Abstract
Mutation in superoxide dismutase-1 (SOD1), which is a cause of ALS, alters the folding patterns of this protein. Accumulation of misfolded mutant SOD1 might activate endoplasmic reticulum (ER) stress pathways. Here we show that transgenic mice expressing ALS-linked SOD1 mutants exhibit molecular alterations indicative of a recruitment of ER's signaling machinery. We demonstrate by biochemical and morphological methods that mutant SOD1 accumulates inside the ER, where it forms insoluble high molecular weight species and interacts with the ER chaperone immunoglobulin-binding protein. These alterations are age- and region-specific, because they develop over the course of the disease and occur in the affected spinal cord but not in the nonaffected cerebellum in transgenic mutant SOD1 mice. Our results suggest a toxic mechanism for mutant SOD1 by which this ubiquitously expressed pathogenic protein could affect motor neuron survival and contribute to the selective motor neuronal degeneration in ALS.
Keywords: neurodegeneration, protein misfolding
ALS is the most common adult-onset paralytic disease characterized by a loss of motor neurons in the cerebral cortex, brainstem, and spinal cord. Insights into the neurodegenerative mechanisms followed the discovery that dominant mutations in the gene for superoxide dismutase-1 (SOD1) cause familial ALS. Overexpression of SOD1 mutants in rodents recapitulates ALS clinical and pathological hallmarks through a toxic gain of function (1). Many mutations in SOD1 decrease its stability and increase its unfolding rates and propensity to aggregate (2). High molecular weight complexes of SOD1 are observed in mammalian cells and spinal cords of transgenic mice expressing this mutant protein (3). In these animals, intracellular ubiquitin-positive proteinaceous inclusions are also often seen in spinal cord motor neurons and, in some cases, in neighboring astrocytes (3–5). These findings posit that accumulation of misfolded mutant SOD1 could contribute to the demise of motor neurons.
Endoplasmic reticulum (ER) stress signaling, otherwise known as the unfolded protein response (UPR), is triggered by an increased load of misfolded proteins in the organelle (6). Herein, we show that transgenic mice expressing mutant SOD1 exhibit age- and region-specific molecular alterations indicative of a broad recruitment of ER signaling pathways, including caspase-12, a prototypical ER cell death effector (7). We also show that mutant SOD1, and to a lesser extent wild-type SOD1 (SOD1WT), do accumulate in the ER. Within this organelle, mutant, but not SOD1WT, forms high molecular weight species and interacts with the ER chaperone immunoglobulin-binding protein (BiP), which is a key component of the ER misfolded protein recognition machinery (6). The preferential accumulation of mutant SOD1 in the ER in the spinal cord cells and the ensuing stress response may represent novel aspects of motor neuron degeneration in this ALS model.
Results
UPR Transcription Factors Are Activated in Affected Transgenic SOD1G93A Mice.
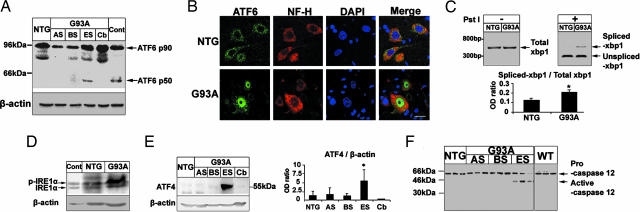
ATF6 is a key transcription factor in the mammalian UPR, synthesized as a membrane-bound precursor and activated by site-directed proteolysis (8). Spinal cord levels of ATF6 mRNA (not shown) or full-length protein (90 kDa) did not differ between transgenic mice expressing the human SOD1 (hSOD1) mutant G93A (SOD1G93A), which is the most widely studied model of ALS (9), and their nontransgenic counterparts (Fig. 1A). Cleaved ATF6 (50 kDa) was detected in spinal cords of transgenic SOD1G93A mice at end stage (5 months old), to a lesser extent at early symptomatic stage (≈3–4 months old), but not at asymptomatic stage (1–2 months old) (Fig. 1A and Fig. 6, which is published as supporting information on the PNAS web site). ATF6 was also cleaved in symptomatic transgenic mice expressing another hSOD1 mutant, i.e., SOD1G85R (Fig. 6), which promotes a rapidly progressive ALS-like phenotype (5). Conversely, cleaved ATF6 was not detected in cerebella of end-stage transgenic SOD1G93A mice or spinal cords of transgenic mice expressing hSOD1WT (Figs. 1A and 6). ATF6 immunofluorescence of spinal cord motor neurons was perikaryal in nontransgenic mice but often nuclear in symptomatic transgenic SOD1G93A mice (Fig. 1B), consistent with the fact that cleaved ATF6 translocates into the nucleus (10).
Fig. 1.
Activation of UPR transcription factors and apoptosis-associated ER factors in transgenic SOD1G93A mice. (A) ATF6 immunoblot in spinal cords from transgenic SOD1G93A mice (G93A) at asymptomatic stage (AS, 1 month old), beginning of symptoms (BS, 3–4 months old), end stage (ES, 5 months old), and from their age-matched nontransgenic littermates (NTG) or from the cerebellum (Cb) of ES-G93A mice. Thapsigargin-treated PC12-cells were used as a positive control (Cont.). Full-length ATF6 is p90, and cleaved ATF6 is p50. (B) Immunolocalization of ATF6 in spinal cord sections: ATF6 (green), neurofilament (red), and DAPI (blue). (Scale bar, 20 μm.) (C) Ratios of IRE1-mediated splicing form of XBP1 (spliced -XBP1) over total XBP1 mRNA. ∗, P < 0.05; Student's t test. (D) Immunoblot for phosphorylated IRE1α (p-IRE1α) and nonphosphorylated IRE1α (IRE1α). (E) ATF4 immunoblot. ∗, P < 0.05; one-way ANOVA, Student–Newman–Keuls test. (F) Immunoblot of procaspase-12 (≈55 kDa) and its cleaved fragment (≈42 kDa). All values are means ± SEM (n = three to six per group). Analyses were performed in ES-G93A and age-matched NTG mouse spinal cords unless indicated otherwise.
