Abstract
The adhesion of bacteria to surfaces plays critical roles in the environment, disease, and industry. In aquatic environments, Caulobacter crescentus is one of the first colonizers of submerged surfaces. Using a micromanipulation technique, we measured the adhesion force of single C. crescentus cells attached to borosilicate substrates through their adhesive holdfast. The detachment forces measured for 14 cells ranged over 0.11 to 2.26 μN, averaging 0.59 ± 0.62 μN. Based on the calculation of stress distribution with the finite element analysis method (dividing an object into small grids and calculating relevant parameters for all of the elements), the adhesion strength between the holdfast and the substrate is >68 N/mm2 in the central region of contact. To our knowledge, this strength of adhesion is the strongest ever measured for biological adhesives.
Keywords: adhesive strength, Caulobacter crescentus, cell mechanics, holdfast, micromanipulation
In the environment, bacteria are typically attached to surfaces as individual cells or as part of a biofilm. Bacterial biofilms are often the cause of biofouling, for example, when they form in implanted catheters, in water distribution systems, or on the hull of ships. The strong attachment of single bacterial cells to a surface provides the critical first step in the biofouling process. Understanding the nature, biosynthesis, and properties of the adhesives that mediate this attachment to surfaces is essential for a full understanding of the mechanisms of biofouling and biofilm formation.
The Gram-negative bacterium Caulobacter crescentus is ubiquitous in aquatic environments (1) and is among the first colonizers of submerged surfaces, initiating the process of biofouling (2). C. crescentus has a dimorphic life cycle with a motile swarmer cell stage, during which the initial attachment to surfaces occurs (3), followed by differentiation of the swarmer cell into a nonmotile cell that contains a polar extension called a stalk. The stalk is tipped by the holdfast, a polysaccharide adhesin that mediates the strong attachment of stalked cells to surfaces. C. crescentus cells attached to a surface are capable of resisting washing with strong jets of water, suggesting that the attachment of single cells is extremely strong (4).
A force of such a magnitude is far outside the range of applicability of laser tweezers, which provide maximum working forces on the order of ≈100 pN. In contrast, atomic force microscopy (AFM) can provide surface adhesion force measurements for cells attached to solid substrates. However, the measured force is typically for the adhesion of the AFM tip to a cell surface, and the coupling is not strong enough to detach the cell completely from its substrate (5, 6). The AFM tip has also been used to push laterally in an attempt to scrape off single cells. The resulting forces were of a few nN to 200 nN in magnitude (7). Flow chamber experiments (8) provided adhesion force measurements for a population of cells by counting the number of cells that remained attached after hydrodynamic shear stress was applied. Micropipette techniques have been previously used in the 5-pN to 20-nN range to investigate receptor–ligand bonding dynamics (9–11). When used in the form of biomembrane force probes, micropipettes delivered forces ranging from 5 to 170 pN (12–15). These forces are still below what is needed for detachment of the single bacterial cells reported here.
In this article, we introduce a method for measuring the adhesion force of single bacterial cells with the ability to measure forces in the micronewton range. We find that the adhesion force of single C. crescentus cells reaches the order of micronewton, the largest ever measured for single bacterial cells to our knowledge.
Results
Force of Attachment of Single C. crescentus Cells.
To measure the strong adhesion of C. crescentus to a solid substrate, we developed a micromanipulation method for measuring forces ranging from tens of nN to tens of μN (Fig. 1). Briefly, cells are allowed to attach to a thin flexible pipette whose force constant has been calibrated by AFM. A suction pipette is used to grab the body of an attached cell and pull the cell perpendicularly away from the flexible pipette. Because of the large force required, the cell body has to be sucked into the pipette, which is then bent away from the pulling direction. Thus the operation here differs from a recent study using the shear stress from continuous aspiration to detach much larger cells, the myotubes, from the substrate, which involves smaller forces generated by the fluid flow (16). In our setup, the force of adhesion is calculated from the amount of bending required to break the cell–pipette contact (see Materials and Methods). Fig. 2 shows an example of successive frames of a pulling operation (also see Movie 1, which is published as supporting information on the PNAS web site). Successful pulls were performed on 14 individual bacterial cells (Table 1). We obtained an average adhesion force of 0.59 ± 0.62 μN that ranged from 0.11 to 2.26 μN. Because each micropipette was reliably calibrated, the variation in adhesion force was most likely caused by the differences in holdfast sizes, different breaking points, and the extent of distortion in the orientation of the stalk with respect to the glass surface as the cell was positioned into the suction trap.
