Abstract
The cellular cytoskeleton is a complex dynamical network that constantly remodels as cells divide and move. This reorganization process occurs not only at the cell membrane, but also in the cell interior (bulk). During locomotion, regulated actin assembly near the plasma membrane produces lamellipodia and filopodia. Therefore, most in vitro experiments explore phenomena taking place in the vicinity of a surface. To understand how the molecular machinery of a cell self-organizes in a more general way, we studied bulk polymerization of actin in the presence of actin-related protein 2/3 complex and a nucleation promoting factor as a model for actin assembly in the cell interior separate from membranes. Bulk polymerization of actin in the presence of the verprolin homology, cofilin homology, and acidic region, domain of Wiskott-Aldrich syndrome protein, and actin-related protein 2/3 complex results in spontaneous formation of diffuse aster-like structures. In the presence of fascin these asters transition into stars with bundles of actin filaments growing from the surface, similar to star-like structures recently observed in vivo. The transition from asters to stars depends on the ratio [fascin]/[G actin]. The polarity of the actin filaments during the transition is preserved, as in the transition from lamellipodia to filopodia. Capping protein inhibits star formation. Based on these experiments and kinetic Monte Carlo simulations, we propose a model for the spontaneous self-assembly of asters and their transition into stars. This mechanism may apply to the transition from lamellipodia to filopodia in vivo.
Keywords: actin self-assembly, asters, cellular protrusions, Monte Carlo simulations, stars
During cellular migration, regulated actin assembly takes place at the plasma membrane (1–3), with continuous disassembly deeper in the cell interior. The site-directed actin polymerization at the plasma membrane results in the extension of cellular protrusions in the form of lamellipodia and filopodia. Although most cultured animal cells assemble both lamellipodia and filopodia, some cells, like keratocytes, emphasize lamellipodia, whereas dendritic cells are dominated by filopodia. One open question is how does a cell “choose” between the formation of lamellipodia and filopodia and what dictates the preference of one structure over the other. For that, it is essential to understand the self-assembly of these actin-based structures and the factors controlling these processes.
Although protrusions of lamellipodia and filopodia are tightly coupled to actin polymerization, the distinct organization and generation of filaments in each structure uses a different mechanism to produce mechanical force. In the lamellipodia, the actin filaments organize into a flat 2D branched network (2, 4), whereas bundles of parallel actin filaments support filopodia (5). In the lamellipodia, the branched nucleation is driven by activation of the actin-related protein (Arp) 2/3 complex (6) by the Wiskott-Aldrich syndrome protein family (7–8), followed by filament elongation and barbed-end capping by capping protein (CP) (9). Formin and Ena/vasodilator-stimulated phosphoprotein proteins concentrated at the tips of filopodia (3, 5, 10) are believed to protect the barbed ends of the filaments from capping and thus to enable persistent filament elongation (3, 11). Competition between branching, capping, and bundling rates governs the type of protrusion. These rates are expected to be regulated by the local abundance of certain regulatory proteins including Arp2/3 complex, CP, and bundling proteins such as fascin (12).
Biomimetic systems provide a simplified controlled environment for characterizing the role of constituent proteins and identifying the fundamental physical principles underlying cellular motility. Most in vitro experiments explore phenomena in the vicinity of surfaces (13–18). To understand how the nano machinery of a cell self-organizes in a more general way, we studied self-assembly in a bulk sample of purified proteins isolated from the influence of cell membranes. The reconstituted medium contained the major proteins participating in the assembly of lamellipodia and filopodia: (i) the constitutively active verprolin homology, cofilin homology, and acidic region (VCA) (16) (or WA6) domain of the Wiskott-Aldrich syndrome protein, which activates Arp2/3 complex; (ii) Arp2/3 complex; (iii) heterodimeric CP, which blocks the barbed ends of actin filaments; and (iv) fascin, which crosslinks actin filaments of the same polarity into bundles in filopodia. This biomimetic system enabled us to control the concentrations of the various proteins and observe the time course of self-assembly by light microscopy. We studied the formation and maturation of various mesoscopic structures, particularly asters and stars, similar to those recently observed in vivo (11).
Results
Arp2/3 Complex Controls Self-Organization of Asters.
