Abstract
Predicting at the time of discovery the prognosis and metastatic potential of cancer is a major challenge in current clinical research. Numerous recent studies searched for gene expression signatures that outperform traditionally used clinical parameters in outcome prediction. Finding such a signature will free many patients of the suffering and toxicity associated with adjuvant chemotherapy given to them under current protocols, even though they do not need such treatment. A reliable set of predictive genes also will contribute to a better understanding of the biological mechanism of metastasis. Several groups have published lists of predictive genes and reported good predictive performance based on them. However, the gene lists obtained for the same clinical types of patients by different groups differed widely and had only very few genes in common. This lack of agreement raised doubts about the reliability and robustness of the reported predictive gene lists, and the main source of the problem was shown to be the small number of samples that were used to generate the gene lists. Here, we introduce a previously undescribed mathematical method, probably approximately correct (PAC) sorting, for evaluating the robustness of such lists. We calculate for several published data sets the number of samples that are needed to achieve any desired level of reproducibility. For example, to achieve a typical overlap of 50% between two predictive lists of genes, breast cancer studies would need the expression profiles of several thousand early discovery patients.
Keywords: DNA microarray gene expression data, outcome prediction in cancer, probably approximately correct sorting, predictive gene list, robustness
One of the central challenges of clinical cancer research is prediction of outcome, i.e., of the potential for relapse and for metastasis. Identification of aggressive tumors at the time of diagnosis has direct bearing on the choice of optimal therapy for each individual. The need for sensitive and reliable predictors of outcome is most acute for early discovery breast cancer patients. Adjuvant chemotherapy is recognized to be useless for ≈75% of this group (1); it is believed that after surgery a large majority of these patients would remain disease free without any treatment. Nevertheless, they are often submitted to the same therapeutic regimen as the small fraction of those who really need chemotherapy and benefit from it.
Considerable effort has been devoted recently to outcome prediction for several kinds of cancer on the basis of gene expression profiling (2–8), with special emphasis on breast carcinoma (9–13). Several of these studies reported considerable predictive success. These successes were, however, somewhat thwarted by two problems: (i) when one group’s predictor was tested (G. Fuks, L.E.-D., and E.D., unpublished data) on another group’s data (for the same type of cancer patients), the success rate decreased significantly; and (ii) comparison of the predictive gene lists (PGLs) discovered by different groups revealed very small overlap. These problems indicate that the currently used PGLs suffer from instability of their membership and of their predictive performance. These statements are well illustrated by two prominent studies of survival prediction in breast cancer. Wang et al. (11), using the Affymetrix technology, analyzed expression data obtained for a cohort of 286 patients with early discovery. They identified and reported a PGL of 76 genes. van’t Veer et al. (9) used Rosetta microarrays to study 96 patients and produced their own list of 70 genes, which were subsequently tested successfully on a larger cohort of 295 patients (10). Each group achieved, using its own genes on its own samples, good prediction performance. However, the overlap between the two lists was disappointingly small: only three genes appeared on both!§ Furthermore, the discriminatory power of the two classifiers, as found on their own data sets, was not reproduced when testing them on the samples of the other study (G. Fuks, L.E.-D., and E.D., unpublished data).
These intriguing problems have received great attention by the community of cancer research and have been addressed in several topical studies. The obvious and most straightforward explanation of these apparent discrepancies is to attribute them to (i) different groups using cohorts of patients that differ in a potentially relevant factor (such as age), (ii) the different microarray technologies used, and (iii) different methods of data analysis. Ein-Dor et al. (14) have shown that the inconsistency of the PGLs cannot be attributed only to the three trivial reasons mentioned above. To this end, they focused on a single data set (9) and repeated many times precisely the analysis performed by van’t Veer et al., thereby eliminating all three differences listed above. Generating many different subsets of samples for training, they showed that van’t Veer et al. (9) could have obtained many lists of equally prognostic genes and that two such lists (obtained by using two different training sets generated from the same cohort of patients) share, typically, only a small number of genes. This discovery was supported by Michiels et al. (15), who did not limit their attention to breast cancer and investigated the stability of seven PGLs published by seven large microarray studies. They showed that the prediction performances that were reported in each study on the basis of its published gene list were overoptimistic in comparison with results obtained by reanalysis of the same data performed (using different training sets) by Michiels et al. (15). Furthermore, they showed, much in the same way as in ref. 14, that the PGLs reported by the various groups were highly unstable and depended strongly on the selection of patients in the training sets. Ioannidis (16), in a comment to ref. 15, and Lonning et al. (17), in a review on genomic studies in breast cancer, cast doubt on the maturity of the published lists to implementation in a routine clinical use and suggest that small sample sizes might actually hinder identification of truly important genes. Similar criticism was expressed in two recent reviews (18, 19), which raise several methodological problems in the process determining the prognostic signature. They conclude that further research is required before applying the identified markers in a routine clinical use.
