Abstract
We present a common allosteric mechanism for control of inflammatory and apoptotic caspases. Highly specific thiol-containing inhibitors of the human inflammatory caspase-1 were identified by using disulfide trapping, a method for site-directed small-molecule discovery. These compounds became trapped by forming a disulfide bond with a cysteine residue in the cavity at the dimer interface ≈15 Å away from the active site. Mutational and structural analysis uncovered a linear circuit of functional residues that runs from one active site through the allosteric cavity and into the second active site. Kinetic analysis revealed robust positive cooperativity not seen in other endopeptidases. Recently, disulfide trapping identified a similar small-molecule site and allosteric transition in the apoptotic caspase-7 that shares only a 23% sequence identity with caspase-1. Together, these studies show a general small-molecule-binding site for functionally reversing the zymogen activation of caspases and suggest a common regulatory site for the allosteric control of inflammation and apoptosis.
Keywords: procaspase, allosteric regulation, apoptosis, fragment-based discovery, inflammation
Caspases are dimeric thiol endopeptidases that cleave specific proteins after aspartic acid residues and drive apoptosis or inflammation (for review, see refs. 1 and 2). Although of considerable medical interest, the active sites of caspases have been very difficult to target with drug-like compounds owing to the enzyme’s preference for negatively charged chemotypes with electrophilic warheads that engage the catalytic cysteine (3).
In past work to develop alternative small-molecule inhibitors to the active site of the apoptotic caspase-7, we used a fragment-based discovery approach called disulfide trapping. This is a site-directed small-molecule-capture approach in which natural or engineered cysteines on the surface of the protein are screened with a library of disulfide-containing small molecules in a redox buffer (4). This approach has been broadly applied in fragment-based drug discovery for both enzymes and challenging protein–protein interface targets (for review see ref. 5). Although the active-site cysteine of caspase-7 failed to yield strong hits from a library of ≈10,000 disulfide-containing compounds, several strong inhibitors were found for another cysteine in a cavity at the dimer interface.
Procaspase-7, the inactive precursor or zymogen, is known to undergo a large structural transition after auto- or transproteolysis. This transition results in cleavage of the N-terminal 23 residues and an internal cut that yields two large and two small subunits of mature caspase-7. Structural studies of caspase-7 containing the disulfide-trapped inhibitors at the dimer-interface cavity showed that the compounds reversed this structural transition and induced a conformation virtually identical to the zymogen form (6).
We wondered whether other caspases could be inhibited by a similar mechanism. The class of human inflammatory caspases-1, -4, and -5 is involved in driving innate immune responses after pathogen stimulation and has been of considerable interest for basic and medicinal purposes (7, 8). Although caspase-1 shares only a 20–30% amino acid sequence identity with the apoptotic caspases, it too contains a cavity at the dimer interface. Caspase-1 lacks a cysteine in the same position as caspase-7 but contains a nearby cysteine in the central cavity, Cys-331. Using this site as an anchor for disulfide trapping, we sought to identify small molecules binding at the dimer interface that can be used to test whether allosteric regulation is common among caspases. This information would provide us with an opportunity for drug discovery that circumvents the challenges imposed by the requirements for active-site ligands and may help elucidate important structure/function relationships in caspases that relate to their evolution and regulation.
Results
Disulfide Trapping and Structural Analysis of Allosteric Inhibitors.
We screened a library of ≈8,000 thiol-containing compounds by mass spectrometry and found compounds that conjugated to Cys-331 of caspase-1 and inhibited the activity of the enzyme. The screen identified 14 neutral compounds that conjugated exclusively to Cys-331 to >30%, a hit rate of ≈0.2%. The conjugation and inhibition was fully reversed in the presence of higher concentrations of reducing agent, such as β-mercaptoethanol [β-ME] and in the caspase-1 mutant C331A, reflecting the importance of the disulfide conjugate for inhibition.