First, we explored the role of the Arp2/3 complex on the dynamical self-assembly of actin in bulk. When activated by VCA, Arp2/3 complex induces the spontaneous self-assembly of actin into diffuse 3D aster-like structures (Fig. 1a and b). Asters formed over a wide range of concentrations of actin (0.52–7.28 μM), Arp2/3 complex (20–200 nM), and VCA (40–850 nM). Fully grown asters appeared in <2 min after initiating a reaction with 100 nM Arp2/3 complex, 200 nM VCA, and 7.28 μM actin monomers. This time is similar to the time required for actin polymerization as measured by biochemical assays (6), therefore nucleation of highly branched microstructures by Arp2/3 complex and their reorganization into asters occurs simultaneously
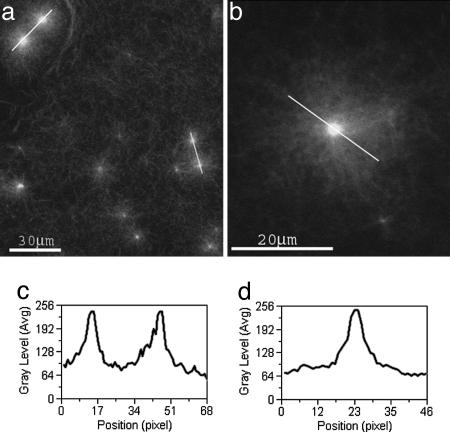
Fig. 1.
Confocal images of asters. Conditions were actin monomer (3.78 μM), GST-VCA (50 nM), and Arp2/3 complex (25 nM). (a and b) Individual asters and aster pairs (marked by white lines in a) are observed. (c) The intensity profile of actin fluorescence along an aster pair (right aster pair) is plotted. Typically, the two asters are almost identical in size and actin content. (d) The intensity profile of the fluorescence, I(r), is plotted along an aster (marked by a black line) versus the radial distance r from the aster center. I(r) decays symmetrically as a function of r from the aster center to the periphery.
We studied the effect of Arp2/3 complex on the number of asters formed per unit area within the 1-μM section of the bulk sample that we observed. The density of asters, ρ̄, was evaluated by measuring the characteristic distance between two asters ξ, where ρ̄ ∼ ξ−2. We tested the dependence of ρ̄ on the ratio x = [Arp2/3 complex]/[G actin] keeping the [VCA]-to-[Arp2/3 complex] ratio constant. The density ρ̄ increases monotonically (roughly linearly) with x (data not shown), implying that the number of asters generated per unit area depends linearly on [Arp2/3 complex], for a constant actin concentration.
Fig. 1d shows the intensity fluorescence profile I(r) along an aster. The intensity decreases monotonically and symmetrically from the aster’s center toward its periphery, reflecting the decreasing density of actin filaments. The next section discusses this behavior of the density profile and the polarity of the growing filaments along the growing aster.
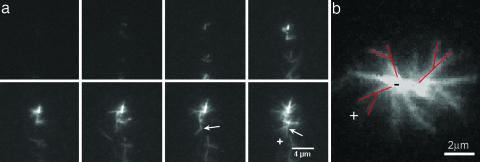
Using total internal reflection fluorescence microscopy, we followed the evolution of asters close to a glass surface, where we could observe the formation of individual asters and filament branches over time (Fig. 2a). These experiments were conducted at low concentrations of actin and Arp2/3 complex, 1 μM and 10 nM, respectively, along with 200 nM activator. Asters first appeared as fluorescent points, which subsequently grew by elongation and formation of branches (6) (Fig. 2a). On average, filaments grow with their barbed ends facing outward from the aster center, as can be assessed by analyzing the growth direction of the branches. The branch marked by an arrow in Fig. 2a grows away from the aster’s center. The angle between this branch and its mother filament is ≈70°, typical to an Arp2/3 branching angle (6). In Fig. 2b we can clearly observe an aster with the branches (marked in red) pointing outward, reflecting the tendency of barbed ends to grow outward.
Fig. 2.
