). Two siRNAs that targeted different regions in the coding sequence of the pabp gene were used (Supplementary Figure S1A). HeLa cells were transfected with either control (PABP#2 inverted sequence) or two different PABP-specific siRNAs. Following a 72-h incubation period, the expression level of PABP was determined by Western blotting (Supplementary Figure S1B). PABP levels dramatically decreased (∼90%) in cells transfected with siPABP#1, whereas siPABP#2 was less effective.
We first examined the effect of PABP-depletion on the levels of different translation factors in HeLa cells by Western blotting (Figure 1A). Unexpectedly, the amount of Paip2 was significantly reduced in PABP-depleted cells, whereas other factors (e.g. eIF4AI, eIF4E and 4E-BP1) remained unchanged. In metazoans, several binding partners of PABC have been identified. These include Paip1 and Paip2, eRF3/GSPT and Tob (Craig et al, 1998; Hoshino et al, 1999; Khaleghpour et al, 2001b; Uchida et al, 2002; Okochi et al, 2005). We therefore examined the effects of PABP depletion on PAM2-containing proteins, Paip1, Paip2 and eRF3. Total cell extracts were prepared at 24, 48 and 72 h post-transfection, and the levels of Paip1, Paip2 and eRF3 proteins were analyzed by Western blotting. Paip2 protein levels decreased in a time-dependent manner by silencing PABP, but the levels of Paip1 and eRF3 were not affected (Figure 1B). Similar results were obtained using 293T cells (data not shown). Thus, PABP depletion caused specific reduction of Paip2 protein levels without any effect on other factors, which also bind to PABP. Possible explanations for this specificity will be addressed in the Discussion.
Figure 1.
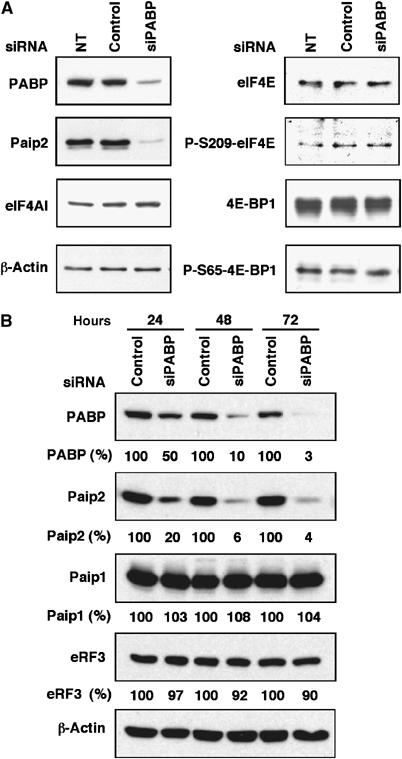
siRNA-mediated PABP depletion results in reduced Paip2 levels. (A) HeLa cells were transfected with control siRNA or PABP siRNA (siPABP#1) for 72 h, and extracts were subjected to Western blot analysis with the indicated antibodies. NT, nontransfected. (B) Extracts were collected at the indicated time points and subjected to Western blot analysis with the indicated antibodies. Data are expressed as the percentage of PABP, Paip1, Paip2, and eRF3 amounts relative to those in control siRNA-transfected cells (set at 100%). The results are representative of two independent experiments.
Paip2 protein is reduced in PABP-depleted cells
To determine the level at which Paip2 protein amount is affected, Northern blot analysis was first performed. Treatment of cells with siRNA against PABP had no effect on the amount of Paip2 mRNA as compared to either untransfected cells or control siRNA-transfected cells (Figure 2A), indicating that the regulation is not of the mRNA level. Next, the effect of siRNA against PABP on both exogenous and endogenous Paip2 was examined. HeLa cells were transiently transfected with the HA-Paip2 plasmid under the control of the cytomegalovirus (CMV) promoter. After 24 h, cells were re-transfected either with control siRNA or with two different PABP-specific siRNAs. Cells were harvested 24 h later, and equal amounts of extract were subjected to SDS–PAGE followed by Western blot analysis (Figure 2B). In agreement with the results described above (Supplementary Figure S1B), siPABP#1 effectively lowered the level of PABP as compared to siPABP#2-transfected cells (Figure 2B, both panels). Also, the extent of decrease in both endogenous Paip2 and HA-Paip2 paralleled the changes in PABP (Figure 2B, right panel). This effect on Paip2 is specific, as PABP depletion failed to affect the level of HA-eIF4E (Figure 2B, left panel). Thus, depletion of PABP specifically downregulates the steady-state amount of Paip2 protein.
Figure 2.
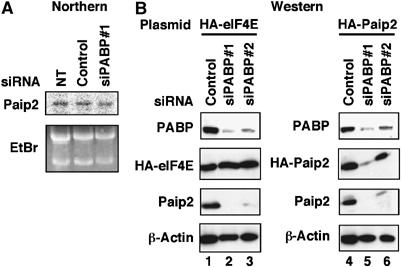
Paip2 is reduced at the protein level in PABP-depleted cells. (A) Northern blot analysis of Paip2 mRNA was performed using total RNA isolated from HeLa cells after 72 h of transfection with the indicated siRNAs (top panel). Ribosomal RNAs (28S and 18S) served as loading controls (bottom panel). NT, nontransfected. (B) HeLa cells were transfected with either HA-eIF4E (lanes 1–3) or HA-Paip2 (lanes 4–6) plasmids. At 24 h after transfection, cells were transfected with control siRNA (lanes 1 and 4), PABP siRNA (siPABP#1; lanes 2 and 5) or PABP siRNA (siPABP#2; lanes 3 and 6). Cells were harvested 48 h later, and extracts were subjected to Western blot analysis with the indicated antibodies. The results are representative of two independent experiments.