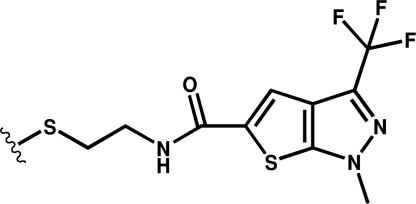
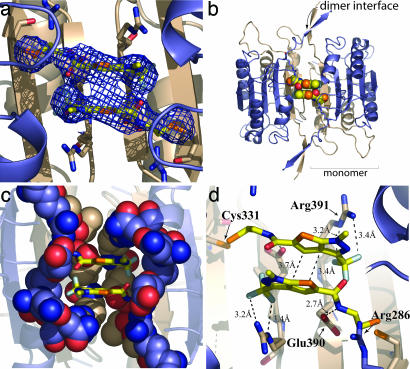
An x-ray structure of caspase-1 in complex with one of these compounds (Compound 34, Scheme 1), a thienopyrazole, was determined to 3.3-Å resolution (see Table 1). Despite the modest resolution, there is strong electron density for Compound 34 (Fig. 1a) and for residues that connect the allosteric and active-site regions. The refined structure shows two molecules of Compound 34 bound symmetrically at the dimer interface (Fig. 1 b and c). The trapped compounds appear close enough for van der Waals and aromatic stacking interactions with each other. The molecules bind in a transorientation where each molecule is tethered at Cys-331 in one subunit but makes most of its noncovalent interactions with the residues of the neighboring subunit. The triflouromethyl group of Compound 34 is packed tightly against the side chain of Arg-391 and, presumably, stabilized by polar interactions. The pyrazole is tightly packed against the Leu-258 side chain located in the large subunit of the adjacent dimer. The rim of the binding pocket is composed of large-subunit residues Gln-257, Glu-241, and Arg-286, and the bottom of the pocket is lined by Thr-288, Glu-390, and Arg-391 from the small subunit (Fig. 1c). A close interaction is seen between the amide linker portion of Compound 34 and the carboxylate of Glu-390 in the small subunit of the monomer (Fig. 1d).
Scheme 1.
Compound 34.
Table 1.
Crystallographic data and refinement statistics
| Mutation | Cys-285, -362, -364, -397 → Ala | WT | Arg-286 → Ala | Glu-390 → Ala | Glu-390 → Ala Arg-286 3 Ala |
|---|---|---|---|---|---|
| PDB ID code | 2FQQ | 2FQR | 2FQS | 2FQU | 2FQV |
| Space group | P3221 | P43212 | P43212 | P43212 | P43212 |
| Cell constants | a = b = 71.04 Å, c = 117.76 Å | a = b = 63.3 Å, c = 161.7 Å | a = b = 62.7 Å, c = 160.0 Å | a = b = 63.3 Å, c = 160.9 Å | a= b = 63.2 Å, c = 142.2 Å |
| X-ray source | Rigaku RU-3R | Rigaku RU-3R | Rigaku RU-3R | Rigaku RU-3R | Rigaku RU-3R |
| Wavelength, Å | 1.54 | 1.54 | 1.54 | 1.54 | 1.54 |
| Resolution, Å* | 20–3.3 (3.41–3.30) | 20–1.8 (1.86–1.80) | 20–2.2 (2.28–2.20) | 20–2.1 (2.18–2.10) | 20–1.9 (1.97–1.90) |
| No. of observations | 11,608 | 109,228 | 61,759 | 77,477 | 73,883 |
| No. of reflections | 5,490 | 31,366 | 16,955 | 19,871 | 23,540 |
| Completeness, %* | 99.3 (99.8) | 97.9 (83.7) | 99.9 (99.8) | 99.9 (99.9) | 91.7 (96.3) |
| Mean l/(σl) | 14.4 (1.5) | 5.6 (1.6) | 5.2 (1.7) | 6.1 (1.2) | 4.2 (2.0) |
| Rmerge on l† | 0.125 (0.495) | 0.064 (0.370) | 0.073 (0.403) | 0.078 (0.377) | 0.093 (0.343) |
| Cutoff criteria | I < −3σ(I) | I < −3σ(I) | I < −3σ(I) | I < −3σ(I) | I < −3σ(I) |
| Model and refinement statistics | |||||
| Resolution range, Å | 20–3.3 | 20–1.8 | 20–2.2 | 20–2.1 | 20–1.9 |
| No. of reflections‡ | 5,225 (247) | 28,476 (1,508) | 15,986 (855) | 18,692 (1,009) | 20,408 (1,109) |
| Completeness, % | 99.5 | 95.6 | 99.3 | 99.1 | 91.4 |
| Cutoff criterion | |F| > 0.0 | |F| > 0.0 | |F| > 0.0 | |F| > 0.0 | |F| > 0.0 |
| No. of residues | 235 | 256 | 256 | 258 | 250 |
| No. of water molecules | 18 | 228 | 147 | 160 | 183 |
| rmsd bond lengths, Å | 0.006 | 0.006 | 0.006 | 0.005 | 0.006 |
| rmsd bond angles, ° | 1.119 | 1.301 | 1.252 | 0.822 | 0.899 |
| Luzzati error, Å | 0.667 | 0.251 | 0.366 | 0.319 | 0.279 |
| Correlation factor§ | 0.792 | 0.935 | 0.904 | 0.924 | 0.915 |
| Rcryst¶ | 24.38 | 20.84 | 23.27 | 22.68 | 22.34 |
| Rfree | 28.02 | 23.57 | 27.01 | 25.85 | 24.91 |
| Ramachandran plot statistics‖ | |||||
| Most favored | 183 (88.4) | 202 (90.2) | 205 (91.5) | 204 (90.3) | 197 (89.5) |
| Additional allowed | 24 (11.6) | 21 (9.4) | 18 (8.0) | 22 (9.7) | 22 (10.0) |
| Generously allowed | 0 (0) | 1 (0.4) | 1 (0.4) | 0 (0.0) | 1 (0.5) |
| Disallowed | 0 (0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Overall G factor** | 0.2 | 0.2 | 0.2 | 0.2 | 0.1 |
rmsd, rms deviation.