An obvious question is: Why does one need a short list of predictive genes? There are at least three reasons for this need. The first reason is technical and goes back to a problem well known in machine learning. In general, the number of genes on the chip is in the ten thousands, and the number of samples is in the hundreds. Hence, by using all genes to classify the samples into good and bad outcome, we take a high risk of overtraining, i.e., of fitting the noise in the data, which may increase the generalization error (the error rate of the resulting predictor on samples that were not used during the training phase). The second reason has to do with our desire to gain some biological insight about the disease: One hopes that the genes that are the most important and relevant for control of the malignancy also will appear on the list of the most predictive ones. Third (and least important), a relatively small number of predictive genes will allow inexpensive mass usage of a custom-designed prognostic chip. The second of these points was questioned by Weigelt et al. (20); these authors addressed further the instability of PGLs and concluded that the membership of a gene in a prognostic list is not necessarily indicative of the importance of that gene in cancer pathology.
These findings raise another question: Why should one worry about the diversity of the derived short PGLs? Clearly, had the predictor based on one group’s genes worked well on patients of other studies, one would not have had to worry about list diversity. However, the observed lack of transferability of predictive power may well be because of the same reason that causes instability of the gene lists. Because one hopes that by generating more stable PGLs one will obtain more robust predictors as well, and in light of their tremendous potential for personalized therapy, assessing the stability of these lists is crucial to guarantee their controlled and reliable utilization.
So far, the lack of stability of these PGLs has been either ignored or demonstrated for a particular experiment by reanalysis of the data. Here, we propose a mathematical framework to define a quantitative measure of a PGL’s stability. Furthermore, we present a method that uses existing data of a relatively small number of samples to project the expected stability one would obtain for a larger set of training samples, thereby helping to design an experiment that generates a list that has a desired stability.
To this end, we introduce a previously undescribed mathematical method for evaluating the stability of outcome PGLs for different cancer types. To measure list stability, we introduce a figure of merit f, which varies between 0 and 1; the higher its value, the more stable the PGL. We show how this figure of merit increases with the number of training samples and determine the number of training samples needed to ensure that the resultant PGL meets a desired level of stability. We perform a comparative study of list quality in several cancer types, using a collection of gene expression data sets supplemented by outcome information for the patients.
Overview and Notation
Denote by Ng the number of genes from which a PGL is to be selected: either the total number of genes on the chip or the number of those that pass a relevant filter (such as significant expression in at least a few samples or variance above a threshold). Either way, Ng ≈ 10,000. The expression levels measured in n samples are used for gene selection and, subsequently, for construction of a predictor of outcome. These samples are routinely referred to as the “training set”; usually n is on the order of a few tens, up to a few hundreds. For each gene, a predictive score is calculated on the basis of its expression over the training set, and the genes are ranked according to this score. The NTOP top-ranked genes are selected as members of the PGL. Usually (9) NTOP is determined by incrementing the number of genes on the list and monitoring the success rate of the resulting predictor, using cross-validation. Broadly speaking, the success rate increases, peaks, and decreases (21, 22), and the optimal number of genes is used as NTOP. Because determination of NTOP is outside the scope of our work, we use a free parameter, α = NTOP/Ng, and calculate our results as a function of α. In typical studies NTOP ≈ n; hence, for our problem α ≈ 0.01.
