Abstract
Most genes are conserved in mammals, but certain gene families have acquired large numbers of lineage-specific loci through repeated rounds of gene duplication, divergence, and loss that have continued in each mammalian group. One such family encodes KRAB-zinc finger (KRAB-ZNF) proteins, which function as transcriptional repressors. One particular subfamily of KRAB-ZNF genes, including ZNF91, has expanded specifically in primates to comprise more than 110 loci in the human genome. Genes of the ZNF91 subfamily reside in large gene clusters near centromeric regions of human chromosomes 19 and 7 with smaller clusters or isolated copies in other locations. Phylogenetic analysis indicates that many of these genes arose before the split between the New and Old World monkeys, but the ZNF91 subfamily has continued to expand and diversify throughout the evolution of apes and humans. Paralogous loci are distinguished by divergence within their zinc finger arrays, indicating selection for proteins with different regulatory targets. In addition, many loci produce multiple alternatively spliced transcripts encoding proteins that may serve separate and perhaps even opposing regulatory roles because of the modular motif structure of KRAB-ZNF genes. The tissue-specific expression patterns and rapid structural divergence of ZNF91 subfamily genes suggest a role in determining gene expression differences between species and the evolution of novel primate traits.
Most genes are conserved as 1:1 orthologs in different mammalian species (Waterston et al. 2002; Lindblad-Toh et al. 2005), but certain gene families contain different numbers and types of genes in each lineage due to an ongoing series of gene duplications, divergence, and gene-loss events. Two of the best-known examples are the genes encoding olfactory receptors and immune receptors, and both lineage-specific and individual differences in these genes are clearly linked to significant variation in phenotype (Nei et al. 1997; Trowsdale et al. 2001; Young et al. 2002; Gilad et al. 2005; Hao and Nei 2005; Sambrook et al. 2005).
Another dramatic example of lineage-specific expansion and divergence is provided by a family of transcription factor genes encoding proteins, in which a chromatin-interaction domain called KRAB (Krüppel-associated box) is associated with tandem arrays of Krüppel-type (C2H2) zinc finger motifs (Thiesen 1990; Thiesen et al. 1991; Shannon et al. 1998, 2003; Mark et al. 1999; Looman et al. 2002). The KRAB domain confers a potent transcriptional repressor function by specific interactions with a corepressor protein, KAP1 (encoded by TRIM28 and also known as TIF1-β or KRIP-1) that serves to recruit chromatin deacetylation machinery (Kim et al. 1996; Moosmann et al. 1996; Schultz et al. 2002, Ayyanathan et al. 2003). KRAB-zinc finger (ZNF) genes are a recent invention, first arising around the time of tetrapod divergence and duplicating aggressively to comprise a family of more than 400 active members in the human genome (Huntley et al. 2006). Most KRAB-ZNF genes reside in large familial clusters, many of which contain substantial numbers of lineage-specific genes. Studies focused on specific subfamilies have shown that paralogs typically diverge through structural changes within the ZNF arrays that are likely to affect DNA-binding properties and, potentially, target choice for the duplicated transcription factors (Looman et al. 2002; Hamilton et al. 2003; Shannon et al. 2003; Krebs et al. 2005).
We have speculated that the rapid expansion and lineage-specific diversification of KRAB-ZNF genes allow the “fine-tuning” of transcriptional regulation and diversification of regulatory networks in evolution (Hamilton et al. 2003), which could contribute to population differences and play a role in speciation. One large cluster of KRAB-ZNF genes located near the centromere of human chromosome 19 (HSA19) is especially interesting because it has long been known to be unique to primates (Bellefroid et al. 1995; Eichler et al. 1998). Here, we present a detailed analysis of this large subfamily of genes and investigate patterns of evolutionary divergence in clades of recently duplicated primate paralogs. Due to their function as transcriptional repressors, the generation and rapid divergence of these genes may help to explain some of the transcriptome differences that have been documented between humans and our closest relatives among the apes (Enard et al. 2002; Khaitovich et al. 2004, 2005; Preuss et al. 2004) and, potentially, individual differences within our own species.
Results
The HSA19p12-13.1 cluster and the ZNF91 subfamily
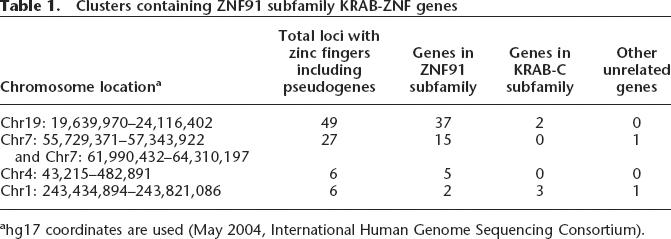
Functional genes and pseudogenes in the HSA19p12-13.1 KRAB-ZNF cluster were identified from the human KRAB-ZNF gene catalog (Huntley et al. 2006; http://znf.llnl.gov). This large primate-specific gene cluster contains over 70 ZNF loci, including 39 intact genes as well as pseudogenes (Fig. 1). All but two of the 39 protein-coding loci are closely related to the previously described gene, ZNF91 (Table 1); for simplicity, we will hereafter refer to this group as the “ZNF91 subfamily.” To identify additional subfamily members, we searched the human genome sequence using KRAB-A, spacer, and zinc finger regions of ZNF91 and several other related genes as BLAST and BLAT queries. This search identified related loci at several chromosomal locations, including a large group clustered around the centromere of HSA7, smaller clusters on HSA4p16.3 and 1q44.1 (Table 1; Fig. 2), and other loci scattered across 17 chromosomes (Supplemental Table S1). All told, the ZNF91 subfamily includes 64 protein-coding genes, 62 of which encode both a KRAB and an array of zinc finger motifs. While ZNF91-subfamily genes are by no means the only primate-specific KRAB-ZNF loci found in the human gene set, this subfamily represents by far the largest clade of genes in the recently expanded group.
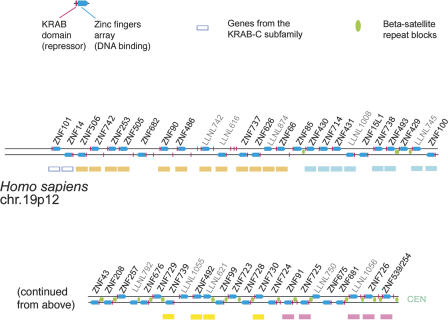
Figure 1.
A diagram of the KRAB-ZNF cluster on HSA19p12 containing genes related to the ZNF91 subfamily. The map shows the physical order of the KRAB-zinc finger loci in the cluster, but is not scaled for genomic distances or relative size. Each finger-containing locus is represented by a blue arrow pointing 5′ to 3′ (left-to-right above the line, opposite strand, below). Loci that contain a KRAB have a red bar at the 5′ end of the symbol. Isolated red bars are KRAB-only pseudogene fragments (not labeled). Locus designations with the format “LLNLxxx” are from the new zinc finger catalog (Huntley et al. 2006) and are here used for pseudogene loci (light-gray text). Colored boxes beneath loci correspond to clade membership in the phylogeny (Fig. 3). In some cases, labeled genes in the cluster maps were not included in the phylogeny in Figure 3 because the spacer region used to create the phylogeny was modified or deleted from the locus, or because they were very divergent and not members of the ZNF91 subfamily, notably the “KRAB-C subfamily” loci (blue outline boxes). The relative positions of β-satellite repeat blocks between loci in this cluster are indicated by green ovals between the lines.
Table 1.
Clusters containing ZNF91 subfamily KRAB-ZNF genes
ahg17 coordinates are used (May 2004, International Human Genome Sequencing Consortium).
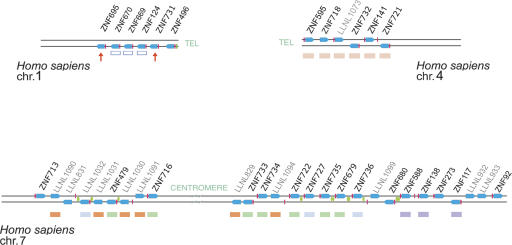
Figure 2.
