Abstract
Krüppel-type zinc finger (ZNF) motifs are prevalent components of transcription factor proteins in all eukaryotes. KRAB-ZNF proteins, in which a potent repressor domain is attached to a tandem array of DNA-binding zinc-finger motifs, are specific to tetrapod vertebrates and represent the largest class of ZNF proteins in mammals. To define the full repertoire of human KRAB-ZNF proteins, we searched the genome sequence for key motifs and then constructed and manually curated gene models incorporating those sequences. The resulting gene catalog contains 423 KRAB-ZNF protein-coding loci, yielding alternative transcripts that altogether predict at least 742 structurally distinct proteins. Active rounds of segmental duplication, involving single genes or larger regions and including both tandem and distributed duplication events, have driven the expansion of this mammalian gene family. Comparisons between the human genes and ZNF loci mined from the draft mouse, dog, and chimpanzee genomes not only identified 103 KRAB-ZNF genes that are conserved in mammals but also highlighted a substantial level of lineage-specific change; at least 136 KRAB-ZNF coding genes are primate specific, including many recent duplicates. KRAB-ZNF genes are widely expressed and clustered genes are typically not coregulated, indicating that paralogs have evolved to fill roles in many different biological processes. To facilitate further study, we have developed a Web-based public resource with access to gene models, sequences, and other data, including visualization tools to provide genomic context and interaction with other public data sets.
The human genome contains ∼30,000 genes (Lander et al. 2001; Venter et al. 2001) including at least 2000 loci encoding transcription factor proteins (TFs) (Messina et al. 2004). The C2H2, or Krüppel-type zinc finger (ZNF), is the most common DNA-binding motif found in eukaryotic TF proteins; ZNF proteins typically contain multiple C2H2 motifs joined together in tandem arrays. Proteins of this type are ancient and numerous, encoded by large and diverse gene families in all eukaryotic genomes. Subfamilies of distinct structure and function have arisen in different evolutionary lineages defined by the types of chromatin interaction modules, or effector motifs, included in the protein (Knochel et al. 1989; Bellefroid et al. 1991; Chung et al. 2002; for reviews, see Collins et al. 2001; Huntley et al., in press).
At least one-third of mammalian ZNF proteins include an effector motif called the Krüppel-associated box, or KRAB, which serves to recruit histone deacetylase complexes to regions surrounding the DNA-binding sites (Bellefroid et al. 1991; Friedman et al. 1996; Pengue and Lania 1996; Abrink et al. 2001; Ayyanathan et al. 2003). KRAB-associated ZNF (KRAB-ZNF) proteins thus function as potent transcriptional repressors (Margolin et al. 1994; Friedman et al. 1996; Ayyanathan et al. 2003). Proteins combining KRAB and ZNF domains are specific to tetrapod vertebrates, but the family has expanded dramatically to include hundreds of members in mammals (Bellefroid et al. 1995; Looman et al. 2002; Hamilton et al. 2003). Many KRAB-ZNF loci reside in familial gene clusters, indicating that the family has evolved primarily through tandem in situ duplication (Bellefroid et al. 1995; Collins et al. 2001; Huntley et al., in press). Recent studies have shown that paralogs diversify through structural changes in the zinc finger arrays that are driven by positive selection (Hamilton et al. 2003, 2006; Shannon et al. 2003; Schmidt and Durrett 2004); since even subtle alterations in zinc-finger array structure can yield proteins with distinct DNA recognition specificities (Miller and Pabo 2001; Krebs et al. 2005), this pattern of divergence suggests an active selection for novel transcription factors with altered regulatory properties. However, since most KRAB-ZNF genes remain completely uncharacterized, it has not been possible to examine the evolutionary histories or functional diversity of this large gene family in depth.
To derive a complete catalog of the KRAB-ZNF gene family, we computationally analyzed and manually curated all segments of the human genome containing KRAB and ZNF domains. These efforts revealed 423 protein-coding genes with alternative transcripts that predict the existence of at least 742 distinct proteins, as well as 341 pseudogene sequences. Analyses of this gene set and comparisons to predicted genes in other mammalian genomes permitted a genome-wide assessment of the mechanisms through which this gene family has evolved. Results of this study can be accessed from a public Web site (http://znf.llnl.gov/) that will be updated as additional data becomes available.
Results
Assembling the human KRAB-ZNF catalog and Web-based resource
KRAB-ZNF genes exist as simple modular structures with one or more KRAB-effector domains and a tandem array of zinc-finger motifs encoded within distinct 5′ and 3′ exons, respectively (Shannon and Stubbs 1998). In addition to the functionally dominant KRAB-A motif, many genes also encode modulating motifs such as KRAB-B (Bellefroid et al. 1991), a novel KRAB-B variant we will refer to as KRAB-BL (KRAB-BL exons are 30 bp larger than KRAB-B exons, extending in the 3′ direction), KRAB-b (Mark et al. 1999), or KRAB-C (Looman et al. 2004). A small number of KRAB-ZNF genes also encode a second vertebrate-specific effector called SCAN (Sander et al. 2003). To catalog the complete gene family, we employed profile Hidden Markov Model (HMM) software (HMMER 2.3, http://hmmer.wustl.edu/) to generate profile HMMs for KRAB, SCAN, and Krüppel-type finger motifs and to scan the human genome for matching sequences.
Based on RNA evidence and HMM-identified motifs, we generated models for alternate transcripts arising from each locus and identified overlaps with publicly available known and predicted genes. Three hundred thirty-four HMM-based models overlapped known loci; manual annotation produced 669 transcript models for these genes, including 495 models that we extended or corrected and 157 public models that were not modified. In addition to the known genes, we identified 89 KRAB-ZNF loci capable of encoding full-length proteins that are not described in public databases (see Methods). Altogether we annotated 423 loci encoding proteins with both effector (KRAB and/or SCAN) and zinc-finger domains (Table 1; Supplemental Table S1), for simplicity we will hereafter refer to the collection as KRAB-ZNF genes.
Table 1.
Characteristics of KRAB-ZNF gene family members
aIncludes all SCAN-KRAB-A genes with and without additional modulating motifs.
In addition to KRAB-ZNF loci, we also identified 254 genes with noncanonical structures, e.g., encoding ZNF-only, KRAB-only, or SCAN-only proteins as their only potential protein product. Loci of this type were annotated as genes only when supported by mRNA evidence; these genes may indeed correspond to functional family members. However, in the following discussions we will focus primarily on the 423 loci capable of encoding proteins with both effector (SCAN, KRAB, or both) and zinc-finger domains. To publicly share the data arising from this analysis, we created a Web-based resource (http://znf.llnl.gov/) that provides access to full descriptions and sequences of all curated KRAB-ZNF gene models and pseudogene loci. Interfaces for searching and browsing the database, including an added track within the UCSC Genome Browser (Kent et al. 2002), are also provided, along with a set of downloadable data files describing the models, motifs identified, and HMMER matrices. We will continue to expand and update the Web resource as new data regarding these loci are made available.