*Numbers in parentheses indicate high-resolution shells.
†Rmerge = ΣhklΣi|I(hkl)i − 〈I(hkl)〉|/ΣhklΣi〈I(hkl)i〉.
‡Numbers in parentheses indicate the numbers of reflections used to calculate the Rfree factor.
§Correlation factor between the structure factors and the model as calculated by sfcheck.
¶Rcryst = Σhkl|Fo(hkl) − Fc(hkl)|/Σhkl|Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
‖Computed with procheck (27). Numbers in parentheses are percentages.
**Overall G factor is a measure of the overall normality of the structure and is obtained from an average of all the different G factors for each residue in the structure. It is essentially a log-odds score based on the observed distributions of these stereochemical parameters (27).
Fig. 1.
Structure of an allosteric inhibitor bound to caspase-1. (a) 2Fo − Fc electron density for Compound 34 used for compound model building, contoured at 1σ, is shown as a blue mesh at the dimer interface (PDB ID code 2FQQ). (b) Two molecules of Compound 34 are shown as spheres in the central cavity at the dimer interface of caspase-1. (c) Residues involved in forming the binding pocket for Compound 34 are shown as spheres. Residues from the large subunit (Glu-241, Gln-257, and Arg-286) are colored blue, and residues from the small subunit (Thr-388, Glu-390, and Arg-391) are colored tan. (d) Residues likely involved in the mechanism of inhibition of caspase-1 by allosteric compounds are displayed. Arg-286 adjacent to the catalytic Cys-285 is located >12 Å from Glu-390, to which it is salt-bridged in the active conformation. The amide nitrogen of the linker group of Compound 34 is within a hydrogen-bonding distance of the Glu-390 carboxylate. Arg-391 is shown to indicate the boundary of the binding pocket.
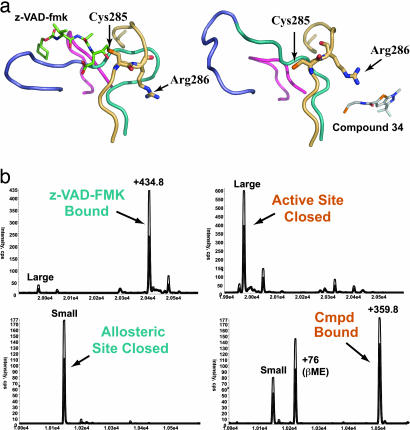
The conjugation with Compound 34 produced three functionally important outcomes when compared with the active form of caspase-1 labeled with an active-site inhibitor (z-VAD-FMK): (i) the catalytic Cys-285 rotated ≈5 Å away from its normal position for hydrolyzing peptide bonds; (ii) the substrate-binding loops collapsed and were no longer positioned to interact productively with a substrate; and (iii) the side chain of Arg-286 rotated toward the substrate-binding cleft but retained a high degree of flexibility, as evidenced by higher B factors (B factor of 52 for all protein atoms versus 95 for the side chain of Arg-286) (Fig. 2a). Given the dramatic and concerted changes that occur during ligand binding at the active site versus the allosteric site, we examined whether these changes were mutually exclusive. Reacting the active-site Cys-285 with the irreversible active-site inhibitor z-VAD-FMK in the large subunit blocked subsequent reaction with Compound 34 on the small subunit (Fig. 2b). Conversely, labeling the enzyme with Compound 34 at Cys-331 prevented labeling by the active-site inhibitor. Thus, the sites appear to work in a reciprocal and mutually exclusive manner.
Fig. 2.
Comparison of the caspase-1 active site in active- and allosteric-site-bound structures. (a) Four loop regions that change position between the active- and allosteric site-bound conformations are shown as ribbons. The active-site residue Cys-285 and the adjacent residue Arg-286 are displayed as sticks. (Left) The active-site-bound structure is shown in complex with the active-site inhibitor z-VAD-FMK (green sticks). (Right) The allosteric-site-bound structure in complex with the allosteric inhibitor Compound 34. (b) Mass-spectrometry results of competitive labeling of the active and allosteric sites with z-VAD-FMK and Compound 34 to large and small subunits, respectively. Either inhibitor was added first to the enzyme for 30 min, followed by addition of the second inhibitor and a 30-min incubation.