X-box-binding protein (XBP1) protein is another transcription factor in the mammalian UPR whose activation relies on protein kinase endoribonuclease 1 (IRE1)-mediated splicing of its mRNA (11). XBP1 mRNA levels in symptomatic transgenic SOD1G93A and age-matched nontransgenic spinal cords did not differ (Fig. 1C). However, spliced XBP1 mRNA was more abundant in symptomatic transgenic SOD1G93A than in age-matched nontransgenic spinal cords (Fig. 1C). Splicing of XBP1 mRNA requires activation of IRE1, which implies IRE1 oligomerization and transautophosphorylation (12). We found that the spinal cord contents of phosphorylated IRE1α were higher in transgenic SOD1G93A mice, whereas the levels of unphosphorylated IRE1α were similar between symptomatic transgenic SOD1G93A and age-matched nontransgenic mice (Fig. 1D).
Activation of PKR (the double-stranded RNA-activated protein kinase)-like ER kinase (PERK) induces transcription factor ATF4 through phosphorylation of eIF2α. To assess the involvement of the PERK-mediated UPR pathway, we investigated ATF4 protein expression in SOD1G93A mouse spinal cords. Immunoblotting revealed more ATF4 in spinal cords from symptomatic transgenic SOD1G93A mice compared to earlier stages of the disease in these mice as well as in nontransgenic controls (Fig. 1E). Increased ATF4 expression was specific to spinal cords (Fig. 1E). Collectively, the above investigations indicate that a broad ER stress response occurs in affected regions such as the spinal cord but not in unaffected regions such as the cerebellum over the course of the disease in transgenic mice expressing mutant but not SOD1WT.
Apoptosis-Associated ER Factors Are Activated in Transgenic SOD1G93A Mice.
Caspase-12 is an ER stress-related cell death effector (7). Levels of procaspase-12 in spinal cord did not typically differ among the groups of mice (Fig. 1F and Fig. 7A, which is published as supporting information on the PNAS web site), although it was reduced in some end-stage transgenic SOD1G93A mice (Fig. 1F). Active caspase-12 was detected in spinal cord of both symptomatic transgenic SOD1G93A and SOD1G85R mice but not in that of the other groups (Figs. 1F and 7A). Spinal cord caspase-12 catalytic activity was also higher in paralyzed transgenic SOD1G93A mice compared with their nontransgenic controls (Fig. 7B). These findings show that the broad ER stress-related transcriptional response occurs in the spinal cord of transgenic mice expressing mutant SOD1 mice in concert with the activation of caspase-12.
SOD1WT and Mutant SOD1 Are Present in the ER.
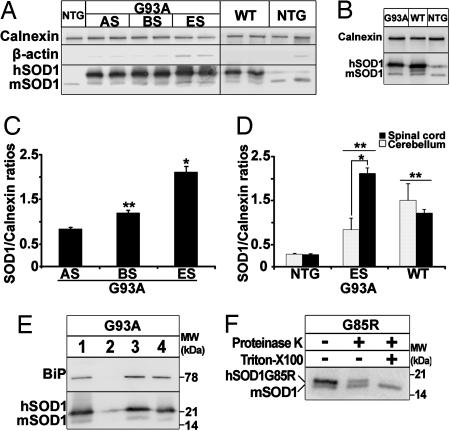
We next sought to investigate whether the observed UPR could be linked to an accumulation of SOD1 in the ER. Purified microsomal fractions enriched in ER membranes were isolated from the spinal cord of different mouse genotypes. Upon equal loading of protein extracts from spinal cord microsomal fractions of transgenic SOD1G93A and SOD1WT mice, both hSOD1 and mouse SOD1 (mSOD1) were detected by immunoblot (Fig. 2A). ER integral membrane protein calnexin was abundant in the microsomal fractions, whereas synaptosomal protein synaptotagmin, mitochondrial protein cytochrome oxidase, and cytosolic/nuclear protein β-actin were virtually absent (Fig. 2A and Fig. 8, which is published as supporting information on the PNAS web site), ruling out that the SOD1 detection was merely a contamination of the microsomal fractions.
Fig. 2.
SOD1WT and mutant SOD1 are present in the ER. (A) Immunoblot of spinal microsomal fractions of SOD1G93A mice at different disease stages and 5-month-old transgenic SOD1WT and nontransgenic mice. The membranes were also probed for the ER resident protein calnexin as internal standard and β-actin. (B) Immunoblot of cerebellar microsomal fractions from mice of the different genotypes. (C) Quantification of microsomal SOD1 content over the course of disease relative to calnexin by optical density analysis (n = three to six per group; one-way ANOVA; ∗, P < 0.01; ∗∗, P < 0.05). (D) Comparison of microsomal SOD1 content in spinal cord and cerebellum. In nontransgenic and transgenic SOD1WT mice, there is no difference in the SOD1:calnexin ratios between spinal and cerebellar microsomal fractions (P > 0.05). In transgenic SOD1G93A mice, the microsomal SOD1:calnexin ratios are higher in the spinal cord than in the cerebellum (n = three to five per group, two-way ANOVA, Student–Newman–Keuls test; ∗, P < 0.001). SOD1:calnexin ratios in microsomal fractions from transgenic SOD1WT and SOD1G93A mice are higher than in nontransgenic mice in both areas (n = three to five per group; two-way ANOVA; Student—Newman–Keuls method; ∗∗, P < 0.05). (E) SOD1 is protected from surface biotinylation under conditions of intact microsomal membranes. Luminal protein BiP is used as positive control. Lane 1, total input; lane 2, eluate from intact membranes. Little SOD1 and no BiP are biotinylated in the absence of detergent. Lane 3, flow-through from intact membrane. Abundant SOD1 and BiP are found among the nonbiotinylated proteins. Lane 4, eluate from solubilized membranes. Both SOD1 and BiP are now biotinylated. (F) SOD1 is protected from proteinase K proteolysis in intact microsomal membranes and digested after addition of detergent. The mSOD1 is resistant to proteolysis in any condition (lower band).