Alternate splicing and pseudogenes
Based on gene models, we identified 818 different transcripts encoding 742 structurally distinct proteins from the 423 KRAB-ZNF genes. The most prevalent class of transcripts includes KRAB-A, KRAB-B, and ZNF-encoding exons, but proteins with many different structures were also predicted. For example, alternate transcripts encoding KRAB-only or fingers-only proteins, or proteins that include only one of several encoded effectors, are frequently generated from the KRAB-ZNF gene sets, as has been described previously for specific genes (Table 1; Supplemental Table S1; Bellefroid et al. 1993; Wu et al. 2003; Oh et al. 2005). Predicted KRAB-ZNF proteins with 2–40 tandem zinc-fingers are encoded by the collection of human genes, with a median number of 12 ZNF motifs per gene. SCAN-containing proteins typically include a smaller number of zinc finger motifs than other proteins in this family (Table 1). This observation suggests that the SCAN proteins might typically recognize shorter DNA-binding motifs, an interpretation that is interesting in light of the fact that many SCAN-ZNF proteins bind DNA as homodimers (Sander and Morris 2002).
We also identified 227 gene fragments and 39 full-length pseudogenes, based on evidence of multiple stop codons, frameshifts, and lack of proper splice junctions. Sequence comparisons confirmed that most pseudogenes arose from neighboring loci by partial-gene duplication events (data not shown), although gene remnants may also be left behind after lineage-specific deletions (Hamilton et al. 2003; Shannon et al. 2003). We also found evidence of 75 processed KRAB-ZNF pseudogene sequences. Three of these processed pseudogenes maintain open reading frames potentially capable of encoding functional proteins: LLNL1071 (HSA3, 32Mb), LLNL1040 (HSA9, 35Mb), and LLNL973 (HSA12, 132Mb). All other processed loci correspond to degraded, nonfunctional copies.
Gene clustering and evolution
We counted 65 KRAB-ZNF, SCAN-ZNF, and mixed gene clusters in the human genome (for cluster definition and criteria, see Methods) (Supplemental Table 1). A total of 384 KRAB-ZNF genes reside in these clusters; the remaining 39 loci were classified as isolated singleton genes (Table 1). Most of the genes are concentrated in 25 major clusters, the largest of which are located on HSA19 (Table 2). A few KRAB-ZNF gene clusters are well conserved in mammals; e.g., a cluster located in HSA5q35.3 contains six tandem genes arranged identically to orthologous genes in human, chimp, dog, and mouse (Table 2; Supplemental Table S6). However, several human clusters are primate specific and most conserved gene clusters display clear evidence of lineage-specific expansion, with different numbers of genes in each species and relatively few orthologous groups.
Table 2.
Properties of major human KRAB-ZNF clusters and related regions in other mammalian genomes
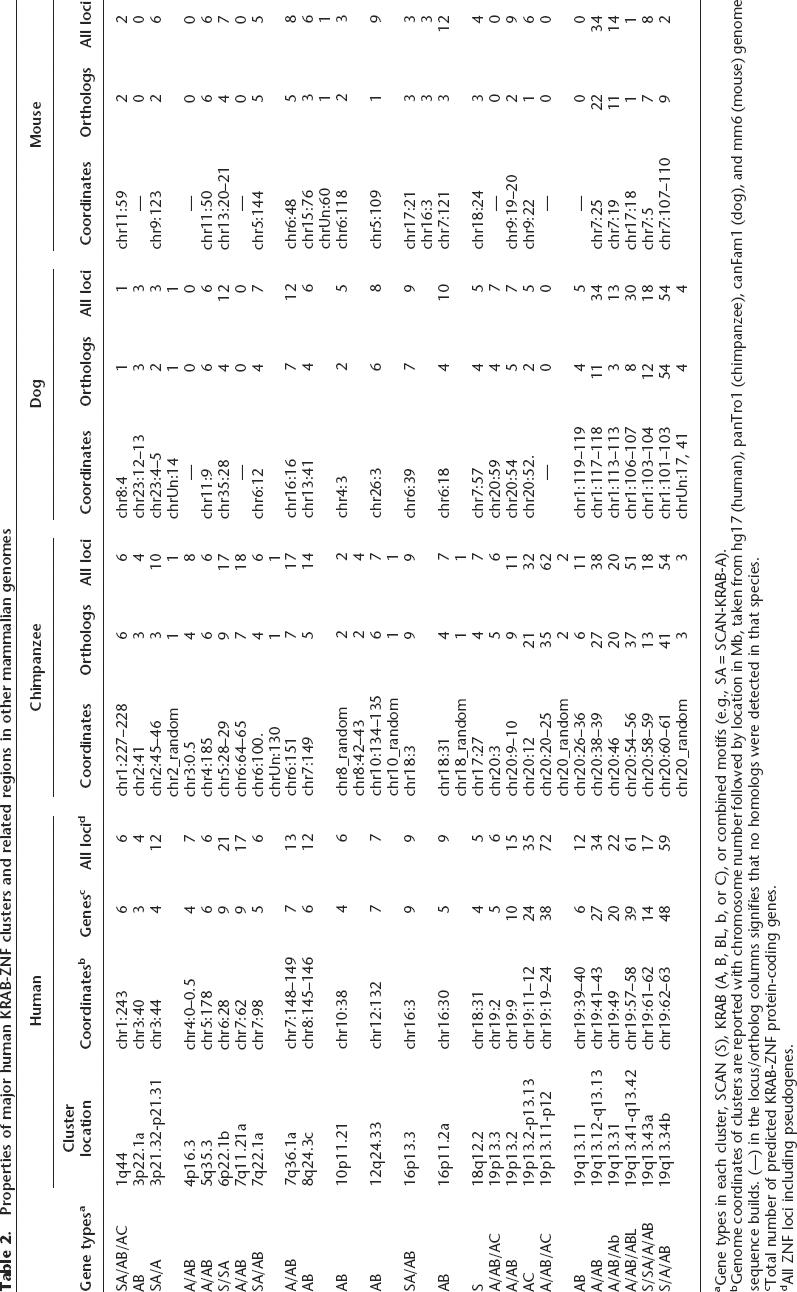
aGene types in each cluster, SCAN (S), KRAB (A, B, BL, b, or C), or combined motifs (e.g., SA = SCAN-KRAB-A).
bGenome coordinates of clusters are reported with chromosome number followed by location in Mb, taken from hg17 (human), panTro1 (chimpanzee), canFam1 (dog), and mm6 (mouse) genome sequence builds. (—) in the locus/ortholog columns signifies that no homologs were detected in that species.
cTotal number of predicted KRAB-ZNF protein-coding genes.
dAll ZNF loci including pseudogenes.