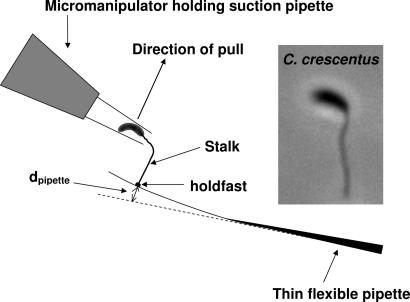
Fig. 1.
Diagram of force measurement by micromanipulation. A single C. crescentus cell is attached to a thin flexible pipette. The bacterium is trapped in place at the tip of the suction pipette. The movement of the suction pipette is controlled by a micromanipulator, which pulls the bacterium up until the cell is detached. The suction pressure is applied by a syringe (not shown). (Inset) Image of a cell with an elongated stalk. (Magnification: ×3,000.)
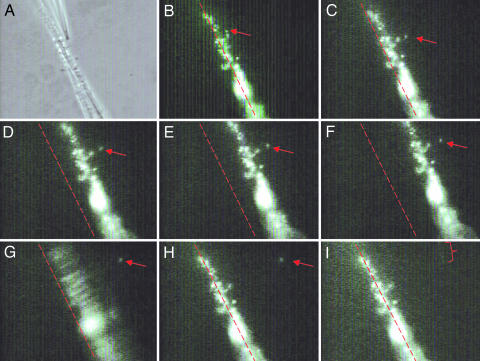
Fig. 2.
Successive frames of a pulling operation. The red arrow indicates the position of a single cell being pulled. (A) Phase contrast showing the tip of a suction pipette holding a C. crescentus cell in place, which is to be pulled from the flexible thin pipette. (B–I) Fluorescence images using NanoOrange to label the cell body, stalk, and holdfast. (C–F) The cell is pulled, and the thin flexible pipette increasingly deviates from its original position (indicated by the red dotted line). (F) The instant right before the cell is pulled off from the thin flexible pipette. (G) The cell is successfully pulled off the thin flexible pipette; the movement of the pipette is easily seen as a blurring of the attached cells. (H) The thin flexible pipette has returned to its original position. (I) The red bracket outlines the cell stalk taken inside the suction pipette following the cell body, a moment after their detachment from the substrate. (Magnification: ×500.) See Movie 1 for a demonstration of the pulling experiment.
Table 1.
Adhesion force for 14 individual cells
| Force constant, N/m | Pipette deviation, μm | Adhesion force, μN |
|---|---|---|
| 0.0236 | 62 | 1.46 |
| 0.0244 | 8.24 | 0.201 |
| 0.0245 | 22.3 | 0.547 |
| 0.0236 | 7.5 | 0.186 |
| 0.0248 | 8.0 | 0.198 |
| 0.0260 | 8.5 | 0.211 |
| 0.0247 | 8.0 | 0.198 |
| 0.0228 | 5.0 | 0.114 |
| 0.0128 | 176 | 2.26 |
| 0.0128 | 88 | 1.13 |
| 0.0139 | 24 | 0.333 |
| 0.0116 | 42 | 0.486 |
| 0.0116 | 56 | 0.647 |
| 0.0115 | 28 | 0.324 |
| Average force, μN | 0.59 ± 0.62 |
Effect of Pulling Rate on Detachment Force.
The detachment force of molecular bonds is known to depend on the rate of pulling (14). Such a rate dependence is expected to diminish with the increasing size and energy scale of the system. We tested the rate dependence in our experiment but found it to be rather small. Specifically, we tested the rate dependence for three cells. For cell one, the pulling force was first raised to 0.33 μN in 3–5 s, and then the cell was kept in the fixed position; the detachment occurred ≈30 min later. Cells two and three were first pulled with the force raised to 0.20 and 0.26 μN, respectively. No detachment occurred in 2 h. The pulling forces for these two cells were then raised to 0.5 and 0.78 μN, respectively. Both cells were detached within 5 s. These results suggest that the detachment force varies within a factor of three (possibly much smaller not knowing what the maximal forces cells two and three could tolerate before detachment) between vanishingly small loading rate (held in position) and a large loading rate on the order of 0.1 μN/s, typical in our detachment experiments. These tests were only semiquantitative, because the setup was manually operated. Nevertheless, the results suggest that the rate dependence in detaching C. crescentus cells is rather different from pulling single molecular bonds, for which case the detachment force varies orders of magnitude as the loading rate varies from pN/s to μN/s (14).