Kinetic and Structural Analysis of Allosteric Site Mutants.
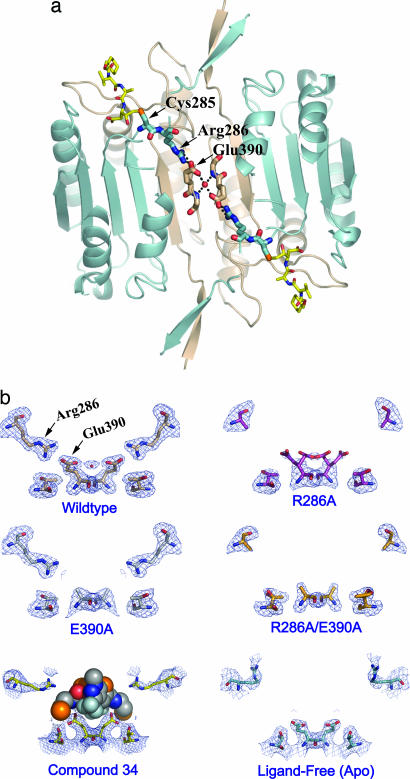
Inspection of the residues that connect the allosteric and active sites reveals a network of residues involved in hydrogen-bonding interactions (Fig. 3a). One of the most conspicuous interactions is a salt bridge between Glu-390 and Arg-286 present in the active form of caspase-1 (9). Compound 34 prevents the formation of this salt bridge and the associated conformational transition (Fig. 1d). Site-directed mutagenesis was used to probe the importance of the Arg-286–Glu-390 salt bridge for enzyme activity. Mutations of Arg-286 to Ala and Glu-390 to Ala separately resulted in a 400- and 460-fold reduction in catalytic efficiency (kcat/KM), respectively (Table 2). The reductions in kcat/KM result from both reductions of kcat and increases in KM, as shown in Table 2. The R286A/E390A double mutant caused only an additional 10-fold decrease in kcat/KM, supporting the notion that these two residues work together to help stabilize the transition state of the reaction. The effects of these mutations 15 Å away from the active site are as dramatic, for example, as those observed for mutating catalytic residues in serine or thiol hydrolases directly involved in oxyanion stabilization (10, 11). The fact that the KM values are very different between the wild-type and mutant enzymes indicates that the reduced activity is not due to destabilization creating mostly misfolded protein with a trace of wild-type enzyme (12). X-ray structures of the mutant enzymes labeled with the active-site inhibitor z-VAD-FMK were solved to >2.2-Å resolution (Table 1). The structures showed that the mutant enzymes were virtually identical to the wild-type protein but simply lacked the key salt-bridge interactions in the central cavity (Fig. 3 a and b).
Fig. 3.
Structural analysis of mutations in the allosteric circuit of caspase-1. (a) A network of interactions across the dimer interface of caspase-1 in the z-VAD-FMK-inhibited protein. The inhibitor is shown as yellow sticks in the upper left and lower right. The active-site Cys-285 and Arg-286 are displayed as blue sticks, Glu-390 at the dimer interface as tan sticks, and a water molecule mediating the interaction between the two Glu residues is shown as a red sphere. (b) The x-ray crystal structure of each allosteric-circuit mutant was determined in the presence of the active-site inhibitor z-VAD-FMK. All structures (PDB ID codes 2FQS, R286A; 2FQU, E390A; and 2FQV, R286A/E390A) adopted a dimeric structure very similar to that of the wild-type enzyme in complex with an active-site inhibitor (PDB ID code 2FQR). No significant conformational changes were observed in the enzymes except for those involving residues in the allosteric circuit. The 2Fo − Fc electron density for residues Arg-286, Glu-390, and Thr-388 is displayed. (Bottom Left) The position of Compound 34 displayed as spheres. (Bottom Right) The ligand-free (apo) conformation of caspase-1.
Table 2.
Kinetic analysis of caspase-1 salt-bridge mutants
| Enzyme | KM | kcat, sec−1 | kcat/KM, M−1·sec−1 | Ratiokcat/KM | nHILL |
|---|---|---|---|---|---|
| WT | 4.0 | 0.77 | 1.9 × 105 | 1 | 1.5 |
| R286A | 370 | 0.17 | 4.7 × 102 | 400 | 1.6 |
| E390A | 180 | 0.07 | 4.1 × 102 | 460 | 1.0 |
| R286A/E390A | 600 | 0.03 | 4.7 × 10 | 4,000 | ND |
ND, not determined.