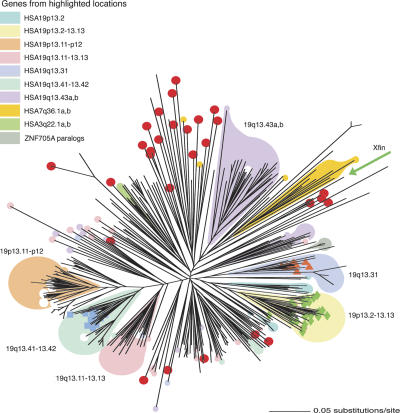
To identify evolutionary relationships, we constructed a phylogenetic tree based on KRAB-A–encoding nucleotide sequences (Fig. 1; Supplemental Table S2). As suggested by previous studies focused on subsets of genes, evolutionary relatedness is typically associated with physical proximity in this family. However, the complete family tree also shows that unrelated KRAB-ZNF genes are physically intermixed at several clustered sites. Therefore, although tandem in situ duplication events have represented the major mechanism of new gene creation in the KRAB-ZNF family, distributed duplication and, possibly, post-duplication rearrangement events have also played a prominent role. It is uncertain at present what effect gene conversion may have had on the evolution of these genes.
Figure 1.
Phylogenetic tree of human KRAB-A motifs. This neighbor-joining phylogenetic tree represents 418 human KRAB-A nucleotide sequences from KRAB-ZNF and SCAN-KRAB-ZNF genes (including some with noncanonical structures). Gene designations are removed from this unrooted phylogram for clarity; a list of the genes including location and aligned KRAB sequences is presented in Supplemental Table S2. Genes from several major physical clusters are colored to show comparisons between physical location and sequence similarity. Loci within a highlighted phylogenetic group that do not map to the same physical cluster as related genes appear as superimposed circles in the appropriate color (or in white for genes that do not belong to any labeled cluster). Genes that also encode SCAN, KRAB-b, KRAB-BL, or KRAB-C motifs are indicated as follows: SCAN, red circles; KRAB-b, orange triangles; KRAB-BL, blue squares; and KRAB-C, green diamonds. The green arrow notes the position of the Xenopus Xfin KRAB sequence, added as a potential outgroup, although the tree is shown unrooted.
Most genes containing the KRAB-A motif also include the KRAB-B modulator, or less common modulators KRAB-b, KRAB-BL, or KRAB-C (Table 1). These associations appear within separate clades in the KRAB-A–based tree, indicating that these distinct motifs arose and were expanded within specific families (Fig. 1). Unlike genes with specific types of KRAB modulators, the SCAN-containing genes do not group together in one evolutionary clade (Fig. 1, red circles). This pattern could be explained if the SCAN-KRAB-ZNF combination is ancient, with a history of frequent loss of one or the other effector domains during the expansion of the gene family. This kind of history would be consistent with the comingling of related genes with different combinations of SCAN and KRAB effector motifs we observed in several clusters (Table 2). However, it is also possible that the SCAN-KRAB combination arose more than once, as has recently been proposed (Looman et al. 2002).
Phylogenetic analyses also highlighted relatedness between clusters and among cluster members and isolated loci distributed at distant chromosomal sites. For example, genes from the large 19p12 cluster, which is known to be primate specific (Bellefroid et al. 1995; Eichler et al. 1998), group together in the tree with a clade of genes from a separate cluster located in 19q13.41. Unlike the 19p12 cluster, this latter clade includes genes with clear mouse and dog orthologs (Table 2, see below) and therefore may be related to progenitor genes for the expanded primate group.
Relationships in certain groups show that some distributed duplicates may subsequently give rise to tandem copies, suggesting one way that new lineage-specific clusters may have been seeded over evolutionary time. For example, seven KRAB-ZNF genes and one pseudogene sequence distributed in HSA4, 8, 11, and 12 show >96% nucleotide sequence identity over >70-kb duplications (ZNF705A paralogs) (highlighted in gray in Fig. 1; Supplemental Table S2). The high degree of similarity between these large segments indicates that most of the duplication events occurred ≤15 million years ago (Mya), and some events may be human specific. Most notably, two adjacent pairs of HSA8 paralogs arose from very recent tandem duplications (ZNF705C and LLNL1103 at 11.9 and 12.2 Mb with 98.5% identity, and LLNL1035 and ZNF705B, at 7.2 and 7.6 Mb with >99% identity); although the human specificity of these duplications must be verified, the chimpanzee genome contains only one ZNF gene at each locus (chr7 at 12.7 and 7.5 Mb, respectively).
Paralogs, orthologs, and recent primate duplications
To identify putative orthologs of human KRAB-ZNF genes, we also generated HMM-based gene models from the chimpanzee, mouse, and dog genomes and searched for reciprocal best BLAST matches between these models and the curated human coding gene set (Supplemental Tables S3–5). We manually inspected alignments in the variable DNA-binding helix regions of the ZNF domains (Choo and Klug 1994; Kim and Berg 1995; Shannon et al. 2003) to confirm the accuracy of identified orthologs (see Methods). Although the draft status of mouse, dog, and chimp genomes makes it impossible to determine the KRAB-ZNF gene repertoires of those species completely, these data provide a clear overall view of KRAB-ZNF gene conservation in mammals. Three hundred ninety-nine of the 423 human KRAB-ZNF proteins detect a clear chimpanzee ortholog, and the human and chimpanzee genes are organized very similarly within related clustered groups (Table 2). We identified 112 genes that are conserved as 1:1 orthologs in mouse, including 103 genes that are conserved as 1:1 reciprocal pairs in all four mammals (Supplemental Table S6). In addition to 1:1 orthologous pairs, we found many human proteins that matched multiple mouse and dog homologs with high similarity and, conversely, sets of multiple human genes that detect the same reciprocal best-matching gene in other species (data not shown). A total of 226 human KRAB-ZNF protein-coding loci detected at least one clear homolog in one or both nonprimate species (Supplemental Table 6); the remaining 197 loci represent potential primate-specific genes (Supplemental Tables S6, S7).