Determination of the Breaking Point of Detached Cells.
The cell-to-surface contact could be broken in a number of positions during a pulling experiment: the cell body–stalk junction, along the stalk, at the stalk–holdfast junction, within the holdfast, and at the holdfast–surface interface. To determine the position of breakage, we grew C. crescentus on glass coverslips and then compared fluorescent holdfast images before and after vigorous rinsing with water. Coverslips where the cell bodies were washed away had the same density of holdfast labeling as undisturbed coverslips (data not shown). This finding suggests that the point of breakage is most likely above the holdfast–substrate junction. AFM images of the coverslips described above after rinsing showed numerous holdfasts, mostly without stalks but some with short stalk fragments. The images of the holdfasts from which the stalks were completely removed were similar to those left on a glass surface by holdfast attachment mutants that are unable to keep the tip of the stalk attached to the holdfast (17) (data not shown). These holdfasts did not have the appearance of craters, suggesting that the holdfast material to which the stalk tip was attached remained with the holdfast on the surface. We conclude that the breaking position was typically either at the holdfast–stalk junction or somewhere along the stalk.
AFM Measurement of the Size of Stalks and Holdfasts.
We performed AFM imaging for 12 stalks to obtain an average diameter of ≈119 ± 10 nm after subtracting the AFM tip broadening effect of ≈60 nm for dried stalks with an average detected height ≈40 nm (based on the geometry for the tip known from the manufacturer). Using a stalk diameter of 119 nm and an average detachment force of 0.59 μN, we found that the average threshold stress the stalk could endure was estimated to be 53 N/mm2, assuming the stalk to be a solid rod. If the stalk is considered a hollow wall, the stress that can be endured by the wall material ought to be larger, and inversely proportional to the cross-sectional area.
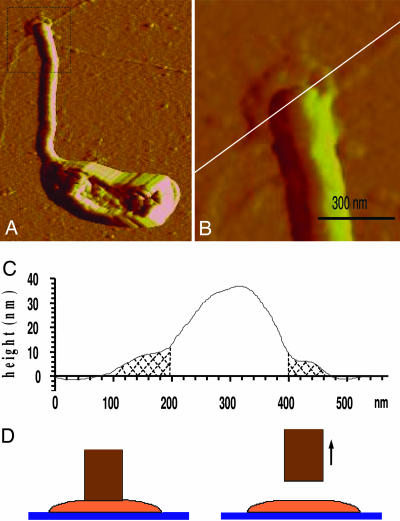
To determine the strength of the adhesion between the holdfast and the substrate, we needed to determine the area over which the force was transmitted. The size of 18 holdfasts was measured with AFM imaging as illustrated in Fig. 3; the diameter varied from 254 to 579 nm with an average of 411 nm (after subtracting the AFM tip size broadening of ≈26 nm on holdfasts with heights of ≈10 nm). The size of the holdfast has previously been shown to vary from cell to cell (17).
Fig. 3.
Measurement of holdfast size and thickness by AFM. (A) AFM image of a C. crescentus stalked cell, grown on glass coverslip, with the holdfast at the end of the stalk. (Total scan area: 3 μm × 2.4 μm.) (B) Enlarged image of the holdfast area. The height is measured along the white cross cut. (C) Plot of the height of the dried holdfast. Shaded regions in the plot indicate the visible holdfast area. For 18 sampled cells, the area of the holdfast ranged from 254 to 579 nm in diameter, and the average value obtained was 411 nm. (D) Schematics showing that when a cell is pulled off the glass substrate the stalk (brown) detaches from the holdfast (orange).
Finite Element Analysis of the Adhesion Strength of the Holdfast.