Diagrams of the KRAB-ZNF clusters on HSA1, HSA4, and HSA7 containing genes related to the ZNF91 subfamily. Maps show the physical order of the KRAB-zinc finger loci in the clusters, but are not scaled for genomic distance or relative size. Due to the spacing between loci, the HSA7 map shown could be defined as more than one cluster, but several clades of closely related loci (colored to match Fig. 3) have copies distributed across the region. Symbols and labels for genes and β-satellite repeats are the same as in Figure 1. The red arrows under the HSA1 cluster denote the two ZNF91 subfamily members in that cluster (the blue outline boxes denote KRAB-C subfamily genes). The HSA7 region shown also has a divergent gene at the p-telomeric end (ZNF713) included in the diagram although relationships between this gene and other KRAB-ZNF loci are unclear. Not shown are four clusters containing a total of 15 pseudogene loci on HSAY and three divergent loci (ZNF267, ZNF720, LLNL1128), which may be distant relatives of the ZNF91 subfamily, clustered together with unrelated genes on HSA16.
Phylogenetic analysis
To examine the evolutionary history of the ZNF91 subfamily, we generated phylogenetic trees based on coding and noncoding sequences surrounding the “spacer” regions of each locus (see Methods). This analysis revealed multiple well-supported clades or subgroupings of loci (Fig. 3). Most notably, genes residing in the HSA19p12 and HSA7 clusters are intermingled throughout the phylogeny. This pattern suggests a history in which one cluster was spawned from another, probably through the duplication or translocation of multiple loci after initial episodes of gene duplication expanded the number of genes in the subfamily during early primate evolution. Because the relationship between HSA7 and HSA19 genes has relatively distant roots, unraveling the early history of these clusters may be complicated by subsequent events, including tandem in situ duplication events (resulting in clades present in only one cluster), gene loss, gene conversion, or “colonization” of one cluster by gene copies from another.
Figure 3.
Neighbor-joining phylogeny of the ZNF91 subfamily based on the spacer and flanking sequences. The tree includes 116 loci (including outgroups); the single best tree is shown, and bootstrap values above 70% based on 1000 bootstrap replicates are indicated on branches. Gene names without a leading number are from the chromosome 19p12 cluster; in all other cases, the initial number indicates the chromosome from which the locus derives (e.g., 7_ZNF588 is from HSA7). Loci with the ZNF catalog designation LLNLxxx (Figs. 1, 2; Supplemental Table S1) are abbreviated “Lxxx.” Colored boxes define certain well-supported clades as designated to the left. For the clades including genes from the main clusters, the box colors match the colored bars under the individual member genes on the cluster maps in Figures 1 and 2. Note that the “Y-chromosome related clade” contains genes from the cluster on HSA7, but these are outnumbered by the pseudogene loci on HSAY. The red arrows indicate the ZNF91 subfamily members in the cluster on HSA1; the gray arrows indicate two nonclustered genes mentioned in the results section. Pseudogenes are marked with a ψ. Processed pseudogenes are indicated by “r” ahead of the locus name.
As also revealed by the analysis, the HSA19p12 and HSA1q44.1 clusters contain a mix of genes from two distinct KRAB-ZNF subfamilies. Specifically, two genes located at the p-telomeric end of the large HSA19p12 cluster, ZNF101 and ZNF14 (open boxes, Fig. 1) have very divergent spacer sequences and cannot be aligned reliably with ZNF91 subfamily loci. ZNF101 and ZNF14 also encode proteins containing KRAB-C motifs (Looman et al. 2004), a distinguishing trait that is shared with genes located in a separate KRAB-ZNF cluster located 8 Mb away (chr19: 11–12Mb). The HSA1q44.1 gene cluster also contains ZNF101-related genes (open boxes, Fig. 2) intermixed with ZNF91 subfamily loci (red arrows, Fig. 2). Because no other clusters mix genes from these two subfamilies, it is uncertain whether the progenitors for both subfamilies were originally linked. However, the ZNF91 subfamily members that reside in the HSA1 cluster are highly divergent from their HSA19 relatives (red arrows, Fig. 3) suggesting that these sets of genes arose by duplication in relatively distant primate history.
Also of note are several clades comprising genes that reside in locations other than HSA1, HSA7, or HSA19. For example, the ‘Y chromosome related clade’ (marked blue-gray, Figs. 2, 3) includes members on four different chromosomes, although the only intact loci are on HSA7. The HSAY pseudogenes are in four clusters that may be undergoing concerted evolution (Rozen et al. 2003; Skaletsky et al. 2003). The distributed singleton ZNF91 subfamily members appear to be the result of both ancient duplication events (e.g., ZNF678 on HSA1 and ZNF195 on HSA11 are divergent in sequence from any relatives; gray arrows, Fig. 3) and very recent, perhaps ape- or human-specific, duplication episodes (the dispersed LLNL1037 clade of pseudogenes share up to 99% sequence identity over variable segmental duplication lengths of <30–160 kb). This mode of duplication, which appears to have been utilized much less frequently than tandem in situ events, has the potential to seed gene clusters at new chromosomal sites (Ohno 1970; Eichler et al. 1998). A possible example of this is a monophyletic group of genes including ZNF141 that are clustered together at the telomere of HSA4p (Figs. 2, 3) and may have been derived from a single progenitor. In addition to segmental duplications, the ZNF91 subfamily includes six scattered, intronless loci. These copies correspond to KRAB-ZNF processed pseudogenes and most can be related to an intact ancestral locus or to a clade of related loci.
Timing and patterns of duplication events
Although expansion of the ZNF91 subfamily created gene copies on many chromosomes, most genes are concentrated in the centromeric regions of HSA19 and HSA7. There are several well-supported clades within the HSA19 and HSA7 clusters that do not have members in the other location, probably representing loci that were generated after the clusters diverged. Members of most related subgroups of HSA19 genes are physical neighbors, indicating that each group has expanded with new copies landing close to their progenitors (Figs. 1, 3). Gene conversion may also have acted on some loci post-duplication, and could reduce the apparent divergence of affected loci through homogenization. In contrast to this relatively simple HSA19 pattern, members of well-supported HSA7 groups are intermingled in physical location. Several HSA7 clades contain loci mapping to both chromosome arms (Figs. 2, 3), indicating that subgroup expansion was followed by a series of duplications or inversions around the centromere, as occurred on HSA10 (Tunnacliffe et al. 1993; Guy et al. 2003).
The pericentromeric HSA19 cluster contains a large number of β-satellite repeats, and unequal crossing-over between blocks of these repeats has been suggested as a factor in expansion of the cluster (Eichler et al. 1998). The β-satellite repeat (BSR) blocks are abundant near the centromere but absent from the telomeric end of the cluster (green ovals, Fig. 1). BSR blocks are also present in the HSA7 clusters, but are sparse among genes on the q arm near the telomeric end (Fig. 2), which contains genes most closely related to those found in HSA19 BSR-dense regions. BSR blocks are not found in the HSA1 and HSA4 clusters. This variation in repeat organization may mean that the expansion of the different clusters, or even sections of the same cluster, may have been influenced by different repeat types (Bailey et al. 2003), with β-satellites potentially associated with the growth of some of the clades in the ZNF91 subfamily but not, for instance, the ZNF505 clade (Figs. 1, 3).
Alignment of noncoding sequences from within the duplicated regions permitted an approximate age to be determined for specific duplication events, assuming a low rate of homogenizing gene conversion. Pairwise comparisons between intron and 3′ UTR sequences of intact loci of different clusters showed divergence levels consistent with a major expansion of much of the ZNF91 gene subfamily occurring before the split between the Catarrhini (Old World monkeys and hominoids [apes and humans]) and Platyrrhini (New World monkeys), around 35–40 million years ago (Li et al. 1987; Li 1997; Goodman et al. 1998; Glazko and Nei 2003). For instance, the most similar HSA19 and HSA7 loci show ∼85% noncoding nucleotide sequence identity, indicating the major clusters were probably separated before this time. This pattern is consistent with conclusions from previous studies on members of the ZNF91 subfamily (Bellefroid et al. 1995; Eichler et al. 1998). However, noncoding sequence similarity was above 90% in alignments of many ZNF91 subfamily gene pairs within clusters, and at least 15 coding genes from the ZNF91 subfamily are included in known recent segmental duplications, not counting duplications producing pseudogenes (Supplemental Table S2; Bailey et al. 2001, 2002). These data indicate that some members of the ZNF91 family have continued to duplicate throughout the evolution of apes and humans.