The figure of merit we introduce and use here, f, is the overlap between two PGLs, obtained from two different training sets of n samples in each. That is, 0 ≤ f ≤ 1 is the fraction of shared genes (out of NTOP) that appear on both PGLs; the closer f is to 1, the more robust and stable are the PGLs obtained from an experiment.
Our central point is that because the n samples of the training sets are chosen at random from the very large population of all patients, the figure of merit f is a random variable. The aim of our work is to calculate Pn,α(f), the probability distribution of f.
Once this distribution has been determined, we are able to answer the following question: For given n and α, what is the probability that the robustness f of the PGL exceeds a desired minimal level? This question is related to the classical concept of probably approximately correct (PAC) learning (23), which we generalize here to “PAC sorting.” Alternatively, we can answer a question such as: How many training samples are needed to construct a PGL whose expected f exceeds a desired value?
Results
Analytical Derivation of Pn,α(f).
Our central result, correct to order 1/Ng, is that this probability distribution has the form
Hence, the probability distribution of f is a Gaussian; We have calculated (see Supporting Text, which is published as supporting information on the PNAS web site) the mean f*n and variance Σn2 as functions of Ng, α, and n We have found that Σn ∼ 1/Ng, and hence in the limit of infinite Ng the overlap between any two PGLs is fixed at f*n and does not depend on the specific realization of the training set, but only on its size n.
Testing the Validity of Our Assumptions.
As described in Materials and Methods, our analytical calculation is based on several assumptions on the model generating the data we have at hand. The extent to which any of these assumptions is fulfilled for real-life data sets varies from case to case and may affect the extent to which our analytical results can be used for a particular data set. Importantly, one can test the correctness of the assumptions by using the real data. A detailed analysis of our assumptions for each of the data sets we have investigated, presented in Supporting Text, shows that our assumptions hold. Excellent agreement between simulations and the analytic calculation was found in five of the six data sets studied.
Breast Cancer Expression Data.
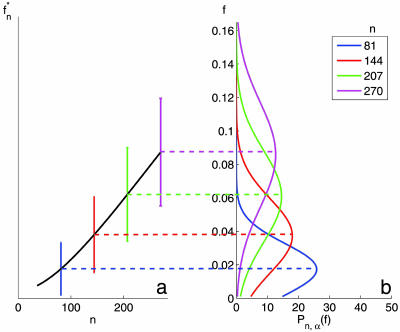
Fig. 1b shows the probability distributions of f, estimated from the data of ref. 10. We set α to 0.0046, which corresponds to a PGL of size 70, and present Pn,α(f) for several values of n. Note that the analytical calculation can be performed for any n irrespective to the number of samples used in the actual experiment (10). As n increases, the typical overlap f*n increases as well; for the range of n shown, the width of the distribution, Σn, also increases (for large n it will start to decrease). In Fig. 1a, we show the variation of f*n with n. Importantly, we see that for these moderate values of n the typical overlap between two PGLs, obtained from two training sets, is of the order of a few percents! For two randomly selected lists of αNg genes, one expects f ≈ α. In Table 1, we show the number of samples needed to achieve a desired level of overlap between a pair of PGLs produced from two randomly chosen training sets. Suppose that we wish to know how many samples are needed to guarantee, with confidence level 1 − δ, that the PGL produced by these samples has an overlap f (with a hypothetical PGL obtained from an independent cohort with the same number of samples) that exceeds a desired level fc = 1 − ε. The number of samples n, needed to achieve this goal is given in column 3 of Table 1 for the data of ref. 10, for a PGL of 70 genes, and in column 4 for ref. 11, for a PGL of 76 genes. When δ = 0.5, we have fc = f*n so that to achieve a typical overlap of f*n = 0.50, n = 2,300 samples are needed for the data of ref. 10 and 3,142 for the data of ref. 11.
Fig. 1.