Included in this set of 197 loci are KRAB-ZNF genes located in the HSA19p12 cluster, which dates back to early primate evolution and underwent a significant expansion ∼40 Mya (Bellefroid et al. 1995; Li 1997; Eichler et al. 1998; Goodman et al. 1998). Subsequent duplications have created new HSA19 paralogs and related genes at several distributed sites (Supplemental Table S7; Hamilton et al. 2006). Altogether, this expanded subfamily includes 62 of the 197 candidate primate-specific KRAB-ZNF protein-coding genes. At least 15 genes in this subfamily are spanned by recent segmental duplications (Bailey et al. 2001) providing support for their recent advent. Recent segmental duplications also span an additional 24 nonconserved KRAB-ZNF coding genes of other subfamilies (Supplemental Table S7). To identify older primate duplications, we used BLAST to identify all locus pairs with >80% sequence identity within noncoding DNA sequences. A total of 126 genes were identified with BLAST matches at this level or above to other human KRAB-ZNF genes, indicating involvement in duplications that occurred in the anthropoid lineage (Supplemental Table S7). Combined evidence of >80% noncoding sequence similarity, involvement in recent segmental duplications, and membership in the known primate-specific subfamily together provide a compelling argument for the primate specificity of at least 136 protein coding loci (32% of all human KRAB-ZNF genes).
KRAB-ZNF gene expression, cluster position, and evolutionary history
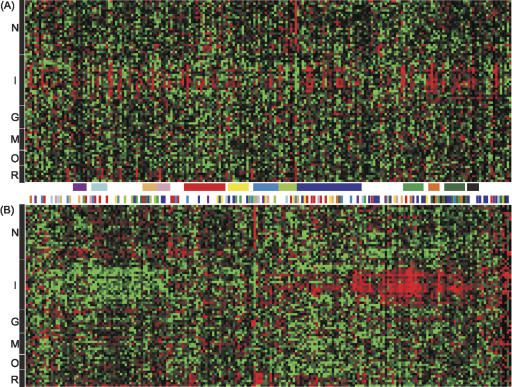
Are clustered KRAB-ZNF genes coexpressed, or is tissue-specific expression of tandem duplicates separately regulated and free to diverge? To address this question, we used the Cluster software package (Eisen et al. 1998) to analyze publicly available microarray data (see Methods) from unique probes sets that could be selected for 211 loci (Supplemental Table S8) and examined potential correlations among genome location, evolutionary history, and microarray expression patterns. Although the 211 genes display diverse patterns of transcriptional activity, some clustering of expression patterns was observed. Most notably, expression data for 51 of the human KRAB-ZNF genes analyzed clustered with >50% correlation into one major group, with highest levels of expression in lymphoid tissues. A second group of 50 genes displayed the opposite pattern, i.e., significantly reduced expression levels in immune-related tissues (Fig. 2; Supplemental Table S8). However, groups of genes with most similar expression patterns were generally derived from different chromosomal regions. In addition, a plot of Pearson's correlation coefficient values for all pairwise comparisons of expression profiles resembles a normal distribution with a median at 0.02 for this probe set (data not shown), indicating no evidence of coregulation. Although, in specific cases, recently duplicated gene pairs have been shown to display overlapping patterns of expression (Shannon et al. 2003), on a global scale related genes were only infrequently grouped as best matches on the basis of expression similarity. Reliable expression data are not available for most of the primate-specific genes, but several sets of close relatives, such as ZNF585A and ZNF585B (with 85% noncoding sequence identity) (Supplemental Table S7), display very different patterns of expression (Fig. 2; Supplemental Table S8). The wide divergence of gene expression patterns and lack of global expression correlation suggest that even recent paralogs can diverge to give rise to genes with distinct patterns of tissue-specific expression.
Figure 2.
Analysis of expression patterns for KRAB and SCAN-KRAB ZNF genes. Expression levels of members of the human KRAB and SCAN-KRAB genes are represented by red and green boxes (denoting higher and lower expression levels, respectively). Vertical columns of boxes represent different genes, and horizontal rows represent different tissues. We have grouped tissues into the following categories as labeled on the left of each panel: N indicates neural; I, immune; G, glandular; M, muscle; O, other organ; and R, reproductive. In panel A, the 211 analyzed genes are arranged in chromosomal order. Selected genomic clusters are indicated by colored boxes beneath the expression pattern. From left to right, the clusters are (as listed in Supplemental Table S1): 16p13.3, 16p11.2a, 19p13.2, 19p13.2-p13.13, 19q13.12-q13.13, 19q13.31, 19q13.41-q13.42, 19q13.43a, 19q13.43b, 6p22.1b, 7q22.1a, 7q36.1a, and 8q24.3c. In panel B, all selected expression profiles (described in text) have been clustered based on expression pattern similarity. Colored hash marks above the expression profiles indicate the chromosomal cluster from panel A with which each profile corresponds.
Discussion
We have identified and curated 423 human loci capable of encoding complete KRAB-ZNF proteins, including 89 novel loci; many of the genes are alternatively spliced to encode predicted protein isoforms with potentially very different functional properties. For example, inclusion of a KRAB-B domain has been shown to enhance repressor activity of KRAB-A proteins (Vissing et al. 1995), whereas the inclusion or exclusion of a SCAN domain may facilitate or abolish dimer formation, respectively (Williams et al. 1999). The prevalence of these alternative transcripts therefore predicts a large array of KRAB-ZNF proteins with several different types of gene regulatory roles. Human KRAB-ZNF proteins typically include a large number of ZNF motifs, with a median number of 12 tandem fingers, predicting a preference for relatively long and specific DNA recognition sequences (Choo and Klug 1994; Berg 1997; Moore et al. 2001). This prediction is confirmed by known binding sites of several such “polydactyl” ZNF proteins (Schoenherr and Anderson 1995; Zheng et al. 2000; Gebelein and Urrutia 2001; Tanaka et al. 2002). However, for certain proteins at least, only subsets of zinc fingers are required for DNA binding (e.g., ZBRK1) (Zheng et al. 2000) and different fingers may be used to recognize targets of distinct sequence composition and types (e.g., ZAC, Hoffmann et al. 2003; and CTCF, Ohlsson et al. 2001). The possibility that these polydactyl proteins might interact with multiple distinct DNA recognition sequences through different subsets of zinc fingers suggests the potential to further amplify the functional diversity of the KRAB-ZNF family.