Evolutionary divergence in two clades of recent KRAB-ZNF gene duplicates
Several HSA19p12 clades show evidence of recent duplication involving intact genes. Multiple pairwise comparisons of aligned intronic sequence between loci showed several pairs of genes with divergence <5%–7%, indicating that some of these loci may be more recent than the hominoid/Old World monkey split (Li et al. 1987; Li 1997; Liu et al. 2003). Analysis of these recently duplicated genes and their homologs in nonhuman primate species provides a clear illustration of the patterns of diversification for new KRAB-ZNF gene copies.
The ZNF431 clade (highlighted in blue-green, Figs. 1, 3) includes six protein-coding genes and two pseudogenes, one of which contains a deleted spacer sequence and was excluded from the phylogenetic analysis (Fig. 3). As confirmed by PCR of primate genomic DNA samples, several genes in this clade date back at least to the Old World monkeys (Fig. 4A), an observation also supported by BLAST and BLAT searches of the draft chimpanzee and rhesus genome sequences (data not shown). No ortholog for ZNF714 was found in the rhesus genome by PCR or in sequence searches. Here, we should note that failure to detect a gene copy by PCR could result from divergence in primer sites, and specific genes may be missing from the draft primate genome sequences; these analyses, therefore, only estimate a minimum age for each locus. However, the high degree of sequence similarity between ZNF431 and ZNF714 provides support for the notion that ZNF714 arose relatively recently in evolutionary time.
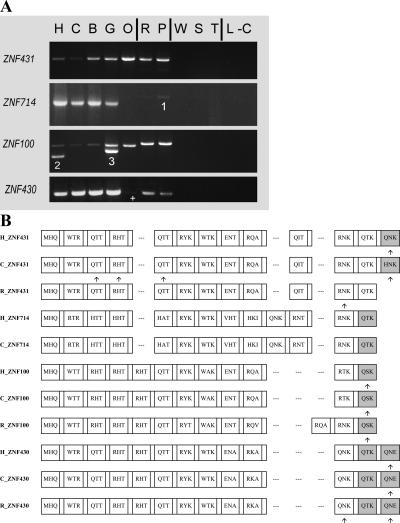
Figure 4.
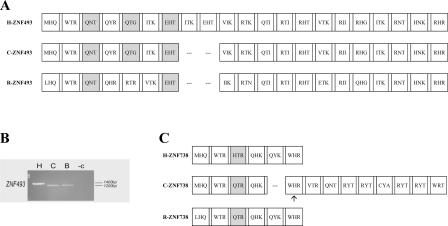
(A) PCR results using primers for selected members of the ZNF431 clade across a panel of primate genomic DNA. The samples are (H) human (Homo sapiens); (C) chimpanzee (Pan troglodytes); (B) bonobo (Pan paniscus); (G) gorilla (Gorilla gorilla); (O) orangutan (Pongo pygmaeus); (R) rhesus macaque (Macaca mulatta); (P) pigtailed macaque (Macaca nemestrina); (W) common woolly monkey (Lagothrix lagotricha); (S) black-handed spider monkey (Ateles geoffroyi); (T) red-chested mustached tamarin (Saguinus labiatus); (L) ring-tailed lemur (Lemur catta); and (−C) the negative control. Vertical lines distinguish the apes, Old World monkeys (the macaques), New World monkeys, and the lemur (a prosimian); see Supplemental Figure S1 for a simple phylogeny of the primates using the same letter codes. Unusual PCR products are numbered as follows: (1) a non-ZNF sequence; (2) LLNL745 (a pseudogene whose closest paralog is ZNF100); (3) a LLNL745-like sequence in gorilla. The + indicates a weak band that was confirmed to be present. Primers were designed from the known genomic sequences and therefore may not identically match all species, resulting in variation in PCR efficiency. LLNL745 may have been deleted in chimpanzee (Newman et al. 2005) and is not in the current chimpanzee genome assembly. (B) Zinc finger alignment hypothesis for the ZNF431 clade. The alignment includes the zinc finger array of the predicted protein for selected human paralogs (H) and their respective orthologs (if found) in chimpanzee (C) and rhesus (R) genomic data. Each box represents a zinc finger; gaps are added when one locus has added or deleted a zinc finger so that flanking fingers remain aligned. The amino acid codes inside each box (for positions −1, 3, and 6, relative to the start of the α-helix) are variable positions involved in sequence-specific DNA target recognition and binding. Degenerate fingers are shaded. Frameshifts in the genomic sequences are indicated by arrows under the fingers, but for the nonhuman primate sequences these are “repaired” to match the orthologous sequences due to the incomplete nature of the nonhuman sequence data. ZNF493 and ZNF738 are not shown due to high-sequence divergence and difficulty in alignment with the other members of the clade; these genes are depicted in Figure 5.
To identify structural changes in the orthologous and paralogous genes, we aligned zinc finger sequences from closely related human and nonhuman primate genes. “Fingers-code” diagrams of the related gene sets, which highlight changes in DNA-binding amino acids at positions −1, 3, and 6 in the α-helix region of the aligned zinc fingers (Choo and Klug 1994), provide a simplified but informative view of structural similarities and differences between the proteins (Fig. 4B). These alignments revealed striking changes in the number and arrangement of zinc fingers in paralogous genes of this clade. For example, predicted ZNF431 and ZNF714 proteins differ by the insertion or deletion of an extra zinc finger within the ZNF array, and nonsynonymous amino acid changes have altered DNA-binding amino acids in many of the remaining fingers (Fig. 4B). Two other closely related genes, ZNF738 and ZNF493, display even more substantial structural differences (Fig. 5). In contrast, orthologous proteins typically have similar zinc finger arrays (Fig. 4B). However, there are also exceptions; the rhesus ZNF100, human ZNF493, and chimpanzee ZNF738 proteins each has structural differences compared with orthologs in the other primate species (Figs. 4, 5).
Figure 5.
Zinc finger exon analysis for ZNF493 and ZNF738, two divergent genes from the ZNF431 clade. The alignments include the human predicted proteins and their orthologs determined from the chimpanzee and rhesus genomic data. Finger boxes, amino acid codes, predicted frameshifts, and degenerate finger shading are as in Figure 4B. The alignment for the predicted ZNF493 proteins (A) shows that the human version has two additional zinc finger motifs compared with both its chimpanzee and rhesus orthologs, indicating the change is an addition of fingers in the human. (B) The relative sizes of ZNF493 zinc finger exon PCR products from genomic DNA of human, chimpanzee, and bonobo is consistent with the human/chimpanzee difference in finger number; species designations are as in Figure 4A. (C) The chimpanzee ZNF738 protein’s differences with the human version, based on draft genomic sequence, include a deleted finger motif, a substitution that eliminates the stop codon seen in human, and a possible frameshift after the deleted finger. Although these differences have not been confirmed, any would change the predicted translation of the chimpanzee protein compared with the human ortholog.
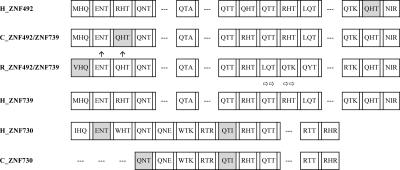
A second example of rapidly evolving genes, the ZNF492 clade (highlighted in yellow, Figs. 1, 3) includes three full-length KRAB-ZNF genes, one partial pseudogene (LLNL621), and one retroposed copy with an intact ORF (LLNL1040) (Fig. 3). ZNF492 and ZNF739 are close relatives that display >97% sequence identity in alignments of noncoding sequences (Supplemental Table S2); these paralogs share the same number of zinc fingers (Fig. 6). However, one zinc finger in human ZNF492 has a mutation disrupting the structurally critical C2H2 pattern of the zinc finger motif. Such “degenerate” fingers cannot bind DNA and potentially affect the overall functional properties of the ZNF array (Gebelein and Urrutia 2001). Although the chimpanzee and rhesus draft genome assemblies contain only a single sequence corresponding to this human gene pair, both genomes are incompletely assembled in surrounding regions and the duplication status is therefore uncertain. The remaining ZNF492 clade members, ZNF730, the pseudogene LLNL621, and the retroposed copy LLNL1040 are present in the chimpanzee genome but are not represented in the available rhesus scaffolds at the time of this submission.
Figure 6.