The overlap f of two top-gene lists derived from data of van de Vijver et al. (10), with α = 0.0046 (corresponding to predictive lists of 70 genes). (a) The mean and standard deviation (represented by vertical bars) of f for various values of n. (b) The probability distribution of f for the same values of n.
Table 1.
The number of samples n needed to get an overlap f ≥ fc = 1 − ε, with confidence 1 − δ, for two PGLs of size αNg
| fc = 1 − ε | δ | n (10) | n (11) |
|---|---|---|---|
| 0.02 | 0.5 | 87 | 104 |
| 0.05 | 0.5 | 170 | 218 |
| 0.10 | 0.5 | 290 | 383 |
| 0.20 | 0.5 | 553 | 743 |
| 0.50 | 0.5 | 2,300 | 3,142 |
| 0.02 | 0.1 | 178 | 195 |
| 0.05 | 0.1 | 270 | 319 |
| 0.10 | 0.1 | 412 | 507 |
| 0.20 | 0.1 | 736 | 930 |
| 0.50 | 0.1 | 3,026 | 3,883 |
We use α = 0.0046 (corresponding to a PGL of 70 genes) and α = 0.0068 (corresponding to a PGL of 76 genes) for refs. 10 and 11, respectively. For δ = 0.5, fc = f*n, and hence n represents the number of samples needed for an average overlap of 1 − ε. The effective number of genes used here (after preprocessing) was Ng = 15,125 for ref. 10 and Ng = 11,130 for ref. 11.
Comparing Data from Several Groups and Types of Cancer.
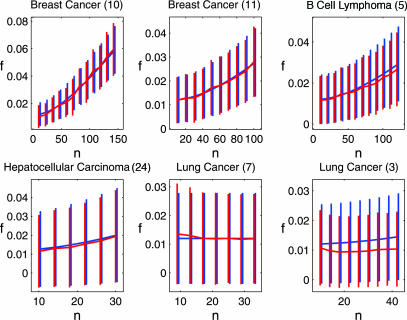
We analyzed the stability of PGLs obtained from gene-expression data from various cancer types: breast (10, 11), lung (3, 7), lymphoma (5) and hepatocellular carcinoma (24). First, we checked the agreement between simulations and our analytic prediction. The results, presented in Fig. 2, show that simulations coincide with the analytical results for the mean and the standard deviation of the overlap between two measured lists, for five of the six data sets studied. Once agreement between simulations and the analytic calculation is established for a particular study, we can rely on the analytic results for values of n that exceed the range that is currently experimentally available. If no agreement is found, we can extrapolate the results of the simulations.
Fig. 2.
The mean overlap f*n as a function of the number of samples, for six different data sets, for α = 0.012. The vertical bars indicate one standard deviation. Analytic estimations are in blue, and the results of simulations are in red. For each data set, the range of n for which results are presented reflects the number of samples of the particular experiment. Numbers in parentheses refer to the reference from which the data were taken.
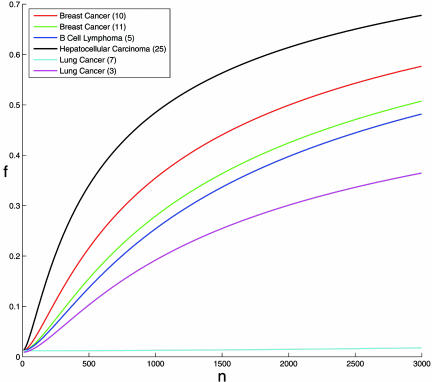
The results are shown in Fig. 3, which presents the expected overlap f*n for all data sets, thus enabling one to compare the robustness of the PGLs obtained using different microarrays and for different cancers. One can see a clear hierarchy of cancers with respect to their PGLs robustness, where hepatocellular carcinoma is most stable, breast cancer ranked second, acute lymphocytic leukemia is third, and lung cancer is characterized by the lowest stability. This result may reflect the difficulty of outcome prediction in the different cancers where survival of hepatocellular carcinoma patients is the easiest to predict, whereas survival prediction in lung cancer is most difficult.
Fig. 3.