Because the holdfast spreads over a much larger area than the tip of the stalk, the stress within the holdfast and at the interfaces is no longer given by the simple formula of force/area. Instead, the stress and strain have to be calculated as tensors (each having up to 3 × 3 components) based on the known geometry and by assuming certain material properties of the holdfast. The deformation of the holdfast was simulated by means of the numerical finite element method. The material was assumed to be elastic based on our previous results (17), with a Young’s modulus of 100 MPa (108 N/m2) and a Poisson ratio of 0.4. This value of Young’s modulus was chosen to be comparable to a strong matrix of biological origin such as the byssal thread of marine mussels (18). At the average detachment stress, the calculated strain is 0.53. A much lower value of Young’s modulus, which might be more suitable for the holdfast material, would lead to a highly nonlinear strain field, which is impossible to calculate without knowing the form of the stress–strain relationship. In our calculation, a linear relationship between stress and strain is assumed to estimate the stress distribution. The Poisson ratio of 0.4 is commonly used for a wide range of materials with low compressibility, including glass and silk. The holdfast was modeled as a disk of diameter 411 nm and thickness 40 nm. This thickness was chosen on the basis of our previous measurements of wet holdfasts, showing that the holdfast thickness shrinks ≈3-fold upon drying (17). The stalk was attached over a circular patch on the top surface of initial radius 59.5 nm (Fig. 3D), imposing uniform displacement in the normal or z-direction where the force is applied. The bottom of the disk was constrained against displacement in the z-direction.
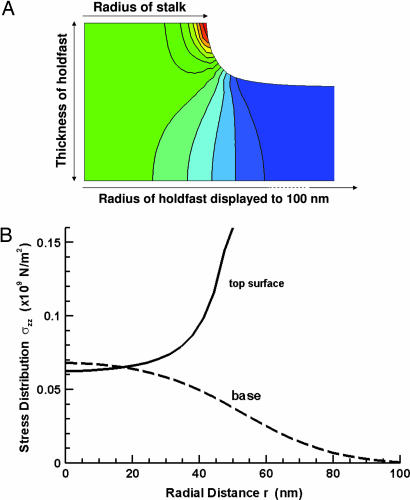
Fig. 4 shows the deformation at a total force of 0.59 μN for the central portion of the disk of radius 59.5 nm. The contour plot in Fig. 4A shows level curves of the tensile stress component σzz throughout the axially symmetric configuration. The graph in Fig. 4B shows in detail the distribution of this stress component at the interface of the holdfast with the stalk (solid curve) and substrate (dashed curve). The solid curve in Fig. 4B, labeled top surface, shows the normal stress exerted in the disk by the stalk over the planar contact surface. The stress is much larger at the interface between the holdfast and the stalk, especially at the edge, which is likely the reason it is easier for detachment to occur at this interface rather than between the holdfast and substrate. The dashed curve in Fig. 4B shows the normal stress exerted by the rigid glass substrate on the base of the disk. The maximum stress at the holdfast–substrate interface is 68 N/mm2 in the central region. The interface is not broken under this force, indicating that the strength of adhesion of the holdfast to the substrate is larger than this value. The integrated force of either distribution is equal to the pulling force.
Fig. 4.
Numerical finite element method simulation of stress and deformation on holdfast material. (A) The contour plot shows level curves of the tensile strength component σzz throughout the axially symmetric configuration with red to blue representing magnitudes from highest to lowest. (B) The solid curve (labeled top surface) shows the normal stress exerted in the disk by the stalk over the planar contact surface. The dashed curve (labeled base) shows the normal stress exerted by the rigid glass substrate on the base of the disk from equilibrium. The resultant forces of these two distributions are equal.