Total internal reflection fluorescent images of an aster formation. Conditions were 1 μM actin monomer, 200 nM GST-VCA, and 10 nM Arp2/3 complex. (a) Images in the time-lapse series are spaced 30 s apart for a total time of 3.5 min. The experiments begin with buffer alone in the chamber. Actin, Arp2/3 complex, and activator are quickly mixed and pulled through the chamber. High concentrations of Arp2/3 complex lead to the formation of asters. Most likely, very small spontaneously nucleated seeds act as nucleation points from which Arp2/3 complex rapidly forms branches autocatalytically. The initial seeds are smaller than the optical resolution and thus appear as points at the center of the aster. Formation of a new branch (≈70° is characteristic to Arp2/3 branching) marked by a white arrow in the two last frames shows that the actin filaments barbed ends (+) point outward. (b) An image of the aster part that is close to the coverslip surface clearly shows branches pointing away from the aster center, three of them are marked in red. The growth direction is thus from the pointed (−) end to the barbed (+) end.
Kinetic Monte Carlo Simulation of Aster Formation.
Fig. 3 shows 2D projections of 3D snapshots from a kinetic Monte Carlo simulation of actin aster growth in the presence of Arp2/3 complex. In these simulations the aster evolves from a nucleus consisting of a single actin filament. After several generations of branching mediated by Arp2/3 complex, the aster is a roughly spherical globule of some radius r0, which may be referred to as the aster’s core. We shall refer to the filaments emanating from this core as “primary” filaments. As time evolves, filaments elongate and branch, resulting in structures similar to those observed experimentally (Figs. 1 and 2).
Fig. 3.
Kinetic Monte Carlo simulations. (a–c) Snapshots taken 10 (a), 13 (b), and 16 (c) s after nucleation. (d and e) Magnified regions from c and b, respectively. All filament tips are barbed ends and labeled in red. (f) (Lower) The volume fraction, ρ, of polymerized actin, and the orientational order parameter, η, as a function of the distance, r, from the nucleation center, after all actin monomer has been consumed and cluster growth completed. (Upper) The average number of polymerized subunits, d, between adjacent branching points on the same filament, as a function of r. The plots represent averages over eight simulation runs. The initial concentrations are similar to those used experimentally: 7.47 μM G actin and 100 nM Arp2/3 complex.
The calculated volume fraction, ρ, occupied by polymerized actin after all actin monomer has been consumed and cluster growth was completed (Fig. 3f Lower) is similar to the fluorescence profile in Fig. 1d. The farthest regions of the aster can be reached only by primary filaments, whose barbed ends point radially, and possibly also by a few, newly born branches. Because branching takes place along all, old and new filaments, the actin density is maximal in the oldest regions of the aster, i.e., near and around its core, and gradually decreases toward its periphery.
The polarity of the growing filaments can be expressed by the “orientational order parameter” η(r)= 〈cosθ〉. The averaging here is over all filament branches originating at r, with θ denoting the angle between the filament direction and the radius vector connecting the aster’s center and the filament’s origin (i.e., its pointed end). Consistent with the qualitative analysis above, Fig. 3f shows that as r increases, the actin density ρ(r) decreases, whereas η(r) increases monotonically, as discussed in more detail below.
At any moment during an aster’s growth, the tips (i.e., the barbed ends) of the primary filaments reached the farthest distance, r*, from the core’s center. There are no pointed ends at r*. Slightly inward, however, we find a few “young” first-generation branches (daughter filaments emerging from the primary filaments). On average, these branches are still oriented radially, and the contribution of their pointed ends to η(r) is about cos(70o) ≈ 0.3. Indeed, the highest-order parameter is at the aster’s periphery, reaching η(r) ≈ 0.3 (Fig. 3f). Further inward, as r decreases, we always find the primary filaments and, in addition, an increasing number of daughter filament generations. The fraction of uncorrelated filament orientations thus that contributes to η(r) increases as r decreases, resulting in smaller η(r).
Fig. 3f shows how the average spacing, d (in monomer units), between adjacent branches on the same mother filament, varies with r (calculated at the end of an aster’s growth). The spacing is calculated as d(r) = NF(r)/2NP(r), where NF(r) is the number of polymerized actin subunits and NP(r) is the number of branching points (or, equivalently, of pointed ends, because all pointed ends in our model emerge as a result of branching events) within a spherical shell at distance r from the aster’s nucleation center. The factor of 2 arises because each new branch on the side of an existing filament creates one new end but results in two F-actin segments originating at this branch point. As expected, d(r), increases rapidly with r.