Paip2 is regulated by the ubiquitin–proteasome system
The simplest explanation of our results is that Paip2 binds to both PABP and EDD via their shared PABC domain and, consequently, these proteins would compete for binding to Paip2. PABP is a very abundant protein (4 μM cellular concentration) (Gorlach et al, 1994). Paip2 was estimated to be about five-fold less abundant than PABP in HeLa cells (Khaleghpour et al, 2001b). Immunoprecipitation of EDD from 293 cell extracts followed by SDS–PAGE and silver staining to estimate the quantity of EDD suggests that EDD might account for ∼0.001% of cell protein (data not shown). Thus, the enzyme appears to be a low abundance protein as would be expected for an E3 ligase. Therefore, under physiological conditions, most of Paip2 is predicted to be bound to PABP. However, when PABP is depleted from cells, Paip2 is expected to interact with EDD and become targeted for degradation. In subsequent experiments we addressed this hypothesis. We first examined the effect of proteasome inhibitors on the steady-state level of Paip2. HeLa cells were transfected with siRNA against PABP. After incubation for 24 h, cells were treated with several proteasome inhibitors for 6 h (we note that although MG132 at a concentration of 20 μM could be fairly toxic, after 6 h the cells were still attached to the plate), and the levels of PABP, Paip2 and β-actin were assessed by Western blotting. Treatment of cells with the proteasome inhibitors MG132, Lactacystin and PSI (proteasome inhibitor I) markedly stabilized (21- to 49-fold) the amount of Paip2 as compared to control (Figure 3A). This effect was specific because treatment of cells with the cysteine protease inhibitor E64d had no effect on Paip2 levels. None of the inhibitors stabilized Paip2 in control siRNA-transfected cells. Paip2 expression in PABP-knockdown cells was restored, albeit incompletely (44%), upon the addition of MG132 in a dose-dependent manner (Figure 3B, left panel). Addition of Velcade, another proteasome inhibitor (Adams, 2001; Xu et al, 2004; Nencioni et al, 2005) restored Paip2 levels to 58% of control (Figure 3B, right panel). These results demonstrate that the proteasome is required for the degradation of more than half of Paip2 by siRNA-mediated PABP depletion.
Figure 3.
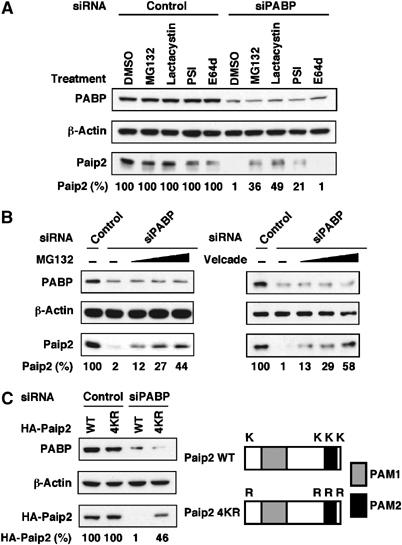
Paip2 levels are regulated by the ubiquitin-proteasome system. (A) Proteasome inhibition causes Paip2 accumulation in PABP-depleted cells. HeLa cells were transfected with control or PABP siRNAs. At 24 h after transfection, cells were treated with either DMSO or with 20 μM each of MG132, Lactacystin, PSI or E64d for 6 h. Cell extracts were then subjected to Western blot analysis with anti-PABP and anti-Paip2 antibodies. Data are expressed as the percentage of Paip2 amount relative to that in control siRNA-transfected cells (set at 100%). (B) At 24 h after siRNA transfection, cells were treated with either DMSO or with increasing concentrations (10, 20 and 40 μM) of MG132 (left panel) or (50, 100 and 200 nM) of Velcade (right panel) for 6 h and then analyzed as in (A). (C) Degradation of Paip2 in PABP knockdown cells is diminished by lysine mutations. Schematic representation of Paip2 WT and Paip2 4KR in which all the lysine residues were replaced by arginines (K2R, K108R, K116R and K123R). Gray and black boxes represent PAM1 and PAM2, respectively. HeLa cells were transfected with the indicated Paip2 plasmids. At 24 h after transfection, cells were transfected with siRNAs and then analyzed as in (A). The results are representative of two independent experiments.
Ubiquitin is conjugated to its target protein in most cases by the formation of an isopeptide bond between its carboxyl group of glycine 76 (the carboxy terminus) and the epsilon amino group of one or more lysine residues or N-terminal amino group of the target protein (Hershko and Ciechanover, 1998; Ciechanover and Ben-Saadon, 2004). Many, but not all, of such modified proteins are subsequently degraded by the proteasome (Weissman, 2001). Paip2 has four lysine residues (Figure 3C), which could be potential targets for ubiquitination. To examine the role of Paip2 lysine residues in ubiquitin-directed degradation, either wild-type (WT) Paip2 or a mutant, in which all four lysines were mutated to arginines (4KR), were expressed in HeLa cells. After incubation for 24 h to allow for expression of Paip2 proteins, cells were re-transfected with siRNAs for PABP, and harvested 24 h later. Paip2 mutant (4KR) levels were markedly increased (46-fold) in the PABP-depleted cells as compared to WT Paip2 reaching 46% level of untransfected cells (Figure 3C). Thus, lysine residues play an important role in the ubiquitin-mediated degradation of Paip2.
PABP depletion stimulates Paip2 ubiquitination
To determine directly whether ubiquitin is conjugated to Paip2, histidine-tagged ubiquitin (His-Ub) (Treier et al, 1994) was expressed in HeLa cells. After incubation for 24 h to allow for expression of His-Ub proteins, cells were re-transfected with siRNA against PABP. At 24 h following transfection, cells were treated with the proteasome inhibitor MG132 for 6 h prior to lysis. Paip2 ubiquitination was measured by capturing His-Ub from cell extracts with TALON metal affinity resin under denaturing conditions, and detecting ubiquitinated Paip2 by an anti-Paip2 antibody. This assay revealed a stronger ubiquitination of Paip2 (that barely entered the gel) in PABP-depleted cells (Figure 4). These results indicate that endogenous Paip2 is polyubiquitinated in response to PABP depletion prior to its degradation by the proteasome.
Figure 4.
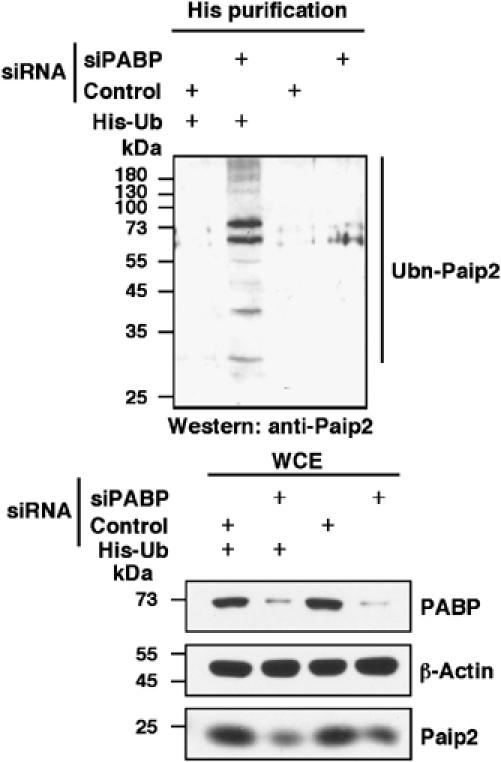
PABP depletion increases Paip2 ubiquitination. HeLa cells were transfected with plasmid encoding His-ubiquitin (His-Ub) or empty vector. At 24 h after transfection, cells were transfected with control or PABP siRNA, and treated with the proteasome inhibitor MG132. His-tagged ubiquitinated proteins were purified using TALON metal affinity resin and anti-Paip2 antibody was used to detect ubiquitinated Paip2 (Ubn-Paip2). Whole-cell extracts (WCE) were subjected to Western blot analysis with the indicated antibodies.