Given that conformational transitions clearly propagate between the central cavity and the active sites of caspase-1, we tested to see whether the binding of substrate at one active site positively enhanced catalysis at the second active site. Indeed, caspase-1 showed strong positive cooperativity at both low and high concentrations of enzyme, with a Hill constant of 1.5 (Table 2; and see Fig. 5, which is published as supporting information on the PNAS web site). Interestingly, the Hill coefficient reduced to 1.0 in the E390A mutant, suggesting that the interaction across the dimer interface between Glu-390 and the central water is a key element of communication between the two active sites. The Hill coefficient for wild-type caspase-1 is comparable to that seen in other positively cooperative dimeric enzymes, such as phosphorylase (13–15). This unique observation of positive cooperativity for an endopeptidase could provide an additional level of control, in that caspase-1 would be less active at low substrate concentrations and more active at high concentrations when concentrated on inflammasomes (16).
Discussion
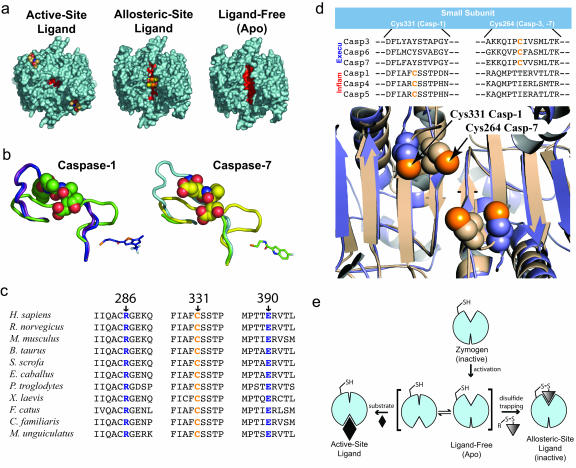
Our screen has identified small-molecular-mass compounds that bind specifically to the allosteric site of caspase-1 and are potent inhibitors of enzyme activity. The mechanism of action of these compounds appears to be to disrupt a network of interactions bridging the active site to the dimer interface. This mechanism is also observed in the allosteric inhibition of caspase-7, suggesting that common structural features have been conserved in these enzymes despite divergence in sequence identity and biological function. The conservation of mechanism is reflected in the structures of inactive conformations of caspases that include ligand-free and zymogen structures. In a comparison of the global structure of caspase-1 in the allosterically inhibited and ligand-free conformations, the allosteric compounds clearly trap the enzyme in the naturally occurring inactive conformation seen in the ligand-free enzyme (Fig. 4a). This finding suggests that the inactive-loop conformation may be more energetically favorable and may be important for reasons such as protecting the active-site cysteine from irreversible oxidation or to allow for a greater degree of regulation over the activity of the enzyme by creating a conformational barrier to catalysis. An overlay of the allosterically inhibited structures of caspase-1 and caspase-7 showed that conjugation of their respective allosteric inhibitors stabilizes virtually identical, zymogen-like conformations (Fig. 4b).
Fig. 4.
A conserved mechanism for dynamic activation of caspases. (a) Comparison of the surface of three structures of caspase-1. The active-site ligand-bound structure of caspase-1 in complex with the active-site inhibitor z-VAD-FMK, the allosteric-site ligand-bound structure of caspase-1 complexed with Compound 34, and the ligand-free apo enzyme structure as reported earlier are shown in aqua (PDB ID code 1SC1). The allosteric cavity between the two monomers is indicated in red. (b) Alignment of three caspase-1 (Left) and three caspase-7 (Right) structures. The ribbon representation of the protein backbone is shown for residues 330–348 for caspase-1, 222–239 for caspase-7 (330–348 for procaspase-7). (Left) Three conformations are shown: allosteric-site inhibitor-bound (blue; PDB ID code 2FQQ), ligand-free (purple; PDB ID code 1SC1), and active-site inhibitor-bound (green; PDB ID code 2FQR). (Right) Three conformations are shown: allosteric-site inhibitor-bound (light green; PDB ID code 1SHL), ligand-free procaspase-7 (light blue; PDB ID code 1GQF), and active-site inhibitor-bound (yellow; PDB ID code 1F1J). The allosteric ligand in caspase-1 (Compound 34) is shown in blue sticks, and the allosteric ligand in caspase-7 (FICA) is shown in green sticks. The active-site inhibitors are shown as spheres. Two rearrangements are observed in these loops when allosteric-site ligands bind that mimic the zymogen or inactive conformation. The first half of the loop moves out of the allosteric pocket, and the second half of the loop moves up into the substrate-binding groove to prevent substrate binding. The upper portion of the caspase-7 loop region in the allosteric structure is not well ordered. (c) Conservation of Arg-286, Cys-331, and Glu-390 in caspase-1 homologs. (d) Structural conservation of a cysteine in the allosteric regulatory region of inflammatory and executioner caspases. (e) Model for the dynamic activation of executioner and inflammatory caspases. After zymogen activation, caspases are in equilibrium between “zymogen-like” and “active-like” conformations. The ligand-free enzyme is subject to trapping into either the active or allosteric conformation by the binding of a site-specific ligand. For simplicity, we show caspase in a monomeric form.
