Abstract
The hrp gene clusters of plant pathogenic bacteria control pathogenicity on their host plants and ability to elicit the hypersensitive reaction in resistant plants. Some hrp gene products constitute elements of the type III secretion system, by which effector proteins are exported and delivered into plant cells. Here, we show that the hrpZ gene product from the bean halo-blight pathogen, Pseudomonas syringae pv. phaseolicola (HrpZPsph), is secreted in an hrp-dependent manner in P. syringae pv. phaseolicola and exported by the type III secretion system in the mammalian pathogen Yersinia enterocolitica. HrpZPsph was found to associate stably with liposomes and synthetic bilayer membranes. Under symmetric ionic conditions, addition of 2 nM of purified recombinant HrpZPsph to the cis compartment of planar lipid bilayers provoked an ion current with a large unitary conductivity of 207 pS. HrpZPsph-related proteins from P. syringae pv. tomato or syringae triggered ion currents similar to those stimulated by HrpZPsph. The HrpZPsph-mediated ion-conducting pore was permeable for cations but did not mediate fluxes of Cl−. Such pore-forming activity may allow nutrient release and/or delivery of virulence factors during bacterial colonization of host plants.
Pathogenic bacteria use highly specialized mechanisms to proliferate in their hosts. Recent work has revealed the importance of a type III protein-secretion system essential for pathogenicity of Gram-negative bacteria (1, 2). Type III secretion systems were reported initially from the mammalian pathogen Yersinia enterocolitica but have been identified also in various phytopathogenic bacteria (2). Specific effector proteins are believed to be injected into the cytosol of eukaryotic host cells by using this secretion apparatus (2). In contrast to the proteins that form the secretion apparatus, the export substrates often are not conserved among pathogenic bacteria (2, 3).
The Yersinia Yop (Yersinia outer proteins) virulon serves as a paradigm for the type III secretion system (1). A total of 25 ysc (Yop secretion) genes encode proteins implicated in protein translocation across the bacterial inner and outer membranes (1, 2, 4). Among 12 secreted Yops are three (YopB, D, and LcrV) that promote translocation of effector proteins across the mammalian-host cell-plasma membrane (1). YopB and YopD are hydrophobic proteins that interact with each other (1, 5). Pore-forming activity ascribed to both proteins (1, 4, 5) has been verified recently by experiments showing that YopB and YopD form an ion-conducting pore after insertion into planar lipid bilayers (6).
Phytopathogenic bacteria harbor a gene cluster (hrp for hypersensitive reaction and pathogenicity) that is essential for pathogenicity in susceptible plants and the ability to elicit the hypersensitive reaction in nonhosts or resistant cultivars of host plants (2, 7). hrp genes are expressed in planta or in media that mimic plant apoplastic conditions (8). Sequence analyses have uncovered a shared subset of hrp genes that were redefined as hrc (for hrp and conserved) (9) and that encode proteins homologous to Yersinia ysc gene products (2, 10–12). This observation suggests evolutionary conservation of molecular mechanisms of pathogenicity used by both mammalian and phytopathogenic bacteria (2). A number of proteins secreted by phytopathogenic bacteria in an hrp-dependent manner have been identified (2), but their molecular modes of action are understood poorly.
Harpins constitute a group of secreted effector proteins traveling along the type III pathway in plant pathogenic bacteria (2, 13). Genes encoding such proteins have been identified in Erwinia amylovora, Erwinia chrysanthemi (hrpN) (14), Pseudomonas syringae pvs. syringae, tomato, or glycinea (hrpZ) (7, 15), and Ralstonia solanacearum (popA) (16). Although not homologous in primary sequence, harpins from different bacterial genera trigger disease resistance-associated responses, such as hypersensitive cell death, and thus activate the plant's surveillance system (2, 13). However, the contribution of harpins to bacterial pathogenicity remains enigmatic. Mutations in hrpN significantly reduced pathogenicity of E. amylovora (14, 17), whereas mutations in hrpZ left the virulence of P. syringae pv. tomato apparently unaffected (18). The presence in P. syringae pathovars of genes functionally redundant to hrpZ, such as hrpW, may account for the unaltered pathogenicity of hrpZ mutants (18). Another open question concerns the site of action of harpins during infection. Immunolocalization studies revealed Ca2+-dependent association of P. syringae pv. syringae harpin with plant cell walls (19). However, whether this interaction identifies a physiological target implicated in colonization of the host plant has yet to be determined.
Here, we report on the analysis of the structure and function of hrpZ in the bean halo-blight pathogen P. syringae pv. phaseolicola (Psph). Structural properties of HrpZPsph resemble those of proteins assumed to interact with membranes such as YopB from Y. enterocolitica. Similar to YopB (6), HrpZPsph and related proteins from P. syringae pvs. tomato or syringae are integrated into bilayer membranes to form an ion-conducting pore in vitro. We propose that pore formation by harpins may either mediate nutrient release from host cells or assist delivery of virulence factors into the plant cell cytoplasm by facilitating movement of proteins through the plasma membrane.
Materials and Methods
Bacterial Strains, Plasmids, and Bacterial Growth.