The typical overlap f*n as a function of the number of samples, for the six different data sets (α = 0.012 was used). All curves except lung cancer (3) were produced using the analytical results. Because no agreement was found between simulation and analytical results for lung cancer (3), this curve was produced using extrapolation of simulation results (see Materials and Methods). Numbers in parentheses refer to the reference from which the data were taken.
Summary and Conclusions
We introduced probably approximately correct (PAC) sorting, a previously undescribed mathematical method for calculating the quality of the PGLs obtained in an experiment, by measuring the overlap f between pairs of gene lists produced from different training sets of size n. We proved that the method can predict with high accuracy Pn,α(f), the probability distribution of f, for any n. Because gene expression profiles are believed to possess high potential of outcome prediction, and such PGLs will be put to clinical prognostic use, obtaining information about their quality and robustness is crucial. Moreover, by discovering the relationship between n, the number of training samples used to obtain a gene list and the quality of the resultant list, our method provides a means for efficient experimental design. Given any desired list quality, we can calculate the number of samples required to achieve it in a particular experiment. Furthermore, by finding the mathematical expressions for Pn,α(f), we gain an insight into the problem of instability of PGLs, discussed at the beginning of this work. In Materials and Methods, we introduce two distributions, q(Ct) and p′n(Z; Zt), and show that the instability is governed mainly by the magnitude of q(Ct) near the values that correspond to the top αNg correlations, and by the variance σn2 of p′n. These two factors determine the sensitivity of the PGL’s composition to random selection of the training set. Our method can be extended to deal with applications to a wide variety of feature selection problems, including pattern recognition and text categorization.
Materials and Methods
The analytical calculations rely on our ability to use available expression data to estimate two distributions, p′n(Z; Zt) and q(Zt), which we define now.
Scoring Genes by Their “Noisy” Correlation with Outcome.
We used one of the accepted ways (9–11, 15) to represent outcome: as a binary variable, with 1 for good and 0 for bad outcome. A sample (patient) is designated as “good outcome” if the metastasis and relapse-free survival time exceeds a threshold, or “bad” if it does not. The simplest score of a gene’s predictive value is the Pearson correlation C of outcome with the gene’s expression levels.¶ Measuring a gene’s correlation with outcome over several different training sets of n samples yields different values of C; i.e., our measurement of a gene’s C is noisy, taken from some distribution pn(C; Ct) around the “true” value Ct.‖ The deviation of a gene’s measured C from Ct, referred to as “noise,” is due to the variation of the measured correlations when the n samples of the training set are randomly selected; it is one of the factors that governs the diversity or lack of robustness of the PGL. Large noise causes large differences in a gene’s correlation when measured over different subsets of samples, potentially inducing large shifts in a gene’s rank, which induce instability of the PGL.
The Distributions pn(C;Ct) Are Different for Each Gene.
A simple transformation on the correlations produces new variables Z = tanh−1(C). Under certain assumptions (25, 26) their distribution, p′n(Z; Zt), is approximately a Gaussian around the true value Zt = tanh−1(Ct), with identical variances for all of the genes, given by σn2 = 1/(n − 3). These assumptions do not necessarily hold for all expression data; we found that the distribution of the noise is Gaussian to a good approximation, and in our analytical calculations we use the same variance for all genes, but this variance has to be estimated for each experiment from the measured data.
The Distribution of the True Correlations,q(Zt), Is Another Important Factor That Affects the Diversity of the PGL.
When the training set changes, the measured Z of each gene changes (by the noise described above). As a result, genes will move in and out of the interval that contains the NTOP highest-ranked ones. Higher density of genes in this interval increases the sensitivity (to noise) of a top gene’s rank, resulting in a more unstable PGL. We estimate Vt, the variance of q(Zt), from the data, as described below, and for the analytic calculation approximate q(Zt) by a Gaussian of variance Vt and mean zero.
Analytical Calculation.
Here we present only an outline of the method; a concise description is in Supporting Text and also Figs. 4–6, which are published as supporting information on the PNAS web site. We do review here the assumptions that are made about the expression data and the approximations taken to carry out the calculation.
Assumption 1.