Common to both mechanisms is the severing of key interactions made by Arg-286 conserved in all of the human inflammatory and executioner caspases. In the inflammatory caspases (Group I when classified by substrate specificity) (17), Arg-286, Glu-390 to which it is salt-bridged, and Cys-331 to which the allosteric compounds are anchored are completely conserved. In fact, a broader sequence alignment that includes all caspase-1 homologs shows complete conservation of Arg-286, Glu-390, and Cys-331 (Fig. 4c). There is a similar salt bridge conserved in the Group-II caspases (caspases with DEXD substrate-specificity, including the executioner caspases-3 and -7); however, the interaction is with Glu-147 that comes from the same large subunit. The configuration of the salt bridge is consistent among the caspases when grouped by substrate specificity (17). In addition, the inflammatory and executioner caspases conserve a thiol near the dimer interface. Although this cysteine arises from very different sites in the gene sequence because of placement on different β-strands, the three-dimensional positions of Cys-331 and Cys-264 are nearly identical (Fig. 4d). Whether there is functional significance to the conservation of a thiol in this site is unknown; however, some reports indicate that an exosite thiol exists in caspase-3 that may be regulated by S-nitrosylation (18). In the Group-III caspases that include the initiator caspases, no salt bridge is conserved in the sequence. The apparent lack of a salt bridge in these caspases suggests that they may be regulated differently. In fact, caspase-9, a Group-III caspase, is inhibited by the bir3 domain of XIAP, a naturally occurring regulator that binds at the dimer interface by displacing one of the monomers, in a manner not known to happen with inhibitors of the other caspases.
The combined data from structures of natural and allosterically inhibited forms of caspase-1 and -7 support a dynamic zymogen-activation model for the inflammatory and executioner caspases (Fig. 4e). After zymogen activation, the mature enzyme exists in a dynamic equilibrium between active and inactive states. Binding of substrate drives the equilibrium to the active form, which explains the positive cooperativity seen in caspase-1. Binding of the allosteric inhibitor traps the ligand-free zymogen-like form, which explains the mutual exclusivity we observe in both caspase-1 and -7.
Identifying and characterizing such “orphan” allosteric sites can provide opportunities for drug discovery. This allosteric site in caspases is less well conserved in detail, offering opportunities for improved selectivity. Moreover, the site binds neutral ligands and does not have the same requirements for a highly charged substrate mimic that impairs cellular transport. Lastly, given the common mechanism for caspase-1 and -7, it is tempting to search for a natural allosteric ligand that could bind and provide a necessary safeguard against spurious and catastrophic caspase activation.
Materials and Methods
Caspase-1 Expression and Mutagenesis.
Recombinant caspase-1 was prepared by expression in Escherichia coli as insoluble inclusion bodies followed by refolding, as described in refs. 9 and 19. Mutagenesis was done by using the QuikChange Site-Directed Mutagenesis kit from Stratagene. For multiple mutations in the same plasmid construct, all sets of primers were included in a single QuikChange reaction with an extension time of 16 min for 18 cycles. This procedure produced ≈1 in 4 multiply mutated clones.
Disulfide Trapping.
Disulfide trapping was performed according to standard procedures documented in several publications (20). Briefly, purified caspase-1 (C285,362,364,397A) was diluted to 5 μM in a buffer containing 50 mM Hepes, 50 mM KCl, and 100 μM β-ME and was incubated at room temperature for 1 h with pools of disulfide-containing compounds in 96-well plates. After the equilibration period, reaction mixtures were analyzed by high-throughput mass spectrometry using a CIT Analytics Autosampler and a QStar Electrospray mass spectrometer (Applied Biosystems). Hits were identified by comparing the molecular masses of covalent complexes bound to the protein’s small subunit to the molecular masses of compounds in the pool. The hits were confirmed by performing a similar reaction with individual compounds. The hits identified by the screen were assayed for caspase-1 inhibition by using a conventional fluorescence-based assay containing caspase-1 at a 10 nM concentration in 50 mM Hepes, 50 mM KCl, 200 mM NaCl, 100 μM β-ME, 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, and the fluorescent substrate Ac-WEHD-AFC. For determination of disulfide dependence, the concentration of the reducing agent was increased by adding β-ME to 10 mM final concentration in the reaction.