Interactions between EDD and Paip2
To examine the physical interaction between endogenous EDD and Paip2, we performed co-immunoprecipitation experiments. Cell lysates were subjected to immunoprecipitation with either preimmune serum or anti-Paip2 antibody, and the immunoprecipitates were analyzed by immunoblotting with anti-EDD, anti-PABP and anti-Paip2 antibodies (Figure 5A). Endogenous EDD was precipitated with anti-Paip2 antibody only in the PABP-depleted cell extract (compare lane 5 to 6). In contrast, endogenous PABP was co-precipitated with anti-Paip2 antibody in both control and PABP-depleted cell extracts (lanes 5 and 6). These results demonstrate that the association between EDD and Paip2 occurs only when PABP is depleted from cells.
Figure 5.
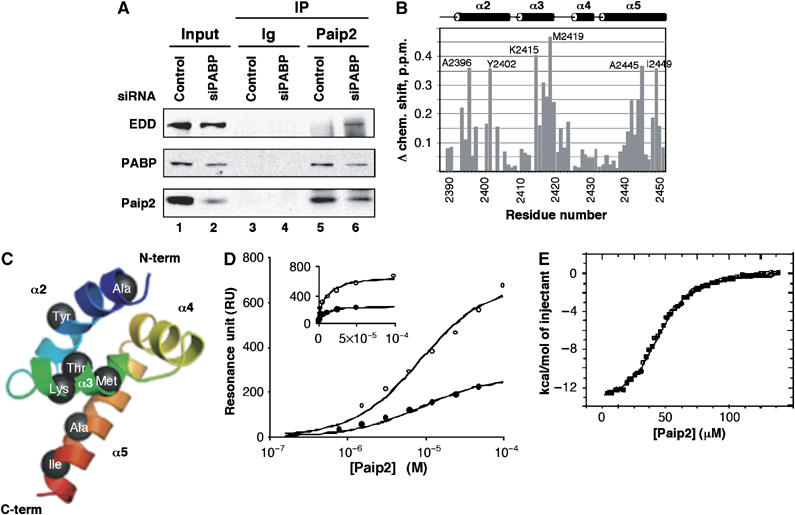
Paip2 interacts with EDD in vivo and in vitro. (A) Interaction of endogenous Paip2 and EDD. Extracts from HeLa cells transfected with control or PABP siRNA for 48 h were subjected to immunoprecipitation (IP) with preimmune rabbit serum (Ig) or anti-Paip2 antibody. The immunoprecipitates were subjected to Western blot analysis with anti-EDD, anti-PABP, and anti-Paip2 antibodies. As a negative control, immunoprecipitates obtained with preimmune rabbit serum (Ig) were used. Input fraction represents 5% of the cell extract used for IP. (B) Paip2-peptide binding to the PABC domain of EDD. Plot of amide chemical shift changes of PABC domain upon addition of the Paip2 peptide. The chemical shift changes were calculated as (ΔHN2+(0.15*ΔN)2)1/2. The most affected amides are labeled. The secondary structure is shown above. (C) Mapping of chemical shift changes on the EDD PABC crystal structure (PDB ID code 1I2 T). Residues with shifts greater than 0.3 ppm are labeled. The figure was generated using MacPyMOL. (D) Measurement of PABC EDD/Paip2 peptide apparent affinity using SPR. The symbols correspond to the average of control-corrected plateau responses (steady-state equilibrium) monitored for Paip2 peptide solutions injected at various concentrations over coupled PABC EDD at two different loadings corresponding to 4000 RU (open symbols) and 1300 RU (closed symbols). Solid lines correspond to the best global fit for binding to a single site. The insert shows the same data plotted with a linear axis for the peptide concentration. (E) Calorimetric titration of EDD (2393–2452) with Paip2 (106–127) at 298 K. The panel shows the integrated heat of each injection after correction for the heat of dilution of Paip2 and normalization for the amount of Paip2 injected. The curve represents the best fit to a model involving a single set of independent sites.
Next, nuclear magnetic resonance (NMR) spectroscopy was used to show that the PABC domain from EDD is sufficient for Paip2 binding and to determine its peptide-binding site. The PABC domain of EDD (residues 2393–2452) was cloned and expressed in media supplemented with 15NH4Cl and 13C-glucose. The 15N-1H correlation spectrum of the unliganded domain was assigned using standard triple resonance experiments (data not shown). Addition of a peptide corresponding to the PAM2 motif of Paip2 (residues 106–127) caused specific chemical shift changes (in slow exchange) in the 15N-1H spectrum of the EDD fragment. The residues displaying the largest spectral changes upon binding of the PAM2 peptide were Met2419, Lys2415, Ala2445, Ala2396, Ile2449, Tyr2402 and Thr2417 (Figures 5B and C). We mapped the chemical shift changes on the crystal structure of EDD PABC domain (Deo et al, 2001) to identify the PAM2 binding site (Figure 5C). Comparison with changes upon Paip2 binding to the PABC domain from PABP (Kozlov et al, 2001) shows an identical pattern of chemical shift changes. This suggests that the PABC domains from PABP and EDD bind peptides in an identical fashion and with overlapping specificity. Therefore, in vivo, PABP and EDD likely compete for Paip2 PAM2 binding.
We next measured the affinity of the Paip2 peptide-PABC domain of EDD interaction using a surface plasmon resonance (SPR)-based sensor, Biacore. The EDD PABC domain was covalently coupled at two different loadings and PAM2 peptides injected to measure binding (Figure 5D). The control-corrected steady-state equilibrium values (Req) corresponding to the various Paip2 peptide injections over the different EDD surfaces were used to determine a global apparent Kd of 8.8 μM for this interaction. This value is 20-fold higher than that observed for the identical peptide binding to the PABC domain from PABP (Kozlov et al, 2004).