Wild-type and mutant strains of Psph race-6 and pilM-deficient Psph strain HB10Y were grown as described (20). Protein secretion was initiated in minimal-growth medium (21). The nonpolar hrpA and hrpZ mutants were constructed as follows: hrpA was deleted from bases 46 to 256 of the coding sequence in pPPY438 by a double digest with Csp45I and HpaI and religated after end filling; hrpZ was deleted from bases 586 to 876 in pPPY438 by digests with MunI and PpuMI and religated. The truncated coding regions were cloned into the suicide vector pOK and introduced into Psph as described (22).
Y. enterocolitica strains were grown as described (5). MRS40(pABL 403) is a derivative of wild-type strain MRS40 lacking yopH, yopO, yopP, yopE, and yopM (23). Strain MRS40(pAB409) is a yopB-deficient derivative of MRS40(pABL403) (23). KNG22703(pSW2276) is a yscN-deficient mutant of wild-type strain KNG22703(pYV227) (24). For complementation of MRS40(pAB409) with yopB, pCNR27-encoding YopB was used (5). For expression of hrpZ in Y. enterocolitica, an XbaI/PstI fragment from vector pT7-7-hrpZ containing the hrpZ ORF (25) was subcloned into pBluescript SK(+) (pSK-hrpZ), facilitating expression from the basal lac promoter. Y. enterocolitica transformation by electroporation, induction of the Yop virulon, and analysis of Yop proteins in culture supernatants were performed as described (26).
Plant Infiltration and Sheep Erythrocyte Hemolysis Assays.
Pathogenicity tests of Psph strains on bean (Phaseolus vulgaris L.) were performed as described (27). Hemolytic activity was assayed as described (5).
Preparation of Recombinant HrpZ.
A 1.38-kb NdeI/HindIII fragment from plasmid pT7-7-hrpZ was introduced into the modified pET vector, pJC40, encoding an N-terminal His10 tag (28). Expression in BL21 (DE3) pLysS Escherichia coli cells was initiated, and expressed proteins were isolated on Ni-nitrilotriacetic acid agarose (Qiagen, Hilden, Germany). Stringent washes with buffer containing 60 mM of imidazole before elution were crucial for obtaining pure protein. Removal of the His10 tag was achieved by Factor Xa (Denzyme, Aarhus, Denmark) treatment for 12 h at 25°C in 50 mM Tris⋅HCl, pH 8.0 and 1 mM CaCl2 containing 1/100 (wt/wt) protease. HrpZPsph was separated by 40% (NH4)2SO4 fractionation and desalted by ultrafiltration with a 30-kDa cut-off concentrator (Amicon). The purity of recombinant HrpZPsph was checked by SDS/PAGE and silver staining. To express fragments of HrpZPsph, the corresponding PCR-generated products were cloned into vector pJC40 as NdeI/BamHI or NdeI/EcoRI fragments, respectively. Recombinant proteins were purified as described above except that ultrafiltration was performed with 10- or 3.5-kDa cut-off filters, respectively. A PCR-generated HindIII/BamHI fragment derived from P. syringae pv. tomato DC3000 DNA was subcloned into vector pJC40 for production of recombinant HrpZPst. Recombinant HrpZPss was produced as described (7) with the vector pSYH10.
Protein Biochemistry.
Phospholipid-binding assays were carried out as described (29). To determine β-glucuronidase activity, plasmid pDGUS (30) carrying the E. coli uidA gene was introduced into Psph strains by triparental mating as described (27). GUS activity was quantified as described (30).
Lipid-Binding and Planar Lipid Bilayer Experiments.
Association of secreted proteins with liposomes and isolation of proteoliposomes was performed as described (6) except that liposomes were added to Psph grown in minimal medium. Lipid-bound proteins were precipitated in 80% (vol/vol) acetone before SDS/PAGE immunoblotting by using the anti-HrpZPsph antiserum. In control experiments, PBS (pH 7.1) without liposomes was added to the bacteria to ensure that hrp gene induction was unaffected. Protein binding to silica beads coated with single lipid bilayers (TRANSIL, NIMBUS Biotechnology, Leipzig, Germany) was performed as described (31). Assays involved incubation of 5 μl of TRANSIL beads and 1 μM of purified recombinant protein for 1 h (room temperature; 10 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, protein:lipid ratio 1:500/1,000). After centrifugation (5 min, 1,500 × g), the beads were washed three times with binding buffer. Unbound proteins and wash fractions were pooled and precipitated by 5% (vol/vol) ice-cold trichloroacetic acid. Bound proteins were eluted with sample buffer for SDS/PAGE and silver staining. Planar lipid bilayer experiments were performed as described (32). The trans compartment of the cuvette is defined to be at ground potential. The sign of the membrane voltage refers to the cis compartment, and a positive current (upward deflections) corresponds to a cation transfer from the cis to the trans compartment. HrpZPsph was added to the aqueous solution of the cis compartment.
Results
HrpZPsph Is Produced and Secreted in an hrp-Dependent, Type III-Specific Manner.