Quantification of SOD1 in spinal cord microsomal fractions revealed that, relative to calnexin, hSOD1 contents rose over the course of disease in transgenic SOD1G93A mice (Fig. 2C). At 5 months of age, spinal microsomal hSOD1 in transgenic SOD1WT mice was ≈4 times higher than spinal microsomal mSOD1 in nontransgenic controls (Fig. 2D). Spinal microsomal hSOD1 in end-stage transgenic SOD1G93A mice was, respectively, ≈7 and ≈1.7 times higher than spinal microsomal mSOD1 in nontransgenic controls and hSOD1 in transgenic SOD1WT mice (Fig. 2D). Spinal cord hSOD1:calnexin ratios did not differ from cerebellar ratios in both 5-month-old transgenic SOD1WT and nontransgenic mice (Fig. 2 B and D). However, these ratios were significantly higher in spinal cords than in cerebella of age-matched symptomatic transgenic SOD1G93A mice (Fig. 2 B and D).
To test whether hSOD1 from transgenic SOD1G93A rodents found in the microsomal fractions resides inside the ER or is merely associated with the cytosolic side of its membranes, we labeled the microsomal surface proteins with a membrane-impermeable N-hydroxysulfosuccinimidobiotin analog and isolated biotinylated proteins with an avidin gel. Microsomal proteins were eluted from the avidin gel, as described in Materials and Methods, and were immunoblotted for SOD1. In the absence of Triton X-100, microsomal biotinylated proteins contained only traces of hSOD1 (Fig. 2E). Conversely, the flow-through (i.e., microsomal nonbiotinylated proteins) contained copious amounts of hSOD1 and some mSOD1 (Fig. 2E). In the presence of 1% Triton X-100, microsomal membranes were permeabilized; biotinylated proteins now comprised abundant amounts of hSOD1 (Fig. 2E), confirming that hSOD1 could react with the N-hydroxysulfosuccinimidobiotin analog. We also used the resident luminal ER protein BiP as an internal control. BiP was completely protected from biotinylation when membranes were intact (i.e., without Triton X-100), but, upon membrane permeabilization by 1% Triton X-100, BiP became highly biotinylated (Fig. 2E).
To confirm the biotinylation results, we used spinal cord microsomal preparations for a proteinase K protection assay; because SOD1G85R is one of the rare SOD1 proteins sensitive to proteinase K digestion (13), we prepared spinal microsomal fractions from transgenic SOD1G85R mice. Without Triton X-100, some SOD1G85R molecules were spared from proteinase K digestion (Fig. 2F), whereas with 1% Triton X-100, they were completely digested by proteinase K (Fig. 2F). mSOD1, which is resistant to proteinase K (13), was unaffected by this treatment even after permeabilization of microsomal membranes (Fig. 2F). Collectively, these data indicate that: (i) SOD1 accumulates within the ER in a genotypic-, time-, and region-dependent manner; (ii) not only SOD1G93A, but at least one other SOD1 mutant, namely SOD1G85R, accumulates in the spinal ER; and (iii) the content of mutant SOD1 in the ER exceeds that of SOD1WT despite the fact that the total cellular expression of the former is lower than of the latter (9).
Microscopical Demonstration of ER Location of SOD1 Within Spinal Motor Neurons.
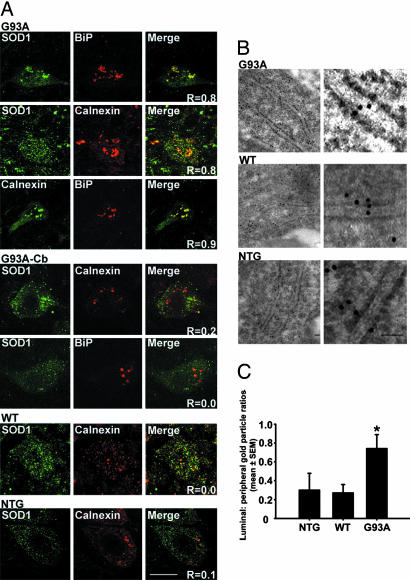
To confirm SOD1 accumulation in the ER, tissue sections from 5-month-old transgenic SOD1G93A and SOD1WT and nontransgenic mice were immunostained for SOD1, calnexin, and BiP (Fig. 3A). Confocal microscopy analysis showed colocalization of calnexin and BiP (overlap coefficient: Rover = 0.9) (14) and of hSOD1 with both calnexin and BiP (Rover = 0.8 and 0.8, respectively) within spared large motor neurons of the spinal anterior horn of paralyzed transgenic SOD1G93A mice (Fig. 3A). Much lower colocalization between SOD1 and calnexin (Rover = 0.2 and 0.0, respectively) was observed within cerebellar dentate nucleus cells of these animals (Fig. 3A). Likewise SOD1 colocalization with calnexin was modest (Rover = 0.0 and 0.1, respectively) within spinal motor neurons of, respectively, 5-month-old transgenic SOD1WT and nontransgenic mice (Fig. 3A).