The distributions of the measured Z values are Gaussian, centered for each gene around its Zt.
Assumption 2.
The variance σn2 is the same for all genes.
Assumption 3.
The noise variables Z − Zt are independent (i.e., uncorrelated noise for different genes).
Assumption 4.
q(Zt), the distribution of the true correlations, can be approximated by a Gaussian with variance Vt. This assumption is easily generalized to represent q(Zt) as a mixture of Gaussians.
Under these assumptions, we can write down an expression for Pn,α(f), which reflects a process of (i) drawing Ng independent true correlations Zt from the distribution q(Zt), (ii) submitting each to a Gaussian noise of variance σn2, and (iii) identifying the αNg top genes. Submitting the Ng true values to another realization of the noise, we obtain another list of αNg genes. Note that for finite n the lists are expected to be different because of noise (nonvanishing σn2). The probability to obtain an overlap f between two PGLs, Pn,α(f), is given by
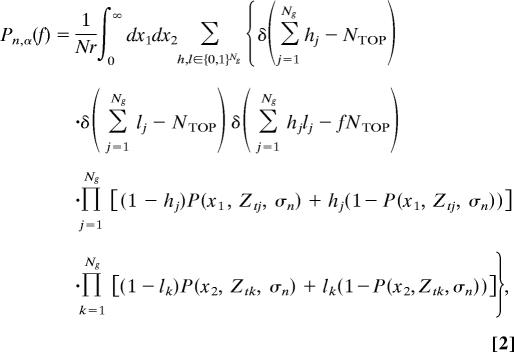 |
where
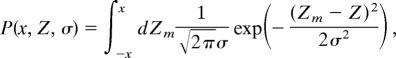 |
δ(.) is the Kronecker delta, Nr is a normalization factor, and Ztj is the true correlation of the jth gene with outcome. h = (h1, . . ., hNg) and l = (l1, . . ., hNg) are binary vectors of size Ng whose nonzero elements correspond to the genes included in the measured NTOP of the first and the second realizations, respectively.
Approximation: 1/Ng expansion:
By using mathematical manipulations, we represent Eq. 2 as a multivariate integral over Ng variables and calculate it using saddle-point integration and expansion (to first order) in 1/Ng, a technique widely used in theoretical physics (27, 28). We have tested and found this to be an excellent approximation, as expected (because Ng ≫ 1/α ≈ 100).
Some single-variable integrations have to be done numerically to obtain the final result, Eq. 1, i.e., that Pn,α(f) is a Gaussian, with mean f *n and variance Σn2 that we know how to calculate on the basis of available data (see Supporting Text).
Derivation of the Variances σn2 and Vt from Real Data.
As mentioned above, the two major components that affect Pn,α(f) are q(Zt), the probability distribution of the true Z values, and the noise variance σn2. Yet, for real data sets, one knows only the Ng measured Z values, obtained for each of the Ng genes on the basis of their expression levels in n samples, n ≤ Ns. Hence, we have access to the measured probability distribution qn(Z), and to the expression data, from which we have to reconstruct the true distribution q(Zt) and the variance of the noise, σn2.
If the noises of the different genes are identical independent Gaussian random variables, the measured qn(Z) is obtained from the true one by adding noise to each Zt, yielding
To determine σn2 and Vt, we randomly select from the full available set of Ns samples, 200 training sets of n samples. For each training set, we calculate the Z values of all genes, and the variance of the resulting “measured” distribution. Thus, we end up with 200 variances obtained from the 200 training sets of n samples. Denote by V(n) the average of these 200 measured variances; this value is our estimate of var[qm(Z)] obtained for n samples. Repeating this procedure for n = n0, …, Ns yields a series of variances V(n). Because the noise is due to the finite number n of samples in a training set, the variance of the noise approaches zero as n → ∞; hence, extrapolation of V(n) to n = ∞ yields our estimate of Vt.