Zinc finger alignment for the loci in the ZNF492 clade. The alignment includes the human paralogs and their orthologs if found in chimpanzee and rhesus genomic data. Finger boxes, amino acid codes, degenerate finger shading, and frameshifts are as in Figure 4B. The chimpanzee ZNF492/ZNF739 homolog finger-exon sequence is in a location and orientation matching human ZNF739 in the panTro1 draft, but there are assembly gaps around it and in the location where ZNF492 (if both are present) would be located. Newman et al. (2005) recorded a possible inversion in this region between human and chimpanzee. The location of the rhesus copy is unknown. Therefore, the presumed nonhuman ortholog sequences are not explicitly identified with either human locus name. The horizontal arrows below the rhesus ZNF492/ZNF739 protein homolog represent a finger in the protein alignment that is divergent from the same-numbered finger in the human and chimpanzee homologs. The rhesus ZNF492/ZNF739 protein’s fingers 8 and 9 are similar enough to fingers 10 and 11 that in this case, as an alternative to mutational change in the finger amino acid sequence, the pattern in this species could also be explained by a loss of fingers followed by an internal duplication of the aforementioned finger motifs, restoring the array to the same number of zinc finger motifs. The ZNF730 gene contains a stop codon after the 12th finger, eliminating several 3′-end ZNF motifs that are included in the ZNF492 and ZNF739 proteins. For chimpanzee ZNF730, the available genomic sequence is missing the first three finger motifs, but the cross-primate PCR results showed a similar band for chimpanzee and human (data not shown).
Zinc finger sequence evolution
ZNF arrays in paralogous genes differ not only in arrangement and number of zinc fingers, but also through nucleotide substitutions that alter the sequence of amino acids involved in nucleic acid binding. We tested the finger array sequences of aligned paralogous and cross-primate orthologous loci in the ZNF431 and ZNF492 clades for evidence of natural selection by comparing the pairwise values for dN (nonsynonymous changes per nonsynonymous site) and dS (synonymous changes per synonymous site). In analyses including the complete ZNF array sequences, most comparisons had a dN/dS ratio <1 (the trend toward purifying selection). However, when only amino acids in the DNA-binding helix regions were analyzed, dN/dS for paralogous alignments (but rarely orthologous alignments) was often >1, indicating positive selection on these regions (Supplemental Table S3). These results reflect the fact that many amino acids comprising the zinc finger motif serve a structural function, and therefore are highly conserved, while there may be much variation in the four amino acid positions per finger motif that are most critical to DNA binding (−1, 2, 3, and 6 relative to the α helix) (Choo and Klug 1994; Kim and Berg 1996). A Z-test indicated significant purifying selection in most comparisons of the whole zinc finger array (P < 0.05), but significant positive selection was only found for the comparison between human sister-genes ZNF431 and ZNF714 (and for five other comparisons including their primate orthologs) when using the Z test on these four coding positions. The Fisher’s exact test gave the same results for purifying selection, but did not reject the null hypothesis of neutral evolution for any of the variable-site comparisons; the comparisons of the DNA-binding sites necessarily will involve smaller numbers of codons per gene, reducing the statistical power of the pairwise analysis. However, Schmidt and Durrett (2004) also found significant evidence for positive selection when they aligned members of the ZNF431 clade with more distantly related genes, although they did not add gaps for tracking finger gain or loss in their alignment.
Expression of full-length genes and alternative splicing
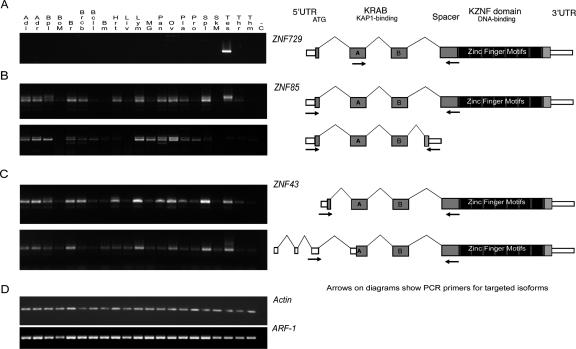
To examine the functional roles of ZNF91 subfamily genes, we analyzed tissue-specific expression of selected loci using RT–PCR. Many genes of this family are widely expressed, but other loci are transcribed in a limited number of tissues. For example, ZNF729 is expressed only in testis (Fig. 7A). Because some KRAB-ZNF genes have splice variants that include or eliminate specific domains (Lovering and Trowsdale 1991; Bellefroid et al. 1993), we also designed primers to detect splicing isoforms of selected primate-specific genes. For example, ZNF85 produces a transcript with the typical KRAB-ZNF structure, as well as an alternate transcript that would be predicted to produce a protein with a KRAB domain but no zinc fingers. This isoform is expressed in multiple tissues, as is the full-length isoform (Fig. 7B). In another example, two confirmed ZNF43 splice variants differ in 5′ UTR exon structure. The longest ORF of the typical splice variant encodes a ZNF protein with a full-length KRAB, whereas the alternate splice variant includes a truncated KRAB-A (Fig. 7C). Both are expressed in multiple tissues and could bind to the same DNA targets, but with different potential to repress transcription.
Figure 7.
RT–PCR gels showing the expression patterns for (A) the KRAB-ZNF gene ZNF729 (a ZNF91 subfamily member on chromosome 19), (B) alternate splice variants of ZNF85 with and without the zinc fingers exon, and (C) alternate splice variants of ZNF43 with different 5′ ends and start sites. Diagrams at right indicate primer locations (arrows) designed to selectively amplify the depicted isoforms for ZNF43 and ZNF85. Notably, each of these isoforms has its first exon within a HERV70/ERV1 LTR repeat. Predicted 5′ ends on many (but not all) of the genes in the cluster overlap with the same type of LTR repeat sequence, which may indicate that the repeats are involved in driving gene expression, as has been shown previously for other genes including related zinc finger genes (Di Cristofano et al. 1995; Abrink et al. 1998). (D, bottom) Gels for two “housekeeping” genes selected as positive controls for the cDNA panels. Guide to the tissues included on the gels: (Adi) Adipose tissue; (Adr) Adrenal gland; (Bpl) Blood, peripheral leukocytes; (BoM) Bone marrow; (Br) Brain; (Brcb) Brain (cerebrum); (Bcll) Brain (cerebellum); (Bm) Brain (medulla oblongata); (Hrt) Heart; (Liv) Liver; (Lym) Lymph node; (MG) Mammary gland; (Pan) Pancreas; (Ov) Ovary; (Pla) Placenta; (Pro) Prostate gland; (Spl) Spleen; (SkM) Skeletal muscle; (Tes) Testis; (Thr) Thyroid; (Thm) Thymus; (−C) negative control. The zinc finger RT–PCR reactions were repeated under varying conditions, and the gels shown are for the 33-cycle experiments, which reveal tissues with present but low-abundance ZNF transcripts.
Discussion
ZNF91 was among the first KRAB-ZNF loci to be described, and the HSA19p12 gene cluster has been known for several years to be primate specific (Bellefroid et al. 1995; Eichler et al. 1998). As elaborated here, the genes have expanded not only through tandem in situ duplications, but also through segmental duplications that have dispersed related genes to distant chromosomal sites. Together with chromosome rearrangement events (Dehal et al. 2001), these distributed duplications may therefore have been a major source for seeding new clusters over evolutionary time. In agreement with previous estimates (Bellefroid et al. 1995, Eichler et al. 1998), data presented here indicate that the initial episodes of expansion of the ZNF91 subfamily occurred before the Catarrhini/Platyrrhini split, but we also identified a substantial number of genes that have continued to duplicate through the rise of apes and humans, creating new lineage-specific genes.
Evolutionary divergence within the zinc finger array of paralogous genes can lead to diversification of gene copies (Looman et al. 2002; Hamilton et al. 2003; Shannon et al. 2003; Krebs et al. 2005); data presented here show that these changes can occur very rapidly, serving to distinguish DNA-binding domains of even very recent duplicates. In the ZNF91 subfamily, we found few examples of KRAB-ZNF paralogs that have retained the same number, arrangement, or sequence of zinc finger motifs. Less frequently, we also identified finger-array deletions and duplications in orthologous sets of primate genes. Because of the intimate relationship predicted between zinc finger arrays and their DNA recognition sites (Choo and Klug 1994; Kim and Berg 1996), these data indicate that the primate-specific KRAB-ZNF genes have diverged specifically in ways that alter DNA-binding properties of the proteins.