Fig. 3.
Microscopical demonstration of ER location of SOD1 in spinal motor neurons. (A) Immunoflourescence analysis of SOD1 colocalization with ER markers calnexin and BiP in spinal cord and cerebellar sections. (Scale bar, 20 μm.) (B) Luminal ER labeling of SOD1 in immunoelectron microscopy. (Scale bar, 50 nm.) (C) Quantification of gold particles inside the ER lumen and in the ER periphery. Values represent means ± SEM (n = 23–30 ER profiles per group). ∗, P < 0.05; one-way ANOVA, Student–Newman–Keuls method.
Immunoelectron microscopy was also performed on these tissue samples by using a rabbit polyclonal anti-SOD1 antibody that recognizes hSOD1WT and hSOD1G93A to the same extent (Fig. 9, which is published as supporting information on the PNAS web site). This ultrastructural analysis revealed SOD1 immunogold labeling over the ER in spinal cord sections from both symptomatic transgenic SOD1G93A and age-matched transgenic SOD1WT mice (Fig. 3B). Only scant ER immunolabeling was seen in spinal cord sections from nontransgenic controls (Fig. 3B) and none upon omission of the anti-SOD1 antibody (not shown). Quantification of SOD1 immunolabeling in spinal cord motor neurons revealed that the number of gold particles over the ER was ≈30% of that within the cytosol in both 5-month-old transgenic SOD1WT and nontransgenic mice (Fig. 3C; Table 1, which is published as supporting information on the PNAS web site). In contrast, the number of gold particles on the ER was ≈73% of that within the cytosol in aged-matched transgenic SOD1G93A mice (Fig. 3C; Table 1). Thus, the SOD1 ER/cytoplasm gold particle ratio is ≈2.7-fold higher in transgenic SOD1G93A than in transgenic SOD1WT mice, although the number of cytosolic gold particles is 15% lower in the former than in the latter (Table 1). Contrasting with spinal cord motor neurons, no significant genotypic difference in ER/cytoplasm SOD1 gold particle numbers was found in cerebellar Purkinje cells (Table 1). These morphological data suggest that, specifically in spinal cord motor neurons, mutant SOD1 accumulates in the ER more than either hSOD1WT or mSOD1.
Microsomal Fractions Contain High Molecular Weight Species of Mutant SOD1.
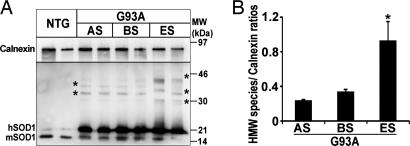
Because SOD1 mutants readily aggregate, we asked whether hSOD1 accumulates in the ER from transgenic SOD1G93A mice as insoluble species. In addition to monomeric hSOD1, high molecular weight complexes were evident in microsomal fractions from transgenic SOD1G93A mouse spinal cords on overexposed immunoblots (Figs. 4A and 8). Like monomeric hSOD1, high molecular weight complexes became more abundant in microsomal fractions over the course of the disease (Fig. 4). Conversely, no high molecular weight SOD1 species were found in the spinal microsomal fractions of either transgenic SOD1WT or nontransgenic mice (Figs. 4A and 8) or in the microsomal fractions extracted from the cerebellum of the three mouse genotypes studied (not shown). The genotypic- and age-dependent increase in the amount of SOD1 aggregates in spinal microsomal fractions was confirmed by size-exclusion filter assay (15) (not shown). The above data indicate that the accumulation of high molecular weight species in microsomal fractions is specific to mutant SOD1 and restricted to affected tissues.
Fig. 4.
High molecular weight aggregates of mutant SOD1 in spinal microsomal fractions. (A) Age-dependent increase of high molecular weight SOD1 species in SOD1G93A spinal microsomal fractions in overexposed immunoblot of spinal microsomal fractions of SOD1G93A mice. Asterisks indicate high molecular weight species. (B) Quantification of high molecular weight species over the course of the disease relative to the ER resident protein calnexin by optical density analysis. Values represent means ± SEM (n = three per group). ∗, P < 0.05; one-way ANOVA, Student–Newman–Keuls method.
Mutant SOD1G93A but Not hSOD1WT or mSOD1 Interacts with BiP.