As noted in previous reports, most of the 423 human KRAB-ZNF genes reside in large familial clusters (Fig. 1, Table 2; Rousseau-Merck et al. 1992; Cannizzaro et al. 1993; Bellefroid et al. 1995). However, we also found a significant number of clusters containing intermixed sets of unrelated genes and distributed singleton copies at many sites. In some cases at least, distributed copies have given rise to tandem duplicates, suggesting a potential means by which new clusters may be seeded at lineage-specific sites. Therefore, both tandem in situ and dispersed segmental duplications have driven the expansion and the genome-wide distribution of this gene family. In contrast to other types of clustered families in which member genes are coexpressed in a limited number of cell types and tissues (e.g., Mombaerts 1999; Zhang et al. 2002; Uhrberg 2005), expression patterns of KRAB-ZNF genes do not correlate with genomic location and neighboring genes often vary widely in transcriptional activity (Fig. 2). These data argue strongly against coregulation of cluster neighbors and indicate that tandem organization is most likely a simple reflection of the KRAB-ZNF family’s duplicative history. Since evolutionary relatedness correlates with genome location in most gene clusters and even very recent paralogs can show distinct patterns of expression, these data also argue that gene expression patterns are free to diverge after paralog duplication. This factor would be likely to enhance the ability of new genes to evolve nonredundant functions (Lynch and Force 2000).
Based on comparisons between the curated human gene set and gene models from draft mouse, dog, and chimpanzee genomes, we present a preliminary classification of human KRAB-ZNF genes according to their degree of conservation or lineage specificity. Although information regarding specific orthologous relationships will change as nonhuman draft sequences are improved, the overall picture of gene repertoire diversity can be clearly discerned. Only 103 of the 423 human KRAB-ZNF genes can be grouped in unambiguous 1:1 orthologous relationships in primate, canine, and rodent lineages; by contrast, at least 136 loci, or nearly one-third of the total human KRAB-ZNF gene set, are primate specific, having arisen since the emergence of Old and New World monkeys. Since regulatory functions are known for only a handful of KRAB-ZNF proteins, the cumulative impact of lineage-specific gain, loss, and divergence of these genes on primate biology remains a matter of conjecture. However, their sheer numbers, their wide range of tissue-specific expression, and their dynamic evolutionary history predict that KRAB-ZNF genes have played a significant role in shaping both primate-specific and deeply conserved traits. A more complete understanding of the functions of the KRAB-ZNF family will be essential for deciphering pathways of vertebrate evolutionary diversity and for building accurate models of gene regulation and its role in human disease.
Methods
Genome searches and initial data analysis
Human KRAB-A, KRAB-B, KRAB-b, KRAB-C, and SCAN protein sequences were collected from RefSeq (the National Center for Biotechnology Information mRNA reference sequence collection) (Pruitt et al. 2000) and trimmed to include only motif residues completely encoded within single exons. Zinc finger protein sequences (X7-C-X2-C-X12-H-X3-H) from HSA19 were collected by a simple pattern-matching script. Sequence alignments for each motif-type were generated by using CLUSTALX (Thompson et al. 1997) and submitted to the HMMER profile HMM matrix building tool HMMBUILD to generate matrices (available for download at http://znf.llnl.gov/). These matrices were used by the HMMER search program to identify all putative motif matches in a full six-frame translation of the hg17 genomic sequences.
In addition, DNA sequences from exons immediately preceding known KRAB-A exons were used to search hg17 chromosomal sequences by using BLAST (Altschul et al. 1990). Output from the HMMER and BLAST searches was compared with the genomic coordinates of publicly available gene models from RefSeq, UCSC Known (known protein-coding genes based on protein data from UniProt, i.e., SWISS-PROT and TrEMBL, and mRNA data from RefSeq), and MGC (Mammalian Gene Collection gene models) (Strausberg et al. 1999) to identify previously characterized loci. The search results were also arranged in chromosomal order and grouped based on proximity and orientation into putative loci.
The human HMM matrices were also used to search the chimp (panTro1), mouse (mm6), and dog (canFam1) six-frame genome translations, and putative loci were generated based on proximity and orientation as above. Crude protein sequences for these nonhuman loci were generated by extending from motif coordinates N- and C-terminally until a translational stop signal was encountered, eliminating overlapping sequences from adjacent motifs, and joining all collected sequences for each locus.
Gene annotation and database curation
Gene model structures were manipulated by using a modified version of APOLLO (http://www.fruitfly.org/annot/apollo/) displaying publicly available mRNA, EST, and other evidence as well as HMM-identified motifs. Public gene models were revised to coincide with RNA evidence. New models were created based on RNA evidence where available but, in some cases, were generated de novo so as to include an open reading frame containing the motifs and maintaining canonical intron splice sites (GT-AG, AT-AG, and GC-AG). Loci were classified as pseudogenes if no gene model could be made that could produce a functional protein. Pseudogenes with limited stop codons or insertions/deletions were re-evaluated for functionality by examining the putative open reading frames of any RNA sequences associated with the locus and identifying discrepancies with the genomic sequence.
Adjacent genes were considered “clustered” if the intergenic sequence separating two KRAB-ZNF genes was <200 kb, even in cases where unrelated genes were found between the ZNF loci (a situation only rarely encountered). This distance cutoff was selected based on a distribution of intergenic distances between KRAB-ZNF genes in the annotation database (for additional details, see legend of Supplemental Fig. S1). Manual inspection of the clustering confirmed that the 200-kb criterion resulted in coclustering of all major groups of neighboring, related genes. Only two pairs of paralogs that could be considered to form familial clusters were omitted by the 200-kb cutoff; these genes—LLNL1035 and ZNF705B, and ZNF705C and LLNL1103—derived from unusually large duplicons were counted as clusters in the final analysis.
Comparative genomic analyses
Protein sequences from all human KRAB and SCAN-KRAB loci were BLASTed against the full set of human ZNF protein sequences and against draft protein sequences predicted from chimp, mouse, or dog ZNF loci. The best match (with expectation value <10−30 and >30% identity) was then BLASTed back against the human protein set; if this second BLAST returned the initial search query, the pair was considered a reciprocal best match. Data were collected for the best six matches for each protein, and reciprocal matches were flagged. The data were then manually checked to identify and correct anomalous results by inspection of the variable helix regions of aligned zinc fingers. Most inaccurate identifications were due to sequencing errors, e.g., those predicting truncated proteins in the draft genome searched; this issue was most acute where very closely related paralogs exist in both species. In cases where a true ortholog is missing from a draft genome sequence, the most similar sequenced paralogous gene will be selected by this method. Chromosomal and relative positions of reciprocal matches were compared both within the human genome and across the four species to evaluate lineage-specific cluster evolution.