The simulation is based on a simple kinetic model involving only two rate processes: monomer addition at barbed ends and branching from nonterminal filamentous subunits. We focus here on the structural evolution of single asters, not the process of nucleating multiple asters, or transition of asters to stars. In this simple simulation, actin monomers and Arp2/3 complexes are the only relevant species. The simulation box consists initially of a single polymerization seed (an actin dimer), embedded in a solution containing NG(0) actin monomers and NArp(0) Arp2/3 complexes. To account for the experimental fact that many asters grow simultaneously in solution, the box volume V is chosen as the average volume per aster, as observed experimentally in a system with the initial concentrations ρG(0) = NG(0)/V and ρArp(0) = NArp(0)/V. This result is equivalent to the assumption that each cluster is embedded in a certain volume containing given total numbers of actin monomers and Arp2/3 complexes. For the results shown in Fig. 3 we used the initial concentrations [actin monomer] = 7.47 μM and [Arp2/3] = 100 nM and set V = 2,500 μm3.
In the simulations, actin filaments are modeled as linear rigid polymers composed of flat-cylindrical monomers stacked one on top of another. The monomer diameter is 9 nm, corresponding to the diameter of an actin filament, and its height is set to 2.57 nm. Branching can take place from any subunit in a filament except the terminal subunit, unless it is already occupied by a daughter branch. The Arp2/3 complex, forming the nucleus of a new branch, is modeled as an actin dimer, initiating a filament at an angle of 70° with the mother filament. We impose an azimuthal angle difference of n(13·2π/28) between a branch originating n monomers away from the first branch along that filament. We explicitly account for excluded volume interactions between actin filaments. To ensure that filaments do not intersect each other, we allow actin monomers and Arp2/3 complex to add only if the new particle does not intersect an existing filament. Monomeric actin and Arp2/3 complex are assumed to be uniformly distributed in solution, with time-dependent concentrations ρG(t) and ρArp(t). This approximation is justified in view of the slow rates of actin polymerization and branching as compared with monomer diffusion (19, 20).
In the simulations the probabilities of polymerization and branching are proportional to the rates of these processes, Rpol(t) = kpol(t)·Nb(t) and Rbr(t) = kbr(t)·Nm(t), respectively, where Nb(t) and Nm(t) stand for the numbers of barbed ends and middle (i.e., nonterminal) F-actin monomers available for branching, respectively. Their time rate of change is governed by the kinetic equations
Here kpol(t) = kpol(2)ρG(t) and kbr(t) = k̃brρArp(t)ρG(t)2 are (pseudo unimolecular) rate constants for polymerization and branching. More explicitly, kpol(t) = kpol(2)ρG(t), where kpol(2) is the bimolecular rate constant for actin polymerization at the barbed end. For the branching rate constant we use the commonly accepted form kbr(t) = k̃brρArp(t)ρG(t)2, which assumes that branching can occur only after binding of Arp2/3 with two G-actin monomers (21); k̃br is the rate constant corresponding to the attachment of this complex to an F-actin monomer. To facilitate the simulations we set kpol(0) = 1/τMC and scale all rates relative to this (largest) rate constant; τMC denotes the simulation (Monte Carlo) time step, i.e., the average time interval between successive polymerization events at the barbed end, corresponding to the actin monomer concentration at t = 0. Experiments suggest that for the initial concentrations considered here, namely [actin monomer] = 7.47 μM and [Arp2/3 complex] = 100 nM, kbr(0) = 1.05 × 10−4 kpol(0)21. In the simulations we thus set kbr(0) = 1.05 × 10−4 τMC−1. We do not include depolymerization of barbed ends, because it is slow. We do not include debranching, because it is relatively slow and any dissociated branches will be constrained by the meshwork and continue to grow at their barbed ends. Aster growth ends when all actin monomers are consumed. For the conditions used in our simulations 99% of actin monomers were consumed after ≈100 s of growth, similar to the experiments described above. Also, ≈40% of the initial Arp2/3 complex remains in solution at the end of aster growth.
Star Formation, Role of Fascin, and CP.