The integrity of the holdfast GlcNac polymers is critical for the strong adhesion of C. crescentus cells. The only characterized component of the holdfast is polymers of GlcNac (19). We have previously shown that the force constant that quantifies the elastic properties of the holdfast dropped to <10% of its original value after treatment with lysozyme, which cleaves oligomers of GlcNac (17). To demonstrate the dependence of the adhesion force on the holdfast GlcNac, we digested the GlcNac polysaccharide of the holdfast with lysozyme as described (17). The lysozyme treatment might also affect the cell wall, which would soften the cell body and the stalk, but may not affect the adhesion as directly as the holdfast. At concentrations of 2.3, 0.1, and 0.01 mg/ml of lysozyme, all cells (five or more cells tested at each concentration) were readily pulled off by syringe suction without making any observable deflections of the pipettes (data not shown), indicating that the GlcNac polysaccharide is a critical component of the strong adhesive force of the holdfast. These lysozyme-treated cells were still too strongly attached to be pulled off the surface with our laser tweezers (data not shown), which generate forces up to the order of 10 pN. The lowered adhesion force of lysozyme-treated holdfasts is probably in a range that is more amenable to a force probe, which works in the range of 100 pN to 10 nN, such as by a controlled flow or by applying an electric field to the bacterium, which is typically negatively charged in aqueous environment.
Discussion
We have demonstrated that the adhesion force of single bacterial cells can be measured directly and conveniently by micromanipulation. As far as we know, the adhesive force of C. crescentus is the strongest measured for microbial cells. Abu-Lail and Camesano (20) showed by using AFM that the adhesion force between bacteria and a number of substrates ranged from 26 pN to 5 nN, and between yeast/fungi and various AFM tips it ranged from 35 pN to 9 nN (20). Sulfate-reducing bacteria produce adhesion forces of ≈6.8 nN onto an AFM tip of radius 50 nm, resulting in an adhesive strength of ≈0.87 N/mm2 (5). Burkholderia cepacia G4 was measured to have an adhesion force of ≈2 nN onto an AFM tip with a ≈50-nm tip radius, which gives an adhesion strength of 0.25 N/mm2 (6). Bacillus mycoides spores on hydrophobic-coated glass surface were measured to have a maximal adhesion force of 56.5 nN, giving 0.032 N/mm2 for a typical spore diameter of ≈1.5 μm (21). Adhesion measurements of yeast cells using ≈1-μm colloidal probes yielded adhesion forces as large as 240 nN, which gave an adhesion strength of 0.3 N/mm2 (22).
One of the strongest adhesion strength known in nature is that of the setae on the toes of geckoes (23, 24). A single seta can generate up to 200 μN of force on an area of ≈20 μm2, which is equivalent to 10 N/mm2. At 68 N/mm2, the adhesion strength of the holdfast is stronger than gecko’s toes setae and far stronger than all other known cellular attachments. C. crescentus holdfast covering 1 cm2 would have the potential to hold a weight of 680 kg on a wet surface, making it an excellent candidate as a biodegradable or even surgical adhesive. The lower limit of the attachment strength measured for the holdfast in this study is stronger than the commercial dentin adhesive, which can provide a bond strength up to 30 N/mm2 (25).
Why would C. crescentus need to adhere to surfaces with such strength? Surface tension detachment forces caused by the passage of an air–liquid interface can be up to 200 nN for a microsized object (26). This surface tension exerts by far the dominant detachment force on microorganisms. C. crescentus has to contend with the passage of air–liquid interfaces, for example, through the action of waves when they are attached near the surface of an aquatic environment. The measured adhesion force of C. crescentus appears to be safely beyond the estimated threshold value, but within an order of magnitude.
The challenge now is to understand the chemical and biophysical basis for the impressive force of the holdfast adhesin. Because polymers of GlcNac play a critical role in the adhesive force of the holdfast, and because such polymers are important adhesins in many systems, including biofilms (27), understanding the basis for the adhesive properties of the holdfast will serve as a useful model system. The discrete location of the holdfast at the tip of the stalk and the fact that its sole function is for adhesion provide clear advantages for the investigation of the mechanisms of bacterial adhesion.
Materials and Methods
Sample Preparation.
C. crescentus CB15 cells were grown in a peptone yeast extract medium (28) at 30°C overnight without agitation in the presence of a thin flexible micropipette, which was fabricated by using a Sutter Instruments (Novato, CA) Micropipette Puller P-97. The pipette has a long tapered thin tip of ≈15 mm in length and ≈2 μm in diameter at the end. Cells were observed by microscopy to have attached to the micropipette. The medium was then changed to Hutner imidazole glucose glutamate medium (29) with a reduced phosphate concentration of 0.03 mM PO4, and the cells on the micropipette were grown for 2 days. This process promoted elongation of the stalks (30).