To further validate the quality of the hits, we compared percent covalent labeling and percent inhibition. For this determination, a disulfide-trapping reaction was performed for 1 h in 50 mM Hepes, pH 7.5, 50 mM KCl, 100 μM β-ME, and 50 μM of the discrete compound by using 5 μM caspase-1. After 1 h, 5 μl of the mixture was transferred to the fluorescence assay buffer and assayed for activity, and the remainder of the mixture was analyzed by mass spectrometry. In reaction with 51 compounds, a linear correlation between inhibition and labeling was observed. The compounds that did not show this relationship were eliminated from further analysis.
Compound Synthesis and Protein Labeling.
Compounds used for more detailed analysis were resynthesized in 100-mg quantity by using a standard amide-coupling reaction. First, a methionine-sulfonyl-containing linker was coupled to the acid or base by amide coupling, followed by displacement of the MTS group by the free thiol cysteamine. The disulfide-containing compounds were purified by reverse-phase HPLC, lyophilized, and solubilized at 100 mM in DMSO.
For labeling of caspase-1 with Compound 34 (C285,362,364,397A) for crystallization, the protein was buffer-exchanged by using prepacked NAP-25 gel filtration columns into 50 mM Hepes, pH 8.0, 50 mM KCl, 200 mM NaCl, and 200 μM β-ME. Approximately 200 μM compound was added to the protein mixture and reacted overnight at 4°C. After incubation, precipitate was removed by centrifugation, and the extent of protein labeling was determined by mass spectrometry. Protein labeled to homogeneity was buffer-exchanged again into 50 mM Hepes, pH 7.5, 50 mM KCl, and 200 mM NaCl and concentrated by using Amicon spin concentrators. Concentrated protein (≈10 mg/ml) was kept on ice or frozen in 20-μl aliquots at −80°C. Crystal screens were set up with Hampton Research HT screens in 24-well plates, and caspase-1-inhibitor complexes were crystallized by hanging-drop vapor diffusion.
For crystallization of wild-type and mutant caspases (R286A, E390A, and R286A/E390A) the protein was refolded around the covalent tripeptide inhibitor z-VAD-FMK (Calbiochem). The purified complexes were analyzed by mass spectrometry to confirm the correct mass, concentrated, and used in Hampton HT screens (see below for crystal growth conditions).
Crystallization, Data Collection, and Structure Determination.
Crystals of caspase-1 in complex with Compound 34 were obtained by hanging-drop vapor diffusion at 22°C against a reservoir of 10 mM Hepes, pH 7.5, 25% PEG 3350, 250 mM (NH4)2SO4, and a drop buffer consisting of 10 mM Hepes, pH 7.5, 8% PEG 3350, and 250 mM (NH4)2SO4. Crystals of caspase-1 in complex with active-site inhibitors were obtained by hanging-drop vapor diffusion at 4°C against a reservoir of 0.1 M Pipes, pH 6.0, 200 mM (NH4)2SO4, 25% PEG 2000 MME, 10 mM DTT, 3 mM NaN3, 2 mM MgCl2 (wild-type protein); 0.1 M Hepes pH 7.0, 2 M (NH4)2SO4, and 25 mM DTT (R286A); 0.1 M Pipes pH 6.0, 350 mM Li2SO4, 20% PEG 2000 MME, 10 mM DTT, 3 mM NaN3, and 2 mM MgCl2 (E390A); and 0.1 M Pipes, pH 6.0, 100 mM Li2SO4, 25% PEG 2000 MME, 10 mM DTT, 3 mM NaN3, and 2 mM MgCl2 (R286A/E390A). All crystals for data collection were cryoprotected in mother liquors supplemented with 20% (vol/vol) glycerol for 1–2 min and immersion in liquid nitrogen.
Diffraction data were collected under standard cryogenic conditions by using a Rigaku RU-3R rotating anode generator and an RAXIS-IV detector, processed and scaled with CrystalClear from Rigaku/Molecular Structure (21). The structures were determined from single-wavelength native diffraction experiments by molecular replacement with amore (22) by using search models from previously determined structures (PDB ID 1SC1 for the allosterically inhibited enzyme, and 1SC3 for the active-site-bound enzyme). The refinement of the initial solutions with refmac (23–25) yielded experimental electron density maps suitable for model building with o (26). The following residues were not visible in the electron density maps for the indicated protein–inhibitor complexes and were omitted from refinement of the final atomic models: 120–150 (C285,362, 64,397A); 120–124 and 145–149 (wild-type enzyme); 120–124 and 145–149 (R286A); 120–124 and 146–148 (E390A); and 120–131, 146–148, and 317 (R286A/E390A). procheck (27) revealed no disallowed (φ, ψ) combinations and excellent stereochemistry (see Table 1 for a summary of crystallographic data and refinement statistics).
Enzyme Kinetic Analysis.