Inclusion of fascin in a reaction mixture containing actin monomers, Arp2/3 complex, and VCA resulted in spontaneous self-assembly of actin into structures with radial spokes that we call stars (Fig. 4). In the early stages of this process the spokes condensed out of a low density cloud of actin surrounding the dense core (Fig. 4a). Under similar conditions, but without fascin, fully grown asters formed in <2 min, as described above. Stars grew outward from the original aster center, indicating that the polarity is preserved, with the barbed ends pointing outward. We thus conclude that on short time scales actin monomers organize into asters (the process is too rapid for observation with our current setup). Bundles appear only at longer time scales, after the actin filaments have elongated significantly, thus allowing their crosslinking by fascin with only a relatively small bending energy penalty (22) δE ≈ 8 kBT (Supporting Text, which is published as supporting information on the PNAS web site). About 5 min later the cloudy corona turned into a fully developed star (Fig. 4b); the diffuse actin cloud disappeared, turning into long actin bundles emanating from the dense Arp2/3 branched network core. Experiments with fluorescent VCA showed that it was present in the core but not in the star bundles (data not shown), so the concentration of Arp2/3 complex is likely to be low in bundles relative to the core.
Fig. 4.
Star formation is observed when fascin is added to a solution of actin, VCA, and Arp2/3 complex. (a and b) Initial conditions were 7.28 μM actin monomer, 200 nM GST-VCA, 100 nM Arp2/3 complex, and 3 μM fascin; the [fascin]/[G actin] = 1/2. (a) After mixing for 2.5 min the star appears to be composed of a dense core surrounded by a low density cloud of actin. It is already possible to observe actin bundles in the cloudy regions. (b) After 7.5 min the cloudy corona turns into a fully developed star; the diffuse actin cloud disappears, turning into long actin bundles (10–30 μm) emanating from the dense core. (c) Addition of 40 nM CP results in inhibition of star formation. The number of stars is decreased and their bundles are shorter (≈2 μm). (d) Stars are not formed in the absence of Arp2/3 complex; instead, an entangled network of filament bundles is observed. Short white arrows mark branching points of actin bundles, and the long white arrow marks the bundling/splitting of several bundles. (e) The dynamic of star growth is followed in time steps of 20 s. Initial protein concentrations were 3.64 μM actin monomer, 200 nM GST-VCA, 100 nM Arp2/3 complex, and 1.5 μM fascin; [fascin]/[actin] = ½ as in a–d. The initial average growth rate of bundles is v̄ = 2.27 ± 0.80 μm/min. (f) Star pairs also form. Their bundles growth rate is the same as for individual stars.
Inclusion of CP in the reaction mixture inhibited the generation and growth of bundles (Fig. 4c). The addition of nM levels of CP to the solution of actin, Arp2/3 complex, and VCA resulted in the generation of condensed actin asters instead of large diffuse asters (data not shown). Adding nM levels of CP to the solution of actin, Arp2/3 complex, VCA, and fascin inhibited star growth. Stars still appeared for [CP] = 15 nM (data not shown) but the spokes were shorter than in a system free of CP, where typical bundle lengths were L ≈ 10–30 μm (Fig. 4b). Upon increasing [CP] to 40 nM the stars became even smaller and developed only few short and straight bundles of typical length L ≈ 2 μm (Fig. 4c). Inhibition of star growth by CP confirmed that the barbed ends of filaments are at the ends of the spokes (Fig. 4c). Under the conditions of Fig. 4c reducing the concentration of fascin to 25% that of actin completely abolished the formation of stars (data not shown).
The Role of Arp2/3 Complex in Star Formation.
Polymerization of actin under our standard conditions but without Arp2/3 complex generated an isotropic mesh of entangled filament bundles rather than stars (Fig. 4d). This fact shows that Arp2/3 is required for star formation. Polymerization of actin without Arp2/3 complex but with fascin produced long actin filament bundles (tens of micrometers long, Fig. 4d). In the presence of fascin, branching points of actin bundles were routinely observed (short white arrows, Fig. 4d). The bundling and splitting process is, of course, not a unique property of single bundles, but also occurred when several bundles were involved (long white arrow, Fig. 4d).
We performed the quantitative data analysis of star growth at half the concentrations of actin and fascin used in Fig. 4 a and b. Under such conditions, the stars are smaller and overlapping effects between neighboring stars are reduced.
At the rapid growth step, which typically lasts up to 5 min (Fig. 4e), the bundles elongate at an average velocity v̄ = 2.27 ± 0.80 μm/min (14 subunits per s). Elongation would be faster with the initial actin monomer concentration of 3.64 μM; however, much of the free actin is consumed during the first step of aster formation. In fact, this velocity is comparable to the growth rate of filopodia in vivo (11, 23). As more actin monomer was consumed, elongation of the bundles slowed and eventually halted. Addition of fresh actin and fascin to the solution restarted the nucleation and growth of new bundles and the elongation of preformed ones (unpublished results).