Microscopy.
For fluorescence microscopy, the holdfast was labeled with 20 μg/ml FITC-conjugated lectin (Molecular Probes) and cells were labeled with 2 μg/ml NanoOrange (Molecular Probes). To image the stalk and holdfast with AFM (Dimension 3100 AFM, Digital Instruments, Santa Barbara, CA), cells were grown in the same conditions as for the force measurement experiments, but were allowed to attach to glass coverslips. After rinsing, the coverslips were dried and imaged in air by using contact mode.
Measurement of Detachment Force by Micromanipulation.
To detach a C. crescentus cell from the flexible micropipette, a suction micropipette was approached to the attached cell. The suction pipette was mounted on a NanoControl micromanipulator (Kleindiek Nanotechnik, Reutlingen, Germany). This micromanipulator can move in incremental steps of 50 nm to 1 μm. Suction pressure was produced by a Stoelting 51222 Manual Microsyringe Driver with a Hamilton 710LT 100-μl syringe. Once a cell was held in place just inside the tip of the suction pipette, we began videotaping while pulling the cell perpendicularly away from the thin flexible pipette to which the cell was attached. The position at which the cell detachment event was observed yielded the cell’s adhesion force.
Deflection measurements were made by using a Nikon TE-2000U inverted microscope with a ×100 objective designed for phase contrast and fluorescence imaging. Cells were labeled with NanoOrange. Pulling movies were recorded by using a Marshall V-1070 black and white charge-coupled device camera and a Sony GV-D800 digital video recorder. Cells attached to the pipette through their holdfast with their body aligned in the direction of the suction pipette were used. Let the deflection of the flexible micropipette to which a cell was attached be dpipette, and the force constant or elastic stiffness of the pipette be kpipette. The pulling force, F, which was counterbalanced by the bending pipette, was obtained by Hooke’s law, F = kpipette · dpipette.
Measuring the Force Constant of the Flexible Micropipette.
To calibrate the force constant at the position of cell detachment, we used AFM (Dimension 3100, Digital Instruments) in contact mode and tipless calibration cantilevers with force constant, kcantilever, of 0.71 N/m. At equilibrium, the force of the thin flexible glass pipette balanced the elastic force of the cantilever,
The elastic deflections of the pipette, dpipette, and cantilever, dcantilever, were generated by the AFM’s piezo electric tube such that for small angles,
Knowing the deviation of the AFM tube, dtube, and deviation of the cantilever, dcantilever, we obtained the deviation of our pipette, dpipette. With a known force constant of the cantilever, kcantilever, we obtained the force constant of the pipette, kpipette. Hence,
Supplementary Material
Acknowledgments
We thank members of our laboratories for critical reading of the manuscript. This work was supported by National Science Foundation Awards DMR 0320676 and DMR 0405156 (to J.X.T.), National Science Foundation Materials Research Science and Engineering Center Award DMR 0079964 (to Brown University), National Institutes of Health Grant GM51986 (to Y.V.B.), Indiana University, and Brown University.