Evolutionary analysis
A tree of phylogenetic relationships was generated by using KRAB-A motifs from KRAB-ZNF and KRAB-SCAN-ZNF loci, including several genes with noncanonical structures. Several loci contained two KRAB-A motifs; in these cases both KRAB-A motifs were used. KRAB-A nucleotide sequences were retrieved from the catalog Web site, and alignments of the sequences were made by using CLUSTALX 1.81 (Thompson et al. 1997). The alignment was manually checked by using SeAl (Rambaut 1996). The PAUP 4.0b10 package (Swofford 2002) was used to generate trees by using mean character differences and the neighbor-joining (NJ) method (Saitou and Nei 1987). A Xenopus KRAB-A (Xfin) sequence was added as a potential outgroup.
Selection of microarray probe sets and expression clustering
Affymetrix-based GNF Atlas expression data pre-analyzed using the MAS5 algorithm (Su et al. 2004) was obtained from the UCSC Genome Bioinformatics Web site (http://genome.ucsc.edu/, Kent et al. 2002), and GNF1H and U133A probe sets relevant to genes in our data set were selected. We included data only from probes that could reasonably be expected to detect expression of a single gene; due to the high degree of sequence similarity between ZNF paralogs, these represented a very small subset of the probe sets designed against KRAB-ZNF genes. To identify relatively unique probe sets, the individual sequences for all probe sets from both chip designs were BLASTed against all catalog transcript sequences (including noncanonical genes). A pool of gene-specific probe sets was then identified based on two criteria: (1) all sequences within a given probe set perfectly aligned to some portion of a single target locus; (2) the ratio of the number of target-specific alignments over the total number of alignments to any locus (i.e., target and nontarget loci to which probes had ≥20/25 matches) was >0.8. This second requirement eliminates probe sets that interrogate additional target(s) well or numerous targets poorly. In instances where multiple candidate probe sets were identified for a single gene, a representative probe set was manually chosen, selecting probes aligning with 3′ regions of the transcripts wherever possible.
By use of Cluster v. 2.11 (Eisen et al. 1998), the expression profiles for the selected probe sets were hierarchically clustered using an uncentered correlation metric. The resulting cdt files were visualized by using TreeView v. 1.60 (Eisen et al. 1998) to generate Figure 2B. Profiles were also manually arranged in chromosomal order and visualized in TreeView to generate Figure 2A. These expression profiles were also used to analyze possible global coregulation by using Pearson’s correlations coefficient.
Acknowledgments
We thank Ivan Ovcharenko for advice on programming and genome searching methods, and David Goodstein and Astrid Terry at the Joint Genome Institute for advice on Apollo and gene annotation. We also thank Colleen Elso, Jutta Kollet, Jason Raymond, and Alice Yamada for critical reviews of the manuscript and Web site. This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory (LLNL) under contract no. W-7405-Eng-48. The project (04-ERD-084) was funded by the Laboratory Directed Research and Development Program at LLNL.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4842106
References
- Abrink M., Ortiz J.A., Mark C., Sanchez C., Looman C., Hellman L., Chambon P., Losson R., Ortiz J.A., Mark C., Sanchez C., Looman C., Hellman L., Chambon P., Losson R., Mark C., Sanchez C., Looman C., Hellman L., Chambon P., Losson R., Sanchez C., Looman C., Hellman L., Chambon P., Losson R., Looman C., Hellman L., Chambon P., Losson R., Hellman L., Chambon P., Losson R., Chambon P., Losson R., Losson R. Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1β. Proc. Natl. Acad. Sci. 2001;98:1422–1426. doi: 10.1073/pnas.041616998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ayyanathan K., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Yamada Y., Tanaka K., Torigoe K., Rauscher III F.J., Tanaka K., Torigoe K., Rauscher III F.J., Torigoe K., Rauscher III F.J., Rauscher III F.J. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes & Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Massa H.F., Trask B.J., Eichler E.E., Trask B.J., Eichler E.E., Eichler E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid E.J., Poncelet D.A., Lecocq P.J., Revelant O., Martial J.A., Poncelet D.A., Lecocq P.J., Revelant O., Martial J.A., Lecocq P.J., Revelant O., Martial J.A., Revelant O., Martial J.A., Martial J.A. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc. Natl. Acad. Sci. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid E.J., Marine J.C., Ried T., Lecocq P.J., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Marine J.C., Ried T., Lecocq P.J., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Ried T., Lecocq P.J., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Lecocq P.J., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Coulie P.G., de Jong P., Szpirer C., de Jong P., Szpirer C., Szpirer C., et al. Clustered organization of homologous KRAB zinc-finger genes with enhanced expression in human T lymphoid cells. EMBO J. 1993;12:1363–1374. doi: 10.1002/j.1460-2075.1993.tb05781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid E.J., Marine J.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Marine J.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Martial J.A., Ihle J.N., Ward D.C., Ihle J.N., Ward D.C., Ward D.C. Emergence of the ZNF91 Kruppel-associated box-containing zinc finger gene family in the last common ancestor of anthropoidea. Proc. Natl. Acad. Sci. 1995;92:10757–10761. doi: 10.1073/pnas.92.23.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J.M. Letting your fingers do the walking. Nat. Biotechnol. 1997;15:323. doi: 10.1038/nbt0497-323. [DOI] [PubMed] [Google Scholar]
- Cannizzaro L.A., Aronson M.M., Thiesen H.J., Aronson M.M., Thiesen H.J., Thiesen H.J. Human zinc finger gene ZNF23 (Kox16) maps to a zinc finger gene cluster on chromosome 16q22, and ZNF32 (Kox30) to chromosome region 10q23-q24. Hum. Genet. 1993;91:383–385. doi: 10.1007/BF00217362. [DOI] [PubMed] [Google Scholar]