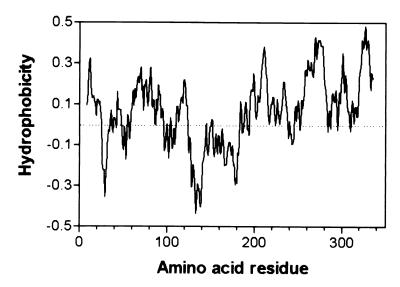
The clone pPPY430 contains the hrp gene cluster from Psph race 6 except for part of hrpRS (33). Comparison of the region of PPY430 containing hrpZ (subclone pPPY438; GenBank accession no. AF 268940) with the hrpZ operon described for other pathovars of P. syringae (15) revealed Psph genes homologous to hrpA, hrpZ, hrpB, hrcJ, hrpD, and hrpE. In contrast to other pathovars of P. syringae (15, 34), Northern blot analysis of RNA prepared from Psph grown in minimal medium revealed separate hrpA and hrpZ + hrpB-specific transcripts (not shown). The deletion of basepairs 46–356 of hrpZ abolished accumulation of the gene product in bacteria grown under hrp-inducing conditions, but no alteration in pathogenicity on bean was associated with the hrpZ mutation (not shown). hrpZPsph encodes a 345-aa protein (35.2 kDa), which shows 53 and 77% protein sequence similarity to the respective orthologs from P. syringae pv. tomato and syringae. Hydropathy plots (Eisenberg algorithm; ref. 35) revealed the amphipathic nature of the encoded protein, which comprises a hydrophilic central region flanked by two hydrophobic terminal domains (Fig. 1).
Figure 1.
Hydrophobicity plot of HrpZPsph based on the Eisenberg algorithm (35).
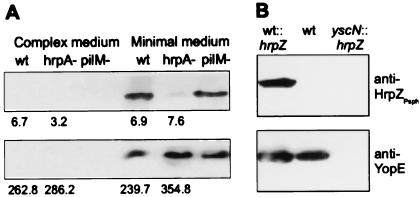
Expression of hrpZ from other P. syringae pvs. is hrp gene dependent (15). By using an antiserum raised against recombinant HrpZPsph, we detected this protein in minimal, but not in complex, media (Fig. 2A). This was because of secretion rather than cell lysis, because no leakage of a constitutively expressed cytoplasmic marker protein β-glucuronidase was observed (Fig. 2A). The hrpA gene of P. syringae pv. tomato, which encodes the major constituent of the Hrp pilus (21), was shown recently to be required for full expression and secretion of putative virulence proteins such as HrpW and AvrPto (36). The Psph hrpA mutant, which was nonpathogenic on bean (not shown), accumulated HrpZPsph intracellularly to the same levels as in wild-type bacteria, but secretion was reduced strongly (Fig. 2A). By contrast, a mutation in pilM, which is required for type IV pilus production (20), did not affect the secretion of HrpZPsph. An hrcJ mutant of Psph also did not secrete HrpZPsph (not shown). Thus, HrpZPsph is produced and secreted in an hrp-dependent and type III-specific manner.
Figure 2.
Analysis of HrpZPsph secretion by Psph and by Y. enterocolitica. (A) hrp-dependent secretion of HrpZPsph. Psph race-6 wild-type (wt), race-6 hrpA− mutant (hrpA−), and a type IV pili mutant of strain HB10Y (pilM−) were grown in complex media or hrp-inducing minimal media (8). Bacteria were pelleted by centrifugation, and secreted proteins were precipitated from the culture supernatant by 5% (vol/vol) trichloroacetic acid. Proteins prepared from the supernatant (Upper) and pellet (Lower) were analyzed by SDS/PAGE and immunoblotting by using an antiserum raised against recombinant HrpZPsph (1:5,000 dilution). Numbers below individual lanes represent β-glucuronidase activity (nmol 4-methylumbelliferone released per minute per bacterium × 1010), which was determined in uidA-transformed Psph strains grown in complex and minimal growth medium (30). (B) Type III-dependent secretion of HrpZPsph. Y. enterocolitica wild-type strain KNG22703(pYV227) (wt), this strain carrying pBluescript SK(+)-hrpZ (wt∷hrpZ), and Y. enterocolitica type III secretion-deficient yscN- mutant KNG22703(pYV2276) transformed with plasmid pBluescript SK(+)-hrpZ (yscN∷hrpZ) were grown under permissive conditions. Analysis of secreted proteins was performed by using antisera raised against HrpZPsph or YopE, respectively.
The apparent evolutionary conservation of type III secretion systems prompted us to investigate whether Psph export substrates can be secreted by the mammalian pathogen Y. enterocolitica. Therefore, we introduced hrpZ under control of the basal lac promoter into strain KNG22703(pYV227). After induction of the yop virulon (5), the transformants produced and secreted HrpZPsph (Fig. 2B). However, secretion of HrpZPsph was not observed in its secretion-deficient yscN mutant KNG22703(pSW2276) (Fig. 2B). Thus, a putative phytopathogenic virulence factor, HrpZPsph, is exported from a mammalian pathogen, Y. enterocolitica, in a type III secretion-dependent manner.
The PROPSEARCH algorithm (37) allows the identification of proteins that may be dissimilar in primary sequence but share common structural properties. Comparison of HrpZPsph predicted similarity to several proteins known to interact with membranes and to form protein complexes, among them Yersinia YopB (1, 5, 6) (reliability score 41%; ref 37). To test for a possible functional similarity between the two proteins, we attempted complementation by hrpZPsph of yopB-deficient Y. enterocolitica strain MRS40(pABL409) lacking virulence factors yopH, yopO, yopP, yopE, and yopM in addition to yopB. Deletion of these genes was designed to eliminate adverse effects of other secreted virulence factors in the sheep erythrocyte hemolysis assay (1, 4). The mutant strain secreted HrpZPsph (not shown) but failed to cause hemolysis (OD570 < 0.01). By contrast, the same strain expressing yopB [MRS40(pABL403)], or the yopB-deficient strain complemented by plasmid-borne yopB, possessed significant hemolytic activity (OD570 = 0.161). Despite structural similarities, HrpZPsph is unable, therefore, to replace YopB in Y. enterocolitica.