We also applied isothermal titration calorimetry (ITC) to determine thermodynamics parameters of the EDD/Paip2 binding (Figure 5E). Addition of the Paip2 peptide to the EDD PABC domain resulted in heat release allowing for measurements of the enthalpy, entropy and Gibbs free energy of binding. The interaction is dominated by favorable enthalpic effects (ΔH of −14.5 kcal mol−1) reflecting van der Waals, hydrogen bonds, and electrostatic interactions. The negative entropy value of −25.58 cal mol−1 indicates a loss of disorder likely due to the immobilization of the peptide backbone. The change in heat capacity upon binding (ΔCp) measures changes in the degree of surface hydration and has been used to estimate the ratio of polar to nonpolar surface buried upon complex formation. The negative ΔCp of −58 cal mol−1 K−1 indicates significant hydrophobic interactions upon Paip2 binding to EDD. Comparison with the value for peptide binding to the homologous domain from PABP (−253 cal mol−1 K−1) suggests that ionic interactions are relatively more important in the case of the Paip2–EDD complex. The Kd value determined by ITC, 6.0 μM, is in excellent agreement with the value from SPR. In both cases, the binding affinity of Paip2 to the EDD PABC domain is significantly weaker than Paip2 binding to the homologous domain from PABP (0.3 μM; Kozlov et al, 2004). This difference may be due to the absence of the first α-helix in the PABC domain from EDD as compared to the PABP domain (Deo et al, 2001; Kozlov et al, 2001). In vivo, the higher affinity of Paip2 for PABP likely protects Paip2 from degradation by EDD and serves to coordinately control the level of Paip2.
Ubiquitination and degradation of Paip2 is mediated by EDD
Next, we examined whether EDD directly catalyzes the ubiquitination of Paip2 in an in vitro ubiquitination system. Immunopurified EDD was incubated with either bacterially expressed recombinant GST-Paip2 or GST, and other required ubiquitination components (including E1 and E2). The products were isolated by using glutathione-agarose beads, resolved by SDS–PAGE and then probed with anti-GST, anti-Paip2 or anti-ubiquitin antibodies (Figure 6A). Omission of E1 (lane 1) or E2 (lane 2) resulted in no ubiquitination. When EDD was omitted (lane 3), one slower migrating than Paip2 molecular weight species was detectable, which probably corresponds to monoubiquitinated Paip2. No ubiquitination was detected in the absence of His-Ub (lane 4). In the presence of all components, GST-Paip2 underwent polyubiquitination (lanes 6 and 7), but GST was not affected (lanes 9 and 10). Probing with the anti-ubiquitin antibody confirmed that the higher molecular weight conjugates of GST-Paip2 contained ubiquitin (lane 7). The anti-ubiquitin antibody detected even higher molecular weight conjugates. These conjugates likely contained multiple ubiquitin molecules per moiety of GST-Paip2 and were therefore readily detected with the anti-ubiquitin antibody (lane 7), but not detected with the anti-Paip2 antibody (lane 6). This result demonstrates that EDD is an E3 ubiquitin ligase for Paip2. Consequently, it is anticipated that EDD depletion should result in increased levels of Paip2. To address this possibility, HeLa cells were transfected consecutively at a 24-h interval, first with either control siRNA or EDD siRNA, and 24 h later re-transfected with either control siRNA or PABP siRNA. At 24 or 48 h after the second siRNA transfection, cell lysates were subjected to immunoblotting with antibodies against EDD, PABP and Paip2 (Figure 6B). Endogenous EDD was barely detectable after EDD siRNA transfections (lanes 2, 4, 6 and 8). Importantly, transfection of EDD siRNAs significantly reduced (1.9- to 3.1-fold), the loss of Paip2 caused by PABP depletion (compare lanes 3 to 4 and 7 to 8; Figures 6B and C). Moreover, Paip2 levels were increased 1.5-fold following transfection with EDD siRNA (lanes 1 and 2, and 5 and 6; Figures 6B and C). Thus, endogenous EDD specifically mediates the degradation of Paip2.
Figure 6.
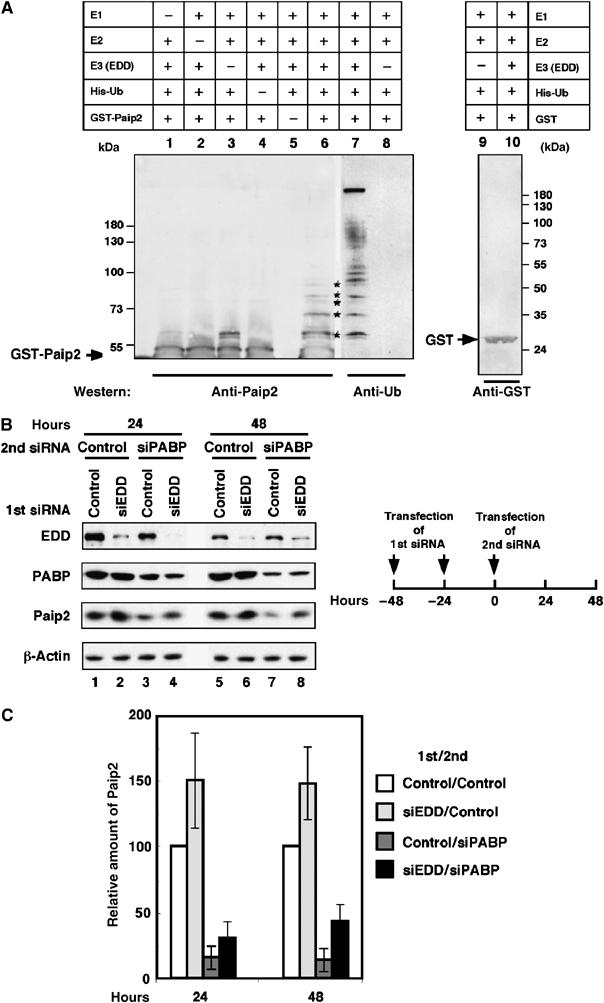
Ubiquitination of Paip2 is mediated by EDD. (A) Ubiquitination of Paip2 by EDD in vitro. EDD, isolated by immunoprecipitation with anti-EDD antibodies from rat testis lysate, was incubated with E1, E2 (UBC4-1), His-ubiquitin, ubiquitin aldehyde, AMP-PNP and either GST-Paip2 or GST. Products were then isolated by using glutathione coupled beads. Higher molecular weight ubiquitinated forms of GST-Paip2 (lane 6, asterisks) were detected by immunoblotting with anti-Paip2. As negative controls, immunoprecipitates obtained with preimmune rabbit IgG were used in the in vitro ubiquitination assay (lane 3) or reactions were carried out with GST as substrate and detected with anti-GST antibodies (lanes 9 and 10). To confirm that the higher molecular weight bands in lane 6 contained ubiquitin, replicates of samples used in lanes 3 and 6 were analyzed by immunoblotting with anti-ubiquitin antibody (lanes 7 and 8). Bands of ubiquitinated GST-Paip2 are marked with asterisks. The positions of GST and GST-Paip2 are indicated by arrows. (B) Depletion of EDD by RNAi prevents Paip2 degradation. HeLa cells were transfected consecutively at a 24-h interval with control or EDD siRNA. At 24 h following the second transfection, cells were transfected with control siRNA or PABP siRNA. At 24 and 48 h after the second siRNA transfection, cell extracts were subjected to Western blot analysis with anti-EDD, anti-PABP and anti-Paip2 antibodies. (C) Histogram of the relative amount of Paip2 from (B). Data are expressed as the percentage of Paip2 amount relative to that in control siRNA-transfected cells (set at 100%). Error bars denote the standard error of four independent experiments.