Alternative splicing offers another way to increase the total number and diversity of zinc finger proteins. Alternate transcripts that skip the KRAB-A, KRAB-B, or both effector-encoding exons are present in the ZNF91 subfamily, encoding isoforms with different transcription factor activity (e.g., Lovering and Trowsdale 1991; Bellefroid et al. 1993; Chong et al. 1995; Vissing et al. 1995; Chen et al. 1998). Also, several genes of the ZNF91 subfamily appear to use alternative 5′ ends, which may place specific isoforms under different types of transcriptional regulation. Due to the structure of these genes, splice variants and paralogous loci may both be selected for simultaneously (an exception to the inverse relationship proposed by Kopelman et al. 2005) as they facilitate different types of diversity.
What kinds of functional roles do these primate-specific transcriptional repressors play? A potential connection between ZNF91 subfamily genes and immune system function (Bellefroid et al. 1991, 1993; Lovering and Trowsdale 1991; Mark et al. 1999, 2001; Nishimura et al. 2001) suggests a role in the “arms race” between the body’s defense mechanisms and the ever-changing suite of threats. However, RT–PCR data presented here and information from other sources (Gonzalez-Lamuno et al. 2002; Shin et al. 2002; Liu et al. 2004; Su et al. 2004) indicate a wider range of expression patterns and functions for ZNF91 subfamily genes. The testis-specific expression of family member ZNF729 provides an excellent example, and suggests a role in reproduction, another pathway that is potentially operating under intense evolutionary selection (Nielsen et al. 2005). In contrast, some ZNF91 subfamily members are expressed in nearly ubiquitous patterns. Here we should note, however, that widespread patterns of expression do not necessarily imply nonspecific or housekeeping functions for genes that encode transcriptional repressors. For example, the widely expressed transcriptional repressor NRSF exerts a specific and profound role in neurological development by inhibiting expression of target genes in all but neural cells (Chong et al. 1995; Chen et al. 1998). Tissues in which certain KRAB-ZNF genes are not expressed may therefore yield the most important clues to biological function.
Taken together, these data highlight the continuing growth and the rapid, prolific diversification of the KRAB-ZNF gene family in primate lineages. The acquisition of lineage- and species-specific transcriptional repressors potentially provides a potent mechanism for evolution and speciation by serving as a means for fine-tuning regulatory networks and may hold a key to understanding the gene expression and biological differences that make humans distinct from our closest relatives.
Methods
Gene identification and phylogenetic analysis
Evolutionary analyses used the spacer or “tether” region (the section of the zinc finger array-containing exon between the splice junction and the first canonical finger motif) of each locus, a region that has been shown to be diagnostic for ZNF91 subfamily members and likely to vary in sequence among KRAB-ZNF genes (Bellefroid et al. 1993; Shannon and Stubbs 1998). The spacer region was extracted from genes in the 19p12 cluster using the batch sequence retrieval capability of the LLNL Biosciences zinc finger gene catalog Web site (Huntley et al. 2006; http://znf.llnl.gov). Related genes in other locations were found via BLAST and BLAT searches of genome databases using the spacer sequences of 19p12 cluster genes, and by examining a KRAB-A sequence phylogeny (Huntley et al. 2006). The KRAB data was also used to select outgroup loci from another subfamily, genes in the cluster on HSA19q13.41-42.
For each ZNF91-related locus, additional sequence flanking the spacer was included (pre-spacer intron on the 5′ side, initial zinc fingers on the 3′ side). After the removal of repetitive elements defined by RepeatMasker (A.F.A Smit, R. Hubley, and P. Green, unpubl., http://www.repeatmasker.org), an alignment of the spacer + flanking regions was made using ClustalX 1.81 (Thompson et al. 1997). The alignment was manually checked using SeAl (Rambaut 1996). The total length of each sequence differed due to repeat removal and sequence insertions or deletions; a typical example, ZNF431, had 300 bp of pre-spacer intronic sequence and 465 bp of exonic sequence comprising the spacer (which contains the remnants of what may have been a degenerate zinc finger motif) and the first two canonical zinc fingers (although in some loci one or both of these may have also become degenerate).
To gain a fully informative view of the evolutionary history of this subfamily, we included all genes and finger-containing pseudogene sequences from the ZNF91 subfamily in the analysis. Related processed pseudogene sequences were also included. Aligned sequence positions where not all loci had matching sequence due to indels (or, in the case of the processed pseudogenes, no intron sequence at all) were rated as “missing data” for those loci. The spacer + flanking sequence alignment contained 116 loci including the outgroups (Supplemental Table S1 lists all of the ingroup loci, plus other finger-containing loci in the major illustrated clusters). The PAUP 4.0b10 package (Swofford 2002) was used to generate trees using mean character differences and the neighbor-joining (NJ) method (Saitou and Nei 1987). The NJ trees were evaluated with 1000 rounds of bootstrapping (Felsenstein 1985). PAUP was also used to construct maximum parsimony trees from the same data, which did not differ greatly from the neighbor-joining results (the major clades were consistent; however, there was a weak association of the dispersed clade with the chromosome 4 clade; tree not shown). For the aligned intronic sequences, divergence times were estimated based on the neutral evolution rates for pseudogenes or introns (Li et al. 1987; Li 1997; Chen and Li 2001; Liu et al. 2003).
Zinc finger array comparisons between paralogs and orthologs
For two groups of related genes containing recent duplications (the ZNF431 clade and the ZNF492 clade, both on HSA19), a more intense scrutiny of the genes’ zinc finger sequences and interspecies differences was carried out. Alignments of the zinc finger motifs were made using nucleotide and amino acid sequences if paralogs shared enough homologous fingers to permit the tracing of finger-array changes across multiple genes. The zinc finger sequences were also compared with the available chimpanzee and rhesus genomic sequence assemblies or fragments via reciprocal-best BLAT searches on the UCSC Browser (Kent et al. 2002). The draft chimpanzee and rhesus sequences sometimes had frameshift-causing insertions and deletions, but it is probable that these are not all real, because some apparent indels were not confirmed when we did partial sequencing of the gel-extracted primate PCR bands. For the purpose of translating the sequence alignment for protein comparisons and testing for selection, the insertions were “repaired” in silico based on the orthologs in the other species, but the fingers possibly affected are marked on Figures 4, 5 and 6.
The zinc finger motif variable amino acid positions were analyzed for orthologs identified across primate species and between paralogs. Alignments were made using the sequences of the whole ZNF array, as well as with selected amino acid positions (a set of six [−1,1,2,3,5,6], and a set of four [−1,2,3,6]) implicated as the most vital in determining the potential target-specificity of a zinc finger motif (Choo and Klug 1994; Kim and Berg 1996; Greisman and Pabo 1997). The MEGA program (Kumar et al. 2001) was used to calculate the number of nonsynonymous changes per nonsynonymous site and synonymous changes per synonymous site, using the modified Nei and Gojobori (1986) method (Nei and Kumar 2000), with pairwise deletion of missing data and the Jukes and Cantor (1969) distance correction. The dN/dS ratios were calculated for the different subsets of aligned amino acid positions. The Z-test and Fisher’s exact test were used to test all alignments for significant purifying or positive selection. ZNF493 and ZNF738 were not included in the analysis of the ZNF431 clade due to the greater amount of change in the zinc finger sequences, which hindered alignment.
PCR using primers designed for the zinc finger-containing exon of selected loci was performed across a panel of primate genomic DNA samples (Coriell phylogenetic panel PRP00001, http://ccr.coriell.org/primates/; note that samples are also obtainable through IPBIR, the Integrated Primate Biomaterials and Information Resource, at www.ipbir.org). The samples included chimpanzee (Pan troglodytes), bonobo (Pan paniscus), gorilla (Gorilla gorilla), orangutan (Pongo pygmaeus), rhesus monkey (Macaca mulatta), pigtailed macaque (Macaca nemestrina), common woolly monkey (Lagothrix lagotricha), black-handed spider monkey (Ateles geoffroyi), red-chested mustached tamarin (Saguinus labiatus), and ring-tailed lemur (Lemur catta). PCR was performed using the Advantage cDNA PCR Kit (Clontech) and the reaction was run according to the following conditions: 2 min at 94°C; then 32 cycles with 30 sec at 94°C, 45 sec at 55–65°C (annealing temperatures were adjusted to each primer set), 2 min 30 sec at 68°C; and a final extension time for 7 min at 70°C. The list of primers for this survey and the expression testing described below is in Supplemental Table S4. The products were separated on a 1.5% agarose gel and selected bands of interest were gel purified. Partial sequencing using the same primers was used to check the identity of selected bands, including any unusual bands, after gel extraction.