Discussion
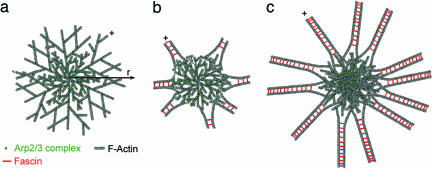
Polymerization of actin in the presence of activated Arp2/3 complex leads to the self-assembly of asters that transition into stars providing that a crosslinking protein (fascin in our experiments) is present (Fig. 5). After the spontaneous nucleation of an actin seed (Figs. 2 and 3), Arp2/3 complexes bind to short actin filaments, initiating an autocatalytic process of branch formation and filament growth (Fig. 5a). After a few generations of branching the cluster growth becomes isotropic, resulting in an aster with roughly spherical symmetry. The growth of the network advances with the barbed ends of the peripheral actin filaments (+ in Fig. 5a) pointing on average outward, as seen in the simulation result of η(r) (Fig. 3f).
Fig. 5.
The transition from asters to stars. (a) The first polymerization step begins by spontaneous nucleation of an actin seed. This step is followed by binding of Arp2/3 complexes to nascent actin filaments, initiating an autocatalytic process of branch formation and filament growth. With time growth of the cluster becomes isotropic, resulting in an aster of roughly spherical symmetry. The growth of the network advances with the barbed ends of the actin filaments (+) pointing outward. (b) The transition of an aster into a star is initiated by the reorganization of the network structure into bundles. The star is now composed of a dense network of actin filaments with short bundles emanating with their barbed ends pointing outward (+) from beyond this core. (c) In the final step of star formation, the bundles elongate by continuous actin polymerization in the outward direction while new bundles continue to emerge close to the star core. During the growth process the density of the star core continues to increase because of the continuous nucleation of new Arp2/3-actin branches.
The transition of an aster into a star in the presence of a crosslinking protein is a natural consequence of the growth of the aster. “Older” regions of the aster are denser than “new” regions, so the actin filament density falls with the distance from the center, r (Fig. 1d) and ρ(r) (Fig. 3f). In addition, the separation of neighboring branches [d(r) in Fig. 3f] increases with r, resulting in longer filaments at the periphery of the growing aster. Longer, less densely packed filaments bend more easily than short filaments. Therefore, fascin bundles actin filaments in regions remote from the aster core, where the gain in fascin binding energy compensates for the actin filament bending energy penalty. At the end of this process, the star is composed of a more or less isotropic core of dense actin filaments network with short bundles emanating outward from this core (+ in Fig. 5b). In the final step of star formation these bundles elongate by continuous actin polymerization while new bundles continue to emerge close to the star core (Figs. 4e and 5c).
In the course of aster-to-star transition the directionality of actin filaments is maintained, with the barbed ends pointing outward. This finding contrasts with another biomimetic system containing latex beads coated with an Arp2/3 activator (17). In this system the generation of the Arp2/3 complex–actin network takes place at the bead’s surface with the filaments’ barbed end pointing toward the bead, whereas the addition of fascin resulted in the growth of bundles with their barbed end pointing away from the bead surface. In living cells, on the other hand, barbed ends of actin filaments in both lamellipodia and filopodia point toward the plasma membrane where they polymerize, meaning that the directionality of actin filaments is maintained during the transition from lamellipodia to filopodia.
The presence of CP in a solution of actin and Arp2/3 complex resulted in the formation of compact asters of dense branched network. Termination of barbed end elongation by CP results in shorter filaments, preserving the pool of actin monomers to make more branches. Deforming these short, constrained actin filaments requires more energy than bending long filaments (Supporting Text), so fascin induces fewer filament bundles around each of these compact asters (Fig. 4c).
The impact of capping on bundle formation illustrates why persistent elongation is an important factor for the assembly of actin filament bundles in cells. In a network of branched actin filaments nucleated by Arp2/3 complex, CP limits filament length, precluding bundling. On the other hand, formins allow persistent growth of barbed ends even in the presence of CP. Formins at the tips of growing filopodia may contribute to the persistent growth of the actin filaments, preventing the CP from blocking their growth (24). In cells the number and the length of filopodia were shown to depend on CP levels (11) and formin expression level (24).