Impact of PABP homeostasis on translation
To address directly the impact of PABP homeostasis on translation, we first determined the effect of PABP knockdown on translation of a reporter mRNA, firefly luciferase (Fluc) (Figure 7A). Depletion of ∼85% (Figure 7B) of PABP resulted in only ∼10% inhibition of translation of the reporter mRNA (Figure 7A). We reasoned that this minimal effect could be explained by the fact that Paip2 WT is degraded under these conditions and, consequently, the activity of the residual PABP would be enhanced. To test this hypothesis, a mutated HA-Paip2 4KR that could not be ubiquitinated, and whose degradation is only partial (Figure 7B), was transfected into the PABP-siRNA treated cells. Under these conditions, treatment of cells with siRNA against PABP caused a significant reduction (50%; Figure 7A) in the translation of the reporter mRNA. Taken together, our data show that PABP activity is subject to homeostatic regulation via ubiquitination of its specific inhibitor, Paip2.
Figure 7.
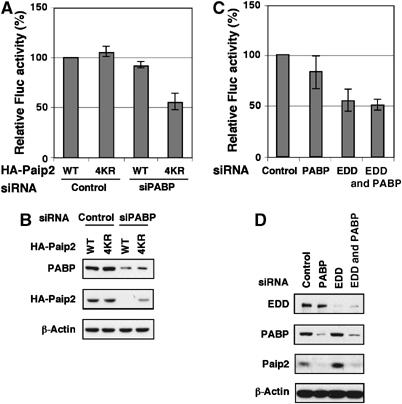
siRNA-mediated reduction of PABP affects translation. (A) HeLa cells were transfected with the indicated HA-Paip2 plasmids and siRNAs as in Figure 3C. At 24 h following the second transfection, cells were transfected with in vitro transcribed capped Fluc mRNA. Cells were harvested 6 h later, and extracts were subjected to Luciferase assay. Data are expressed as the percentage of Fluc activity in cells transfected with control siRNA and WT HA-tagged Paip2 (set at 100%). Error bars denote the standard deviation from three independent experiments. (B) Western blot analysis of the cell extracts from (A) was performed with the indicated antibodies. (C) HeLa cells were transfected with the indicated siRNAs as in Figure 6B. At 24 h following the second siRNA transfection, cells were transfected with Fluc mRNA and then analyzed as in (A). Data are expressed as the percentage of Fluc activity in cells transfected with control siRNA (set at 100%). Error bars denote the standard deviation from three independent experiments. (D) Western blot analysis of the cell extracts from (C) was performed with the indicated antibodies.
To further address the function of EDD in translation, we determined the effect of EDD and/or PABP knockdown on translation of Fluc mRNA (Figure 7C). HeLa cells transfected with siRNAs against EDD or PABP exhibited a strong depletion of the corresponding proteins (Figure 7D). Knockdown of PABP did cause an inhibition of ∼15% in the translation of Fluc mRNA. Knockdown of EDD caused a greater inhibition of translation (∼45%), possibly because Paip2 levels were increased (Figure 7D). However, a combined knockdown of EDD and PABP caused no further inhibition (Figure 7C). The lack of additive effect upon double knockdown of PABP and EDD suggests that EDD and PABP are epistatic, and functioning through the same pathway. These results rule out a model by which EDD knockdown inhibits translation through other substrates or other means. Taken together, our data show that EDD affects translation through Paip2 ubiquitination.
Discussion
In this study, we demonstrate that Paip2 is degraded via the E3 ubiquitin ligase, EDD, and the proteasome following PABP depletion in cells. Based on these observations, we propose a model for the regulation of Paip2 by PABP (Figure 8). PABP binds to Paip2 with a 1:2 stoichiometry (Khaleghpour et al, 2001a). One Paip2 molecule binds to the RRM region of PABP through its PAM1 region while the second Paip2 molecule interacts with the PABC domain of PABP through its PAM2 region. Here, we show that EDD also binds via its PABC domain to Paip2. Significantly, we demonstrate that the same motif in Paip2 (PAM2) interacts with the PABC domain of both PABP and EDD and that the affinity of Paip2 for PABP is higher than for EDD. We thus propose that PABP normally prevents Paip2 ubiquitination by blocking Paip2 interaction with EDD ubiquitin ligase by a direct competition. In PABP-depleted cells, the PAM2 region of Paip2 is free to interact with EDD, which leads to Paip2 ubiquitination and degradation through the 26S proteasome.
Figure 8.
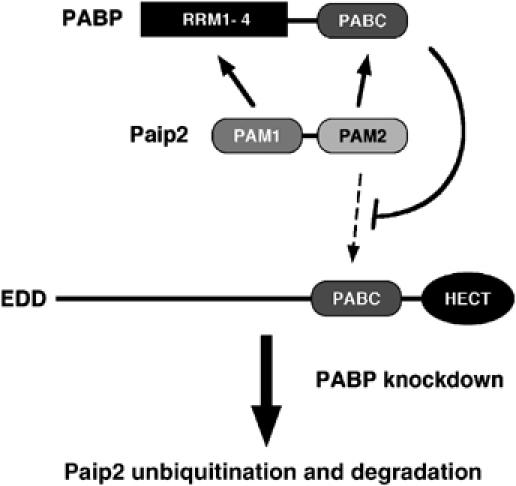
Model of ubiquitin-dependent degradation of Paip2. Two PABC-containing proteins, PABP and EDD, regulate the turnover of Paip2.