Expression panel analysis
A set of total RNA and mRNAs of human tissues were purchased from BD Biosciences and cDNA was generated using Superscript III First-Strand Synthesis System for RT–PCR (Invitrogen). The concentrations for the cDNA samples were measured using a spectrophotometer, and normalized to 200ng/μL. PCR was performed using the Advantage cDNA PCR Kit, and the reaction was run according to the following conditions: 2 min at 94°C; then 25 or 33 amplification cycles with 30 sec at 94°C, 45 sec at 55–65°C (depending on the different primers), 2 min 30 sec at 68°C; and a final extension time for 7 min at 70°C. Gel purification and sequencing were done as described above to confirm transcript identity and exon structure. The RT–PCR reactions were repeated, including with different cycle numbers, to confirm consistent patterns of expression for each primer set (although the tests with lower cycle number did not show bands for tissues in which each ZNF transcript was least abundant). Primers were designed to be unique to the target genes, and in the case of ZNF85 and ZNF43, to detect specific splice variants (Supplemental Table S4). All cDNA samples were tested using control RT–PCR primers that were designed for Human Cytoskeletal Beta Actin and for Human ADP Ribosylation Factor 1 (ARF-1).
Acknowledgments
We thank Colleen Elso and Joomyeong Kim for critical comments on the manuscript. This work was performed under the auspices of the U.S. Department of Energy (DOE) by the University of California, Lawrence Livermore National Laboratory (LLNL) under Contract No. W-7405-Eng-48. The project (04-ERD-084) was funded by the Laboratory Directed Research and Development Program at LLNL.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4843906
References
- Abrink M., Larsson E., Hellman L., Larsson E., Hellman L., Hellman L. Demethylation of ERV3, an endogenous retrovirus regulating the Kruppel-related zinc finger gene H-plk, in several human cell lines arrested during early monocyte development. DNA Cell Biol. 1998;17:27–37. doi: 10.1089/dna.1998.17.27. [DOI] [PubMed] [Google Scholar]
- Ayyanathan K., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher 3rd F.J., Lechner M.S., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher 3rd F.J., Bell P., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher 3rd F.J., Maul G.G., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher 3rd F.J., Schultz D.C., Yamada Y., Tanaka K., Torigoe K., Rauscher 3rd F.J., Yamada Y., Tanaka K., Torigoe K., Rauscher 3rd F.J., Tanaka K., Torigoe K., Rauscher 3rd F.J., Torigoe K., Rauscher 3rd F.J., Rauscher 3rd F.J. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes & Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Yavor A.M., Massa H.F., Trask B.J., Eichler E.E., Massa H.F., Trask B.J., Eichler E.E., Trask B.J., Eichler E.E., Eichler E.E. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bailey J.A., Liu G., Eichler E.E., Liu G., Eichler E.E., Eichler E.E. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid E.J., Poncelet D.A., Lecocq P.J., Revelant O., Martial J.A., Poncelet D.A., Lecocq P.J., Revelant O., Martial J.A., Lecocq P.J., Revelant O., Martial J.A., Revelant O., Martial J.A., Martial J.A. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc. Natl. Acad. Sci. 1991;88:3608–3612. doi: 10.1073/pnas.88.9.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid E.J., Marine J.C., Ried T., Lecocq P.J., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Marine J.C., Ried T., Lecocq P.J., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Ried T., Lecocq P.J., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Lecocq P.J., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Riviere M., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Amemiya C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Poncelet D.A., Coulie P.G., de Jong P., Szpirer C., Coulie P.G., de Jong P., Szpirer C., de Jong P., Szpirer C., Szpirer C., et al. Clustered organization of homologous KRAB zinc-finger genes with enhanced expression in human T lymphoid cells. EMBO J. 1993;12:1363–1374. doi: 10.1002/j.1460-2075.1993.tb05781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid E.J., Marine J.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Marine J.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Matera A.G., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Bourguignon C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Desai T., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Healy K.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Bray-Ward P., Martial J.A., Ihle J.N., Ward D.C., Martial J.A., Ihle J.N., Ward D.C., Ihle J.N., Ward D.C., Ward D.C. Emergence of the ZNF91 Kruppel-associated box-containing zinc finger gene family in the last common ancestor of anthropoidea. Proc. Natl. Acad. Sci. 1995;92:10757–10761. doi: 10.1073/pnas.92.23.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.C., Li W.H., Li W.H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.F., Paquette A.J., Anderson D.J., Paquette A.J., Anderson D.J., Anderson D.J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Chong J.A., Tapia-Ramirez J., Kim S., Toledo-Aral J.J., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G., Tapia-Ramirez J., Kim S., Toledo-Aral J.J., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G., Kim S., Toledo-Aral J.J., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G., Toledo-Aral J.J., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G., Zheng Y., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G., Boutros M.C., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G., Altshuller Y.M., Frohman M.A., Kraner S.D., Mandel G., Frohman M.A., Kraner S.D., Mandel G., Kraner S.D., Mandel G., Mandel G. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Choo Y., Klug A., Klug A. Toward a code for the interactions of zinc fingers with DNA: Selection of randomized fingers displayed on phage. Proc. Natl. Acad. Sci. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Predki P., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Olsen A.S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Kobayashi A., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Folta P., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Lucas S., Land M., Terry A., Ecale Zhou C.L., Rash S., Land M., Terry A., Ecale Zhou C.L., Rash S., Terry A., Ecale Zhou C.L., Rash S., Ecale Zhou C.L., Rash S., Rash S., et al. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science. 2001;293:104–111. doi: 10.1126/science.1060310. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A., Strazullo M., Longo L., La Mantia G., Strazullo M., Longo L., La Mantia G., Longo L., La Mantia G., La Mantia G. Characterization and genomic mapping of the ZNF80 locus: Expression of this zinc-finger gene is driven by a solitary LTR of ERV9 endogenous retroviral family. Nucleic Acids Res. 1995;23:2823–2830. doi: 10.1093/nar/23.15.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E.E., Hoffman S.M., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Hoffman S.M., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Adamson A.A., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., Gordon L.A., McCready P., Lamerdin J.E., Mohrenweiser H.W., McCready P., Lamerdin J.E., Mohrenweiser H.W., Lamerdin J.E., Mohrenweiser H.W., Mohrenweiser H.W. Complex β-satellite repeat structures and the expansion of the zinc finger gene cluster in 19p12. Genome Res. 1998;8:791–808. doi: 10.1101/gr.8.8.791. [DOI] [PubMed] [Google Scholar]
- Enard W., Khaitovich P., Klose J., Zollner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., Khaitovich P., Klose J., Zollner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., Klose J., Zollner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., Zollner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., Muchmore E., Varki A., Ravid R., Varki A., Ravid R., Ravid R., et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gebelein B., Urrutia R., Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol. Cell. Biol. 2001;21:928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y., Man O., Glusman G., Man O., Glusman G., Glusman G. A comparison of the human and chimpanzee olfactory receptor gene repertoires. Genome Res. 2005;15:224–230. doi: 10.1101/gr.2846405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko G.V., Nei M., Nei M. Estimation of divergence times for major lineages of primate species. Mol. Biol. Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lamuno D., Loukili N., Garcia-Fuentes M., Thomson T.M., Loukili N., Garcia-Fuentes M., Thomson T.M., Garcia-Fuentes M., Thomson T.M., Thomson T.M. Expression and regulation of the transcriptional repressor ZNF43 in Ewing sarcoma cells. Pediatr. Pathol. Mol. Med. 2002;21:531–540. doi: 10.1080/15227950290112789. [DOI] [PubMed] [Google Scholar]