Caspase-1 wild-type protein or caspase-1 mutant enzymes were produced as described. Active-site titrations using the active-site inhibitors z-VAD-FMK and Ac-WEHD-CHO were used to determine the concentration of active caspase-1 for kinetic analysis. Substrate titrations using optimal enzyme concentrations were done by using the fluorescent substrate Ac-WEHD-AFC at room temperature in an assay buffer containing 50 mM Hepes, pH 7.5, 50 mM KCl, 200 mM NaCl, 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, and 10 mM DTT. Determination of KM and Vmax was done by plotting the logarithm of substrate concentration on the x axis against the initial velocity v measured in relative fluorescence units per unit time. In GraphPad prism, the kinetic constants were determined by curve fitting using a sigmoidal dose–response with variable slope; Y = Bottom + (Top − Bottom)/(1 + 10‸((LogEC50 − X) × Hillslope)). The Hill constant was determined as the slope of a plot of the logarithm of the substrate concentration on the x axis against the logarithm of the partial Vmax.
Supplementary Material
Acknowledgments
We thank J. W. Lam, T. O’Brien, and M. M. Sopko for advice and assistance. Special thanks to colleagues at Sunesis Pharmaceuticals, Inc., for supportive interactions and generous research funding. This work was supported by National Institutes of Health Postdoctoral Fellowship 7F32AR052602-02 (to J.M.S.) from the National Institute of Arthritis and Musculoskeletal and Skin Diseases. This research was supported in part by the Sandler Family Supporting Foundation.
Abbreviations
- ME
mercaptoethanol.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2FQQ, 2FQR, 2FQS, 2FQU, 2FQV).
References
- 1.Fuentes-Prior P., Salvesen G. S. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y. Protein Sci. 2004;13:1979–1987. doi: 10.1110/ps.04789804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien T., Lee D. Mini Rev. Med. Chem. 2004;4:153–165. doi: 10.2174/1389557043487448. [DOI] [PubMed] [Google Scholar]
- 4.Erlanson D. A., Braisted A. C., Raphael D. R., Randal M., Stroud R. M., Gordon E. M., Wells J. A. Proc. Natl. Acad. Sci. USA. 2000;97:9367–9372. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arkin M. R., Wells J. A. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 6.Hardy J. A., Lam J., Nguyen J. T., O’Brien T., Wells J. A. Proc. Natl. Acad. Sci. USA. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinon F., Tschopp J. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Talanian R. V., Brady K. D., Cryns V. L. J. Med. Chem. 2000;43:3351–3371. doi: 10.1021/jm000060f. [DOI] [PubMed] [Google Scholar]
- 9.Romanowski M. J., Scheer J. M., O’Brien T., McDowell R. S. Structure (Cambridge, MA) 2004;12:1361–1371. doi: 10.1016/j.str.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Menard R., Carriere J., Laflamme P., Plouffe C., Khouri H. E., Vernet T., Tessier D. C., Thomas D. Y., Storer A. C. Biochemistry. 1991;30:8924–8928. doi: 10.1021/bi00101a002. [DOI] [PubMed] [Google Scholar]
- 11.Braxton S., Wells J. A. J. Biol. Chem. 1991;266:11797–11800. [PubMed] [Google Scholar]
- 12.Fersht A. Enzyme Structure and Mechanism. New York; San Francisco: Freeman; 1985. pp. 104–105. [Google Scholar]
- 13.Buchbinder J. L., Guinovart J. J., Fletterick R. J. Biochemistry. 1995;34:6423–6432. doi: 10.1021/bi00019a023. [DOI] [PubMed] [Google Scholar]
- 14.Wang J. H., Tu J. I. J. Biol. Chem. 1970;245:176–182. [PubMed] [Google Scholar]
- 15.Madsen N. B., Shechosky S. J. Biol. Chem. 1967;242:3301–3307. [PubMed] [Google Scholar]
- 16.Petrilli V., Papin S., Tschopp J. Curr. Biol. 2005;15:R581. doi: 10.1016/j.cub.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 17.Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V., Nordstrom P., Roy S., Vaillancourt J., et al. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto A., Comatas K. E., Liu L., Stamler J. S. Science. 2003;301:657–661. doi: 10.1126/science.1079319. [DOI] [PubMed] [Google Scholar]
- 19.Scheer J. M., Wells J. A., Romanowski M. J. Protein Expr. Purif. 2005;41:148–153. doi: 10.1016/j.pep.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Erlanson D. A., Wells J. A., Braisted A. C. Annu. Rev. Biophys. Biomol. Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 21.Pflugrath J. W. Acta Crystallogr. D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 22.Navaza J. Acta Crystallogr. D. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. Acta Crystallogr. D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. Acta. Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Pannu N. S., Murshudov G. N., Dodson E. J., Read R. J. Acta Crystallogr. D. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- 26.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. J. Appl. Crystallogr. D. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.