Association of HrpZPsph with Lipid Membranes.
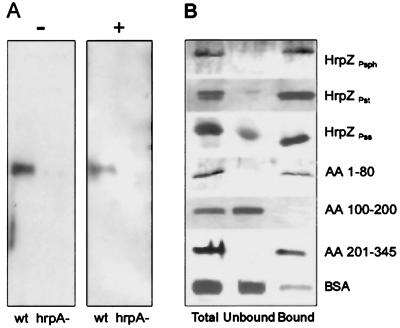
Hemolysis of sheep erythrocytes is an extreme measure of membrane-disintegrating activity. To pursue analysis of the predicted membrane-interacting activity of HrpZPsph further, we tested whether the protein would associate with membrane-constituting lipids. In contrast to control experiments performed with human annexin (29), HrpZPsph did not show Ca2+-dependent binding to phospholipids (not shown). However, the phospholipid mixtures used were of amorphous structure rather than ordered membranes. To study the interaction of secreted native HrpZPsph with lipid membranes, liposomes prepared from asolectin (6) were added to Psph grown in minimal medium. After 12 h of incubation, bacteria were pelleted, and the proteoliposomes were concentrated and purified by sucrose-density gradient centrifugation. Purified proteoliposomes were washed to eliminate sucrose and electrostatically bound proteins and were analyzed for HrpZPsph. Fig. 3A shows that HrpZPsph was found in the culture supernatant in the absence of liposomes and in the purified proteoliposomes. When a Psph hrpA mutant was treated identically, neither secretion nor membrane association of HrpZPsph was observed.
Figure 3.
HrpZPsph interacts with lipid membranes. (A) Psph race-6 wild-type (wt) and race-6 mutant hrpA− were grown in hrp-inducing minimal media (8) in the absence (−) or presence (+) of liposomes. Proteins prepared from the supernatant or proteoliposomes were separated and analyzed by SDS/PAGE immunoblotting with an anti-HrpZPsph antiserum. (B) TRANSIL beads coated with 1-hexadecanoyl-2-(cis-9-octadecenoyl)-sn-glycero-3-phosphocholine (POPC)/1-hexadecanoyl-2-(cis-9-octadecenoyl)-sn-glycero-3-phosphoethanolamine (POPE) (80:20) were incubated for 1 h with 1 μM of the proteins indicated (Total). After separation of lipid-bound (Bound) from unbound material, proteins were analyzed by SDS/PAGE and silver staining. HrpZPst, HrpZ from P. syringae pv. tomato DC3000; HrpZPss, HrpZ from P. syringae pv. syringae; AA 1–80, AA 100–200, AA 201–345, HrpZPsph fragment encompassing the N-terminal 80 amino acid (AA) residues, amino acids 100–200, or the C-terminal amino acids 201–345, respectively.
To verify membrane association of HrpZPsph further, we used silica beads coated with a single phospholipid bilayer (TRANSIL), which allow measurement of protein binding to membranes (31). Because the lipid molecules are not covalently linked to the support and are separated by an ultrathin layer of water molecules, this material has characteristics reminiscent of the lipid bilayer of biological membranes (38). When purified recombinant HrpZPsph was added to TRANSIL beads coated with POPC/POPE (80:20), approximately 80% of the protein applied was found to bind (Fig. 3B, Table 1). Interestingly, a mixture of phosphocholine alone esterified with various fatty acids yielded only 55% binding (Table 1). However, supplementing phosphocholine-coated beads carrying a neutral net charge with 2% of negatively charged 1,2-ditetradecanoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] increased binding to 95%.
Table 1.
Binding* of HrpZPsph to TRANSIL beads coated with single lipid membrane bilayers
| Membrane composition | Bound protein (% of total) |
|---|---|
| 1,2-diacyl-sn-glycero-3-phosphocholine | 55 ± 5 |
| POPC/POPE (80∶20) | 80 ± 5 |
| 1,2-di(trans-9-octadecenoyl)-sn-glycero-3- phosphocholine/1,2-ditetradecanoyl-sn- glycero-3-[phospho-rac-(1-glycerol)] (98∶2) | 95 ± 5 |
Binding of HrpZPsph to TRANSIL beads coated with single lipid membrane bilayers was performed as described in Materials and Methods. Total protein applied, lipid-bound, and unbound protein were separated by SDS/PAGE, and Coomassie-stained proteins were quantified by light densitometry.
Importantly, purified recombinant HrpZ from P. syringae pv. tomato or syringae bound to POPC/POPE (80:20) beads to a similar extent as HrpZPsph (Fig. 3B). To identify regions within HrpZPsph potentially involved in binding to membranes, we tested fragments of the protein. In contrast to the hydrophilic central core of the protein, both the hydrophobic N-terminal 80-mer fragment and the hydrophobic C-terminal 144-mer fragment bound to POPC/POPE (80:20) TRANSIL beads (Fig. 3B). Importantly, BSA, a major lipid and fatty acid carrier protein of the circulatory system (39), bound only very little to the TRANSIL beads. Thus, binding of HrpZ of different origin to lipid membranes is apparently specific. In addition, binding of 1 μM of different HrpZ suggests high affinity of the proteins to membranes.
HrpZPsph Has Ion Pore-Forming Activity.