Our observations in vitro are consistent with the effect of depleting CP from cultured cells (11). Depletion of CP resulted in spontaneous reorganization of the actin cytoskeleton with loss of the leading edge, redistribution of the Arp2/3 complex into the cell interior, and explosive formation of filopodia and star-like structures composed of actin filaments. Mejillano et al. (11) found that the same proteins complex (Ena/vasodilator-stimulated phosphoprotein/formin) at the bundle tips in filopodia and stars. In both cases, bundles of actin filaments emerged from branched networks preserving the actin filament polarity during the transition. Thus, in vivo stars and filopodia are very similar except for their symmetry. Filopodia elongate from a more or less planar surface (i.e., cell membrane), whereas star bundles grow radially from their nucleation center.
The stars that self-assemble in vitro highly resemble those generated in vivo; the star core is composed of a branched network and the filaments in bundles emerging from this core have their barbed ends outward. Lamellipodia are 2D asters with the barbed ends of the filaments directed toward the membrane. If the filaments grow long enough, they can be bundled by crosslinking proteins, spontaneously forming parallel bundles similar to filopodia.
Our minimal model system of pure proteins confirms that actin, activated Arp2/3 complex, fascin, and CP can mimic complex events at the leading edge of cells. Compared with a cell this system is simple and controlled, making it possible to characterize the roles of a limited set of proteins in the higher-order assembly of actin filaments. The asters and stars in the in vitro system are equilibrium self-assembled structures. This fact is in contrast to the reorganization of asters and vortices by motor proteins (25–28), which is considered to be an “active” process leading to steady-state structures that depend on ATP hydrolysis to maintain their shape.
Materials and Methods
Protein Purification.
Actin was purified from rabbit skeletal muscle acetone powder (29). Recombinant Wiskott-Aldrich syndrome protein–VCA (16) and fascin (30) were expressed in Escherichia coli as GST fusion proteins. Recombinant mouse CP was purified from E. coli (31). Arp2/3 complex was purified from bovine thymus (32). Actin labeled on Cys-374 with Alexa 568 or 488 was purchased from Molecular Probes.
Motility Assay.
The motility medium contained 10 mM Hepes (pH 7.7), 1.7 mM Mg-ATP, 5.5 mM DTT, 0.12 mM 1,4-diazabicyclo[2.2.2]octane (an antibleaching reagent), 0.1 M KCl, 1 mM MgCl2, 1% BSA, and various concentrations of G actin, Arp2/3 complex, VCA, CP, and fascin. No vasodilator-stimulated phosphoprotein was needed.
Microscopy.
Actin assembly was monitored for ≈1 h by fluorescence with an Olympus IX-71 microscope using intermittent illumination from a mercury lamp. The labeled actin fraction was 1/26 and the temperature was 22°C. Time-lapse images were acquired by using a Coolsnap HQ camera (Photometrics, Tucson, AZ). Data analysis was performed with metamorph (Universal Imaging, Downingtown, PA).
Total Internal Reflection Fluorescence Microscopy.
We observed actin polymerization, aster generation, and aster growth on the surface of a glass slide with total internal reflection evanescent wave illumination on an Olympus IX70 microscope (33). The protein concentrations were 10 nM Arp2/3 complex, 200 nM GST-VCA, and 1 μM actin with 30% labeled at Cys-374 with Oregon green (34).
Supplementary Material
Acknowledgments
We thank A. Atayeva for the purification of fascin, actin, and VCA; D. Vignjevic (Curie Institute, Paris) for providing the fascin plasmid; and D. Groswasser for careful reading of the manuscript. This work was supported by the Reimund Stadler Minerva Center, Israel Cancer Association Grant 20040070 (to A.B.-G.), Israel Science Foundation Grant 551/04 (to A.B.-G.), Israel Science Foundation Grant 227/02 (to A.B.-S.), U.S.–Israel Binational Science Foundation Grant 2002-75 (to A.B.-S.), and National Institutes of Health Grants GM26338 and GM26132 (to T.D.P.).