- Goodman M., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Porter C.A., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Czelusniak J., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Page S.L., Schneider H., Shoshani J., Gunnell G., Groves C.P., Schneider H., Shoshani J., Gunnell G., Groves C.P., Shoshani J., Gunnell G., Groves C.P., Gunnell G., Groves C.P., Groves C.P. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Greisman H.A., Pabo C.O., Pabo C.O. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- Guy J., Hearn T., Crosier M., Mudge J., Viggiano L., Koczan D., Thiesen H.J., Bailey J.A., Horvath J.E., Eichler E.E., Hearn T., Crosier M., Mudge J., Viggiano L., Koczan D., Thiesen H.J., Bailey J.A., Horvath J.E., Eichler E.E., Crosier M., Mudge J., Viggiano L., Koczan D., Thiesen H.J., Bailey J.A., Horvath J.E., Eichler E.E., Mudge J., Viggiano L., Koczan D., Thiesen H.J., Bailey J.A., Horvath J.E., Eichler E.E., Viggiano L., Koczan D., Thiesen H.J., Bailey J.A., Horvath J.E., Eichler E.E., Koczan D., Thiesen H.J., Bailey J.A., Horvath J.E., Eichler E.E., Thiesen H.J., Bailey J.A., Horvath J.E., Eichler E.E., Bailey J.A., Horvath J.E., Eichler E.E., Horvath J.E., Eichler E.E., Eichler E.E., et al. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10p. Genome Res. 2003;13:159–172. doi: 10.1101/gr.644503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A.T., Huntley S., Kim J., Branscomb E., Stubbs L., Huntley S., Kim J., Branscomb E., Stubbs L., Kim J., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L. Lineage-specific expansion of KRAB zinc-finger transcription factor genes: Implications for the evolution of vertebrate regulatory networks. Cold Spring Harb. Symp. Quant. Biol. 2003;68:131–140. doi: 10.1101/sqb.2003.68.131. [DOI] [PubMed] [Google Scholar]
- Hao L., Nei M., Nei M. Rapid expansion of killer cell immunoglobulin-like receptor genes in primates and their coevolution with MHC Class I genes. Gene. 2005;347:149–159. doi: 10.1016/j.gene.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Huntley S., Baggot D.M., Hamilton A.T., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Baggot D.M., Hamilton A.T., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Hamilton A.T., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Tran-Gyamfi M., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Yang S., Kim J., Gordon L., Branscomb E., Stubbs L., Kim J., Gordon L., Branscomb E., Stubbs L., Gordon L., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L.2006A comprehensive catalogue of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes T.H., Cantor C.R., Cantor C.R.1969. Evolution of protein molecules. In Mammalian protein metabolism (ed. N.H. Munro) pp. 21–123. Academic Press; New York [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P., Muetzel B., She X., Lachmann M., Hellmann I., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W., Muetzel B., She X., Lachmann M., Hellmann I., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W., She X., Lachmann M., Hellmann I., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W., Lachmann M., Hellmann I., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W., Hellmann I., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W., Steigele S., Do H.H., Weiss G., Enard W., Do H.H., Weiss G., Enard W., Weiss G., Enard W., Enard W., et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Hellmann I., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Enard W., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Nowick K., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Leinweber M., Franz H., Weiss G., Lachmann M., Pääbo S., Franz H., Weiss G., Lachmann M., Pääbo S., Weiss G., Lachmann M., Pääbo S., Lachmann M., Pääbo S., Pääbo S. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Kim C.A., Berg J.M., Berg J.M. A 2.2 A resolution crystal structure of a designed zinc finger protein bound to DNA. Nat. Struct. Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- Kim S.S., Chen Y.M., O’Leary E., Witzgall R., Vidal M., Bonventre J.V., Chen Y.M., O’Leary E., Witzgall R., Vidal M., Bonventre J.V., O’Leary E., Witzgall R., Vidal M., Bonventre J.V., Witzgall R., Vidal M., Bonventre J.V., Vidal M., Bonventre J.V., Bonventre J.V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc. Natl. Acad. Sci. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman N.M., Lancet D., Yanai I., Lancet D., Yanai I., Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat. Genet. 2005;37:588–589. doi: 10.1038/ng1575. [DOI] [PubMed] [Google Scholar]
- Krebs C.J., Larkins L.K., Khan S.M., Robins D.M., Larkins L.K., Khan S.M., Robins D.M., Khan S.M., Robins D.M., Robins D.M. Expansion and diversification of KRAB zinc-finger genes within a cluster including Regulator of sex-limitation 1 and 2. Genomics. 2005;85:752–761. doi: 10.1016/j.ygeno.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I.B., Nei M., Tamura K., Jakobsen I.B., Nei M., Jakobsen I.B., Nei M., Nei M. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Li W.-H. Molecular evolution. Sinauer Associates; Sunderland, MA.: 1997. [Google Scholar]
- Li W.H., Tanimura M., Sharp P.M., Tanimura M., Sharp P.M., Sharp P.M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J. Mol. Evol. 1987;25:330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas III E.J., Zody M.C., Chang J.L., Kulbokas III E.J., Zody M.C., Kulbokas III E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Liu G., Zhao S., Bailey J.A., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E., Zhao S., Bailey J.A., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E., Bailey J.A., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E., Sahinalp S.C., Alkan C., Tuzun E., Green E.D., Eichler E.E., Alkan C., Tuzun E., Green E.D., Eichler E.E., Tuzun E., Green E.D., Eichler E.E., Green E.D., Eichler E.E., Eichler E.E. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–368. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhu C., Luo J., Wang Y., Li D., Li Y., Zhou J., Yuan W., Ou Y., Liu M., Zhu C., Luo J., Wang Y., Li D., Li Y., Zhou J., Yuan W., Ou Y., Liu M., Luo J., Wang Y., Li D., Li Y., Zhou J., Yuan W., Ou Y., Liu M., Wang Y., Li D., Li Y., Zhou J., Yuan W., Ou Y., Liu M., Li D., Li Y., Zhou J., Yuan W., Ou Y., Liu M., Li Y., Zhou J., Yuan W., Ou Y., Liu M., Zhou J., Yuan W., Ou Y., Liu M., Yuan W., Ou Y., Liu M., Ou Y., Liu M., Liu M., et al. ZNF411, a novel KRAB-containing zinc-finger protein, suppresses MAP kinase signaling pathway. Biochem. Biophys. Res. Commun. 2004;320:45–53. doi: 10.1016/j.bbrc.2004.05.130. [DOI] [PubMed] [Google Scholar]
- Looman C., Abrink M., Mark C., Hellman L., Abrink M., Mark C., Hellman L., Mark C., Hellman L., Hellman L. KRAB zinc finger proteins: An analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 2002;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- Looman C., Hellman L., Abrink M., Hellman L., Abrink M., Abrink M. A novel Kruppel-Associated Box identified in a panel of mammalian zinc finger proteins. Mamm. Genome. 2004;15:35–40. doi: 10.1007/s00335-003-3022-0. [DOI] [PubMed] [Google Scholar]
- Lovering R., Trowsdale J., Trowsdale J. A gene encoding 22 highly related zinc fingers is expressed in lymphoid cell lines. Nucleic Acids Res. 1991;19:2921–2928. doi: 10.1093/nar/19.11.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark C., Abrink M., Hellman L., Abrink M., Hellman L., Hellman L. Comparative analysis of KRAB zinc finger proteins in rodents and man: Evidence for several evolutionarily distinct subfamilies of KRAB zinc finger genes. DNA Cell Biol. 1999;18:381–396. doi: 10.1089/104454999315277. [DOI] [PubMed] [Google Scholar]
- Mark C., Looman C., Abrink M., Hellman L., Looman C., Abrink M., Hellman L., Abrink M., Hellman L., Hellman L. Molecular cloning and preliminary functional analysis of two novel human KRAB zinc finger proteins, HKr18 and HKr19. DNA Cell Biol. 2001;20:275–286. doi: 10.1089/104454901750232472. [DOI] [PubMed] [Google Scholar]