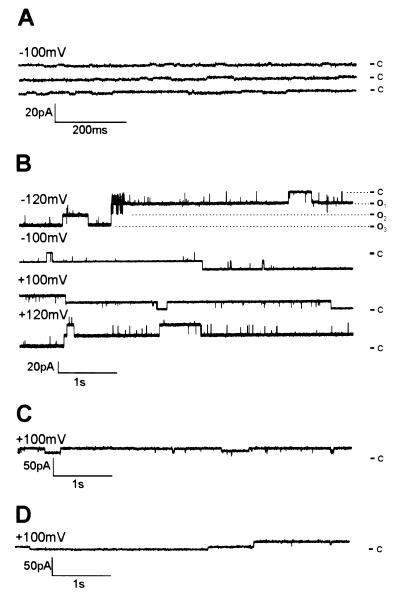
The predicted structural similarity between YopB and HrpZPsph as well as its association with membranes prompted an investigation of the formation of ion-conducting pores. We used the planar lipid bilayer technique, a protein-free membrane system widely used for isolation and characterization of membrane-active compounds (32, 40). His-tagged HrpZPsph was engineered for purification to apparent homogeneity by Ni-nitrilotriacetic acid agarose ligand affinity chromatography. Removal of contaminating proteins was crucial to these assays, in particular the abundant E. coli porins (41). When control E. coli proteins prepared by an identical procedure as His-tagged HrpZPsph were applied to bilayer membranes consisting of POPC/POPE (80:20), only marginal ion fluctuations were detectable (Fig. 4A). By contrast, addition of purified HrpZPsph to the cis compartment of lipid bilayers induced distinct current fluctuations (Fig. 4B). Events induced by HrpZPsph occurred without apparent lag phase and comprised rapid successions of open and closed states as well as prolonged phases of either state. HrpZPsph concentrations required to induce the consistent channel-like ion fluxes were 2 nM. Raising this concentration to 22 nM resulted in increased fluctuation frequency, which eventually caused bilayer instabilities. Control experiments showed that BSA did not cause pore formation, demonstrating specificity of the observed event (data not shown; ref. 32). However, addition of HrpZ protein encoded by the hrpZ genes of P. syringae pv. tomato (HrpZPst) or syringae (HrpZPss), respectively, evoked ion currents of very similar unitary conductance to those stimulated by HrpZPsph (Fig. 4 C and D).
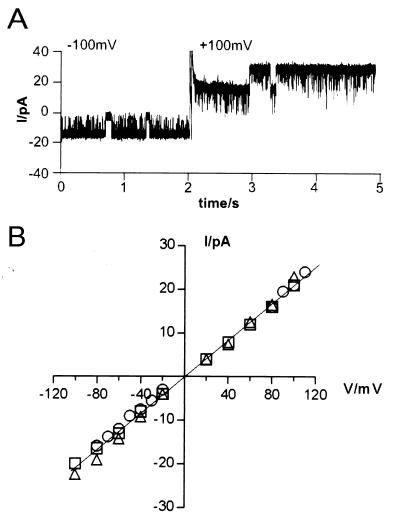
Figure 4.
HrpZPsph and related HrpZPst and HrpZPss trigger ion-current fluctuations in planar lipid bilayers. (A) Protein from E. coli contaminants was prepared as described in Materials and Methods; purified recombinant HrpZPsph (B), HrpZPst (C), or HrpZPss (D) was added to the cis-aqueous solution of the bilayer cuvette, and the induced current traces were recorded. In B, HrpZPsph-induced traces were recorded at different membrane potentials, as indicated. Electrolyte solutions (cis/trans) contained 100 mM KCl and 10 mM Hepes, pH 7.0. Dashed lines indicate different open states. c, closed, o1-o3, open states. Note different y axes in A–D.
Ion current fluctuation patterns induced by HrpZPsph did not alter significantly in response to changes of the membrane potential applied (from −120 mV to +120 mV; Fig. 4B). Experiments in which the membrane potential was switched from negative to positive voltage revealed bidirectional ion currents mediated by the HrpZPsph pore (Fig. 5A). Thus, the ion-conducting HrpZPsph pore does not constitute an ion rectifier. A current–voltage relationship of HrpZPshph-induced ion fluxes was established (Fig. 5B), facilitating determination of the pore unitary conductance as ΛK+ = 207 pS. Such a large ion-transport capacity is consistent with a proposed virulence function of HrpZPsph as a mediator of nutrient release from host plant cells. Ion channel-like pores mediate currents along the electrochemical gradient of a particular ion. Because cytoplasmic K+ and Cl− concentrations of plant cells are much higher than apoplastic concentrations of these ions, both may be potential substrates for such an ion-conducting structure. Therefore, we tested the ion specificity of the pore formed by HrpZPsph and found that cations K+, Na+ (Fig. 5B), and Ca2+ (not shown) permeated in a similar manner. However, when most of the KCl was substituted by K+-gluconate, for which the anion is considered ion-channel impermeable, no apparent change in HrpZPsph-induced ion conductivity was observed. Thus, monovalent anions such as Cl− may not pass the pore formed by HrpZPsph.
Figure 5.