This is the first report describing that an E3 ligase competes with another protein for binding to its target. Several studies reported that protein stabilization can occur through heterodimerization. A prime example is Saccharomyces cerevisiae where MATα2 is protected from degradation by MATa1 (Johnson et al, 1998). The α1 and α2 proteins are mating type specific transcriptional regulators, which are short lived in haploid cells, but stable in diploid cells where they are co-expressed and heterodimerize. Other examples include the inhibition of ubiquitination and stabilization of E2F1 by the pRB tumor suppressor protein (Campanero and Flemington, 1997) as well as the abrogation of targeting of T-cell antigen receptor α (TCRα) for ER degradation by CD3-δ (Bonifacino et al, 1990).
What is the physiological importance of PABP regulation by Paip2? Our results suggest that Paip2 acts as an effector of PABP homeostasis. That is, a decrease in PABP levels augment Paip2 degradation via ubiquitination (Figure 8). As Paip2 levels decrease, the relative amount of PABP, which does not interact with Paip2 increases and the overall PABP activity is restored by a positive feedback loop mechanism. It is conceivable that, in vivo, Paip2 levels are tightly regulated by this feedback compensatory mechanism in order to prevent major deviations in PABP activity. PABP is an essential protein: in S. cerevisiae, deletion of the PAB1 gene is lethal (Sachs et al, 1987) and a P-element insertion in the D. melanogaster PABP gene is embryonic lethal (Sigrist et al, 2000). It will therefore be of interest to investigate how changes in PABP levels correlate with Paip2 levels during development, differentiation and growth. In this respect, EDD was shown to be overexpressed in several cancers including breast and ovarian tumors (Clancy et al, 2003; Fuja et al, 2004). In light of our results, it is likely that overexpression of EDD results in increased ubiquitination and degradation of Paip2. This in turn would result in increased activity of PABP and enhanced translation, which could contribute to oncogenic transformation (Kahvejian et al, 2001; Khaleghpour et al, 2001b). This hypothesis is consistent with the observation that PABP is overexpressed in several tumors (Zhang et al, 1997; Verlaet et al, 2001).
EDD plays an essential role in extraembryonic development as EDD knockout embryos die at E10.5 (Saunders et al, 2004). EDD is also involved in many signaling pathways including hedgehog signaling and DNA damage response (Henderson et al, 2002; Honda et al, 2002; Lee et al, 2002; Saunders et al, 2004). The interaction of EDD with proteins such as calcium- and integrin-binding protein (CIB/KIP/calmyrin) and DNA topoisomerase II-binding protein 1 (TopBP1) suggest a role for EDD in cellular DNA damage response pathways (Henderson et al, 2002; Honda et al, 2002). The binding of these proteins to EDD is mediated via the HECT domain. Therefore, it would be of interest to study the possible implications of these interactions for translational control by Paip2.
A Paip2 mutant, 4KR, which cannot be ubiquitinated by the traditional manner of isopeptide bond formation, only stabilizes the protein to 46% of control levels (Figure 3C). Only one of the four lysine residues lies in the PAM2 domain (Figure 3C). Mutagenesis studies of peptides binding to the PAM2 domain from Paip2 have been published (Kozlov et al, 2004). While this lysine of PAM2 is not important for PABC domain interaction with Paip2, it is expected that ubiquitination of this residue would abrogate PABC binding. We examined the interaction between PABP and HA-Paip2 4KR mutant by co-immunoprecipitation experiments, and found it similar to HA-Paip2 WT (data not shown). Using the in vivo ubiquitination assay, we also examined the ubiquitination of HA-Paip2 4KR mutant, and found that it is still ubiquitinated in 293 cells albeit to a lesser extent as compared to HA-Paip2 WT (data not shown). This raises the possibility that the N-terminal amino group is also a site of ubiquitination (Ciechanover and Ben-Saadon, 2004; Coulombe et al, 2004; Kuo et al, 2004). Interestingly, EDD was recently identified as a member of the UBR family of proteins, which recognize N-terminal degrons (Tasaki et al, 2005). Although the second amino acid of Paip2 is lysine, which is a destabilizing amino acid in the N-end rule pathway and so might be recognized by EDD, the UBR E3s classically ubiquitinate substrates on an internal lysine residue (Chau et al, 1989). It remains possible that there are additional pathways leading to Paip2 degradation. This hypothesis is favored by the observations that (a) proteasome inhibitors do not completely stabilize Paip2; (b) that EDD depletion only partly restores Paip2 stability and (c) that deletion of the PAM2 motif destabilizes Paip2 (3A, B, 6B and 7D; data not shown).
Paip1 and eRF3 have also been reported to interact with PABP through their PAM2 motifs (Hoshino et al, 1999; Khaleghpour et al, 2001b; Roy et al, 2002; Uchida et al, 2002). However, we demonstrate that Paip1 and eRF3 are not degraded in PABP-knockdown cells (Figure 1B). This result was not predicted since the PAM2 motifs within Paip1 and eRF3 are similar to that in Paip2 and would be expected to bind to EDD. It may be that the PAM2 motif is not sufficient for degradation of these proteins or that an additional yet unknown factor is required for specific ubiquitination of Paip1 and eRF3. Alternatively, Paip1 and eRF3 may be stabilized by interacting with other proteins. Indeed, Paip1 is known to interact with eIF4A (Craig et al, 1998) and eRF3 with eRF1 (Hoshino et al, 1999).
In conclusion, we have described a novel mechanism whereby protein stability is mediated by competition between an E3 ubiquitin ligase and a functional partner (PABP) for a specific binding site, such as the PAM2 motif of Paip2. This may be an example of a hitherto unrecognized widespread mechanism for coordinately regulating protein levels, and in the case of PABP, could serve as a feedback mechanism to control its activity in the cell.
Materials and methods
Cell culture and transfection
HeLa S3 and 293T cells were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (Gibco) and penicillin/streptomycin solution (Gibco) in 5% CO2. DNA transfection was performed using LipofectAMINE Plus reagent (Invitrogen) following the manufacturer's protocol.
Plasmids
Full-length Paip2 (Khaleghpour et al, 2001b) was subcloned into pcDNA3 with a hemagglutinin (HA) tag to generate pcDNA3-HA-Paip2. pcDNA3-HA-eIF4E was described previously (Pyronnet et al, 1999). Mutations in Paip2 were generated by site-directed mutagenesis of HA-Paip2 with the QuikChange site-directed mutagenesis kit (Stratagene), according to the manufacturer's protocol. The plasmid encoding the firefly luciferase, pSP72-LUC-A, was described previously (Imataka et al, 1998).