- Choo Y., Klug A., Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc. Natl. Acad. Sci. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.R., Schafer U., Jackle H., Bohm S., Schafer U., Jackle H., Bohm S., Jackle H., Bohm S., Bohm S. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in. Drosophila. EMBO Rep. 2002;3:1158–1162. doi: 10.1093/embo-reports/kvf243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T., Stone J.R., Williams A.J., Stone J.R., Williams A.J., Williams A.J. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E.E., Hoffman S.M., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Hoffman S.M., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., McCready P., Lamerdin J.E., Mohrenweiser H.W., Lamerdin J.E., Mohrenweiser H.W., Mohrenweiser H.W. Complex β-satellite repeat structures and the expansion of the zinc finger gene cluster in 19p12. Genome Res. 1998;8:791–808. doi: 10.1101/gr.8.8.791. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D., Spellman P.T., Brown P.O., Botstein D., Brown P.O., Botstein D., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Fredericks W.J., Jensen D.E., Speicher D.W., Huang X.P., Neilson E.G., Rauscher III F.J., Fredericks W.J., Jensen D.E., Speicher D.W., Huang X.P., Neilson E.G., Rauscher III F.J., Jensen D.E., Speicher D.W., Huang X.P., Neilson E.G., Rauscher III F.J., Speicher D.W., Huang X.P., Neilson E.G., Rauscher III F.J., Huang X.P., Neilson E.G., Rauscher III F.J., Neilson E.G., Rauscher III F.J., Rauscher III F.J. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes & Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Gebelein B., Urrutia R., Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol. Cell. Biol. 2001;21:928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Schneider H., Shoshani J., Gunnell G., Groves C.P., Shoshani J., Gunnell G., Groves C.P., Gunnell G., Groves C.P., Groves C.P. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Hamilton A.T., Huntley S., Kim J., Branscomb E., Stubbs L., Huntley S., Kim J., Branscomb E., Stubbs L., Kim J., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L. Lineage-specific expansion of KRAB zinc-finger transcription factor genes: Implications for the evolution of vertebrate regulatory networks. Cold Spring Harb. Symp. Quant. Biol. 2003;68:131–140. doi: 10.1101/sqb.2003.68.131. [DOI] [PubMed] [Google Scholar]
- Hamilton A.T., Huntley S., Tran-Gyamfi M., Baggott D.M., Gordon L., Stubbs L., Huntley S., Tran-Gyamfi M., Baggott D.M., Gordon L., Stubbs L., Tran-Gyamfi M., Baggott D.M., Gordon L., Stubbs L., Baggott D.M., Gordon L., Stubbs L., Gordon L., Stubbs L., Stubbs L.2006Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Ciani E., Boeckardt J., Holsboer F., Journot L., Spengler D., Ciani E., Boeckardt J., Holsboer F., Journot L., Spengler D., Boeckardt J., Holsboer F., Journot L., Spengler D., Holsboer F., Journot L., Spengler D., Journot L., Spengler D., Spengler D. Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol. Cell. Biol. 2003;23:988–1003. doi: 10.1128/MCB.23.3.988-1003.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S., Hamilton A., Kim J., Branscomb E., Stubbs L., Hamilton A., Kim J., Branscomb E., Stubbs L., Kim J., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L. Tandem gene family expansion and genomic diversity. In Comparative genomics: A guide to the analysis of eukaryotic genomes (ed. M.D. Adams). Humana Press; New York, (in press) [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.A., Berg J.M., Berg J.M. Serine at position 2 in the DNA recognition helix of a Cys2-His2 zinc finger peptide is not, in general, responsible for base recognition. J. Mol. Biol. 1995;252:1–5. doi: 10.1006/jmbi.1994.0468. [DOI] [PubMed] [Google Scholar]
- Knochel W., Poting A., Koster M., el Baradi T., Nietfeld W., Bouwmeester T., Pieler T., Poting A., Koster M., el Baradi T., Nietfeld W., Bouwmeester T., Pieler T., Koster M., el Baradi T., Nietfeld W., Bouwmeester T., Pieler T., el Baradi T., Nietfeld W., Bouwmeester T., Pieler T., Nietfeld W., Bouwmeester T., Pieler T., Bouwmeester T., Pieler T., Pieler T. Evolutionary conserved modules associated with zinc fingers in Xenopus laevis. Proc. Natl. Acad. Sci. 1989;86:6097–6100. doi: 10.1073/pnas.86.16.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C.J., Larkins L.K., Khan S.M., Robins D.M., Larkins L.K., Khan S.M., Robins D.M., Khan S.M., Robins D.M., Robins D.M. Expansion and diversification of KRAB zinc-finger genes within a cluster including regulator of sex-limitation 1 and 2. Genomics. 2005;85:752–761. doi: 10.1016/j.ygeno.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li W.H. Molecular evolution. Sinauer Associates; Sunderland, MA.: 1997. [Google Scholar]
- Looman C., Abrink M., Mark C., Hellman L., Abrink M., Mark C., Hellman L., Mark C., Hellman L., Hellman L. KRAB zinc finger proteins: An analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 2002;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- Looman C., Hellman L., Abrink M., Hellman L., Abrink M., Abrink M. A novel Kruppel-associated box identified in a panel of mammalian zinc finger proteins. Mamm. Genome. 2004;15:35–40. doi: 10.1007/s00335-003-3022-0. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin J.F., Friedman J.R., Meyer W.K., Vissing H., Thiesen H.J., Rauscher III F.J., Friedman J.R., Meyer W.K., Vissing H., Thiesen H.J., Rauscher III F.J., Meyer W.K., Vissing H., Thiesen H.J., Rauscher III F.J., Vissing H., Thiesen H.J., Rauscher III F.J., Thiesen H.J., Rauscher III F.J., Rauscher III F.J. Kruppel-associated boxes are potent transcriptional repression domains. Proc. Natl. Acad. Sci. 1994;91:4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark C., Abrink M., Hellman L., Abrink M., Hellman L., Hellman L. Comparative analysis of KRAB zinc finger proteins in rodents and man: Evidence for several evolutionarily distinct subfamilies of KRAB zinc finger genes. DNA Cell Biol. 1999;18:381–396. doi: 10.1089/104454999315277. [DOI] [PubMed] [Google Scholar]
- Messina D.N., Glasscock J., Gish W., Lovett M., Glasscock J., Gish W., Lovett M., Gish W., Lovett M., Lovett M. An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res. 2004;14:2041–2047. doi: 10.1101/gr.2584104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Pabo C.O., Pabo C.O. Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger–DNA recognition. J. Mol. Biol. 2001;313:309–315. doi: 10.1006/jmbi.2001.4975. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor genes in humans. Curr. Opin. Genet. Dev. 1999;9:315–320. doi: 10.1016/s0959-437x(99)80047-1. [DOI] [PubMed] [Google Scholar]