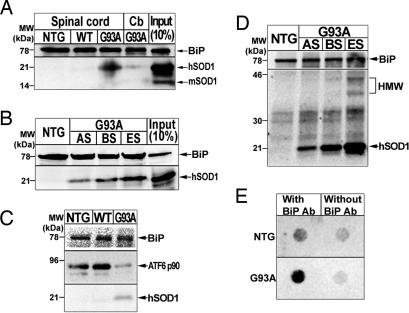
Among the ER molecules, BiP can engage the ER stress response after its binding to a broad range of misfolded proteins (6). Coimmunoprecipitation studies revealed that SOD1G93A, but not SOD1WT, interacts with BiP in spinal microsomal fractions (Fig. 5A and Fig. 10A, which is published as supporting information on the PNAS web site) in an age-dependent manner (Figs. 5B and 10B). No such interaction could be detected in cerebellar microsomal fractions (Figs. 5A and 10B). We also found that coimmunoprecipitation with anti-BiP antibody pulled down more SOD1 and less ATF6 from spinal microsomal fractions of symptomatic transgenic SOD1G93A mice than from those of age-matched transgenic SOD1WT and nontransgenic mice (Fig. 5C). In unstressed cells, BiP binds to the luminal domains of IRE1, PERK and ATF6, keeping them in an inactive state (6). As unfolded proteins accumulate in the ER lumen, BiP dissociates from these ER stress sensors (6). Our data thus suggest that during the course of the disease, BiP is titrated from the luminal domain of ATF6 by the ER accumulation of mutant SOD1G93A in microsomal fractions from spinal cords. Of note, coimmunoprecipitation showed that BiP pulled down not only microsomal SOD1 monomers but also high molecular weight species (Fig. 5D). To confirm these results, we performed a size-exclusion filter assay on spinal microsomal samples immunoprecipitated with BiP antibody. In spinal cords from 5-month-old nontransgenic mice, negligible amounts of SOD1 were trapped, as was the case for samples immunoprecipitated without BiP antibody (Fig. 5E). Conversely, in spinal microsomal fractions from end-stage transgenic SOD1G93A mice, considerable amounts of trapped SOD1 were detected (Fig. 5E). These data indicate that BiP also associates with aggregated SOD1 in the ER of transgenic SOD1G93A mice.
Fig. 5.
Mutant SOD1, but not hSOD1WT or mSOD1, interacts with BiP in the microsomal fraction in vivo. (A) Immunoprecipitation with an anti-BiP antibody followed by immunoblot using anti-BiP and anti-SOD1 antibodies. (B) Immunoprecipitation with an anti-BiP antibody using spinal microsomal fractions of SOD1G93A mice at different disease stages and nontransgenic mice. Input represents 10% of protein from spinal microsomal fraction. (C) Immunoprecipitation with an anti-BiP antibody followed by immunoblot using anti-BiP, -SOD1, and -ATF6 antibodies. (D) High molecular weight (HMW) SOD1 species in microsomal fractions interact with BiP. (E) Immunoprecipitated with/without anti-BiP antibody followed by a size-exclusion filter assay.
To identify the region of BiP responsible for the interaction with mutant SOD1, we used in vitro GST pull-down assays using fusion GST-BiP truncated constructs in which only the 44-kDa N-terminal ATPase domain [N44 fragment (residue 1–391), the 12.4-kDa peptide-binding domain (PBD, BiP residues 392–509), or the 13.7-kDa α-helical C-terminal tail (CTT, BiP residues 516–636)] were deleted (Fig. 11A, which is published as supporting information on the PNAS web site). As shown in Fig. 11B, both the ΔN44 and ΔCTT mutants contained the PBD, and both, like full-length BiP, bound to SOD1G93A; similar results were obtained for SOD1G85R (not shown). However, ΔPBD that retained both the N44 domain and the CTT region exhibited minimal interaction with hSOD1G93A (Fig. 11B), suggesting that the BiP PBD domain is required for optimal binding to mutant SOD1.
Discussion
ALS-linked mutations destabilize the precursor monomer, weaken the dimer interface, or both (16), and promote aggregation (3). In keeping with mutant SOD1 folding abnormalities, herein we show that the constitutive expression of SOD1G93A and SOD1G85R, which are two of the most extensively characterized ALS-linked SOD1 mutants in transgenic rodents, are associated with a recruitment of key mediators of the UPR (6). Among the three ER-resident transmembrane proteins identified as proximal sensors of UPR, we found an age-dependent activation of the basic leucine-zipper transcription factor ATF6 specifically in degenerating areas, such as spinal cord, but not in unaffected areas, such as cerebellum, of transgenic SOD1G93A mice (Figs. 1A and 6). Within the diseased tissues, ATF6 immunostaining was identified in the nuclei of spared motor neurons (Fig. 1B), implying that ATF6 did transit to the Golgi of these cells in response to a buildup of misfolded proteins, where it was cleaved into the active transcription factor (8). Corroborating this interpretation is the demonstration of mutant SOD1 in the immunoisolated transGolgi network of spinal cords from transgenic SOD1G37R mice (17). Among the other ER-resident transmembrane proteins that transduce the UPR, we found evidence of increased IRE1 phosphorylation (Fig. 1D) and ensuing splicing of XBP1 (Fig. 1C). Likewise, eIF2α (not shown) and ATF4 (Fig. 1E), whose phosphorylation and expression are typically modulated by PERK, were both increased in symptomatic transgenic SOD1G93A mice. These data suggest that, over the course of the disease, motor neurons expressing ALS-linked SOD1 mutants are the site of a UPR aimed at improving their capacity to withstand the ER accumulation of misfolded mutant SOD1.
The UPR can also activate apoptosis via activation of caspase-12 (7). In transgenic SOD1G93A and SOD1G85R mice, we (Figs. 1F and 7) and Wootz et al. (18) found that cleaved activated caspase-12 progressively appears in spinal cord lysates as the disease worsens. Caspase-12 activation can be mediated by Fas (CD95) ligand in L929sAhFas fibrosarcoma cells Fas (CD95) (19) and by caspase-7 in human embryonic kidney 293T cells (20). In turn, caspase-12 can activate caspase-9 and -3 (20). Cultured embryonic motor neurons are susceptible to Fas ligand (21), and activation of both caspase-7 and -3 occurs in transgenic SOD1G93A motor neurons (22) after the mitochondrial release of cytochrome c and the activation of caspase-9 (22). Therefore, caspase-12 may function in ALS both as a link between Fas-mediated extrinsic death signals and intrinsic executioner caspases and as a feed-forward amplifying loop for the mitochondrial-dependent apoptotic pathway, which is instrumental in motor neuron death (22).