Electrophysiological properties of HrpZPsph-induced currents. (A) HrpZPsph-induced currents do not exhibit rectifying properties. After addition of HrpZPsph to the cis compartment of the bilayer cuvette, ion currents were recorded at a membrane potential of −100 mV and after immediately switching the membrane potential to +100 mV after 2 sec of total recording. (B) Linear current–voltage relationship of HrpZPsph-induced ion fluxes. Current–voltage relationships of HrpZPsph-induced nonselective cation currents were established in three different electrolyte solutions: 100 mM KCl and 10 mM Hepes, pH 7.0 (○); 100 mM NaCl and 10 mM Hepes, pH 7.0 (▫); and 90 mM K+-gluconate, 10 mM KCl, and 10 mM Hepes, pH 7.0 (▵). The means of three recordings are given.
Discussion
The Psph harpin resembles the hrpZ gene products identified in P. syringae pvs. glycinea, syringae, or tomato (15). HrpZPsph secretion depended on functional hrp genes and on a functional bacterial type III secretion system (Fig. 2). As shown for AvrPto from P. syringae pv. tomato and AvrB from P. syringae pv. glycinea (3), export of HrpZPsph by the type III secretion system was achieved in the mammalian pathogen, Y. enterocolitica. However, whereas avr gene products are effector proteins assumed to be injected directly into host-plant cells (42, 43), our studies show that bacterial proteins that probably target the plant cell surface, such as HrpZPsph, are also export substrates of the type III pathway in Y. enterocolitica.
Like previous studies of hrpZ of other pathovars (18, 44), our research failed to provide unambiguous genetic evidence for a function of HrpZPsph in bacterial pathogenicity. This fact may be explained by the presence of other proteins functionally redundant to HrpZPsph, such as HrpW, of which a crosshybridizing gene was found also in Psph (18). Although hrpZ is not essential for bacterial pathogenicity, maintenance of the gene in the hrp regulon, evolutionary conservation of this and structurally related genes among numerous plant pathogens, as well as the absence of the gene product in saprophytic Pseudomonas fluorescens (7), strongly suggest that there is a significant but as-yet-undefined role for the encoded proteins in colonization of host plants.
Both native and recombinant HrpZPsph associated with lipid bilayer membranes (Fig. 3). Deletion experiments further showed that lipid binding is determined mainly by both termini of the protein, which harbor the most hydrophobic regions of this amphipathic molecule (Figs. 1 and 3B). For many membrane-interacting proteins, binding is mediated by ubiquitous membrane phosphoglycerolipids such as phosphatidic acid or phosphatidylserine [negatively charged lipids or neutral phosphatidylethanolamine (ref. 45 and references therein)]. Interestingly, association of HrpZPsph to membranes was increased strongly in the presence of negatively charged phosphoglycerolipids and phosphatidylethanolamine (Table 1), both of which are constituents of plant plasma membranes (46).
Furthermore, addition of HrpZPsph to protein-free lipid bilayers yielded a stable cation conductance, which is indicative of spontaneous self-organization of the protein into ion-conducting protein structures reminiscent of true ion channels (Fig. 4). This finding identified membranes as a physiological target for HrpZ proteins in addition to the plant cell wall (19). Using protein-free lipid bilayers rather than plant protoplasts to assess the membrane-integrating activity of HrpZPsph allowed distinction between the abilities of proteins to form ion pores themselves or to trigger receptor-mediated activation of ion fluxes. In tobacco, HrpZPss from P. syringae pv. syringae activates a K+/H+ exchange (19), but it is unknown whether this activation is mediated by specific receptors, by direct interaction of HrpZPss with ion channels, or by direct insertion of HrpZPss into membranes. The predicted structural similarity of HrpZPsph to Y. enterocolitica YopB led us to test whether HrpZPsph could functionally replace YopB, but expression of hrpZPsph in a yopB-deficient Y. enterocolitica strain failed to complement mutated yopB. Nevertheless, the electrophysiological properties of the ion-conducting pores formed by HrpZPsph were reminiscent of those formed by YopB, i.e., distinct levels of open states, a large unitary conductance, and poor ion selectivity (6). Interestingly, current fluctuations mediated by YopB alone differed significantly from those monitored after insertion of YopB and YopD into lipid bilayers (6). In addition, genetic and biochemical evidence (1) indicated that both Yop proteins may interact to form a functional pore structure. The failure of HrpZPsph in Yersinia complementation assays may therefore reflect its inability to interact with YopD. It is tempting to speculate that HrpZPsph, although able to form ion-conducting pores itself, may also interact with other bacterial proteins to establish a pore in the plasma membrane of host plant cells.
A direct consequence of HrpZPsph pore formation may be a rapid hyperpolarization of the plant plasma membrane resulting predominantly from K+ efflux. Interestingly, the HrpZPsph channel seems impermeable to another major charge carrier of the plant cytoplasm, Cl−, which may be because of the charge composition within the ion-conducting pore structure. Large unitary conductance and limited ion selectivity are also features reminiscent of bacterial homotrimeric porins (47). However, further investigations are required to elucidate the subunit structure of the HrpZPsph pore and, thus, possible similarities to bacterial porins.