Chemicals
MG132, Lactacystin, PSI and E64d were obtained from Calbiochem. Compounds were dissolved in DMSO and used at the concentrations indicated. Velcade (PS-341/Bortezomib) was obtained from Millennium Pharmaceuticals, through Pierre Laneuville (McGill University).
Antibodies and Western blotting
The following antibodies were used: rabbit polyclonal anti-PABP (Imataka et al, 1998), rabbit polyclonal anti-Paip1 (Craig et al, 1998), rabbit polyclonal anti-Paip2 (Khaleghpour et al, 2001b), mouse monoclonal anti-eIF4E (BD Biosciences Pharmingen), rabbit polyclonal anti-phospho-eIF4E (Ser209) (Cell signaling), rabbit polyclonal 4E-BP (Cell signaling), rabbit polyclonal anti-phospho-4E-BP1 (Ser65) (Cell signaling), rabbit polyclonal anti-eIF4AI (Ferraiuolo et al, 2004), rabbit polyclonal anti-eRF3 (Hoshino et al, 1999), rabbit polyclonal anti-EDD (Oughtred et al, 2002), mouse monoclonal anti-HA (Berkeley Antibody Company), mouse monoclonal anti-β-Actin (Berkeley Antibody Company), anti-ubiquitin (Sigma; U5379), rabbit polyclonal anti-GST (Amersham Pharmacia). Western blot analysis was performed as previously described (Ferraiuolo et al, 2004; Roy et al, 2004).
Northern blotting
Total RNA was isolated using Qiagen RNAeasy kit according to the manufacturer's protocol. For Northern blots, 20 μg of total RNA was resolved on a 1.2% agarose/formaldehyde gel, transferred onto a Hybond-N membrane (Amersham Pharmacia), and probed with a 32P-labeled random-primed DNA probe derived from the Paip2 open reading frame.
Immunoprecipitation
For the crosslinking of anti-Paip2 antibody to protein A beads, 25 μl of anti-Paip2 antibody was incubated with 300 μl of protein A Sepharose (Amersham Biosciences) in a final volume of 10 ml containing 0.2 M triethanolamine–HCl (pH 9.0). Antibody and beads were then crosslinked in 20 mM dimethyl pimelinediimidate (DMP; Sigma) for 1 h at room temperature. The reaction was stopped by washing with 0.2 M ethanolamine–HCl (pH 8.0). Cells were lyzed in buffer A (20 mM HEPES–KOH (pH 7.6), 100 mM KCl, 1 mM DTT, 0.1 mM EDTA, 10% glycerol and 0.1% Nonidet P-40). For immunoprecipitations, cell extracts (0.5–1 mg of protein) were incubated with anti-Paip2 antibody crosslinked to protein A beads (bed volume of 30 μl) in a 300 μl total volume for 3 h at 4°C. Beads were washed four times in 1 ml of cold buffer A. Proteins were eluted with 2 × Laemmli sample buffer, boiled for 5 min, and processed for Western blotting.
Ubiquitination assay
In vivo or in vitro ubiquitination assays were carried out as detailed in Supplementary data.
In vitro transcription and RNA transfection
In vitro transcription of capped firefly luciferase (Fluc) mRNA was described previously (Imataka et al, 1998; Svitkin et al, 2001). For RNA transfections, the Fluc RNA (200 ng) were transfected into HeLa cells using DMRIE-C reagent (Invitrogen) or Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Cells were harvested 6 h post-transfection and luciferase activity was measured using the luciferase assay kit (Promega).
Small interfering RNA (siRNA) transfections
All siRNA duplexes were designed as 21-mers with 3′-dTdT overhangs (Elbashir et al, 2002). siRNAs were purchased from Dharmacon. The sequences of the siRNAs are as follows: siPABP#1, 5′-AAGGUGGUUUGUGAUGAAAAU-3′; siPABP#2, 5′-AACUAAGACCAAGUCCUCGCU-3′; siPABP#2 INV, 5′-AAUCGCUCCUGAACCAGAAUC-3′; siEDD, 5′-AAUUGUGCAACGUAGCAGAGU-3′; The siPABP#2 INV was used as a control. siRNA transfections were performed as described previously (Costa-Mattioli et al, 2004; Ferraiuolo et al, 2004). All experiments were carried out with siPABP#1, except where otherwise indicated.
Protein chemistry
PABC expression and purification, NMR spectroscopy, ITC measurements and SPR-based biosensor analysis were carried out as described in Supplementary data.
Supplementary Material
Supplementary Materials and Methods
Supplementary Figure S1
Supplementary Figure S2
Acknowledgments
We thank M Costa-Mattioli for the design of siRNAs, S Hoshino and T Katada for the eRF3 antibody, P Laneuville for Velcade, CP Park, T Shimogori and T Lubell for help in preparing the manuscript, and Y Svitkin, H Imataka and P Cormier for helpful discussions. We thank C Lister for excellent technical assistance. This research was supported by a Public Health Service Research Grant from the NIH (Grant #5R01 GM66157-03) to NS and the Canadian Institute of Health Research (CIHR) to KG and SSW MY is a postdoctoral fellow supported by Uehara Memorial Foundation Fellowship and CIHR Fellowship. KY is a postdoctoral fellow supported by Uehara Memorial Foundation Fellowship. AK is a recipient of a McGill Faculty of Medicine Internal Fellowship. ZP is a recipient of a McGill University Health Centre Research Institute Studentship. KG and SSW are a Québec Fonds de la recherche en santé Chercheur National award recipients. NS is a CIHR distinguished scientist and a Howard Hughes Medical Institute International Scholar.