Abbreviation
- AFM
atomic force microscopy.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Poindexter J. S. Microbiol. Rev. 1981;45:123–179. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corpe W. A. In: Acker R. F., Brown B. F., DePalma J. R., Iverson W. P., editors. Proceedings of the Third International Congress on Marine Corrosion and Fouling; Evanston, IL: Northwestern Univ. Press; 1972. pp. 598–608. [Google Scholar]
- 3.Bodenmiller D., Toh E., Brun Y. V. J. Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith C. S., Hinz A., Bodenmiller D., Larson D. E., Brun Y. V. J. Bacteriol. 2003;185:1432–1442. doi: 10.1128/JB.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang H. H., Chan K. Y., Xu L. C. J. Microbiol. Methods. 2000;40:89–97. doi: 10.1016/s0167-7012(99)00137-2. [DOI] [PubMed] [Google Scholar]
- 6.Camesano T. A., Logan B. E. Environ. Sci. Technol. 2000;34:3354–3362. [Google Scholar]
- 7.Sagvolden G., Giaever I., Pettersen E. O., Feder J. Proc. Natl. Acad. Sci. USA. 1999;96:471–476. doi: 10.1073/pnas.96.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burmeister J. S., Vrany J. D., Reichert W. M., Truskey G. A. J. Biomed. Mater. Res. 1996;30:13–22. doi: 10.1002/(SICI)1097-4636(199601)30:1<13::AID-JBM3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 9.Francis G., Fisher L., Gamble R., Gingell D. J. Cell Sci. 1987;87:519–523. doi: 10.1242/jcs.87.4.519. [DOI] [PubMed] [Google Scholar]
- 10.Zinkl G. M., Zwiebel B. I., Grier D. G., Preuss D. Development (Cambridge, U.K.) 1999;126:5431–5440. doi: 10.1242/dev.126.23.5431. [DOI] [PubMed] [Google Scholar]
- 11.Kishino A., Yanagida T. Nature. 1988;334:74–76. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- 12.Chesla S. E., Selvaraj P., Zhu C. Biophys. J. 1998;75:1553–1572. doi: 10.1016/S0006-3495(98)74074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans E., Berk D., Leung A. Biophys. J. 1991;59:838–848. doi: 10.1016/S0006-3495(91)82296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merkel R., Nassoy P., Leung A., Ritchie K., Evans E. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 15.Shao J. Y., Hochmuth R. M. Biophys. J. 1996;71:2892–2901. doi: 10.1016/S0006-3495(96)79486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin M. A., Engler A. J., Barber T. A., Healy K. E., Sweeney H. L., Discher D. E. Biophys. J. 2004;86:1209–1222. doi: 10.1016/S0006-3495(04)74195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G., Smith C. S., Brun Y. V., Tang J. X. J. Bacteriol. 2005;187:257–265. doi: 10.1128/JB.187.1.257-265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waite J. H., Vaccaro E., Sun C., Lucas J. M. Philos. Trans. R. Soc. London B. 2002;357:143–153. doi: 10.1098/rstb.2001.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merker R., Smit J. Appl. Environ. Microbiol. 1988;54:2078–2085. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abu-Lail N. I., Camesano T. A. J. Microsc. (Oxford) 2003;212:217–238. doi: 10.1111/j.1365-2818.2003.01261.x. [DOI] [PubMed] [Google Scholar]
- 21.Bowen W. R., Fenton A. S., Lovitt R. W., Wright C. J. Biotechnol. Bioeng. 2002;79:170–179. doi: 10.1002/bit.10321. [DOI] [PubMed] [Google Scholar]
- 22.Bowen W. R., Hilal N., Lovitt R. W., Wright C. J. Colloids Surf. A. 1998;136:231–234. [Google Scholar]
- 23.Autumn K., Sitti M., Liang Y. A., Peattie A. M., Hansen W. R., Sponberg S., Kenny T. W., Fearing R., Israelachvili J. N., Full R. J. Proc. Natl. Acad. Sci. USA. 2002;99:12252–12256. doi: 10.1073/pnas.192252799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Autumn K., Liang Y. A., Hsieh S. T., Zesch W., Chan W. P., Kenny T. W., Fearing R., Full R. J. Nature. 2000;405:681–685. doi: 10.1038/35015073. [DOI] [PubMed] [Google Scholar]
- 25.Nakajima M., Sano H., Burrow M. F., Tagami J., Yoshiyama M., Ebisu S., Ciucchi B., Russell C. M., Pashley D. H. J. Dent. Res. 1995;74:1679–1688. doi: 10.1177/00220345950740100901. [DOI] [PubMed] [Google Scholar]
- 26.Gomez-Suarez C., Busscher H. J., van der Mei H. C. Appl. Environ. Microbiol. 2001;67:2531–2537. doi: 10.1128/AEM.67.6.2531-2537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadovskaya I., Vinogradov E., Flahaut S., Kogan G., Jabbouri S. Infect. Immun. 2005;73:3007–3017. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poindexter J. S. Bacteriol. Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poindexter J. S. J. Bacteriol. 1978;135:1141–1145. doi: 10.1128/jb.135.3.1141-1145.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonin M., Quardokus E. M., O’Donnol D., Maddock J., Brun Y. V. J. Bacteriol. 2000;182:337–347. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.