SOD1 is a cytosolic protein that lacks organelle-targeting sequences and is synthesized by free ribosomes (23). Nonetheless, SOD1 is secreted by multiple cell types by a brefeldin-A-sensitive mechanism (17, 24) and colocalizes with ER markers in transfected COS7 and Neuro2A cells (17, 25). Our study shows that both hSOD1WT and mSOD1, as well as mutant G93A and G85R, can be identified in the ER of rodent spinal cords (Figs. 2–4). The association of mutant SOD1 with the ER of the spinal cord is not a simple consequence of overexpression, because accumulation of SOD1G93A in the ER exceeds that of SOD1WT (Fig. 2 A and D) despite its lower cellular expression; SOD1G93A, 4.1 ng/μg total protein vs. SOD1WT, 6.7 ng/μg total protein (9). This genotypic difference was not observed in unaffected tissues such as cerebellum (Fig. 2 B and D) nor was it seen for other abundant cytoplasmic protein such as GAPDH (not shown). Thus, mutant SOD1 association with the ER does not reflect an indiscriminate import, residual cytoplasmic contamination of the purified microsomal fractions, or a passive entry through damaged microsomal membranes.
How SOD1 is posttranslationally imported within these organelles remains enigmatic. Proteins are transported across the ER membrane through a channel formed by the heterotrimeric Sec61p complex in partnering with the Sec62/63 complex and the ER-resident Hsp70 chaperone BiP (26). In this capacity, BiP regulates the channel opening (27) and, upon binding to the incoming protein, it acts as a molecular ratchet, optimizing the inward movement of the substrate through the opened channel (28). Our demonstration of mutant SOD1 binding to the Hsp70-like domain of BiP (Fig. 11) is consistent with the possibility that BiP-associated translocon complex mediates SOD1 import in the ER. Correlatively, mutant SOD1 may be more efficiently imported inside the ER than SOD1WT, because BiP interacts preferentially with misfolded proteins. Thus, more mutant SOD1 may be detected in the ER by virtue of a greater import. Alternatively, a lower disposition of mutant SOD1 by the ER, perhaps because of its tendency to form insoluble species, could also account for the greater abundance of the mutant protein in this organelle. In addition to its role in the import of proteins, BiP negatively regulates ATF6, IRE1, and PERK involved in the UPR by binding to their luminal domains (6). Upon ER stress, these proteins are freed and activated, because BiP preferentially binds to accumulating misfolded proteins. In agreement with this molecular model, we found in spinal microsomal preparations more BiP bound to mutant SOD1 than to ATF6 in symptomatic transgenic SOD1G93A compared to age-matched nontransgenic littermates and transgenic SOD1WT mice (Fig. 5C). Based on our results, we propose that BiP promotes both the preferential translocation of misfolded SOD1 into the ER and the activation of the ER stress response in the mutant SOD1 model of ALS.
Compared with its wild-type counterpart, mutant SOD1 in the spinal ER was present both as monomers and insoluble high molecular weight complexes (Figs. 4A and 8). Although there are indications that neurodegeneration in ALS may not strictly result from a cell-autonomous process, we propose that the part of ALS pathogenesis that takes place inside motor neurons is multifactorial, revolves around protein aggregation, and includes ER stress. Minimal misfolded protein-mediated stress may suffice to cause a slow demise of motor neurons, which may explain the relentless progressive nature of the disease. The preferential accumulation of mutant SOD1 inside the ER of spinal cords, together with a selective activation of the UPR in spinal cord, may explain why expression of mutant SOD1, which occurs in all cells, would inflict greater damage on spinal motor neurons in familial ALS. Poorly addressed by this scenario, however, are the questions why most of the reported molecular alterations become conspicuous in end-stage animals only, and how this mutant SOD1-related deleterious cascade could apply to sporadic ALS. ER stress could indeed be a late pathogenic event that exacerbates, but does not initiate, ALS neurodegeneration. However, the use of whole tissue extracts may render its detection at the earlier stages of the disease uncertain. Thus, our late detection of ER stress may reflect more a caveat of the method used rather than its true time course within motor neurons. As for sporadic ALS, proteinaceous inclusions containing cystatin C or neurofilament are widely seen in spinal cord motor neurons from sporadic ALS cases. It is therefore worth considering that misfolded proteins other than mutant SOD1 might accumulate in the ER and recruit the UPR machinery in sporadic ALS.
Materials and Methods
Animals.
Both transgenic G1H that expressed ≈18 copies of hSOD1G93A gene and N1029 mice (The Jackson Laboratory), which expressed >10 copies of hSOD1WT gene, were used. On postnatal day 14, all mice were genotyped. For selected investigations, transgenic mice expressing hSOD1G85R as well as transgenic SOD1G93A rats were used (5, 29). For all experiments, the nontransgenic animals were strict littermates.
Protein Preparation and Immunoblot.
Total proteins from spinal cord and cerebellum were isolated as described (30). Positive control for ER stress-associated proteins was obtained by exposing differentiated PC12 cells to 1 μM thapsigargin (31). All Western blots were performed as described (30); the various primary antibodies used are listed in Supporting Text, which is published as supporting information on the PNAS web site.
RT-PCR and Detection for XBP1 mRNA Splicing Form.
Extraction of total RNA and RT-PCR were performed as described (30). The analysis of the splicing form of XBP1 mRNA was performed as described by Calfon et al. (11). Detailed information is provided in Supporting Text.
Subcellular Fractionation.