The electrophysiological properties of the HrpZPsph pore are similar also to those of ion channels formed by toxins from plant pathogens, syringomycins, and syringopeptins from P. syringae pv. syringae and beticolins from the fungus Cercospora beticola (48, 49). This similarity raises the question whether HrpZPsph may act simply as a toxin during infection of host plants. However, in contrast to syringomycins, harpins seem not to be toxic directly to plant cells. Hypersensitive cell death observed after infiltration of HrpZPss into tobacco leaves was prevented completely by inhibitors of RNA and protein biosynthesis or ion channel antagonists and is thus caused by plant metabolic activities associated with programmed cell death (7, 18). More likely, pore formation benefits the bacterium by releasing cellular metabolites into the apoplast, thus providing an environment more advantageous to bacterial growth. Interestingly, a rapid efflux of sucrose was shown previously to accompany K+ efflux in both susceptible and resistant responses of Phaseolus vulgaris plants to P. syringae pv. syringae infection (50).
Besides compromising the integrity of the plant plasma membrane to enable nourishment of the attacking pathogen, another role of HrpZPsph during infection can be envisaged. HrpZPsph, like YopB, may constitute an element of the protein secretion/translocation apparatus, and the membrane-integrating activity of HrpZPsph may serve to facilitate delivery of virulence factors into host plant cells via the type III secretion/translocation machinery. This view is supported by studies that revealed HrpZPss to be essential for delivery of putative virulence factors from saprophytic P. fluorescens or E. coli expressing the P. syringae pv. syringae hrp regulon (42, 51). Because Psph hrpZ mutants were not impaired in pathogenicity, other functional pore-forming proteins may exist. Future studies may entail isolation and identification of such proteins by use of liposomes and lipid-coated TRANSIL beads. Furthermore, it will be crucial to determine the size of the HrpZPsph pore to identify substrates likely to be translocated into or out of plant cells through such a channel.
Acknowledgments
We thank Ulla Bonas (Martin-Luther-Universität, Halle, Germany), Dierk Scheel (Institut für Pflanzenbiochemie, Halle, Germany), and Dirk Nennstiel (Martin-Luther-Universität) for critical reading of the manuscript, Ulla Bonas for providing plasmid pDGUS, and Elina Roine and Martin Romantschuk (University of Helsinki, Finland) for the kind gift of Psph pilM-strain and P. syringae pv. tomato DC3000. We are grateful to Véronique Cabiaux (Université Libre de Bruxelles, Belgium) and Alan Collmer (Cornell University, Ithaca, New York) for providing freshly prepared liposomes and plasmid pSYH10, respectively. This work was supported by grants from the European Community (BIO-CT97–2244) (to J.L., G.T., C.S., A.T.), and by the U.K. Biotechnology and Biological Sciences Research Council (to J.W.M.).
Abbreviations
- POPC
1-hexadecanoyl-2-(cis-9-octadecenoyl)-sn-glycero-3-phosphocholine
- POPE
1-hexadecanoyl-2-(cis-9-octadecenoyl)-sn-glycero-3-phosphoethanolamine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF 268940).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011265298.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011265298
References
- 1.Cornelis G R. J Bacteriol. 1998;180:5495–5504. doi: 10.1128/jb.180.21.5495-5504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galan J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D M, Fouts D E, Collmer A, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12839–12843. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Håkansson S, Schesser K, Persson C, Galyov E E, Rosquist R, Homble F, Wolf-Watz H. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 5.Neyt C, Cornelis G R. Mol Microbiol. 1999;33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 6.Tardy F, Homble F, Neyt C, Wattiez R, Cornelis G R, Ruysschaert J M, Cabiaux V. EMBO J. 1999;18:6793–6799. doi: 10.1093/emboj/18.23.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He S Y, Huang H-C, Collmer A. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 8.Huynh T V, Dahlbeck D, Staskawicz B J. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 9.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H C, Hutcheson S W, Panopoulos N J, Van Gijsegem F. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang H C, He S Y, Bauer D, Collmer A. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gough C L, Genin S, Zischek C, Boucher C A. Mol Plant–Microbe Interact. 1992;5:384–389. doi: 10.1094/mpmi-5-384. [DOI] [PubMed] [Google Scholar]
- 12.Fenselau S, Balbo I, Bonas U. Mol Plant–Microbe Interact. 1992;5:390–396. doi: 10.1094/mpmi-5-390. [DOI] [PubMed] [Google Scholar]
- 13.Kjemtrup S, Nimchuk Z, Dangl J L. Curr Opin Microbiol. 2000;3:73–78. doi: 10.1016/s1369-5274(99)00054-5. [DOI] [PubMed] [Google Scholar]
- 14.Wei Z M, Laby R J, Zumoff C H, Bauer D W, He S Y, Collmer A, Beer S V. Science. 1992;257:85–88. doi: 10.1126/science.1621099. [DOI] [PubMed] [Google Scholar]