- Moore M., Klug A., Choo Y., Klug A., Choo Y., Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc. Natl. Acad. Sci. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.J., Li Y., Lau Y.F., Li Y., Lau Y.F., Lau Y.F. Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol. Reprod. 2005;72:407–415. doi: 10.1095/biolreprod.104.034447. [DOI] [PubMed] [Google Scholar]
- Ohlsson R., Renkawitz R., Lobanenkov V., Renkawitz R., Lobanenkov V., Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Pengue G., Lania L., Lania L. Kruppel-associated box-mediated repression of RNA polymerase II promoters is influenced by the arrangement of basal promoter elements. Proc. Natl. Acad. Sci. 1996;93:1015–1020. doi: 10.1073/pnas.93.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Katz K.S., Sicotte H., Maglott D.R., Katz K.S., Sicotte H., Maglott D.R., Sicotte H., Maglott D.R., Maglott D.R. Introducing RefSeq and LocusLink: Curated human genome resources at the NCBI. Trends Genet. 2000;16:44–47. doi: 10.1016/s0168-9525(99)01882-x. [DOI] [PubMed] [Google Scholar]
- Rambaut A.1996. Se-Al: Sequence Alignment Editor. http://iubio.bio.indiana.edu/soft/iubionew/molbio/dna/analysis/Pist/main.html [Google Scholar]
- Rousseau-Merck M.F., Tunnacliffe A., Berger R., Ponder B.A., Thiesen H.J., Tunnacliffe A., Berger R., Ponder B.A., Thiesen H.J., Berger R., Ponder B.A., Thiesen H.J., Ponder B.A., Thiesen H.J., Thiesen H.J. A cluster of expressed zinc finger protein genes in the pericentromeric region of human chromosome 10. Genomics. 1992;13:845–848. doi: 10.1016/0888-7543(92)90166-p. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sander T.L., Morris J.F., Morris J.F. Characterization of the SCAN box encoding RAZ1 gene: Analysis of cDNA transcripts, expression, and cellular localization. Gene. 2002;296:53–64. doi: 10.1016/s0378-1119(02)00792-8. [DOI] [PubMed] [Google Scholar]
- Sander T.L., Stringer K.F., Maki J.L., Szauter P., Stone J.R., Collins T., Stringer K.F., Maki J.L., Szauter P., Stone J.R., Collins T., Maki J.L., Szauter P., Stone J.R., Collins T., Szauter P., Stone J.R., Collins T., Stone J.R., Collins T., Collins T. The SCAN domain defines a large family of zinc finger transcription factors. Gene. 2003;310:29–38. doi: 10.1016/s0378-1119(03)00509-2. [DOI] [PubMed] [Google Scholar]
- Schmidt D., Durrett R., Durrett R. Adaptive evolution drives the diversification of zinc-finger binding domains. Mol. Biol. Evol. 2004;21:2326–2339. doi: 10.1093/molbev/msh246. [DOI] [PubMed] [Google Scholar]
- Schoenherr C.J., Anderson D.J., Anderson D.J. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Shannon M., Stubbs L., Stubbs L. Analysis of homologous XRCC1-linked zinc-finger gene families in human and mouse: Evidence for orthologous genes. Genomics. 1998;49:112–121. doi: 10.1006/geno.1998.5230. [DOI] [PubMed] [Google Scholar]
- Shannon M., Hamilton A.T., Gordon L., Branscomb E., Stubbs L., Hamilton A.T., Gordon L., Branscomb E., Stubbs L., Gordon L., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13:1097–1110. doi: 10.1101/gr.963903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg R.L., Feingold E.A., Klausner R.D., Collins F.S., Feingold E.A., Klausner R.D., Collins F.S., Klausner R.D., Collins F.S., Collins F.S. The mammalian gene collection. Science. 1999;286:455–457. doi: 10.1126/science.286.5439.455. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. PAUP: Phylogenetic analysis using parsimony. Sinauer Associates; Sunderland, MA.: 2002. [Google Scholar]
- Tanaka K., Tsumaki N., Kozak C.A., Matsumoto Y., Nakatani F., Iwamoto Y., Yamada Y., Tsumaki N., Kozak C.A., Matsumoto Y., Nakatani F., Iwamoto Y., Yamada Y., Kozak C.A., Matsumoto Y., Nakatani F., Iwamoto Y., Yamada Y., Matsumoto Y., Nakatani F., Iwamoto Y., Yamada Y., Nakatani F., Iwamoto Y., Yamada Y., Iwamoto Y., Yamada Y., Yamada Y. A Kruppel-associated box-zinc finger protein, NT2, represses cell-type-specific promoter activity of the α2(XI) collagen gene. Mol. Cell. Biol. 2002;22:4256–4267. doi: 10.1128/MCB.22.12.4256-4267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Plewniak F., Jeanmougin F., Higgins D.G., Jeanmougin F., Higgins D.G., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrberg M. The KIR gene family: Life in the fast lane of evolution. Eur. J. Immunol. 2005;35:10–15. doi: 10.1002/eji.200425743. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Smith H.O., Yandell M., Evans C.A., Holt R.A., Yandell M., Evans C.A., Holt R.A., Evans C.A., Holt R.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vissing H., Meyer W.K., Aagaard L., Tommerup N., Thiesen H.J., Meyer W.K., Aagaard L., Tommerup N., Thiesen H.J., Aagaard L., Tommerup N., Thiesen H.J., Tommerup N., Thiesen H.J., Thiesen H.J. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995;369:153–157. doi: 10.1016/0014-5793(95)00728-r. [DOI] [PubMed] [Google Scholar]
- Williams A.J., Blacklow S.C., Collins T., Blacklow S.C., Collins T., Collins T. The zinc finger-associated SCAN box is a conserved oligomerization domain. Mol. Cell. Biol. 1999;19:8526–8535. doi: 10.1128/mcb.19.12.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Yu L., Bi G., Luo K., Zhou G., Zhao S., Yu L., Bi G., Luo K., Zhou G., Zhao S., Bi G., Luo K., Zhou G., Zhao S., Luo K., Zhou G., Zhao S., Zhou G., Zhao S., Zhao S. Identification and characterization of two novel human SCAN domain-containing zinc finger genes ZNF396 and ZNF397. Gene. 2003;310:193–201. doi: 10.1016/s0378-1119(03)00551-1. [DOI] [PubMed] [Google Scholar]
- Zhang H.B., Liu D.P., Liang C.C., Liu D.P., Liang C.C., Liang C.C. The control of expression of the α-globin gene cluster. Int. J. Hematol. 2002;76:420–426. doi: 10.1007/BF02982807. [DOI] [PubMed] [Google Scholar]
- Zheng L., Pan H., Li S., Flesken-Nikitin A., Chen P.L., Boyer T.G., Lee W.H., Pan H., Li S., Flesken-Nikitin A., Chen P.L., Boyer T.G., Lee W.H., Li S., Flesken-Nikitin A., Chen P.L., Boyer T.G., Lee W.H., Flesken-Nikitin A., Chen P.L., Boyer T.G., Lee W.H., Chen P.L., Boyer T.G., Lee W.H., Boyer T.G., Lee W.H., Lee W.H. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol. Cell. 2000;6:757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]