Tissue from selected CNS regions was homogenized in 1:10 (wt/vol) volumes of buffer containing 250 mM sucrose, 20 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, 2 mM EDTA, and a protease inhibitor (Complete mini, Roche Diagnostics) (pH 8) and centrifuged (800 × g, 10 min). This supernatant (10,000 × g, 20 min) and the subsequent one were centrifuged (100,000 × g, 1 h). The resulting supernatant was designated as cytosolic fraction. The pellet was resuspended in homogenization buffer and designated as microsomal fraction. Protein concentrations were determined by the bicinchoninic acid method (Pierce). Samples were analyzed by SDS/PAGE and immunoblot, as described above.
Biotinylation Assay.
Microsomal fractions of SOD1G93A transgenic rats and nontransgenic controls from fresh spinal cords were used in a cell surface biotinylation kit according to the manufacturer's instructions (Pierce). Detailed information is provided in Supporting Text. The processed samples were analyzed by immunoblot.
Proteinase K Protection Assay.
The microsomal fractions of the spinal cord homogenates of symptomatic SOD1G85R mice were incubated with or without 100 μg/ml proteinase K (Roche Diagnostics) in the presence or absence of 1% Triton X-100 for 20 min on ice. Proteolysis was terminated by addition of 2 mM phenylmethylsulfonyl fluoride (final concentration), and the products were analyzed by SDS/PAGE and immunoblot.
Immunofluorescence Analysis.
The immunohistochemical analyses were performed on 4% paraformaldehyde-fixed tissue by using our standard protocol (32). The various primary and secondary antibodies used are listed in Supporting Text. Nuclei were stained with DAPI (Molecular Probes) at a dilution of 1:3,000 in PBS for 30 min. Sections were examined by using confocal microscopy (Zeiss LSM 510 NLO Multiphoton Confocal Microscope). Rover values were calculated by using the colocalization analysis tool in the lsm software (14).
Immunoelectron Microscopy.
Spinal cords and cerebella of SOD1G93A, SOD1WT, and nontransgenic mice (two per group) were processed for immuno-EM as described in Supporting Text. Sections were examined in a JEOL 1200EX electron microscope. Gold particles within the ER and the cytosol were counted. Using a similar reasoning as Liu et al. (33), the cytosol was defined as the area within 30 nm of either side of the ER membrane.
Filter Trap Assay.
Microsomal fractions of spinal cord homogenates from SOD1G93A, and nontransgenic mice were prepared as described above. Total protein (150 μg) was immunoprecipitated with anti-BiP antibody, diluted with 20 vol of PBS containing 0.1% SDS, and filtered under vacuum through a prewetted 0.2-μm cellulose acetate membrane (Osmonics, Minnetonka, MN) using a dot blot device (Bio-Rad). After two washes under vacuum with PBS, the membranes were incubated in 5% dry milk in PBS for 1 h and subsequently analyzed by immunoblot.
Immunoprecipitations.
Microsomal fractions were resuspended in (2-[N-morpholino]ethanesulfonic acid (Mes)-buffered solution containing 25 mM Mes (pH 6.0), 75 mM NaCl, and 1% Triton X-100 with protease inhibitor mixture (Roche Diagnostics) and processed for immunoprecipitation as described in Supporting Text.
Statistics.
All values are expressed as the mean ± SEM unless stated otherwise. Differences between means were analyzed by Student's t test, differences among means were analyzed by using one- or two-way ANOVA. When ANOVA showed significant differences, pair-wise comparisons were performed by the Student–Newman–Keuls post hoc test. The null hypothesis was consistently rejected at the 0.05 level.
Supplementary Material
Acknowledgments
We thank Dr. David Ron (New York University School of Medicine, New York) for his advice on the study and his insightful comments on the manuscript, Dr. Susan Brown (Weill Medical College of Cornell University, New York) for the kind gift of transgenic SOD1G85R mice, and Dr. Linda M. Hendershot (St. Jude Children's Research Hospital, Memphis, TN) for providing an aliquot of the rabbit anti-BiP antibody. We are also grateful to Dr. Masashi Aoki and Prof. Yasuto Itoyama (Tohoku University, Sendai, Japan) for providing transgenic SOD1 G93A rats and to Dr. Elizabeth J. Ryu (Columbia University, New York) for providing the thapsigargin-treated PC12 cell extract. This study is supported by National Institutes of Health/National Institute of Neurological Disorders and Stroke Grants R01 NS38586 and NS42269; P50 NS38370; and P01 NS11766-27; National Institutes of Health/National Institute on Aging Grant R01 AG21617-01; National Institutes of Health/National Institute on Environmental Health Sciences Grant R21 ES013177; U.S. Department of Defense Grant DAMD 17-03-1; the Parkinson's Disease Foundation (New York); the Lowenstein Foundation; and the Muscular Dystrophy Association/Wings Over Wall Street. H.K. is a recipient of a fellowship from the Naito Foundation Subsidy for Inter-Institute Research. G.A. is recipient of the Charlotte Buehler Stipend from the Austrian Research Funds. M.N. is a recipient of the Gardner Fellowship from the Muscular Dystrophy Association.