Motivated by the form σn2 = 1/(n − 3) given by Fisher (25, 26), we fit the measured V(n) to
where b < 0, and hence Vt = c. We see from Eqs. 3 and 4 that if the noise is uncorrelated, a·(n − 3)b = σn2. Indeed, we get for most data sets a ≈ 1 and b ≈ −1 (see Table 2, which is published as supporting information on the PNAS web site). Note that our analytical calculation assumes uncorrelated noise. To test this assumption, we estimate the variance of the noise in an independent, more direct way (see below); deviation of this estimate, σ̂n2, from a·(n − 3)b implies that the noise is correlated, and Assumption 3 of our analytical method does not hold. We claim that when this assumption breaks down, by setting
we create an “effective problem” with uncorrelated noise, which provides a good approximation to the original problem. To prove our claim we used Eq. 5 to get our analytical prediction of f as a function of n. Comparison with simulations (see below) reveals good agreement, which supports our claim.
Simulations to Measure the Distribution of f.
To perform these simulations, we created a model that enables us to generate an unlimited number of samples. The motivation for generating the samples in this particular way is described in Supporting Text. Simulations were performed by measuring, for each gene i, the mean and variance of its expression values, first over all good prognosis samples, yielding μg(i) and σg(i), and then over all poor prognosis samples, yielding μp(i) and σp(i). These means and variances were used to create, for each gene, two Gaussians, G(μg(i), σg(i)) and G(μp(i), σp(i)), approximating, for n = Ns, the probability distribution of the gene expression over the good- and poor-prognosis samples, respectively. Note that the true distributions were those corresponding to n = ∞. Therefore, the aforementioned Gaussians had to be rescaled to approximate the true distributions. This rescaling was done by adjusting the difference between the means of each pair of Gaussians, μg(i) − μp(i), so that the resulting distribution of Z values would fit the true one (see details in Supporting Text). The ultimate set of Ng pairs of Gaussians was used to create artificial good- and poor-prognosis samples in the following way. An artificial poor (good) prognosis patient was generated by drawing Ng gene expression values from the Ng Gaussians of the poor (good) prognosis population. In this way, we were able to generate an unlimited number of samples (training cohorts), which allowed us to obtain simulation results for any desired n. We generated the PGL of a given training cohort of n samples by calculating for each gene its Z, ranking the genes according to this score and selecting the αNg top-ranking genes as members of the PGL. We repeated this procedure for 1,000 different cohorts, ending up with 1,000 PGLs. We then calculated the overlap f between 500 independent pairs of PGLs to obtain the distribution of f.
Independent Estimate of the Noise.
To test directly the validity of Assumption 2, an independent estimate of the noise was obtained by randomly drawing 100 cohorts of n samples (see Simulations to Measure the Distribution of f) and for each gene measuring Z for every cohort. We then calculated the variance of these 100 values of Z and averaged it over all genes. The result is our independent estimate σ̂n2 (see Fig. 5).
Extrapolation.
By using the data of a particular experiment, we determined Pn,α(f) analytically and by simulations. The advantages of the analytic method are obvious; the disadvantage is that the calculation relies on the validity of certain assumptions, which need to be tested for each data set. Discrepancy between the analytic results and simulations (that are based on the data) indicates that some of the assumptions do not hold; in such a case we rely on the simulations, which are extrapolated to the regime (e.g., number of training samples n) of interest, which usually lies beyond the current experiment’s range. To extrapolate simulation results, the analytical function was multiplied by the factor that yielded the best fit to the simulation curve.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ridgefield Foundation and the Wolfson Foundation, by European Community FP6 funding, and by the European Community’s Human Potential Program under Contract HPRN-CT-2002-00319, Statistical Physics of Information Processing and Combinatorial Optimisation.
Abbreviations
- PGL
predictive gene list.
Footnotes
Conflict of interest statement: No conflicts declared.
§Because different platforms were used, the maximal possible number of shared genes is 55.
Actually, the absolute values |C| matter for ranking genes, because negative correlation is as informative as positive.
Ct corresponds to measuring C over an infinite number of samples (here, over all of the early discovery cancer patients in the world). Obviously, the numbers Ct are not known, but they exist.