References
- Adams J (2001) Proteasome inhibition in cancer: development of PS-341. Semin Oncol 28: 613–619 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD (1990) Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell 63: 503–513 [DOI] [PubMed] [Google Scholar]
- Callaghan MJ, Russell AJ, Woollatt E, Sutherland GR, Sutherland RL, Watts CK (1998) Identification of a human HECT family protein with homology to the Drosophila tumor suppressor gene hyperplastic discs. Oncogene 17: 3479–3491 [DOI] [PubMed] [Google Scholar]
- Campanero MR, Flemington EK (1997) Regulation of E2F through ubiquitin–proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci USA 94: 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Ben-Saadon R (2004) N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol 14: 103–106 [DOI] [PubMed] [Google Scholar]
- Clancy JL, Henderson MJ, Russell AJ, Anderson DW, Bova RJ, Campbell IG, Choong DY, Macdonald GA, Mann GJ, Nolan T, Brady G, Olopade OI, Woollatt E, Davies MJ, Segara D, Hacker NF, Henshall SM, Sutherland RL, Watts CK (2003) EDD, the human orthologue of the hyperplastic discs tumour suppressor gene, is amplified and overexpressed in cancer. Oncogene 22: 5070–5081 [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Svitkin Y, Sonenberg N (2004) La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol Cell Biol 24: 6861–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe P, Rodier G, Bonneil E, Thibault P, Meloche S (2004) N-terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol Cell Biol 24: 6140–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AW, Haghighat A, Yu AT, Sonenberg N (1998) Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392: 520–523 [DOI] [PubMed] [Google Scholar]
- Deo RC, Sonenberg N, Burley SK (2001) X-ray structure of the human hyperplastic discs protein: an ortholog of the C-terminal domain of poly(A)-binding protein. Proc Natl Acad Sci USA 98: 4414–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T (2002) Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26: 199–213 [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N (2004) A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci USA 101: 4118–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuja TJ, Lin F, Osann KE, Bryant PJ (2004) Somatic mutations and altered expression of the candidate tumor suppressors CSNK1 epsilon, DLG1, and EDD/hHYD in mammary ductal carcinoma. Cancer Res 64: 942–951 [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW (2004) Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Gorlach M, Burd CG, Dreyfuss G (1994) The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res 211: 400–407 [DOI] [PubMed] [Google Scholar]
- Henderson MJ, Russell AJ, Hird S, Muñoz M, Clancy JL, Lehrbach GM, Calanni ST, Jans DA, Sutherland RL, Watts CK (2002) EDD, the human hyperplastic discs protein, has a role in progesterone receptor coactivation and potential involvement in DNA damage response. J Biol Chem 277: 26468–26478 [DOI] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC (2000) The pathway and mechanism of initiation of protein synthesis. In Translational Control of Gene Expression, Mathews MB (ed), pp 33–88. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (1996) Ubiquitin-dependent protein degradation. Annu Rev Genet 30: 405–439 [DOI] [PubMed] [Google Scholar]
- Honda Y, Tojo M, Matsuzaki K, Anan T, Matsumoto M, Ando M, Saya H, Nakao M (2002) Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J Biol Chem 277: 3599–3605 [DOI] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T (1999) The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680 [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA 92: 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Gradi A, Sonenberg N (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 17: 7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PR, Swanson R, Rakhilina L, Hochstrasser M (1998) Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94: 217–227 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Roy G, Sonenberg N (2001) The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harbor Symp Quant Biol 66: 293–300 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'boutchou MN, Sonenberg N (2005) Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, Imataka H, O'Connor-McCourt M, Sonenberg N (2001a) Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol 21: 5200–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N (2001b) Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7: 205–216 [DOI] [PubMed] [Google Scholar]
- Kozlov G, De Crescenzo G, Lim NS, Siddiqui N, Fantus D, Kahvejian A, Trempe JF, Elias D, Ekiel I, Sonenberg N, O'Connor-McCourt M, Gehring K (2004) Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J 23: 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K (2001) Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci USA 98: 4409–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ (2004) N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev 18: 1862–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR (1997) Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem 272: 16247–16255 [DOI] [PubMed] [Google Scholar]
- Lee JD, Amanai K, Shearn A, Treisman JE (2002) The ubiquitin ligase hyperplastic discs negatively regulates hedgehog and decapentaplegic expression by independent mechanisms. Development 129: 5697–5706 [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A (2003) Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB (2000) Origins and principles of translational control. In Translational Control of Gene Expression, Mathews MB (ed), pp 1–32. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Nencioni A, Hua F, Dillon CP, Yokoo R, Scheiermann C, Cardone MH, Barbieri E, Rocco I, Garuti A, Wesselborg S, Belka C, Brossart P, Patrone F, Ballestrero A (2005) Evidence for a protective role of Mcl-1 in proteasome inhibitor-induced apoptosis. Blood 105: 3255–3262 [DOI] [PubMed] [Google Scholar]
- Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T (2005) Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes Cells 10: 151–163 [DOI] [PubMed] [Google Scholar]
- Oughtred R, Bedard N, Adegoke OA, Morales CR, Trasler J, Rajapurohitam V, Wing SS (2002) Characterization of rat100, a 300-kilodalton ubiquitin-protein ligase induced in germ cells of the rat testis and similar to the Drosophila hyperplastic discs gene. Endocrinology 143: 3740–3747 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras AC, Fukunaga R, Hunter T, Sonenberg N (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J 18: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O'Connor-McCourt M, Sonenberg N (2002) Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol 22: 3769–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy G, Miron M, Khaleghpour K, Lasko P, Sonenberg N (2004) The Drosophila poly(A) binding protein-interacting protein, dPaip2, is a novel effector of cell growth. Mol Cell Biol 24: 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A (2000) Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In Translational Control of Gene Expression, Mathews MB (ed), pp 447–466. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sachs AB, Davis RW, Kornberg RD (1987) A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol 7: 3268–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders DN, Hird SL, Withington SL, Dunwoodie SL, Henderson MJ, Biben C, Sutherland RL, Ormandy CJ, Watts CK (2004) Edd, the murine hyperplastic disc gene, is essential for yolk sac vascularization and chorioallantoic fusion. Mol Cell Biol 24: 7225–7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Thiel PR, Reiff DF, Lachance PE, Lasko P, Schuster CM (2000) Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405: 1062–1065 [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Imataka H, Khaleghpour K, Kahvejian A, Liebig HD, Sonenberg N (2001) Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA 7: 1743–1752 [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ Jr, Sachs AB (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15: 7168–7177 [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT (2005) A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol 25: 7120–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78: 787–798 [DOI] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T (2002) A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J Biol Chem 277: 50286–50292 [DOI] [PubMed] [Google Scholar]
- Verlaet M, Deregowski V, Denis G, Humblet C, Stalmans MT, Bours V, Castronovo V, Boniver J, Defresne MP (2001) Genetic imbalances in preleukemic thymuses. Biochem Biophys Res Commun 283: 12–18 [DOI] [PubMed] [Google Scholar]
- Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2: 169–178 [DOI] [PubMed] [Google Scholar]
- Xu Q, Farah M, Webster JM, Wojcikiewicz RJ (2004) Bortezomib rapidly suppresses ubiquitin thiolesterification to ubiquitin-conjugating enzymes and inhibits ubiquitination of histones and type I inositol 1,4,5-trisphosphate receptor. Mol Cancer Ther 3: 1263–1269 [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW (1997) Gene expression profiles in normal and cancer cells. Science 276: 1268–1272 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Supplementary Figure S1
Supplementary Figure S2