- 15.Preston G, Huang H C, He S Y, Collmer A. Mol Plant–Microbe Interact. 1995;8:717–732. doi: 10.1094/mpmi-8-0717. [DOI] [PubMed] [Google Scholar]
- 16.Arlat M, Van Gijsegem F, Huet J-C, Pernollet J-C, Boucher C A. EMBO J. 1994;13:543–553. doi: 10.1002/j.1460-2075.1994.tb06292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer D W, Wei Z M, Beer S V, Collmer A. Mol Plant–Microbe Interact. 1995;8:484–491. doi: 10.1094/mpmi-8-0484. [DOI] [PubMed] [Google Scholar]
- 18.Charkowski A O, Alfano J R, Preston G, Yuan J, He S Y, Collmer A. J Bacteriol. 1998;180:5211–5217. doi: 10.1128/jb.180.19.5211-5217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyos M E, Stanley C W, He S Y, Pike S, Pu X A, Novacky A. Mol Plant–Microbe Interact. 1996;9:608–616. [Google Scholar]
- 20.Roine E, Nunn D N, Paulin L, Romantschuk M. J Bacteriol. 1996;178:410–417. doi: 10.1128/jb.178.2.410-417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E L, Kalkkinen N, Romantschuk M, He S Y. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huguet E, Hahn K, Wengelnik K, Bonas U. Mol Microbiol. 1998;29:1379–1390. doi: 10.1046/j.1365-2958.1998.01019.x. [DOI] [PubMed] [Google Scholar]
- 23.Boland A, Cornelis G R. Infect Immun. 1998;66:1878–1884. doi: 10.1128/iai.66.5.1878-1884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woestyn S, Allaoui A, Wattiau P, Cornelis G R. J Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tampakaki N, Hatziloukas E, Grimm C, Panopoulos N. In: Pseudomonas syringae Pathovars and Related Pathogens. Rudolph K, Burr T J, Mansfield J W, Stead D, Vivian A, Von Kietzell J, editors. Vol. 9. Dordrecht, The Netherlands: Kluwer; 1997. pp. 392–396. [Google Scholar]
- 26.Michiels T, Wattiau P, Brasseur R, Ruysschaert J M, Cornelis G. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson R W, Athanassopoulos E, Tsiamis G, Mansfield J W, Sesma A, Arnold D L, Gibbon M J, Murillo J, Taylor J D, Vivian A. Proc Natl Acad Sci USA. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clos J, Brandau S. Protein Expression Purif. 1994;5:133–137. doi: 10.1006/prep.1994.1020. [DOI] [PubMed] [Google Scholar]
- 29.Tait J F, Gibson D, Fujikawa K. J Biol Chem. 1989;264:7944–7959. [PubMed] [Google Scholar]
- 30.Rossier O, Wengelnik K, Hahn K, Bonas U. Proc Natl Acad Sci USA. 1999;96:9368–9373. doi: 10.1073/pnas.96.16.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz A A P, Schleiff E, Röhring C, Loidl-Stahlhofen A, Vergeres G. Anal Biochem. 1999;268:343–353. doi: 10.1006/abio.1998.3080. [DOI] [PubMed] [Google Scholar]
- 32.Klüsener B, Weiler E W. FEBS Lett. 1999;459:263–269. doi: 10.1016/s0014-5793(99)01261-2. [DOI] [PubMed] [Google Scholar]
- 33.Mansfield J, Jenner C, Hockenhull R, Bennett M A, Stewart R. Mol Plant–Microbe Interact. 1994;7:726–739. doi: 10.1094/mpmi-7-0726. [DOI] [PubMed] [Google Scholar]
- 34.Huang H C, Lin R H, Chang C J, Collmer A, Deng W L. Mol Plant–Microbe Interact. 1995;8:733–746. doi: 10.1094/mpmi-8-0733. [DOI] [PubMed] [Google Scholar]
- 35.Eisenberg D, Schwarz E, Komarony M, Wall R. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 36.Wei W, Plovanich-Jones A, Deng W L, Qin Q L, Collmer A, Huang H C, He S Y. Proc Natl Acad Sci USA. 2000;97:2247–2252. doi: 10.1073/pnas.040570097. . (First Published February 18, 2000; 10.1073/pnas.040570097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hobohm U, Sander C. J Mol Biol. 1995;251:390–399. doi: 10.1006/jmbi.1995.0442. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt, J., Danner, B. & Bayerl, T. M. (2001) Langmuir16, in press.
- 39.Kragh-Hansen U. Dan Med Bull. 1990;37:57–84. [PubMed] [Google Scholar]
- 40.Mirzabekov T A, Silberstein A Y, Kagan B L. Methods Enzymol. 1999;294:661–674. doi: 10.1016/s0076-6879(99)94038-7. [DOI] [PubMed] [Google Scholar]
- 41.Pratt L A, Hsing W, Gibson K E, Silhavy T J. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- 42.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 44.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thevissen K, Cammue B P A, Lemaire K, Winderickx J, Dickson R C, Lester R L, Ferket K K A, Van Even F, Parret A H A, Broekaert W F. Proc Natl Acad Sci USA. 2000;97:9531–9536. doi: 10.1073/pnas.160077797. . (First Published August 8, 2000; 10.1073/pnas.160077797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staehelin L A, Newcomb E H. In: Biochemistry and Molecular Biology of Plants. Buchanan B B, Gruissem W, Jones R L, editors. Vol. 1. Rockville, MD: Am. Soc. of Plant Physiologists; 2000. pp. 2–50. [Google Scholar]
- 47.Phale P S, Philippsen A, Kiefhaber T, Koebnik R, Phale V P, Schirmer T, Rosenbusch J P. Biochemistry. 1998;37:15663–15670. doi: 10.1021/bi981215c. [DOI] [PubMed] [Google Scholar]
- 48.Bender C L, Alarcon-Chaidez F, Gross D C. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goudet C, Milat M-L, Sentenac H, Thibaud J-B. Mol Plant–Microbe Interact. 2000;13:203–209. doi: 10.1094/MPMI.2000.13.2.203. [DOI] [PubMed] [Google Scholar]
- 50.Atkinson M M, Baker C J. Phytopathology. 1987;77:1573–1578. [Google Scholar]
- 51.Alfano J R, Bauer D W, Milos T M, Collmer A. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]