Abbreviations
- VCA
verprolin homology, cofilin homology, and acidic region
- Arp
actin-related protein.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Welch M. D., Mallavarapu A., Rosenblatt J., Mitchison T. J. Curr. Opin. Cell Biol. 1997;9:54–61. doi: 10.1016/s0955-0674(97)80152-4. [DOI] [PubMed] [Google Scholar]
- 2.Pollard T. D., Borisy G. G. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Schafer D. A. Nature. 2004;430:734–735. doi: 10.1038/430734a. [DOI] [PubMed] [Google Scholar]
- 4.Svitkina T. M., Borisy G. G. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M., Borisy G. G. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchoin L., Amann K. J., Higgs H. N., Marchand J.-B., Kaiser D. A., Pollard T. D. Nature. 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 7.Machesky L. M., Insall R. H. Curr. Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 8.Takenawa T., Miki H. J. Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- 9.Cooper J. A., Schafer D. A. Curr. Opin. Cell Biol. 2000;12:97–103. doi: 10.1016/s0955-0674(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 10.Bear J. E., Svitkina T. M., Krause M., Schafer D. A., Loureiro J. J., Strasser G. A., Maly I. V., Chage O. Y., Cooper J. A., Borisy G. G., et al. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 11.Mejillano M. R., Kojima S., Applewhite D. A., Gertler F. B., Svitkina T. M., Borisy G. G. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 12.Kureishy N., Sapountzi V., Prag S., Anilkumar N., Adams J. C. BioEssays. 2002;24:350–361. doi: 10.1002/bies.10070. [DOI] [PubMed] [Google Scholar]
- 13.Loisel T. P., Boujemaa R., Pantaloni D., Carlier M.-F. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 14.Brieher W. M., Coughlin M., Mitchison T. J. J. Cell Biol. 2004;165:233–242. doi: 10.1083/jcb.200311040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron L. A., Footer M. J., van Oudenaarden A., Theriot J. A. Proc. Natl. Acad. Sci. USA. 1999;96:4908–4913. [Google Scholar]
- 16.Bernheim-Groswasser A., Weisner S., Goldsteyn R., Carlier M. F., Sykes C. Nature. 2002;417:308–311. doi: 10.1038/417308a. [DOI] [PubMed] [Google Scholar]
- 17.Vignjevic D., Yarar D., Welch M. D., Peloquin J., Svitkina T., Borisy G. G. J. Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giardini P. A., Fletcher D. A., Theriot J. A. Proc. Natl. Acad. Sci. USA. 2003;100:6493–6498. doi: 10.1073/pnas.1031670100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham V. C., Krishnamurthi V., Taylor D. L., Lanni F. Biophys. J. 1999;77:1721–1732. doi: 10.1016/S0006-3495(99)77018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard T. D., Blanchoin L., Mullins R. D. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 21.Carlsson A. E., Wear M. A., Cooper J. A. Biophys. J. 2004;86:1074–1081. doi: 10.1016/S0006-3495(04)74182-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mogilner A., Rubinstein B. Biophys. J. 2005;89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schirenbeck A., Bretschneider T., Arasada R., Schleicher M., Faix J. Nat. Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- 24.Pellegrin S., Mellor H. Curr. Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Surrey T., Nedelec F., Leibler S., Karsenti E. Science. 2001;292:1167–1171. doi: 10.1126/science.1059758. [DOI] [PubMed] [Google Scholar]
- 26.Verkhovsky A. B., Svitkina T. M., Borisy G. G. J. Cell Sci. 1997;110:1693–1704. doi: 10.1242/jcs.110.15.1693. [DOI] [PubMed] [Google Scholar]
- 27.Kruse K., Joanny J. F., Julicher F., Prost J., Sekimoto K. Phys. Rev. Lett. 2004;92:07810114. doi: 10.1103/PhysRevLett.92.078101. [DOI] [PubMed] [Google Scholar]
- 28.Cytrynbaum E. N., Rodionov V., Mogilner A. J. Cell Sci. 2003;117:1381–1397. doi: 10.1242/jcs.00919. [DOI] [PubMed] [Google Scholar]
- 29.Spudich J. A., Watt S. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 30.Ono S., Yamakita Y., Yamashiro S., Matsudaira P. T., Gnarra J. R., Obinata T., Matsumura F. J. Biol. Chem. 1997;272:2527–2533. doi: 10.1074/jbc.272.4.2527. [DOI] [PubMed] [Google Scholar]
- 31.Machesky L. M., Atkinson S. J., Ampe C., Vandekerckhove J., Pollard T. D. J. Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgs H. N., Blanchoin L., Pollard T. D. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- 33.Amann K. J., Pollard T. D. Proc. Natl. Acad. Sci. USA. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn J., Pollard T. D. Biophys. J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.