- Moosmann P., Georgiev O., Le Douarin B., Bourquin J.P., Schaffner W., Georgiev O., Le Douarin B., Bourquin J.P., Schaffner W., Le Douarin B., Bourquin J.P., Schaffner W., Bourquin J.P., Schaffner W., Schaffner W. Transcriptional repression by RING finger protein TIF1 β that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nei M., Kumar S., Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York.: 2000. [Google Scholar]
- Nei M., Gu X., Sitnikova T., Gu X., Sitnikova T., Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman T.L., Tuzun E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Tuzun E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Morrison V.A., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Hayden K.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., Ventura M., McGrath S.D., Rocchi M., Eichler E.E., McGrath S.D., Rocchi M., Eichler E.E., Rocchi M., Eichler E.E., Eichler E.E. A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 2005;15:1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Bustamante C., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Tanenbaum D.M., Civello D., White T.J., Civello D., White T.J., White T.J., et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Narita T., Miyazaki E., Ito T., Nishimoto N., Yoshizaki K., Martial J.A., Bellfroid E.J., Vissing H., Taniyama T., Narita T., Miyazaki E., Ito T., Nishimoto N., Yoshizaki K., Martial J.A., Bellfroid E.J., Vissing H., Taniyama T., Miyazaki E., Ito T., Nishimoto N., Yoshizaki K., Martial J.A., Bellfroid E.J., Vissing H., Taniyama T., Ito T., Nishimoto N., Yoshizaki K., Martial J.A., Bellfroid E.J., Vissing H., Taniyama T., Nishimoto N., Yoshizaki K., Martial J.A., Bellfroid E.J., Vissing H., Taniyama T., Yoshizaki K., Martial J.A., Bellfroid E.J., Vissing H., Taniyama T., Martial J.A., Bellfroid E.J., Vissing H., Taniyama T., Bellfroid E.J., Vissing H., Taniyama T., Vissing H., Taniyama T., Taniyama T. Characterization of the human Fc gamma RIIB gene promoter: Human zinc-finger proteins (ZNF140 and ZNF91) that bind to different regions function as transcription repressors. Int. Immunol. 2001;13:1075–1084. doi: 10.1093/intimm/13.8.1075. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer-Verlag; Berlin, New York.: 1970. [Google Scholar]
- Preuss T.M., Caceres M., Oldham M.C., Geschwind D.H., Caceres M., Oldham M.C., Geschwind D.H., Oldham M.C., Geschwind D.H., Geschwind D.H. Human brain evolution: Insights from microarrays. Nat. Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- Rambaut A.1996. Se-Al: Sequence Alignment Editor. http://iubio.bio.indiana.edu/soft/iubionew/molbio/dna/analysis/Pist/ main.html [Google Scholar]
- Rozen S., Skaletsky H., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Skaletsky H., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Marszalek J.D., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Waterston R.H., Wilson R.K., Page D.C., Wilson R.K., Page D.C., Page D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J.G., Bashirova A., Palmer S., Sims S., Trowsdale J., Abi-Rached L., Parham P., Carrington M., Beck S., Bashirova A., Palmer S., Sims S., Trowsdale J., Abi-Rached L., Parham P., Carrington M., Beck S., Palmer S., Sims S., Trowsdale J., Abi-Rached L., Parham P., Carrington M., Beck S., Sims S., Trowsdale J., Abi-Rached L., Parham P., Carrington M., Beck S., Trowsdale J., Abi-Rached L., Parham P., Carrington M., Beck S., Abi-Rached L., Parham P., Carrington M., Beck S., Parham P., Carrington M., Beck S., Carrington M., Beck S., Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., Durrett R., Durrett R. Adaptive evolution drives the diversification of zinc-finger binding domains. Mol. Biol. Evol. 2004;21:2326–2339. doi: 10.1093/molbev/msh246. [DOI] [PubMed] [Google Scholar]
- Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., Rauscher III F.J., Ayyanathan K., Negorev D., Maul G.G., Rauscher III F.J., Negorev D., Maul G.G., Rauscher III F.J., Maul G.G., Rauscher III F.J., Rauscher III F.J. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M., Stubbs L., Stubbs L. Analysis of homologous XRCC1-linked zinc-finger gene families in human and mouse: Evidence for orthologous genes. Genomics. 1998;49:112–121. doi: 10.1006/geno.1998.5230. [DOI] [PubMed] [Google Scholar]
- Shannon M., Kim J., Ashworth L., Branscomb E., Stubbs L., Kim J., Ashworth L., Branscomb E., Stubbs L., Ashworth L., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L. Tandem zinc-finger gene families in mammals: Insights and unanswered questions. DNA Seq. 1998;8:303–315. doi: 10.3109/10425179809034075. [DOI] [PubMed] [Google Scholar]
- Shannon M., Hamilton A.T., Gordon L., Branscomb E., Stubbs L., Hamilton A.T., Gordon L., Branscomb E., Stubbs L., Gordon L., Branscomb E., Stubbs L., Branscomb E., Stubbs L., Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13:1097–1110. doi: 10.1101/gr.963903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.N., Kim I., Lee J.S., Koh G.Y., Lee Z.H., Kim H.H., Kim I., Lee J.S., Koh G.Y., Lee Z.H., Kim H.H., Lee J.S., Koh G.Y., Lee Z.H., Kim H.H., Koh G.Y., Lee Z.H., Kim H.H., Lee Z.H., Kim H.H., Kim H.H. A novel zinc finger protein that inhibits osteoclastogenesis and the function of tumor necrosis factor receptor-associated factor 6. J. Biol. Chem. 2002;277:8346–8353. doi: 10.1074/jbc.M110964200. [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Repping S., Pyntikova T., Ali J., Bieri T., Pyntikova T., Ali J., Bieri T., Ali J., Bieri T., Bieri T., et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. PAUP* Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates; Sunderland, MA.: 2002. [Google Scholar]
- Thiesen H.J. Multiple genes encoding zinc finger domains are expressed in human T cells. New Biol. 1990;2:363–374. [PubMed] [Google Scholar]
- Thiesen H.J., Bellefroid E., Revelant O., Martial J.A., Bellefroid E., Revelant O., Martial J.A., Revelant O., Martial J.A., Martial J.A. Conserved KRAB protein domain identified upstream from the zinc finger region of Kox 8. Nucleic Acids Res. 1991;19:3996. doi: 10.1093/nar/19.14.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Plewniak F., Jeanmougin F., Higgins D.G., Jeanmougin F., Higgins D.G., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J., Barten R., Haude A., Stewart C.A., Beck S., Wilson M.J., Barten R., Haude A., Stewart C.A., Beck S., Wilson M.J., Haude A., Stewart C.A., Beck S., Wilson M.J., Stewart C.A., Beck S., Wilson M.J., Beck S., Wilson M.J., Wilson M.J. The genomic context of natural killer receptor extended gene families. Immunol. Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- Tunnacliffe A., Liu L., Moore J.K., Leversha M.A., Jackson M.S., Papi L., Ferguson-Smith M.A., Thiesen H.J., Ponder B.A., Liu L., Moore J.K., Leversha M.A., Jackson M.S., Papi L., Ferguson-Smith M.A., Thiesen H.J., Ponder B.A., Moore J.K., Leversha M.A., Jackson M.S., Papi L., Ferguson-Smith M.A., Thiesen H.J., Ponder B.A., Leversha M.A., Jackson M.S., Papi L., Ferguson-Smith M.A., Thiesen H.J., Ponder B.A., Jackson M.S., Papi L., Ferguson-Smith M.A., Thiesen H.J., Ponder B.A., Papi L., Ferguson-Smith M.A., Thiesen H.J., Ponder B.A., Ferguson-Smith M.A., Thiesen H.J., Ponder B.A., Thiesen H.J., Ponder B.A., Ponder B.A. Duplicated KOX zinc finger gene clusters flank the centromere of human chromosome 10: Evidence for a pericentric inversion during primate evolution. Nucleic Acids Res. 1993;21:1409–1417. doi: 10.1093/nar/21.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissing H., Meyer W.K., Aagaard L., Tommerup N., Thiesen H.J., Meyer W.K., Aagaard L., Tommerup N., Thiesen H.J., Aagaard L., Tommerup N., Thiesen H.J., Tommerup N., Thiesen H.J., Thiesen H.J. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995;369:153–157. doi: 10.1016/0014-5793(95)00728-r. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Young J.M., Friedman C., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Friedman C., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Williams E.M., Ross J.A., Tonnes-Priddy L., Trask B.J., Ross J.A., Tonnes-Priddy L., Trask B.J., Tonnes-Priddy L., Trask B.J., Trask B.J. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 2002;11:535–546. doi: 10.1093/hmg/11.5.535. [DOI] [PubMed] [Google Scholar]