References
- 1.Early Breast Cancer Trialists’ Collaborative Group. Lancet. 1998;352:930–942. [PubMed] [Google Scholar]
- 2.Bair E., Tibshirani R. PLoS Biol. 2004;2:5011–5022. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer D. G., Kardia S. L., Huang C. C., Giordano T. J., Levin A. M., Misek D. E., Lin L., Chen G., Gharib T. G., Thomas D. G., et al. Nat. Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 4.Khan J., Wei J. S., Ringner M., Saal L. H., Ladanyi M., Westermann F., Berthold F., Schwab M., Antonescu C. R., Peterson C., et al. Nat. Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenwald A., Wright G., Chan W. C., Connors J. M., Campo E., Fisher R. I., Gascoyne R. D., Muller-Hermelink H. K., Smeland E. B., Giltnane J. M., et al. N. Engl. J. Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 6.Yeoh E. J., Ross M. E., Shurtleff S. A., Williams W. K., Patel D., Mahfouz R., Behm F. G., Raimondi S. C., Relling M. V., Patel A., et al. Cancer Cells. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharjee A., Richards W. G., Staunton J., Li C., Monti S., Vasa P., Ladd C., Beheshti J., Bueno R., Gillette M., et al. Proc. Natl. Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramaswamy S., Ross K. N., Lander E. S., Golub T. R. Nat. Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 9.van’t Veer L. J., Dai H., van de Vijver M.J., He Y. D., Hart A. A., Mao M., Peterse H. L., van der Kooy K., Marton M. J., Witteveen A. T., et al. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 10.van de Vijver M. J., He Y. D., van’t Veer L. J., Dai H., Hart A. A., Voskuil D. W., Schreiber G. J., Peterse J. L., Roberts C., Marton M. J., et al. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Klijn J. G., Zhang Y., Sieuwerts A. M., Look M. P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M. E., Yu J., et al. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 12.Sorlie T., Perou C. M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M. B., van de Rijn M., Jeffrey S. S., et al. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West M., Blanchette C., Dressman H., Huang E., Ishida S., Spang R., Zuzan H., Olson J. A., Jr., Marks J. R., Nevins J. R. Proc. Natl. Acad. Sci. USA. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ein-Dor L., Kela I., Getz G., Givol D., Domany E. Bioinformatics. 2005;21:171–178. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 15.Michiels S., Koscielny S., Hill C. Lancet. 2005;365:488–492. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 16.Ioannidis J. P. A. Lancet. 2005;365:454–455. doi: 10.1016/S0140-6736(05)17878-7. [DOI] [PubMed] [Google Scholar]
- 17.Lonning P. E., Sorlie T., Borresen-Dale A. L. Nat. Clin. Pract. Oncol. 2005;2:26–33. doi: 10.1038/ncponc0072. [DOI] [PubMed] [Google Scholar]
- 18.Ahmed A. A., Brenton J. D. Breast Cancer Res. 2005;7:96–99. doi: 10.1186/bcr1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brenton J. D., Carey L. A., Ahmed A. A., Caldas C. J. Clin. Oncol. 2005;23:7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 20.Weigelt B., Peterse J. L., van’t Veer L. J. Nat. Rev. Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 21.Hughes G. F. IEEE Trans. Information Theory. 1969;15:615–618. [Google Scholar]
- 22.Jain A. K., Waller W. G. Pattern Recognit. 1978;10:365–374. [Google Scholar]
- 23.Valiant L. G. Commun. ACM. 1984;27:1134–1142. [Google Scholar]
- 24.Iizuka N., Oka M., Yamada-Okabe H., Nishida M., Maeda Y., Mori N., Takao T., Tamesa T., Tangoku A., Tabuchi H., et al. Lancet. 2003;361:923–929. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- 25.Fisher R. A. Biometrika. 1915;10:507–521. [Google Scholar]
- 26.Fisher R. A. Metron. 1921;1:3–32. [Google Scholar]
- 27.Huang K. Statistical Mechanics. New York: Wiley; 1963. [Google Scholar]
- 28.Bleistein N., Handelsman R. A. Asymptotic Expansions of Integrals. New York: Dover; 1986. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.