Abbreviations
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- SOD1
superoxide dismutase-1
- hSOD1
human SOD1
- mSOD1
mouse SOD1
- SOD1WT
wild-type SOD1
- IRE1
protein kinase endoribonuclease
- XBP1
X-box-binding protein
- PERK
PKR (the double-stranded RNA-activated protein kinase)-like ER kinase
- BiP
immunoglobulin-binding protein.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Julien J. P. Cell. 2001;104:581–591. doi: 10.1016/s0092-8674(01)00244-6. [DOI] [PubMed] [Google Scholar]
- 2.Stathopulos P. B., Rumfeldt J. A., Scholz G. A., Irani R. A., Frey H. E., Hallewell R. A., Lepock J. R., Meiering E. M. Proc. Natl. Acad. Sci. USA. 2003;100:7021–7026. doi: 10.1073/pnas.1237797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston J. A., Dalton M. J., Gurney M. E., Kopito R. R. Proc. Natl. Acad. Sci. USA. 2000;97:12571–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruijn L. I., Houseweart M. K., Kato S., Anderson K. L., Anderson S. D., Ohama E., Reaume A. G., Scott R. W., Cleveland D. W. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 5.Bruijn L. I., Becher M. W., Lee M. K., Anderson K. L., Jenkins N. A., Copeland N. G., Sisodia S., Rothstein J. D., Borchelt D. R., Price D. L., et al. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 6.Schroder M., Kaufman R. J. Annu. Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B. A., Yuan J. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 8.Ye J., Rawson R. B., Komuro R., Chen X., Dave U. P., Prywes R., Brown M. S., Goldstein J. L. Mol. Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 9.Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., Deng H.-X., et al. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 10.Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calfon M., Zeng H., Urano F., Till J. H., Hubbard S. R., Harding H. P., Clark S. G., Ron D. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 12.Bertolotti A., Zhang Y., Hendershot L. M., Harding H. P., Ron D. Nat. Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 13.Ratovitski T., Corson L. B., Strain J., Wong P., Cleveland D. W., Culotta V. C., Borchelt D. R. Hum. Mol. Genet. 1999;8:1451–1460. doi: 10.1093/hmg/8.8.1451. [DOI] [PubMed] [Google Scholar]
- 14.Manders E. M., Verbeek F. J., Aten J. A. J. Microsc. 2006;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang J., Xu G., Gonzales V., Coonfield M., Fromholt D., Copeland N. G., Jenkins N. A., Borchelt D. R. Neurobiol. Dis. 2002;10:128–138. doi: 10.1006/nbdi.2002.0498. [DOI] [PubMed] [Google Scholar]
- 16.Lindberg M. J., Bystrom R., Boknas N., Andersen P. M., Oliveberg M. Proc. Natl. Acad. Sci. USA. 2005;102:9754–9759. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urushitani M., Sik A., Sakurai T., Nukina N., Takahashi R., Julien J. P. Nat. Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 18.Wootz H., Hansson I., Korhonen L., Napankangas U., Lindholm D. Biochem. Biophys. Res. Commun. 2004;322:281–286. doi: 10.1016/j.bbrc.2004.07.118. [DOI] [PubMed] [Google Scholar]
- 19.Kalai M., Lamkanfi M., Denecker G., Boogmans M., Lippens S., Meeus A., Declercq W., Vandenabeele P. J. Cell Biol. 2003;162:457–467. doi: 10.1083/jcb.200303157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao R. V., Hermel E., Castro-Obregon S., del Rio G., Ellerby L. M., Ellerby H. M., Bredesen D. E. J. Biol. Chem. 2001;276:33869–33874. doi: 10.1074/jbc.M102225200. [DOI] [PubMed] [Google Scholar]
- 21.Raoul C., Estevez A., Nishimune H., Cleveland D., deLapeyriere O., Henderson C., Haase G., Pettmann B. Neuron. 2002;35:1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 22.Guégan C., Przedborski S. J. Clin. Invest. 2003;111:153–161. doi: 10.1172/JCI17610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirano K., Fukuta M., Adachi T., Hayashi K., Sugiura M., Mori Y., Toyoshi K. Biochem. Biophys. Res. Commun. 1985;129:89–94. doi: 10.1016/0006-291x(85)91407-x. [DOI] [PubMed] [Google Scholar]
- 24.Turner B. J., Atkin J. D., Farg M. A., Zang d. W., Rembach A., Lopes E. C., Patch J. D., Hill A. F., Cheema S. S. J. Neurosci. 2005;25:108–117. doi: 10.1523/JNEUROSCI.4253-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tobisawa S., Hozumi Y., Arawaka S., Koyama S., Wada M., Nagai M., Aoki M., Itoyama Y., Goto K., Kato T. Biochem. Biophys. Res. Commun. 2003;303:496–503. doi: 10.1016/s0006-291x(03)00353-x. [DOI] [PubMed] [Google Scholar]
- 26.Osborne A. R., Rapoport T. A., van den, Berg B. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 27.Alder N. N., Shen Y., Brodsky J. L., Hendershot L. M., Johnson A. E. J. Cell Biol. 2005;168:389–399. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matlack K. E., Misselwitz B., Plath K., Rapoport T. A. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- 29.Nagai M., Aoki M., Miyoshi I., Kato M., Pasinelli P., Kasai N., Brown R. H., Jr., Itoyama Y. J. Neurosci. 2001;21:9246–9254. doi: 10.1523/JNEUROSCI.21-23-09246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Almer G., Guégan C., Teismann P., Naini A., Rosoklija G., Hays A. P., Chen C., Przedborski S. Ann. Neurol. 2001;49:176–185. [PubMed] [Google Scholar]
- 31.Ryu E. J., Harding H. P., Angelastro J. M., Vitolo O. V., Ron D., Greene L. A. J. Neurosci. 2002;22:10690–10698. doi: 10.1523/JNEUROSCI.22-24-10690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guégan C., Vila M., Rosoklija G., Hays A. P., Przedborski S. J. Neurosci. 2001;21:6569–6576. doi: 10.1523/JNEUROSCI.21-17-06569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J., Lillo C., Jonsson P. A., Vande Velde C., Ward C. M., Miller T. M., Subramaniam J. R., Rothstein J. D., Marklund S., Andersen P. M., et al. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.